Abstract
Background
Bacteria employ multiple mechanisms to control gene expression and react to their constantly changing environment. Bacterial growth in rich laboratory medium is a dynamic process in which bacteria utilize nutrients from simple to complex and change physical properties of the medium, as pH, during the process. To determine which genes are differentially expressed throughout growth from mid log to stationary phase, we performed global transcript analysis.
Results
The S. agalactiae transcriptome is dynamic in response to growth conditions. Several genes and regulons involved in virulence factor production and utilization of alternate carbon sources were differentially expressed throughout growth.
Conclusion
These data provide new information about the magnitude of plasticity of the S. agalactiae transcriptome and its adaptive response to changing environmental conditions. The resulting information will greatly assist investigators studying S. agalactiae physiology and pathogenesis.
Background
Bacteria employ multiple mechanisms to control gene expression and react to their constantly changing environment. These processes are especially critical for bacterial pathogens to survive and cause disease in humans and other hosts. Global control of gene expression is achieved using alternative sigma factors, two-component systems (TCSs), small regulatory RNAs, regulators such as RelA and LuxS, or concerted action of regulons (for a review see [1-6] and references therein). Gram positive pathogens such as group A Streptococcus (S. pyogenes, GAS) and group B Streptococcus (S. agalactiae, GBS) lack (or have limited number) of alternative sigma factors of fully confirmed function [7-9]. Analyses of global transcription in GAS under various growth conditions including saliva, blood, and tissue has shown that environmental response regulation is achieved using other mechanisms such RNA stability [10], "stand alone" regulators such as mga [11], or TCSs [12-15]. These transcriptome analyses have been especially useful in providing new information about microbial physiology and leads for pathogenesis research. However, the transcriptional response of GBS to changing growth conditions has not been fully analyzed, only single reports were recently published [16]. GBS is an important human and cow pathogen, responsible for thousands of severe invasive infections in man and large economic loss attributable to bovine mastitis (see [17,18] and references therein).
One of the best examples of sequential gene regulation is bacterial growth in complex medium and activation of stationary phase genes. During growth, bacteria utilize available nutrients, presumably from simple to more complex, and alter their environment (e.g. decrease or increase in pH) as a result of metabolic byproduct release. Therefore, stationary phase can be considered the acid/alkali stress, depending on the type of metabolism and nutrients utilized. GAS grown to stationary phase sequentially expresses genes involved in various aspects of GAS physiology, metabolism and virulence, many genes activated or repressed during the transition to stationary phase have also been shown to play a role in GAS virulence [19]. The purpose of the present study was to identify growth phase-regulated genes in GBS, with a special interest in providing new information about virulence factor gene expression.
Methods
Sample collection for microarray analysis
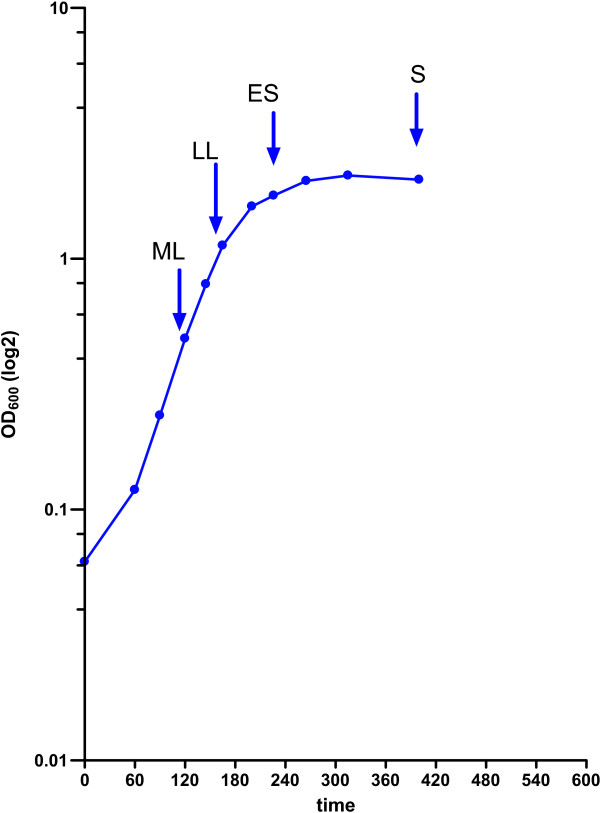
GBS strain NEM316 [7] was grown as three static cultures (3 biological replicates) in liquid Todd Hewitt medium with 0.5% yeast extract in the 5% CO2 atmosphere at 37°C [12]. Samples were collected at OD600 approximately 0.5, 1.0, 2.0 and 2.5, representing mid-logarithmic (ML), late-logarithmic (LL), early stationary (ES) and stationary (S, about 3 h from entering the phase) growth phases, respectively. Growth curve of bacterial cultures used for data collection is presented as Figure 1. Five ml of each sample were immediately mixed after collection with 10 ml of RNAProtect (Qiagen), centrifuged and stored at -80°C until processing.
