Abstract
The malarial parasite Plasmodium falciparum possesses a functional thioredoxin and glutathione system comprising the dithiol-containing redox proteins thioredoxin (Trx) and glutaredoxin (Grx), as well as plasmoredoxin (Plrx), which is exclusively found in Plasmodium species. All three proteins belong to the thioredoxin superfamily and share a conserved Cys-X-X-Cys motif at the active site. Only a few of their target proteins, which are likely to be involved in redox reactions, are currently known. The aim of the present study was to extend our knowledge of the Trx-, Grx-, and Plrx-interactome in Plasmodium. Based on the reaction mechanism, we generated active site mutants of Trx and Grx lacking the resolving cysteine residue. These mutants were bound to affinity columns to trap target proteins from P. falciparum cell extracts after formation of intermolecular disulfide bonds. Covalently linked proteins were eluted with dithiothreitol and analyzed by mass spectrometry. For Trx and Grx, we were able to isolate 17 putatively redox-regulated proteins each. Furthermore, the approach was successfully established for Plrx, leading to the identification of 21 potential target proteins. In addition to confirming known interaction partners, we captured potential target proteins involved in various processes including protein biosynthesis, energy metabolism, and signal transduction. The identification of three enzymes involved in S-adenosylmethionine (SAM) metabolism furthermore suggests that redox control is required to balance the metabolic fluxes of SAM between methyl-group transfer reactions and polyamine synthesis. To substantiate our data, the binding of the redoxins to S-adenosyl-L-homocysteine hydrolase and ornithine aminotransferase (OAT) were verified using BIAcore surface plasmon resonance. In enzymatic assays, Trx was furthermore shown to enhance the activity of OAT. Our approach led to the discovery of several putatively redox-regulated proteins, thereby contributing to our understanding of the redox interactome in malarial parasites.
Author Summary
Protection from oxidative stress and efficient redox regulation are essential for malarial parasites which have to grow and multiply rapidly in various environments. As shown by glucose-6 phosphate dehydrogenase deficiency, a genetic variation protecting from malaria, the parasite–host cell unit is very susceptible to disturbances in redox equilibrium. This is the major reason why redox active proteins of Plasmodium currently belong to the most attractive antimalarial drug targets. The dithiol-containing redox proteins thioredoxin (Trx) and glutaredoxin (Grx), as well as plasmoredoxin (Plrx), which is exclusively found in Plasmodium species, represent central players in the redox network of malarial parasites. To extend our knowledge of interacting partners and the functions of these proteins, we carried out pull-down assays with immobilized active site mutants of Trx, Grx, and Plrx and whole cell parasite lysate. After elution of bound proteins and mass spectrometric identification, about 20 interacting partners were identified for each of the redox proteins. Data was supported using BIAcore surface plasmon resonance. The identified interacting proteins, which are likely to be redox-regulated, are involved in important cellular processes including protein biosynthesis, energy metabolism, polyamine synthesis, and signal transduction. Our results contribute to our understanding of the redox interactome in malarial parasites.
Introduction
The malarial parasite Plasmodium falciparum (Pf) possesses two major functional redox systems: The thioredoxin system [1],[2] comprising NADPH, thioredoxin reductase, thioredoxin (Trx) [2],[3], and thioredoxin-dependent peroxiredoxins [4]–[8], and a glutathione system comprising NADPH, glutathione reductase [9], glutathione, glutathione S-transferase [10],[11], glutaredoxin (Grx) [12], and the glyoxalases I and II [13],[14].
Dithiol Trx and Grx belong to the thioredoxin superfamily whose members characteristically share the ‘thioredoxin-fold’ consisting of a central four-stranded β-sheet surrounded by α-helices [15], and an active site with two conserved cysteine residues that specify the biological activity of the protein. Besides Trx with the classical active site sequence CGPC and Grx possessing a CPYC-motif, also tryparedoxin, protein disulfide isomerase, glutathione peroxidase, glutathione S-transferase, and DsbA (a disulfide bond forming protein of bacteria) belong to the thioredoxin superfamily [15]–[17]. In addition, a redox-active protein named plasmoredoxin (Plrx), which is unique and highly conserved among Plasmodium species, has been identified and analyzed by Becker et al. [18]. Homology modeling of Plrx indicated a characteristic thioredoxin fold including the active site sequence WCKYC which results in assigning also Plrx to the thioredoxin superfamily [18]. The gene encoding Plrx was found to be non-essential for Plasmodium berghei and Plrx knock out parasites did not reveal a significant phenotype throughout the complete life-cycle [19].
The regulation of a number of phenomena in the cell has been linked to the reversible conversion of disulfides to dithiols thereby modulating the activities of the respective proteins [20]. Several recent articles have described the identification of Trx- and Grx-interacting proteins in plants and other organisms. The putative target proteins are involved in many processes, including oxidative stress response (e.g. peroxiredoxins (Prxs), ascorbate peroxidase, catalase), nitrogen, sulfur, and carbon metabolisms (e.g. S-adenosylmethionine synthetase, S-adenosyl-L-homocysteine hydrolase, phosphoglycerate kinase), protein biosynthesis (e.g. several elongation factors), and protein folding (e.g. heat shock proteins, protein disulfide isomerase) [21]–[26].
For Plasmodium, it has been shown that Trx as well as Grx and Plrx may operate as dithiol reductants on ribonucleotide reductase in vitro [1]. All three disulfide oxidoreductases are probably involved in redox regulation and/or antioxidant defense of the parasite: Trx serves as an electron donor for Plrx, oxidized glutathione disulfide and Prxs in vitro [1],[27],[28]. For example, the cytosolic 2-Cys peroxiredoxin of Plasmodium involved in the detoxification of reactive oxygen and nitrogen species and has been shown to be also fueled by plasmoredoxin [29]. Another peroxiredoxin, the so-called antioxidant protein, represents a further electron acceptor of Grx and Plrx in vitro [28].
The reduction of protein disulfides by Trx, Grx and Plrx is based on a dithiol exchange mechanism. The N-terminal cysteine residue of the Cys-X-X-Cys motif initiates a nucleophilic attack on the disulfide target resulting in the formation of a mixed disulfide. The intermolecular disulfide bond is subsequently cleaved by the C-terminal resolving cysteine residue of the active site motif, yielding reduced substrate and the oxidoreductase disulfide [20],[30].
Here we present redox-affinity chromatographical studies in order to gain further insight into the interactome of Plasmodium Trx, Grx, and Plrx by capturing potential target proteins. Our approach led to the identification of around 20 binding partners for each of the proteins applied. Furthermore, the interaction of S-adenosyl-L-homocysteine hydrolase (SAHH) and ornithine aminotransferase (OAT) with the redoxins was studied in more detail by surface plasmon resonance experiments.