Figure 1.
Growth curve of NEM316 in THY medium. Arrows denote points of sample collection.
Glucose content of the medium at the beginning and end of the culture was measured using Optium Xido glucometer (Abbot) and pH was checked using pH test strips (Macherey Nagel).
RNA isolation
GBS cells were mechanically opened by shaking with glass beads (Lysing Matrix B, MPBio) and TRIZOL (Invitrogen). RNA was isolated according to Chomczynski and Sacchi [20], with an additional purification step using RNeasy columns (Qiagen). Targets for hybridization with the array were prepared according to array manufacturer (Affymetrix) as described previously [12].
Array hybridization and data acquisition
The custom expression array manufactured by Affymetrix [21] contained redundant sets of probes representing 1,994 open reading frames (ORFs) of previously sequenced GBS strain NEM316 [7]. Arrays were hybridized and scanned according to the manufacturers instructions. The results of hybridization were normalized to mean of total intensity of GBS probes to allow multiple time point comparison. Array hybridization results are presented as Additional file 1 and are deposited in GEO database http://www.ncbi.nlm.nih.gov/projects/geo/ under GSE12238 accession number.
Results and Discussion
General trends in transcription
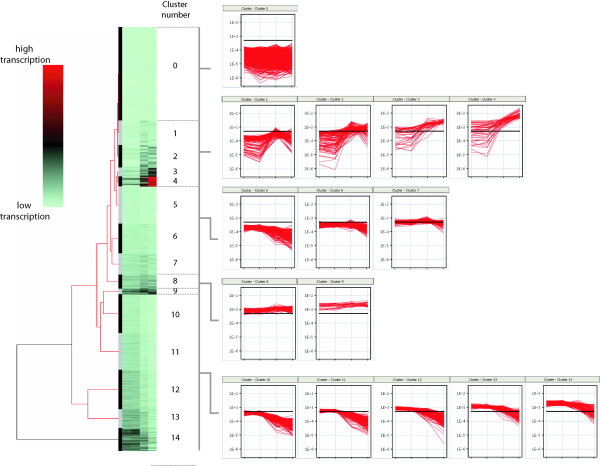
After determining transcript levels for all probe sets, the 1,994 transcripts were grouped into 15 clusters based on their behavior during growth (Figure 2) (self organizing map algorithm; Array Assist 5.1.0 package, Stratagene). The clusters were grouped into five main categories. The first 3 categories contain genes whose transcription did not correspond to growth phase, and were either expressed at low (cluster 0), medium (clusters 6, 7), or high (clusters 8, 9) levels in all phases of growth. Category 4 genes (clusters 1–4) exhibited increased transcription in ES or S phase, and category 5 genes (clusters 5, 10–14) had transcription levels that peaked in ML phase and decreased into S phase.
Figure 2.
Grouping of S. agalactiae transcripts into distinct 15 clusters based on expression profiles from ML to S growth phases. The dendrogram and clusters were generated using a self organizing map algorithm and represent changes in expression of 1,994 transcripts at four consecutive time points: ML, LL, ES, and S phases. Cluster 0 genes had low level of transcription. Clusters 1–4 genes positively correlated with stationary phase of growth transcription level and peaked in the ES (clusters 1 and 2) or S (clusters 3 and 4) phase of growth. Clusters 5 and 10–14 are negatively correlated with the S phase of growth; transcription of genes grouped in these clusters reached their peak in ML phase and decreased in S phase. Genes in clusters 6–9 are expressed relatively steadily during growth although at various levels of expression, ranging from very high (cluster 9) to mid-low (cluster 6). The black horizontal line on the cluster graphs represents average transcription level of the complete dataset. The transcript level in each cluster is plotted using a logarithmic scale. Number of transcripts in clusters: Cluster 0, 440; Cluster 1, 115; Cluster 2, 106; Cluster 3, 42; Cluster 4, 47; Cluster 5, 175; Cluster 6, 140; Cluster 7, 100; Cluster 8, 66; Cluster 9, 26; Cluster 10, 183; Cluster 11, 173; Cluster 12, 185; Cluster 13, 89; Cluster 14, 107.
Genes exhibiting growth phase-independent transcription
Genes in clusters 6, 7, 8, and 9 did not show growth phase-dependent transcriptional regulation. The genes are clustered instead based on their transcript level and general profile. Clusters 6 and 7 contain genes that are expressed at the same level until ES phase to slightly lower expression in S phase. Clusters 8 and 9 contain genes, which the transcript level is steady or slightly increases over time. Cluster 9 is especially interesting in that it contains a group of highly expressed genes that includes fabF, fabZ, fabH, and accBCD (gbs0331, 0336–0341) encoding subunits of beta-ketoacyl-ACP synthase, subunits of acetyl-CoA carboxylase, 3-hydroxydecanoyl-ACP dehydratase, and biotin carboxylase. Other genes in cluster 9 involved in energy production are ATP synthase subunits (atpABEF, gbs 0875–7 and 9). Interestingly, cluster 9 contains a transcript of putative catabolite control protein A (ccpA), and the amount grows steadily to increase about three-fold in S phase in comparison with ML (Table 1). CcpA is a major mediator of carbon catabolite repression – the control mechanism of nutrient utilization. In GAS, CcpA has recently been shown to be a critical direct link between carbohydrate utilization and virulence [21]. Function of CcpA in GBS has been not experimentally confirmed yet. Based on the consensus CcpA binding site (cre sequence), we detected that genome of NEM316 strain contains multiple putative cre sites in promoter sequences of multiple genes (Table 2), what might be correlated with changes in expression of genes involved in arginine and carbohydrate metabolism (see below). The transcript encoding HPr carrier protein, another element of the CcpA regulatory pathway in Gram-positive bacteria, also belongs to cluster 9. HPr kinase, however, is an S phase-related gene (see below).