Results
Identification of potential thioredoxin target proteins in P. falciparum
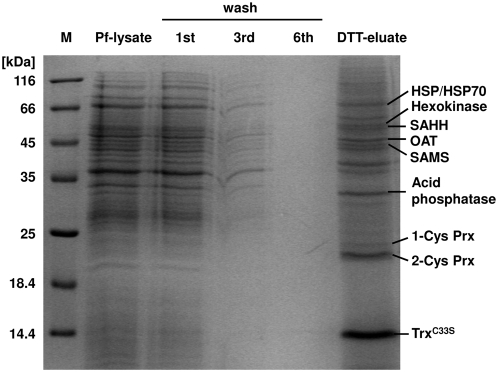
Based on the dithiol exchange mechanism catalysed by Trx, we generated the active site cysteine mutant TrxC33S (Table S1) which is able to catch target proteins as a mixed disulfide intermediate. This method is well established for Trx [31] and has been applied successfully to a whole range of organisms [21],[24],[32],[33]. By using the immobilized mutant as bait, we successfully captured potential target proteins from trophozoite stage P. falciparum cell lysate as shown in Figure 1. Elution of interacting proteins with DTT was carried out after extensive washing with NaCl-containing buffer (see wash in Figure 1) in order to remove unspecifically bound proteins and to increase specificity of the eluate fraction. (For direct comparison of the DTT eluates from Trx, Grx, and Plrx pull downs, please see Figure S1). When comparing the P. falciparum cell lysate fraction on the gel with the one of the eluate, it became evident that this approach facilitates an enrichment of potential target proteins on the column (e.g. see Prxs bands). The strong band for TrxC33S in the eluate is likely to reflect the cleavage of dimeric bait protein (TrxC33S-SS-TrxC33S) and is not a result of inefficient washing. Captured proteins were analyzed by peptide mass fingerprinting with MALDI-MS which enabled us to identify 17 putative Trx-linked proteins that are summarized in Table 1, [34].
Figure 1. SDS-PAGE profile of the captured proteins by Trx-affinity chromatography.
The thioredoxin mutant TrxC33S was immobilized on CNBr-activated Sepharose 4B resin before incubating the column with 7–10 mg of Plasmodium falciparum cell lysate and extensive washing steps with NaCl-containing buffer. Target proteins were eluted with 10 mM DTT. The obtained protein samples were separated on a 12% polyacrylamide gel, and protein bands were identified after tryptic digestion by MALDI-TOF analysis. Some prominent proteins are indicated in the figure. For a complete list of protein bands that could be reproducibly and unambiguously assigned, please see Table 1.
Table 1. Potential thioredoxin target proteins identified in Plasmodium falciparum.
| Protein Name | Accession Number | Protein MW [kDa] | Peak Time Expressiona [Hours] | Predicted Localisation, Expression Levelsb | Protein Isoelectric Point | Protein Coverage [%] | Masses Matched | |
| Swiss-Prot/GenBank | PlasmoDB | |||||||
| Plasmoredoxin (Thioredoxin-like redox-active protein) | Q8I224 | PFC0166w | 21.7 | 16 | c, ++ | 8.94 | 15 | 3 |
| Human peroxiredoxin 2 (Prx 2) | P32119 | 21.8 | — | c | 5.67 | 19 | 4 | |
| 2-Cys peroxiredoxin (2-Cys-Prx) | Q8IL80 | PF14_0368 | 21.8 | 11 | c, +++ | 6.65 | 44 | 5 |
| 1-Cys peroxiredoxin (1-Cys-Prx) | Q8IAM2 | PF08_0131 | 25.2 | 25 | c, +++ | 6.31 | 29 | 6 |
| GTPase, putative | Q8IDL8 | MAL13P1.241 | 26.1 | 36 | c, + | 5.55 | 15 | 3 |
| 14-3-3 protein homologue, putative | Q8IB17 | MAL8P1.69 | 29.5 | 34 | c, +++ | 4.96 | 23 | 4 |
| Acid phosphatase, putative | Q8IM55 | PF14_0036 | 35.8 | 37 | c, ++ | 5.67 | 33 | 9 |
| S-adenosylmethionine synthetase | Q7K6A4 | PFI1090w | 44.8 | 30 | c, +++ | 6.28 | 21 | 6 |
| Ornithine aminotransferase | Q6LFH8 | PFF0435w | 46.1 | 18 | c, +++ | 6.47 | 16 | 5 |
| HAP protein/Plasmepsin III | Q8IM15 | PF14_0078 | 51.7 | 48 | tm (api), +++ | 8.04 | 16 | 8 |
| S-adenosyl-L-homocysteine hydrolase | Q7K6A6 | PFE1050w | 53.8 | 30 | c, +++ | 5.64 | 8 | 3 |
| Hexokinase | Q6LF74 | PFF1155w | 55.3 | 12 | c, +++ | 6.72 | 8 | 3 |
| Pyruvate kinase, putative | Q6LF06 | PFF1300w | 55.7 | 22 | c, +++ | 7.50 | 17 | 5 |
| Fork head domain protein, putative | Q8IEN7 | PF13_0042 | 68.3 | ? | c, (++) | 9.04 | 8 | 4 |
| Heat shock protein | Q8I2X4 | PFI0875w | 72.4 | 33 | sp, +++ | 5.18 | 17 | 8 |
| Heat shock 70 kDa protein | Q8IB24 | PF08_0054 | 73.9 | 01 | c, +++ | 5.50 | 11 | 6 |
| Heat shock protein 86 | Q8IC05 | PF07_0029 | 86.2 | 17 | c, +(+) | 4.94 | 14 | 7 |
Data depicted from PlasmoDB and Ginsburg, Hagai. “Malaria Parasite Metabolic Pathways” (http://sites.huji.ac.il/malaria/), hours represent the 48-hour red blood cycle.
Data depicted from PlasmoDB. If no target sequence or localization signal is predicted, the respective proteins are given as cytosolic (c).
peptide.
tm, predicted transmembrane domain; sp, predicted signal peptide; +++, expression >75%; ++, expression 50%–75%; +, expression less than 50%. Data from PlasmoDB [34].
Identification of potential glutaredoxin target proteins in P. falciparum
The method described above for Trx was also used for the identification of Grx-interacting proteins. The principle of this approach has been previously established by Rouhier et al., 2005, for plant Grx [25]. Grx was mutated (GrxC32S; Table S1) and the mutant was coupled to CNBr-activated Sepharose. We were able to identify 17 target candidates for Grx which are presented in Table 2. These proteins overlapped only partially with the proteins captured by Trx (please compare Tables 1 and 2). For example, the two glycolytic enzymes hexokinase and a putative pyruvate kinase were identified both with Grx and Trx. L-lactate dehydrogenase, however, reacted only with Grx and glyceraldehyde-3-phosphate dehydrogenase only with Grx and Plrx. Several heat shock proteins as well as enzymes involved in SAM metabolism were captured with all three bait proteins. Two ribosomal proteins involved in protein translation were found to interact exclusively with Grx or with Grx and Plrx, respectively. Interestingly, plasmoredoxin was verified as Grx- (as well as Trx-) electron acceptor which had been suggested by former in vitro studies [18]. A putative phosphoethanolamine N-methyltransferase was furthermore identified as a Grx-specific target.
Table 2. Potential glutaredoxin target proteins identified in Plasmodium falciparum.
| Protein Name | Accession Number | Protein MW [kDa] | Peak Time Expressiona [Hours] | Predicted Localization, Expression Levelsb | Protein Isoelectric Point | Protein Coverage [%] | Masses Matched | |
| Swiss-Prot | PlasmoDB | |||||||
| 40S ribosomal protein S12 | O97249 | PFC0295C | 15.4 | 11 | c, ++ | 4.91 | 23 | 3 |
| Ribosomal protein S19s, putative | Q8IFP2 | PFD1055w | 19.7 | 9 | c, +++ | 10.17 | 34 | 6 |
| Plasmoredoxin (Thioredoxin-like redox-active protein) | Q8I224 | PFC0166w | 21.7 | 16 | c, ++ | 8.94 | 20 | 4 |
| 14-3-3 protein homologue, putative | Q8IB17 | MAL8P1.69 | 29.5 | 34 | c, +++ | 4.96 | 20 | 4 |
| Phosphoethanolamine N-methyltransferase, putative | Q8IDQ9 | MAL13P1.214 | 31.0 | 26 | c, +++ | 5.43 | 23 | 4 |
| L-lactate dehydrogenase | Q76NM3 | PF13_0141 | 34.1 | 21 | sp (tm), +++ | 7.12 | 28 | 7 |
| Guanine nucleotide-binding protein, putative/receptor for activated C kinase homolog | Q8IBA0 | PF08_0019 | 35.7 | 11 | c, +++ | 6.24 | 25 | 4 |
| Glyceraldehyde-3-phosphate dehydrogenase | Q8IKK7 | PF14_0598 | 36.6 | 12 | c, +++ | 7.59 | 21 | 5 |
| S-adenosylmethionine synthetase | Q7K6A4 | PFI1090w | 44.8 | 30 | c, +++ | 6.28 | 9 | 3 |
| Ornithine aminotransferase | Q6LFH8 | PFF0435w | 46.1 | 18 | c, +++ | 6.47 | 14 | 6 |
| S-adenosyl-L-homocysteine hydrolase | Q7K6A6 | PFE1050w | 53.8 | 30 | c, +++ | 5.64 | 14 | 4 |
| Hexokinase | Q6LF74 | PFF1155w | 55.3 | 12 | c, +++ | 6.72 | 11 | 4 |
| Pyruvate kinase, putative | Q6LF06 | PFF1300w | 55.7 | 22 | c, +++ | 7.50 | 36 | 12 |
| Heat shock protein | Q8I2X4 | PFI0875w | 72.4 | 33 | sp, +++ | 5.18 | 11 | 6 |
| Heat shock 70 kDa protein | Q8IB24 | PF08_0054 | 73.9 | 01 | c, +++ | 5.50 | 17 | 8 |
| Heat shock protein 86 | Q8IC05 | PF07_0029 | 86.2 | 17 | c, +(+) | 4.94 | 20 | 14 |
| Elongation factor 2 | Q8IKW5 | PF14_0486 | 93.5 | 13 | c, +++ | 6.35 | 17 | 11 |
Data depicted from PlasmoDB and Ginsburg, Hagai. “Malaria Parasite Metabolic Pathways” (http://sites.huji.ac.il/malaria/), hours represent the 48-hour red blood cycle.
Data depicted from PlasmoDB. If no target sequence or localization signal is predicted, the respective proteins are given as cytosolic (c).
tm, predicted transmembrane domain; sp, predicted signal peptide; +++, expression >75%; ++, expression 50%–75%; +, expression less than 50%. Data from PlasmoDB [34].
Identification of potential plasmoredoxin target proteins in P. falciparum
Plrx was first described in 2003 by Becker et al. [18] and is unique for and highly conserved among Plasmodium species. So far, the physiological function of Plrx is not completely understood, and the protein is not essential for the survival of the parasite as shown very recently by Buchholz and co-workers [19]. To gain further insight, we generated an active site mutant (PlrxC63S; Table S1) in analogy to Trx and Grx. The mutant was then used for affinity chromatography resulting in 21 proteins which are potential electron acceptors of Plrx (Table 3). Among several overlapping proteins also captured with Trx or Grx, Plrx was shown to specifically interact with the putative co-chaperone GrpE and a putative disulfide isomerase, both assisting in protein folding, as well as with a putative acyl carrier protein and enzymes involved in DNA synthesis, -repair, and signal transduction.
Table 3. Potential plasmoredoxin target proteins identified in Plasmodium falciparum.
| Protein Name | Accession Number | Protein MW [kDa] | Peak Time Expressiona [Hours] | Predicted Localization, Expression Levelsb | Protein Isoelectric Point | Protein Coverage [%] | Masses Matched | |
| Swiss-Prot/ GenBank | PlasmoDB | |||||||
| 40S ribosomal protein S12 | O97249 | PFC0295C | 15.4 | 11 | c, ++ | 4.91 | 23 | 3 |
| Acyl carrier protein, putative | Q7KWJ1 | PFB0385w | 15.8 | 23 | sp (api), ++ | 8.87 | 19 | 3 |
| 14-3-3 protein homologue, putative | Q8IB17 | MAL8P1.69 | 29.5 | 34 | c, +++ | 4.96 | 20 | 4 |
| Co-chaperone GrpE, putative | Q8IIB6 | PF11_0258 | 34.5 | 19 | n, ++ | 8.8 | 20 | 4 |
| 14-3-3 Protein, putative | Q8ID86 | MAL13P1.309 | 35.1 | 01 | tm, + | 7.08 | 17 | 3 |
| Glyceraldehyde-3-phosphate dehydrogenase | Q8IKK7 | PF14_0598 | 36.6 | 12 | c, +++ | 7.59 | 30 | 7 |
| Hypothetical protein | Q8IJX6 | PF10_0065 | 37.8 | c, + | 8.67 | 16 | 4 | |
| Ribonucleotide reductase small subunit (R2) | Q8IM38 | PF14_0053 | 40.6 | 33 | c, ++ | 5.37 | 14 | 4 |
| S-adenosylmethionine synthetase | Q7K6A4 | PFI1090w | 44.8 | 30 | c, +++ | 6.28 | 20 | 5 |
| Conserved GTP-binding protein, putative | Q8IBM9 | MAL7P1.122 | 45.2 | 11 | c, ++ | 6.88 | 35 | 9 |
| Ornithine aminotransferase | Q6LFH8 | PFF0435w | 46.1 | 18 | c, +++ | 6.47 | 22 | 8 |
| Endonuclease iii homologue, putative | Q6LFC2 | PFF0715c | 49.2 | 26 | c, + | 9.37 | 11 | 4 |
| HAP protein/Plasmepsin III | Q8IM15 | PF14_0078 | 51.7 | 48 | tm (api), +++ | 8.04 | 18 | 8 |
| Hypothetical protein | Q8ILQ4 | PF14_0190 | 54.7 | c, ++ | 5.53 | 9 | 3 | |
| Hexokinase | Q6LF74 | PFF1155w | 55.3 | 12 | c, +++ | 6.72 | 11 | 4 |
| Disulfide isomerase, putative | Q8I6S6 | MAL8P1.17 | 55.5 | 31 | sp (tm),++ | 5.56 | 9 | 3 |
| Pyruvate kinase, putative | Q6LF06 | PFF1300w | 55.7 | 22 | c, +++ | 7.5 | 28 | 9 |
| Heat shock protein | Q8I2X4 | PFI0875w | 72.4 | 33 | sp, +++ | 5.18 | 9 | 6 |
| Heat shock 70 kDa protein | Q8IB24 | PF08_0054 | 73.9 | 01 | c, +++ | 5.5 | 28 | 12 |
| Heat shock protein 86 | Q8IC05 | PF07_0029 | 86.2 | 17 | c, +(+) | 4.94 | 13 | 8 |
| Elongation factor 2 | Q8IKW5 | PF14_0486 | 93.5 | 13 | c, +++ | 6.35 | 10 | 5 |
Data depicted from PlasmoDB and Ginsburg, Hagai. “Malaria Parasite Metabolic Pathways” http://sites.huji.ac.il/malaria/, hours represent the 48-hour red blood cycle.