Table 1.
Fold changes in transcript levels of GBS genes.
| Gene | Fold change in S phase (S/ML ratio) | Putative function |
|---|---|---|
| S phase related genes | ||
| hrcA, grpE, dnaK (gbs0094–96), | +4 to +7.5 | Stress response |
| clpE, and clpL (gbs0535 and gbs1376) | +4.5 and +7.5 | Chaperones |
| gbs1202/1204, gbs1721, gbs1772 | + 30 to +64 | Putative stress response proteins from Gls24 and universal stress response families |
| gbs2083–2085 | +350 to over +1000 | arginine/ornithine antiporter, carbamate kinase, ornithine carbamoyltransferase |
| gbs2122–2126 | +55 to +150 | arginine deiminase ornithine carbamoyltransferase, arginine/ornithine antiporter carbamate kinase |
| glpKDF (gbs0263–5) | +45 to +63 | putative operon responsible for glycerol uptake and utilization. |
| Nutrient utilization and energy metabolism | ||
| fba gbs0125 | +2.2 | fructose-bisphosphate aldolase |
| plr gbs1811 | +3.1 | glyceraldehyde 3P-dehydrogenase |
| pgk gbs1809 | +2.8 | phosphoglycerate kinase |
| eno gbs0608 | +2.5 | enolase |
| acoAB (gbs 0895–0896) | +4 | pyruvate dehydrogenase |
| ldh gbs0947 | +2.8 | L-lactate dehydrogenase |
| Regulators and signal transduction systems | ||
| gbs 1671/2 | -2 | TCS CovR/S |
| gbs1908/9 | +10/14 | TCS, homolog of GAS Spy1106/7 (SF370) |
| gbs1934/5 | +5/+5 | TCS, homolog of Spy1061/2 (SF370) |
| gbs2081/2 | -2.3/-1.7 | TCS, putative arginine utilization regulator |
| gbs2086/7 | 2.5/2.6 | TCS, putative arginine utilization regulator |
| gbs1834/5 | -7.5/-11.7 | TCS |
| gbs1397/8 | -7/-5.8 | TCS |
| gbs0597/8 | -5/-8.5 | TCS |
| gbs0121/2 | -2/1 | TCS |
| gbs0298/9 | -3/-1.8 | TCS |
| gbs0309/10 | -3.3/-3 | TCS |
| gbs0429/30 | -2.4/-1.6 | TCS |
| gbs0963/4 | +1.7/+2 | TCS |
| gbs1019/20 | -1.9/-1.9 | TCS |
| gbs1947/8 | -3/-2.4 | TCS |
| gbs1943/4 | -2.1/-2.7 | TCS |
| gbs0680 | +3.1 | CcpA |
| gbs0191 | +50 | putative transcriptional antiterminator of the BglG family |
| gbs0469 | -34 | Regulator of unknown function |
| relA (gbs1928) | -50 | GTP pyrophosphokinase |
| codY (gbs1719;) | -8 | Global regulator |
| luxS (gbs0294) | -3 | Quorum sensing |
| mecA (gbs0135) | -20 | Global regulator of competence |
| Virulence factors | ||
| gbs1420 | +6.26 | choline-binding protein |
| gbs1539 | +4.67 | cell wall anchored protein |
| gbs1929 | +5.48 | putative nucleotidase |
| gbs1143 | +2.61 | cell wall anchored protein |
| gbs0451 | -2 | paralog of C5A peptidase precursor |
| gbs1104 | -6.15 | cell wall anchored protein |
| gbs1529 | -11 | putative fibronectin binding protein |
| gbs0850 | -3 | putative fibronectin binding protein |
| gbs1307 | -3 | laminin binding protein |
| gbs1926 | -3 | putative laminin binding protein |
| gbs1475/6 | -5.5 | sortases |
| gbs0644–0654 | -1.6 to -2.8 | hemolysin |
| gbs1061–1076 | -2.5 to -12.9 | pathogenicity island IX |
| gbs1233–1247 | -3 to -12.4 | capsule synthesis |
| cpsX gbs1250 | 4.4 | capsule synthesis regulator |
| gbs1478/9, gbs1481, gbs1484/5, gbs1492–1494 | -3 to -12 | Putative group B antigen |
| Cfa gbs2000 | +11.6 | CAMP factor |
The table presents fold change in S phase in comparison with ML phase, classification into functional categories, and putative function.