Data depicted from PlasmoDB. If no target sequence or localization signal is predicted, the respective proteins are given as cytosolic (c).
tm, predicted transmembrane domain; sp, predicted signal peptide; n, predicted nuclear localization with mitochondrial signal sequence, +++, expression >75%; ++, expression 50%–75%; +, expression less than 50%. Data from PlasmoDB [34].
BIAcore interaction analysis of S-adenosyl-L-homocysteine hydrolase with Trx, Grx, and Plrx
To further investigate the interaction between the redoxins and newly captured proteins we cloned, overexpressed and purified S-adenosyl-L-homocysteine hydrolase and ornithine aminotransferase. These two proteins were chosen since they both are involved in SAM metabolism and they could be produced without major problems in recombinant form. In a first step the interaction of SAHH, which was trapped with the Trx and Grx affinity column, and OAT (see below) with the three redox-active proteins was investigated by BIAcore surface plasmon resonance (SPR) analyses. Oxidized SAHH was attached covalently to the sensor chip surface, and various forms of the redox-active proteins, including wild type and active site mutants lacking one or two cysteines, were used as analytes.
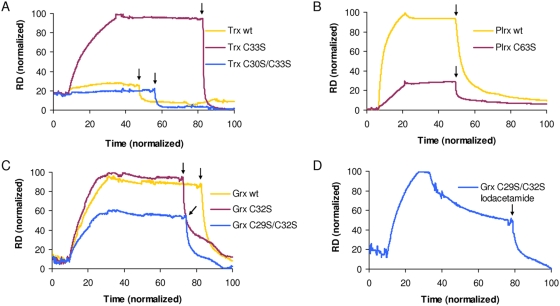
Figure 2A shows the sensorgram of Trx binding to SAHH. TrxC33S, the mutant used in the pull-down assay, associated strongly with SAHH, and the binding was not affected during buffer flow over the chip which is indicative for a covalent interaction. The fact that the protein complex could efficiently be dissociated by DTT confirms the existence of a disulfide bond between the two proteins. In contrast, wild type Trx as well as TrxC30S/C33S which lack both active site cysteines caused no obvious increase in resonance units.
Figure 2. SPR analysis of S-adenosyl-L-homocysteine hydrolase (SAHH) with Trx, Grx, and Plrx.
SAHH on the sensor surface was oxidized with 30 µl of 0.5 mM 5,5′-dithiobis(2-nitrobenzoate) (DTNB) (not shown). Then, 30 µl of the analyte (10 µM in HBS buffer) was injected, followed by buffer flow over the chip. The dissociation phase was initiated by addition of 30 µl of a 2 mM DTT solution as indicated by an arrow. The BIAcore sensorgrams show the response difference (RD) between SAHH-loaded flow chamber 2 and control flow chamber 1 (FC2–FC1). (A–C) Interaction of SAHH with wild type and active site mutants of Trx, Grx, and Plrx. In order to analyze a non-covalent interaction of Grx with SAHH, the GrxC29S/C32S mutant was treated with iodoacetamide prior to SPR analysis (D).
For Plrx, the interaction of the wild type protein with SAHH was much stronger as for the single cysteine mutant. Both forms, however, bound stable to SAHH and dissociated upon addition of DTT (Figure 2B).
The sensorgram in Figure 2C shows the interaction of SAHH with Grx, which was comparable for the wild type protein and the cysteine mutant GrxC32S. Interestingly, also the double mutant of Grx was able to form a covalent complex with SAHH, the interaction, however, was not as intense as for the two other variants. Grx possesses, apart from the two active site cysteines, one additional cysteine in its sequence (Cys 88). In order to verify if the observed interaction is due to a disulfide bridge between residue Cys 88 and SAHH, the double mutant was treated with iodoacetamide resulting in an alkylation of the thiol group. Indeed, this Grx form with a modified Cys 88 reacted with SAHH in a mainly non-covalent manner as shown in Figure 2D. Only a part of the Grx could be eluted with DTT which is presumably due to incomplete alkylation.
Interaction between ornithine amino transferase and thioredoxin
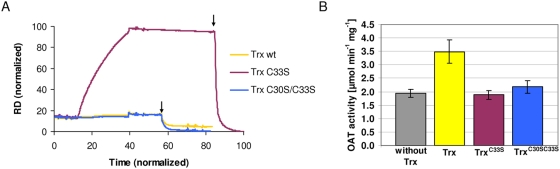
As a second protein captured by the redoxins in the pull down assays, we studied ornithine aminotransferase (OAT) in more detail. Here we focused on SPR and enzyme kinetic analyses with Trx. As described above for the SAHH SPR experiments, OAT was attached covalently to the sensor chip surface and wild type Trx as well as active site single and double mutants, lacking one or two cysteines, were used as analytes. Figure 3A shows the sensorgram of Trx binding to OAT. TrxC33S, the mutant used in the pull-down assay, associated strongly with OAT. This interaction could not be disturbed by washing but could be specifically solved by DTT indicating a disulfide bond formation between OAT and the Trx mutant. Again, as observed for SAHH, wild type Trx as well as TrxC30S/C33S which lacks both active site cysteines showed no obvious interaction.
Figure 3. SPR analysis and specific activity of ornithine aminotransferase (OAT) in the presence of Trx.
(A) SPR analysis of OAT with Trx. The experimental setup was chosen as described in the Figure 2 caption. A strong and specific interaction can be observed with the C33S mutant, whereas wild type Trx and the double mutant C30S/C33S show hardly any interaction. (B) Specific activity of PfOAT in the presence of equimolar concentrations of Trx, TrxC33S, and TrxC30S/C33S.
The biological significance of the interaction between Trx and OAT was further studied in enzymatic assays (Figure 3B). Interestingly, the addition of equimolar concentrations of wild type thioredoxin to the assay resulted in 75% increase in OAT activity. In contrast, the Trx single and double mutants did not induce activity changes which indicates that an intact active site with two cysteine residues is required for the activation of OAT.
Discussion
In the present study, we intended to identify novel target proteins of P. falciparum Trx, Grx, and Plrx in order to extend our knowledge of their physiological functions. An affinity chromatography approach using active site cysteine mutants as bait had already been described for Trx [21], [24], [31]–[33] and Grx [25] from other organisms. Here, we applied this strategy successfully to P. falciparum Trx and Grx as well as to another member of the thioredoxin superfamily, namely Plrx. For Trx and Grx, 17 (partially overlapping) target candidates were detected each (Tables 1 and 2). Most of these proteins had not previously been associated with redox-mediated regulation in P. falciparum. Furthermore, our approach represents the first interactome study carried out with a plasmoredoxin. For Plrx we identified 21 putative target proteins (Table 3). Taken together 33 different putative interacting proteins were identified with these three redoxins, a number which fits to results obtained on other organisms with Trx [31] (Table 1). All target proteins identified are present in asexual red blood cell stages of the parasites (PlasmoDB, see Tables 1– 3) with most proteins predicted to be cytosolic. The same cytosolic localization is predicted for our three bait redoxins.
The different target proteins found are involved in various metabolic processes that can be clustered as summarized in Table 4. The physiological relevance of the observed interactions between the target proteins and our redox-active proteins will be studied in further detail. For S-adenosyl-L-homocysteine hydrolase, a central enzyme of S-adenosylmethionine metabolism, and ornithine aminotransferase the interaction was verified exemplarily and studied in more detail using BIAcore surface plasmon resonance (SPR) analyses. For OAT also activity assays were carried out to show the influence of thioredoxin on the physiological reaction of the enzyme (see below).
Table 4. Functional clusters of PfTrx1, PfGrx1, and PfPlrx target protein candidates captured in the present study.
| Anti-Oxidative Stress System | |||
| Trx | 2-Cys peroxiredoxin (2-Cys-Prx, cytosolic) | ||
| Trx | 1-Cys peroxiredoxin (1-Cys-Prx, cytosolic) | ||
| Trx | Human peroxiredoxin 2 | ||
| Trx | Grx | Plasmoredoxin (Thioredoxin-like redox-active protein) | |
| Transcription/Translation | |||
| Trx | Fork head domain protein, putative | ||
| Grx | Plrx | Elongation factor 2 | |
| Grx | Plrx | 40S ribosomal protein S12 | |
| Grx | Ribosomal protein S19s, putative | ||
| Protein Folding | |||
| Trx | Grx | Plrx | Heat shock protein |
| Trx | Grx | Plrx | Heat shock 70 kDa protein |
| Trx | Grx | Plrx | Heat shock protein 86 |
| Plrx | Co-chaperone GrpE, putative | ||
| Plrx | Disulfide isomerase, putative | ||
| Carbohydrate Metabolism | |||
| Trx | Grx | Plrx | Hexokinase |
| Trx | Grx | Plrx | Pyruvate kinase, putative |
| Grx | Plrx | Glyceraldehyde-3-phosphate dehydrogenase | |
| Grx | L-lactate dehydrogenase | ||
| SAM Metabolism | |||
| Trx | Grx | Plrx | Ornithine aminotransferase |
| Trx | Grx | Plrx | S-adenosylmethionine synthetase |
| Trx | Grx | S-adenosyl-L-homocysteine hydrolase | |
| Lipid Metabolism | |||
| Grx | Phosphoethanolamine N-methyltransferase, putative | ||
| Plrx | Acyl carrier protein, putative | ||
| Hemoglobin Catabolism | |||
| Trx | Plrx | HAP protein/Plasmepsin III | |
| DNA Synthesis and Repair | |||
| Plrx | Endonuclease iii homologue, putative | ||
| Plrx | Ribonucleotide reductase small subunit | ||
| Signal Transduction | |||
| Trx | Grx | Plrx | 14-3-3 protein homologue, putative |
| Plrx | 14-3-3 protein, putative | ||
| Trx | GTPase, putative | ||
| Grx | Guanine nucleotide-binding protein, putative/receptor for activated C kinase homolog, PfRACK | ||
| Plrx | Conserved GTP-binding protein, putative | ||
| Others | |||
| Trx | Acid phosphatase, putative | ||
| Plrx | Hypothetical protein | ||
| Plrx | Hypothetical protein | ||
Antioxidant enzymes and redox control
Peroxiredoxins, repeatedly identified as target proteins in our study, represent central antioxidant and redox-regulatory proteins in Plasmodium. This notion is underlined by the fact that malarial parasites possess neither catalase nor a classical glutathione peroxidase [1]. Many Prxs are present at high intracellular concentrations and are therefore detected in almost every proteomic analysis. From the four P. falciparum Prxs both cytosolic proteins, 2-Cys-Prx and 1-Cys-Prx, were identified in our study using Trx as a bait. On the other hand, no peroxiredoxin was captured with Grx or Plrx (Tables 2 and 3). Since cytosolic 2-Cys-Prx and 1-Cys-Prx are both highly abundant redox proteins, the results suggest an excellent specificity of our method (e.g. with Grx and Plrx as negative controls for potential interacting partners of Trx). Our data support former in vitro studies indicating that Trx is the preferred physiological electron donor of these Plasmodium peroxiredoxins having distinct antioxidant and regulatory functions in vivo [1],[28],[35],[36]. The other two Prxs from P. falciparum as well as a glutathione peroxidase-like enzyme, were not found in our study. This might be due to the lower protein abundance or due to the subcellular localization resulting in different substrate specificities (although the three enzymes were shown to accept electrons from at least one of the bait proteins in vitro) [28], [35]–[37]. Detection of an erythrocytic Prx in the pull-down with Trx (Table 1) might indicate a contamination during lysate preparation. However, very recently we showed that P. falciparum can import the human peroxiredoxin 2 into its cytosol, suggesting that the parasite exploits this antioxidant system of its host (Koncarevic et al., under revision). Non-recombinant plasmoredoxin was captured from parasite lysates with Trx and Grx (Tables 1 and 2). Also this interaction had been described before by biochemical assays and was now verified [18]. It is noteworthy that no other member of the thioredoxin superfamily was found as a target of plasmoredoxin although P. falciparum has many Trx- and Grx-like proteins [1],[28]. One might therefore speculate that Plrx is involved in the cross talk of the thioredoxin and the glutaredoxin system.