Table 2.
Putative CcpA binding sites in promoter regions of genes encoded by S. agalactiae NEM316 genome.
| Match | Start (nt) | End (nt) | Homology with consensus(%) | ORF number | Name | Putative function |
|---|---|---|---|---|---|---|
| TGACAACGGTAAAA | 16111 | 16124 | 92 | gbsr001 | 16S ribosomal RNA | |
| TGAAAACGCTTTAA | 48894 | 48907 | 92 | gbs0032 | N-acetylmannosamine-6-phosphate 2-epimerase | |
| TGACAAGGATGTCA | 65156 | 65169 | 92 | gbs0049 | ruvB | Holliday junction DNA helicase ruvB |
| TTAAAGCGCTTTCA | 69320 | 69333 | 92 | gbs0053 | adh2 | Alcohol dehydrogenase |
| TGTAAACGATTACA | 72238 | 72251 | 100 | gbs0054 | adhA | Alcohol dehydrogenase |
| TGGGAACGGTTTCA | 130310 | 130323 | 92 | gbs0119 | ABC transporter permease protein | |
| TGTAATCGCTTACT | 130334 | 130347 | 92 | gbs0119 | ABC transporter permease protein | |
| TATTAACGTTAACA | 142634 | 142647 | 92 | gbs0130 | Membrane protease protein family | |
| TGTCAACTATATCA | 176297 | 176310 | 92 | gbs0155 | Multimodular transpeptidase-transglycosylase PBP 1B | |
| TGTAATCGTTTACA | 209972 | 209985 | 100 | gbs0189 | PTS system, trehalose-specific IIBC component | |
| TGTAAACGGTTACT | 214120 | 214133 | 92 | gbs0191 | Transcription antiterminator, BglG family | |
| TGAAAAAGGTAACA | 243786 | 243799 | 92 | gbs0227 | pseudogene | |
| TGTTACCGTTTTCA | 284183 | 284196 | 100 | gbs0266 | NADH peroxidase | |
| TGAAAGCGGTTATA | 349577 | 349590 | 92 | gbs0326 | Ribosome-associated factor Y | |
| AGAAAGCGTTAACA | 349601 | 349614 | 92 | gbs0326 | Ribosome-associated factor Y | |
| TTAAAACGTTTTCA | 375767 | 375780 | 92 | gbs0348 | manL | PTS system, mannose-specific IIAB component |
| TGATACCGTTCACT | 480733 | 480746 | 92 | gbs0452 | alpha-L-Rha alpha-1,3-L-rhamnosyltransferase | |
| TAATAACGTTAACA | 515726 | 515739 | 92 | gbs0493 | Hypothetical protein | |
| TGAAAACATTTACA | 540267 | 540280 | 92 | gbs0520 | typA | GTP-binding protein TypA BipA |
| TGACACCGTTTTCA | 592276 | 592289 | 100 | gbs0569 | Acetoin dehydrogenase | |
| AGATAGCGGTCACA | 608177 | 608190 | 92 | gbs0583 | Adenosine deaminase | |
| TGATATCGCTTTCA | 638255 | 638268 | 100 | gbs0615 | Class B acid phosphatase | |
| TGAAAGTGTTGACA | 661185 | 661198 | 92 | gbs0644 | cylX | Hypothetical protein |
| TAAAAGCGTTTACA | 684988 | 685001 | 92 | gbs0669 | SUGAR SODIUM SYMPORTER | |
| AGATAACGGTTACA | 690270 | 690283 | 92 | gbs0673 | 4-Hydroxy-2-oxoglutarate aldolase | |
| TAAAAACGCTAACA | 837159 | 837172 | 92 | gbs0813 | Glycerate kinase | |
| TTAGAGCGTTTTCA | 870536 | 870549 | 92 | gbs0844 | udk | Uridine kinase |
| TGTAAGCCTTGTCA | 879217 | 879230 | 92 | gbs0852 | Hypothetical protein | |
| TGTAAACCATCTCA | 903332 | 903345 | 92 | gbs0875 | atpE | ATP synthase C chain |
| TGAAAACGTAATCA | 903356 | 903369 | 92 | gbs0875 | atpE | ATP synthase C chain |
| TGTTAACGCTATTA | 913902 | 913915 | 92 | gbs0887 | pheT | Acetyltransferase, GNAT family |
| TGAAAACCGTTTCA | 981187 | 981200 | 92 | gbs0940 | 16S rRNA m(2)G 1207 methyltransferase | |
| TGAAAGCGTTTATA | 1145634 | 1145647 | 92 | gbs1100 | pgmA | Phosphoglucomutase |
| AGAAAACGGTATCA | 1157589 | 1157602 | 92 | gbs1112 | apbE | Iron-sulfur cluster assembly repair protein ApbE |
| TAATACCGTTATCA | 1200221 | 1200234 | 92 | gbs1156 | Na+ driven multidrug efflux pump | |
| TGAAATCGATTACA | 1235422 | 1235435 | 100 | gbs1192 | gabD | Succinate-semialdehyde dehydrogenase [NADP+] |
| TGTAAAGGTTTTCA | 1237447 | 1237460 | 92 | gbs1195 | ska | streptokinase |