Protein biosynthesis
Based on our results, some components of the translational machinery seem to be redox-regulated in malarial parasites. Two ribosomal proteins and the elongation factor 2 were identified as redox sensitive targets in Plasmodium (Table 4) and had also been described as Trx or Grx targets in other organisms [21]–[23],[25],[26],[32]. The redox sensitivity of elongation factor 2 has been confirmed in vitro and in vivo. Analyses of the effect of oxidative stress on protein synthesis indicated that elongation factor 2, the main protein involved in the elongation step, is oxidatively modified resulting in lower amounts of active protein [38]. Besides, several redox-dependent chaperones were identified in our work (Table 4). Hsp70 had already been linked to Trx and Grx in plants, Chlamydomonas and Synechocystis [21]–[23],[25],[26],[33]. A recent study demonstrated the formation of a complex between an Arabidopsis Trx-like protein and yeast Hsp70 that is released under oxidative stress. Cysteine 20 which is conserved in virtually all the Hsp70 chaperones has been suggested as target of redox regulation [39]. The existence of a redox-regulated molecular chaperone network has been described by Hoffmann and coworkers focusing on Hsp33 which is reduced in vivo by the glutaredoxin and thioredoxin systems [40]. The present work adds the Plasmodium protein disulfide isomerase to the list of known redox-dependent components involved in protein folding [22],[25],[26]. Thioredoxin was reported to regulate translation via an interaction with a protein disulfide isomerase, namely RB60, in the green algae Chlamydomonas [41].
Carbohydrate metabolism
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was found to interact with Grx and Plrx in our study, had been described previously as a protein undergoing thiol/disulfide redox status changes which affect its enzymatic activity. Molecular modeling studies of plant GAPDH indicated a disulfide bond between an N-terminal and C-terminal cysteine being involved in redox regulation, which alters the geometry of the active site [42],[43]. Two of eight cysteines of PfGAPDH are strictly conserved among prokaryotes and eukaryotes: Cys 153 which is directly involved in catalysis, and the near-neighbor Cys 157 which is located on the same helical segment at a distance of 8.6 Å (PDB entry 2B4T; [44]). Indeed, Cys 149 of rabbit muscle GAPDH has been identified as target for the oxidation by peroxynitrite [45]. A reversible disulfide formation of GAPDH under oxidizing conditions was furthermore observed in vertebrate cells [46],[47], S-glutathionylation, however, did not inactivate the enzyme [48]. Several chloroplast enzymes including GAPDH possess the unique property of being activated by reduced thioredoxins in the light [49],[50]. In a number of former studies on plants, GAPDH had also been identified to be redox-regulated by members of the thioredoxin superfamily [25],[26],[33].
In addition to GAPDH, two other glycolytic enzymes, namely hexokinase and pyruvate kinase, as well as L-lactate dehydrogenase were captured in our study. Most of the enzymes involved in glycolysis are sulfhydryl proteins sensitive to oxidation suggesting that they may be controlled by the redox state of the cells [51],[52]. Possibly, the Plasmodium enzymes are regulated by similar mechanisms involving redox-active proteins. A putative redox interaction, however, remains to be studied with recombinant enzymes in detail. In this context, one should mention that the glycolytic enzyme enolase seems to be redox-regulated by a cytosolic thioredoxin system in a limited number of plant species [53].
Hemoglobin catabolism
Hemoglobin is a major nutrient source during the intraerythrocytic life stage of P. falciparum and is initially processed by four different but homologous proteases, namely plasmepsins I, II, and IV and histo-aspartic protease (HAP; plasmepsin III), located in the acidic food vacuole of the parasite. Plasmepsins possess active site aspartates one of which is substituted in the HAP enzyme by histidine [54]. Among all Plasmodium species, four plasmepsin cysteines are strictly conserved and form two disulfide bonds which are located on the surface of the proteins (PDB entries 2ANL [55], 1MIQ [56], 1LEE [57]). These disulfides are potentially accessible for an interaction with redox-active proteins. Our results indicate an interaction of Trx and Plrx with plasmepsin III. The presence of these two redox-active proteins and their respective reducing backup systems in the food vacuole for regulation of enzyme activities seems to be very unlikely. One might, however, speculate that the HAP protein is putatively redox-regulated by cytosolic dithiol proteins on the way to its destination, the food vacuole. Of course, this hypothesis needs to be substantiated by further detailed studies.
Signal transduction
Increasing attention has been paid to the roles of thioredoxin as a key molecule in redox signaling beyond its intrinsic antioxidant activity. For Plasmodium, an involvement of redox-active proteins in signal transduction has not been elucidated so far. In our study, we detected a GTPase and two guanine nucleotide-binding proteins (Table 4) which had not been described in the context of redox control before. In addition, two 14-3-3 proteins were captured with our redox-active proteins (Table 4) which confirm the findings of Rouhier et al. [25] who first demonstrated the redox-regulation of a 14-3-3 protein in plants. Vice versa, a global proteomic analysis aimed at identifying proteins that bind to 14-3-3 proteins during interphase and mitosis revealed Trx and a peroxiredoxin as binding partners [58]. 14-3-3 is an evolutionarily conserved protein that is most noted as a mediator in signal transduction events and cell cycle regulation [59]. In a very recent investigation conducted by Aachmann et al. [60], the interaction of selenoprotein W with 14-3-3 proteins had been studied using NMR. Selenoprotein W has a thioredoxin-like fold with a CXXU motif located in an exposed loop similar to the redox-active site in thioredoxin. The specific interaction which is suggested to be physiologically relevant involves the active site residues CXXU and indicates that 14-3-3 are redox-regulated proteins. These findings support the possible role of a redox-based signaling network in Plasmodium that needs to be further unraveled.
S-adenosylmethionine (SAM) metabolism and interaction analysis of SAHH with redox-active proteins
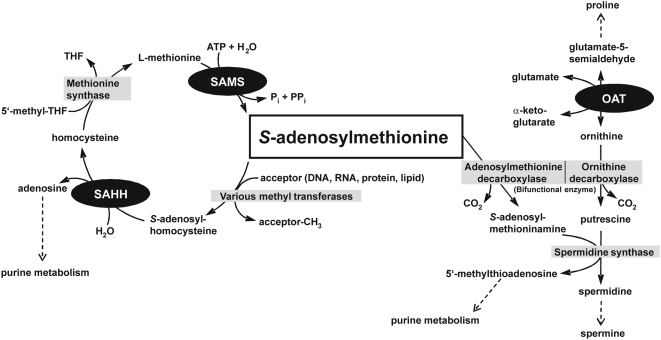
Three enzymes involved in S-adenosylmethionine metabolism, ornithine aminotransferase (OAT), S-adenosylmethionine synthetase (SAMS), and S-adenosyl-L-homocysteine hydrolase (SAHH) have been identified in this study to interact with Trx, Grx and Plrx. The first function of activated methionine in SAM is to serve as a methyl-group donor (Figure 4, left side): SAM is formed from methionine and ATP by SAMS. SAM-dependent methylation reactions lead to formation of S-adenosyl-L-homocysteine which is hydrolyzed into L-homocysteine and adenosine by SAHH. A putative regulation of SAMS by Trx [22],[26], as well as SAHH by Grx had been described before [25]. Methionine synthase catalyzes the reaction step between SAHH and SAMS in the “activated methyl cycle” and also represents an established target for Trx and Grx [25],[26]. Thus it is very likely that regeneration of the methyl-group donor SAM is tightly controlled by redox regulation in P. falciparum and other organisms. OAT which is required for the formation of ornithine has not been described in the context of redox control. However, such a regulation is quite logical considering another function of SAM. The second function of activated methionine in SAM is to serve (after decarboxylation) as the aminoalkyl-donor for the synthesis of polyamines (Figure 4, right side). This is also the case for ornithine which reacts (after decarboxylation) with the SAM-derivative to form spermidine. Thus, we hypothesize that redox regulation of OAT and SAMS is coupled to the tight control of polyamine synthesis. We furthermore suggest that regulation of OAT and the other enzymes is required to balance the metabolic fluxes of SAM between methyl-group transfer reactions and polyamine synthesis (Figure 4).