| TGTAAACGTTTTTA | 1248933 | 1248946 | 92 | gbs1200 | Hydrolase (HAD superfamily) | |
| TTTAAACGCTATCA | 1314589 | 1314602 | 92 | gbs1273 | rmlA | Glucose-1-phosphate thymidylyltransferase |
| TGAAACCGGTTTGA | 1337103 | 1337116 | 92 | gbs1293 | glycerophosphoryl diester phosphodiesterase | |
| TGAAAGCTCTGACA | 1489796 | 1489809 | 92 | gbs1437 | Transcriptional regulators, LysR family | |
| TGACAGCGCAATCA | 1492240 | 1492253 | 92 | gbs1441 | capA | Capsule biosynthesis protein capA |
| TGTAACCGTTTTTA | 1518448 | 1518461 | 92 | gbs1468 | pflC | Pyruvate formate-lyase activating enzyme |
| TGTAACCGCTTTCT | 1742894 | 1742907 | 92 | gbs1684 | Zn-dependent alcohol dehydrogenase | |
| TGTACACGATATCA | 1749143 | 1749156 | 92 | gbs1689 | ABC transporter substrate-binding protein | |
| TGAAAACCCTAACA | 1752507 | 1752520 | 92 | gbs1694 | Dihydroxyacetone kinase | |
| TGACAACGTTAAAA | 1824783 | 1824796 | 92 | gbs1764 | mutS2 | DNA mismatch repair protein mutS |
| TGTAAGCGTTTTAA | 1920050 | 1920063 | 92 | gbs1856 | ulaA | PTS system, 3-keto-L-gulonate specific IIC component |
| TGACACCGGTATAA | 1925222 | 1925235 | 92 | gbs1862 | Amino acid ligase family protein | |
| TTATACCGTTTTCA | 1929838 | 1929851 | 92 | gbs1865 | hslO | 33 kDa chaperonin |
| TGTAAACGTTTTTA | 1939040 | 1939053 | 92 | gbs1874 | ahpC | Peroxiredoxin |
| TGTAATCTCTTACA | 1946899 | 1946912 | 92 | gbs1875 | ahpF | Peroxiredoxin reductase (NAD(P)H) |
| TTATAGCGCTTTCA | 1957716 | 1957729 | 92 | gbs1879 | pepO | Oligoendopeptidase O |
| TGATAACTATGTCA | 1990172 | 1990185 | 92 | gbs1918 | lacA.1 | Galactose-6-phosphate isomerase lacA subunit |
| TGAAAGCGGTTTAA | 2014283 | 2014296 | 92 | gbs1939 | PTS system, mannose fructose family IIA component | |
| TGTAAACGCTTTTA | 2101628 | 2101641 | 92 | gbs2026 | udp | Uridine phosphorylase |
| TGATATCGTAATCA | 2130043 | 2130056 | 92 | gbs2055 | argR2 | Arginine repressor, argR |
| AGATATCGCTTTCA | 2157836 | 2157849 | 92 | gbs2085 | Ornithine carbamoyltransferase | |
| AGAAATCGCTTTCA | 2195324 | 2195337 | 92 | gbs2122 | arcA | Arginine deiminase |
Genome was searched using BLAST algorithm (CLC DNA Workbench 4) with cre consensus sequence: TGWNANCGNTNWCA (N any nucleotide, W indicates adenine or thymine) and accuracy 90%. Start/End coordinates according to genome sequence
Log phase related genes
Almost 50% of all GBS transcripts that were represented on the chip had similar patterns of expression and were classified into clusters 5, 10, 11, 12, 13, and 14. Transcript levels peaked in ML phase and decreased gradually to their lowest levels in S phase. These six clusters differ in their basal level of expression in L phase. The genes assigned to cluster 5 were expressed at low levels in ML phase, whereas genes in cluster 14 had very high transcripts in ML phase. Cluster 5 contains genes involved in multiple cellular and metabolic processes, whereas cluster 14 genes are involved predominantly in synthesis of ribosomal proteins. Clusters 12–14 contain genes encoding RNA polymerase subunits (gbs0084, gbs0105, gbs0156/7, gbs0302) that are down regulated from -2.3 to -10 times, which likely indicates a slowing of gene transcription. RpoD (gbs1496, encoding the major σ70) is also down regulated (~-3×). The RpoE subunit (gbs0105) plays a role in the development of sepsis during GBS infection [22], and its down regulation during growth might have consequences for GBS virulence.
S phase related genes
We identified a group of known stress response genes present in clusters 1–4 that were significantly up-regulated in S phase, including hrcA, grpE, dnaK (gbs0094–96), clpE, and clpL (gbs0535 and gbs1376). Transcription of genes putatively involved in the stress response such as Gls24 and universal stress response family proteins gbs1202/1204, gbs1721, and gbs1778 were also elevated in S phase compared to ML phase (Table 1).
Two apparent operons responsible for arginine/ornithine transport and metabolism were also among the group of highly transcribed S phase genes. One operon (gbs2083–2085) encodes an arginine/ornithine antiporter, carbamate kinase, and ornithine carbamoyltransferase, respectively, and is up-regulated 350 to >1,000 times. The second operon (gbs2122–2126) encodes arginine deiminase, a second ornithine carbamoyltransferase, a second arginine/ornithine antiporter, and another carbamate kinase and is up-regulated ~55 to 150 times. Enzymes encoded by genes in these apparent operons are involved in arginine fermentation via the arginine deiminase pathway. They allow GBS to use arginine as an energy source after simple carbohydrates are exhausted from the medium, as would occur during stationary phase. On the other hand, activation of arginine deiminase pathway might have protective function against acidic conditions in a way similar to oral Streptococci [23] as we observed decrease of pH from about 7.9 to 5.5 between ML and S growth phases.
Metabolic changes toward the utilization of increasingly complex nutrient and carbon sources (see below) can be reflected by changes in utilization of simple carbohydrates (drop in the glucose concentration in the medium from ~300 mg/ml in ML to non detectable level in S) and by changes in transcription of the glpKDF (gbs0263–5, +45 to +63 times), a putative operon responsible for glycerol uptake and utilization.
Trends in expression of genes involved in nutrient utilization and energy metabolism
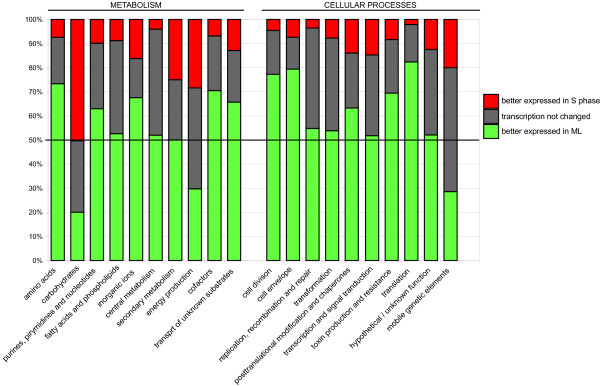
In contrast to genes involved in other aspects of GBS metabolism and physiology, the only genes significantly up-regulated in S phase compared to ML were involved in carbohydrate metabolism (Figure 3). For example, we observed increased levels of certain glycolytic enzymes such as fructose-bisphosphate aldolase (gbs0125), glyceraldehyde 3P-dehydrogenase (gbs1811), phosphoglycerate kinase (gbs1809), enolase (gbs0608), pyruvate dehydrogenase (acoAB), and L-lactate dehydrogenase (gbs0947) (Table 1). This finding is similar to the results reported recently by Chaussee et al [19] showing that transcripts encoding proteins involved in carbohydrate utilization and transport were more abundant in S phase, presumably to maximize carbohydrate utilization. The authors suggested that increased transcription of genes involved in central metabolism and sequential utilization of more complex carbohydrates might be a particularly useful adaptation during infection of tissues where the concentration of carbohydrates is low [19]. In GAS, transcripts of genes involved in transport and metabolism of lactose, sucrose, mannose, and amylase were also more abundant during the stationary phase of growth [19], similar to our findings in GBS (Additional file 2). Similar to links between carbohydrate metabolism and virulence in GAS [21], also carbohydrate metabolism in GBS might be connected to strain invasiveness and strain tissue-disease specificity [24].
Figure 3.
Trends in transcript levels of genes involved in metabolism and cellular processes. 1,994 of GBS transcripts represented on the chip were grouped into functional categories (see Table 1 and Additional file 2). The total number of genes in each category is shown as 100% and the number of transcripts more highly expressed in ML or S phase and transcripts with unchanged expression are presented as a fraction of the 100%.
Changes in expression of regulators and signal transduction systems
TCSs are especially important in the control of global gene expression, especially in the absence of alternative sigma factors. Of the multiple TCSs in GBS, only covR/S (gbs 1671/2) has been well characterized. CovR/S in GBS controls expression of multiple virulence factors, such as hemolysin, CAMP factor, and multiple adhesins [25]. The transcript levels of covR/S are down regulated in S phase, which may be responsible for the observed changes in transcription of virulence factors such as cyl genes encoding hemolysin. However, because the putative effect of CovRS on the camp and cyl genes seems to be opposite to those observed in covRS NEM316 mutant [26] it suggests that these genes are under influence of additional regulators.