Figure 4. Overview of the S-adenosylmethionine metabolism in P. falciparum.
On the left side, the C1 metabolism is shown comprising the putatively redox-regulated proteins SAMS and SAHH identified in the present work. OAT, which is also suggested as being redox-sensitive, is involved in the formation of ornithine, an important precursor for polyamine synthesis, which is shown on the right side. The trans-sulfuration pathway from homocysteine to cysteine has not been included in the figure. See text for details.
SPR analyses were carried out in order to identify the cysteine residue(s) of the three redox-active proteins on which the interaction with oxidized SAHH is based (Figure 2). For Trx, only the TrxC33S mutant was able to bind to SAHH suggesting that a disulfide bond between SAHH and residue Cys 30 at the Trx active site is responsible for the interaction (Figure 2A). The double mutant could probably not be captured due to the lack of a reactive cysteine residue. For wild type Trx strikingly different redox potentials of SAHH and Trx might favor a complete reaction resulting in reduced SAHH and oxidized Trx without trapping the short-lived intermolecular disulfide intermediate. The fact that the intermediate during the reaction of oxidized SAHH with wild type Plrx (Figure 2B) is more pronounced or stable than for wild type Trx might be explained by a more positive redox potential of Plrx. The reduced amount of intermediate seen for PlrxC63S might be due to a significantly altered redox potential of the mutant. The reduced stability of the intermediate might be also the reason why SAHH was not captured with the Plrx affinity column. Besides the redox interaction of the Grx active site cysteine residue(s) with SAHH, Grx is capable of forming a disulfide bond with SAHH via Cys 88 (Figure 2C). The residue is quite conserved (see alignment in ref. [12]) and is located at the active site between the GlyGly-turn ending β-strand four and α-helix four. The homologous residue Cys 117 in yeast Grx5 was also shown to be involved in redox reactions in vitro [61]. Which of the Grx cysteine residues acts mainly as electron donor under physiological conditions remains speculative, although the significance of the N-terminal active site cysteine for disulfide formation was substantiated by the study of Rouhier et al. [25]. Furthermore, the non-covalent interaction observed for Grx (Figure 2D) supports the presence of a specific protein-protein interaction between Grx and SAHH.
In addition to SAHH, SPR experiments were carried out for ornithine amino transferase, another protein captured by the pull down assays, and thioredoxin. Figure 3A shows a strong interaction of OAT with the active site single mutant of Trx. This interaction was based on a disulfide bond formation as proven by the fact that it could be solved rapidly and efficiently by DTT. In contrast to the single mutant, the Trx active site double mutant and the wild type Trx showed hardly any interaction with OAT. The biological significance of these data was studied by activity assays. As indicated in Figure 3B, the addition of equimolar amounts of wild type thioredoxin to the assay resulted in 75% increase in OAT activity. As expected, the Trx single and double mutants did not induce activity changes which indicates that an intact active site motif with two cysteine residues is required for the activation of OAT. Our data substantiate the above hypothesis that in Plasmodium falciparum the activity of OAT is redox-regulated. To our knowledge this is the first time that such a regulation has been proposed for ornithine aminotransferase.
Further putative redox-linked processes in Plasmodium
Class I ribonucleotide reductase has been suggested to be a substrate for Plrx according to in vitro assays [18] and was now also identified as a Plrx interacting partner by affinity chromatography (Table 3). It should be noted that the small subunit (R2) of ribonucleotide reductase identified in our study is not the electron acceptor oxidizing dithiol proteins but generates the radical required for catalysis [62]. Either the interaction between R2 and Plrx was indirect and subunit R1 was overlooked in the mass spectrometric analyses or Plrx interacted directly with one of the eight cysteine residues of R2. The latter possibility is quite speculative but might point to a novel regulatory function of R2. Identification of a phosphoethanolamine N-methyltransferase and the acyl carrier protein suggests a potential redox regulation of lipid metabolism in malarial parasites. Furthermore, a putative redox sensitivity of an endonuclease iii homologue involved in DNA repair has, according to our knowledge, not been described before. The same holds true for an acid phosphatase whose function in Plasmodium remains to be studied in detail.
In order to determine if the interaction of the captured proteins with the members of the thioredoxin family and the resulting putative redox changes are of biological relevance - as started for OAT - further biochemical, biophysical and cell-biological studies will have to be conducted in the next years. Among others, these studies will include cloning, expression, mutagenesis and purification of the interacting proteins, assessment of protein-protein interactions under quasi-physiological conditions, enzyme kinetic studies in different redox environments, modeling, cocrystallization, and x-ray crystallographic or NMR analyses of protein-redoxin complexes, and knock out/knock down experiments. The expected data will further enhance our knowledge on redox regulatory processes in Plasmodium, on host-parasite interactions and on the potential of redox metabolism as antimalarial drug target.
Materials and Methods
All chemicals used were of the highest purity available and were obtained from Roth (Karlsruhe, Germany), Sigma-Aldrich (Steinheim, Germany), or Merck (Darmstadt, Germany). PCR primers were obtained from MWG-Biotech (Ebersberg, Germany). The vectors pHSG398, pET28a and pDrive were obtained from Takara (Japan), Novagen (Darmstadt, Germany) and Qiagen (Hilden, Germany), respectively. The expression system QIAexpress, comprising vector pQE30, E. coli host strain M15, and Ni-NTA agarose resin, was purchased from Qiagen (Hilden, Germany) and E. coli strain BL 21 from Novagen (Darmstadt, Germany). CNBr-activated Sepharose 4B was obtained from GE Healthcare (München, Germany). RPMI 1640 medium was from Gibco (Paisley, UK).