Several other GBS genes encoding putative TCSs and regulators had significant changes in transcript levels during the growth phases studied. For example, transcript levels of gbs1908/9 increased 10/14 times between ML and S phases. The GAS homologs (M5005_Spy_0830/1 in strain MGAS5005 and Spy1106/7 in strain SF370) regulate an operon located downstream that encodes NAD-dependent malic enzyme and malate-sodium symport. In a ΔM5005_Spy_0830 deletion strain, the transcript of these downstream genes is decreased 23/40 times [12], indicating positive regulation. Organization of this chromosomal region in GBS is very similar to GAS, and gbs1906 and gbs1907 encode putative homologues to the GAS NAD-dependent malic enzyme and malate-sodium symport proteins, respectively. Genes gbs1906/7 are 63/81 times up-regulated in S phase; therefore this operon appears to be regulated in a similar manner in both GBS and GAS.
The transcript level of another GAS TCS homolog, gbs1934/5, is also elevated. Gbs1934/5 has close identity (~85%) with GAS M5005_Spy_0785/6 (Spy1061/2 in strain SF370), a TCS that has been implicated in the regulation of the mannose/fructose-specific phosphotransferase (PTS) system [12]. Interestingly, in GBS there is also a homolog of this PTS system located directly downstream of gbs1934/5 that is highly up-regulated (46.5 to 468 times) in S phase. Therefore, based on gene position, homology, and transcription regulation patterns, it is reasonable to speculate that these genes function similarly in GBS and GAS.
The possible functions of other TCSs can be inferred from their position. Two sets of TCSs are located directly upstream (gbs2081/2) and downstream (gbs2086/7) of an operon with arginine catabolism genes that are highly up regulated in S phase (see above). The transcript levels of both TCSs change dynamically during growth (Table 1 and Additional file 2). It is probable that genes encoding arginine catabolism proteins might be under tight control of both or either TCS. However, this needs to be confirmed experimentally. Thus, our transcript profiling results are consistent with the hypothesis that in the absence of global response gene regulation medicated by alternative sigma factors, GBS uses multiple TCSs as key mediators regulating the response to changes in the environment (Table 1).
Among putative regulators of unknown function, the highest changes were observed for gbs0191 encoding a transcriptional antiterminator of the BglG family (+50 times, putative CcpA binding site) and gbs0469 (-34 times). Surprisingly, we observed down regulation of expression of other global regulators that are associated with stress and the stringent response to starvation. These include the gene relA (gbs1928) that encodes a putative GTP pyrophosphokinase (-50), codY (gbs1719; -8), the cell density dependent regulator luxS (-3), and the putative mecA (gbs0135) homolog (-20). This result was unexpected given that relA, codY, and luxS are up-regulated in S phase GAS [19].
Transcripts of proven or putative virulence genes
We observed changes in the transcript level of multiple genes encoding proteins with a carboxyterminus cell-wall anchoring motif. The putative location off the proteins on the cell surface suggests that they may play a role in GBS virulence or pathogen-host interaction. Four transcripts were significantly up-regulated in S phase gbs1420 (+6.3), encoding choline-binding protein, gbs1539 (+4.7) and gbs1929 (+5.5) encoding a putative nucleotidase, and gbs1143 (+2.6). We also observed down regulation in S phase of transcripts for several cell wall anchored proteins including a paralog of C5A peptidase precursor gbs0451 (-2), gbs1104 (-6.2), putative adhesin gbs1529 (-11) and fbp (gbs0850, -3), and putative laminin binding proteins (gbs1307, gbs1926; -3). Down regulation in S phase of proteins involved in bacterial attachment is consistent with results reported for GAS [14,15,19]. It is believed that several cell surface proteins are produced during the initial stages of infection to promote adhesion, and later are down-regulated to avoid immune detection.
Other known virulence factors of GBS that showed decreased transcription in S phase included an operon encoding hemolysin (gbs0644–0654), genes encoded on the putative pathogenicity island IX (gbs1061–1076), the putative group B antigen (gbs1478/9, gbs1481, gbs1484/5, gbs1492–1494), and genes involved in capsule synthesis (gbs1233–1247). The putative kinase cpsX (gbs1250) was upregulated 4.4 times (Table 1). Down regulation of capsule and putative and known surface antigens is known to occur in GAS [14,15,19]. For example, capsule, an antiphagocytic factor, is expressed during establishment of GAS infection and is later down-regulated once the infection is established [14,15]. Our results imply a similar scenario could be occurring in GBS. The only transcript encoding a proven virulence factor that was increased in S phase was CAMP factor (+11.6, cfa, gbs2000).
Conclusion
Our results demonstrate that GBS gene transcript levels are highly dynamic throughout the growth cycle in vitro, likely reflecting exposure to an environment that is altering significantly during growth. The organism activates genes involved in metabolism of nutrients and carbon sources other than glucose such as complex carbohydrates and arginine and protect against changing pH. GBS slows down cell division and decreases transcription and translation. Production of virulence factors involved in establishment of the infection is reduced during growth. The global changes of transcript profiles we identified in GBS grown in rich medium are similar to patterns exhibited by GAS. Our results provide new information useful for the study of pathogen-host interactions and gene regulation in pathogenic bacteria.
Authors' contributions
IS performed the research, IS and JMM analyzed the data and wrote the paper.
Supplementary Material
Supplemental table 1- Normalized hybridization values. File contains normalized hybridization values for each array used in the study. ML-mid logarithmic, LL-late logarithmic, ES-early stationary, S-stationary. P-"present" signal (detected in sample), M-"marginal" signal, A-"absent" signal (not detected).
Supplemental table 2 Changes in transcription of 1,994 transcripts present on array (S/ML ratio). Green- genes down regulated in S phase, Red – genes up regulated in S phase, Gray – P values below 0.05.
Contributor Information
Izabela Sitkiewicz, Email: iza.sitkiewicz@gmail.com.
James M Musser, Email: jmmusser@tmhs.org.
Acknowledgements
Authors would like to thank Kathryn Stockbauer for critical reading of the manuscript.
References
- Commichau FM, Forchhammer K, Stulke J. Regulatory links between carbon and nitrogen metabolism. Curr Opin Microbiol. 2006;9:167–172. doi: 10.1016/j.mib.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. Role of RelA of Streptococcus mutans in global control of gene expression. J Bacteriol. 2008;190:28–36. doi: 10.1128/JB.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/S1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couve E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Scott JR, Moran CP Jr. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol Microbiol. 2001;42:495–502. doi: 10.1046/j.1365-2958.2001.02657.x. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett TC, Bugrysheva JV, Scott JR. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J Bacteriol. 2007;189:1866–1873. doi: 10.1128/JB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- Sitkiewicz I, Musser JM. Expression microarray and mouse virulence analysis of four conserved two-component gene regulatory systems in group a streptococcus. Infect Immun. 2006;74:1339–1351. doi: 10.1128/IAI.74.2.1339-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA III, Sumby P, Sitkiewicz I, Granville C, DeLeo FR, Musser JM. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci USA. 2005;102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Barry WT, Gowen BB, Johnson CR, Wright FA, Musser JM. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Gardner DJ, Long RD, Welty DM, Barry WT, Johnson CA, Parkins LD, Wright FA, Musser JM. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri AK, Margarit I, Broenstrup M, Brettoni C, Hua L, Gygi SP, Telford JL, Grandi G, Paoletti LC. Transcriptional and proteomic profiles of group B Streptococcus type V reveal potential adherence proteins associated with high-level invasion. Infect Immun. 2007;75:1473–1483. doi: 10.1128/IAI.00638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe GP. Streptococcus agalactiae mastitis: a review. Can Vet J. 1997;38:429–437. [PMC free article] [PubMed] [Google Scholar]
- Muller AE, Oostvogel PM, Steegers EA, Dorr PJ. Morbidity related to maternal group B streptococcal infections. Acta Obstet Gynecol Scand. 2006;85:1027–1037. doi: 10.1080/00016340600780508. [DOI] [PubMed] [Google Scholar]
- Chaussee MA, Dmitriev AV, Callegari EA, Chaussee MS. Growth phase-associated changes in the transcriptome and proteome of Streptococcus pyogenes. Arch Microbiol. 2008;189:27–41. doi: 10.1007/s00203-007-0290-1. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Shelburne SA III, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AL, Needham RH, Rubens CE. The Delta subunit of RNA polymerase is required for virulence of Streptococcus agalactiae. Infect Immun. 2003;71:4011–4017. doi: 10.1128/IAI.71.7.4011-4017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivey RG, Kuhnert WL, Hahn K. Genetics of acid adaptation in oral streptococci. Crit Rev Oral Biol Med. 2001;12:301–314. doi: 10.1177/10454411010120040201. [DOI] [PubMed] [Google Scholar]
- Domelier AS, van dM-M, Grandet A, Mereghetti L, Rosenau A, Quentin R. Loss of catabolic function in Streptococcus agalactiae strains and its association with neonatal meningitis. J Clin Microbiol. 2006;44:3245–3250. doi: 10.1128/JCM.02550-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy MC, Zouine M, Fert J, Vergassola M, Couve E, Pellegrini E, Glaser P, Kunst F, Msadek T, Trieu-Cuot P, Poyart C. CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol Microbiol. 2004;54:1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- Jiang SM, Ishmael N, Hotopp JD, Puliti M, Tissi L, Kumar N, Cieslewicz MJ, Tettelin H, Wessels MR. Variation in the group B Streptococcus CsrRS regulon and effects on pathogenicity. J Bacteriol. 2008;190:1956–1965. doi: 10.1128/JB.01677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table 1- Normalized hybridization values. File contains normalized hybridization values for each array used in the study. ML-mid logarithmic, LL-late logarithmic, ES-early stationary, S-stationary. P-"present" signal (detected in sample), M-"marginal" signal, A-"absent" signal (not detected).
Supplemental table 2 Changes in transcription of 1,994 transcripts present on array (S/ML ratio). Green- genes down regulated in S phase, Red – genes up regulated in S phase, Gray – P values below 0.05.