Site-directed mutagenesis of PfTrx1 (PF14_0545), PfGrx1 (PFC0271c), and Plrx (PFC0166w), heterologous expression and protein purification
Cloning of PfTrx1, PfGrx1, and Plrx has been described previously [3],[12],[18]. Mutations of PfTrx1C33S, PfTrx1C30S/C33S, PfGrx1C32S, PfGrx1C29S/C32S, and PlrxC63S were introduced by PCR with Pfu polymerase (Promega Corp.) using mutated primers (Table S1). Methylated non-mutated template plasmids were digested with DpnI, and competent XL1-Blue cells were subsequently transformed. The introduction of the correct mutation was confirmed by sequencing.
pQE30 constructs of wild type and mutant genes were expressed in E. coli strain M15 (Qiagen). Cells containing the respective plasmid were grown at 37°C in LB medium supplemented with carbenicillin (100 µg/ml) and kanamycin (50 µg/ml) to an optical density at 600 nm of 0.5 to 0.6, and expression was subsequently induced for 4 h with 1 mM isopropyl-β-D-1-thiogalactopyranoside. Cells were harvested, resuspended in 50 mM sodium phosphate, 300 mM NaCl, pH 8.0, and sonicated in the presence of protease inhibitors. After centrifugation, the supernatant was applied to a Ni-NTA column, and recombinant proteins were eluted with buffer containing 75 mM imidazole. For coupling to a CNBr-activated Sepharose 4B, the respective proteins were dialysed against 100 mM NaHCO3, 500 mM NaCl, pH 8.3.
Cultivation of Plasmodium falciparum and preparation of parasite cell extract
Intraerythrocytic stages of P. falciparum were grown in continuous culture as described by Trager and Jensen [63], with slight modifications. Parasites were maintained at 1 to 10% parasitemia and 3.3% hematocrit in an RPMI 1640 culture medium supplemented with A+ erythrocytes, 0.5% lipid-rich bovine serum albumin (Albumax), 9 mM (0.16%) glucose, 0.2 mM hypoxanthine, 2.1 mM L-glutamine, and 22 µg/ml gentamicin. All incubations were carried out at 37°C in 3% O2, 3% CO2, and 94% N2. Synchronization of parasites in culture to ring stages was carried out by treatment with 5% (w/v) sorbitol [64]. Trophozoite stage parasites were harvested by suspending the red cells for 10 minutes at 37°C in a 20-fold volume of saponin lysis buffer containing 7 mM K2HPO4, 1 mM NaH2PO4, 11 mM NaHCO3, 58 mM KCl, 56 mM NaCl, 1 mM MgCl2, 14 mM glucose, and 0.02% saponin, pH 7.4. The saponin lysis was repeated twice before washing the parasites with PBS and freezing the parasite pellet at −80°C.
For preparing the parasite cell extract, the pellet was diluted in an equal volume of buffer containing 100 mM Tris, 500 mM NaCl, pH 8.0. Parasites were disrupted by four cycles of freezing in liquid nitrogen and thawing in a waterbath at room temparature followed by sonication at 4°C. After centrifugation at 100,000 g for 30 min at 4°C, the obtained supernatant was used as cell lysate for affinity chromatography columns.
Capturing of the target proteins by immobilized mutants of PfTrx1, PfGrx1, and Plrx
1 mg of the respective pure mutants PfTrx1C33S, PfGrx1C32S, and PlrxC63S in coupling buffer (100 mM sodium carbonate, 500 mM NaCl, pH 8.3) was incubated for 1 h at room temperature under gentle agitation with 10 µl CNBr-activated Sepharose 4B resin, which had been swelled in 1 mM HCl according to the manufacturer's instructions. After termination of the coupling reaction by centrifugation and washing of the resin with coupling buffer, unmodified reactive groups of the resin were blocked by incubation with 100 mM Tris, pH 8.0 for 2 h at room temperature. Plasmodium falciparum cell lysate (∼800 µl) containing 7–10 mg protein was incubated with 10 µl of the liganded resin at room temperature for at least 2 h under gentle stirring, before washing the resin with 100 mM Tris, 500 mM NaCl, pH 8.0 to remove non-specifically bound proteins. The washing steps were repeated until the absorbance of the washing solution at 280 nm became zero. Finally, 10 µl resin were suspended in 22 µl 100 mM Tris, 500 mM NaCl, 10 mM DTT, pH 8.0, and were incubated for 30 min at room temperature. Elution was repeated with 12 µl elution buffer, and eluates were pooled. The eluted proteins were separated by SDS polyacrylamide gel electrophoresis. Bands of interest were excised, and then subjected to tryptic digestion and MALDI-TOF analysis as described below. To verify that the interaction between the redoxins and the captured proteins was specific and based on the proposed disulfide-dependent mechanism a number of control pull down experiments was performed. As bait proteins wild type Trx, Grx and Plrx as well as the double mutants of Trx (C30S/C33S) and Grx (C29S/C32S) were employed. In addition, CNBr-activated Sepharose 4B resin without immobilized protein was used to study unspecific binding of proteins. These control experiments were carried out in analogy to the original pull downs. As shown in Figure S2, hardly any bands could be detected in these controls indicating that the binding to the single mutants of Trx, Grx and Plrx in our pull down experiments was specific.
Protein identification by peptide mass fingerprinting with MALDI-MS
Excised gel pieces were destained with 25 mM NH4HCO3 in 50% acetonitrile by washing them three times for 10 min each. The gel pieces were vacuum-dried and then incubated with modified porcine trypsin (Promega) at a final concentration of 0.1 mg/ml in 25 mM NH4HCO3, pH 8.0 for 16 h at 37°C. The peptides were extracted three times with 30 µl of 5% trifluoroacetic acid in 50% acetonitrile and the extract was concentrated in a speedvac. The obtained solutions were loaded onto the MALDI target plate by mixing 0.3 µl of each solution with the same volume of matrix solution (10 mg/ml α-cyanohydroxycinnaminic acid in acetonitrile/H2O (1∶1, v/v) and allowed to dry. Measurements were performed using a Voyager 4182 MALDI-TOF instrument (Applied Biosystems, Darmstadt, Germany), operating in the positive ion reflector mode with an accelerating voltage of 25 kV. The laser wavelength was 337 nm and the laser repetition rate was 20 Hz. The final mass spectra were produced by averaging 60 laser shots. Each spectrum was internally calibrated with the masses of two trypsin autolysis products. For peptide mass fingerprinting identification, the tryptic peptide mass maps were searched against Swiss-Prot (http://www.expasy.uniprot.org) and PlasmoDB (http://www.plasmodb.org) databases by using the search engine Protein Prospector MS-Fit. Standard search parameters were set to allow a mass accuracy of 15 ppm and two missed tryptic cleavages.
PCR amplification, sequencing, and subcloning of PfSAHH and PfOAT
For cloning procedures of PfSAHH, restriction sites were introduced for EcoRI (underlined) and NcoI (italic), and for EcoRI (underlined) and XhoI (italic), respectively, at the 5′-ends of the respective primers (primer for PfSAHH: N-terminal: 5′-GGGCGAATTC CCATGGTTGAAAATAAGAGTAAGGTC-3′; C-terminal: 5′-CGCGGAATTCCTCGAGATATCTGTATTCGTTACTCT-3′). For cloning procedures of PfOAT the primers contained restriction sites for NcoI (underlined) and XhoI (italic) (primer for PfOAT: N-terminal: 5′-AGCCCATGGATTTCGTTAAAGAATTAAAAAGTAG-3′; C-terminal: 5′-ACGCTCGAGTAAATTGTCATCAAAAAATTTAACAG-3′). The genes for PfSAHH and PfOAT were amplified by PCR using a gametocyte cDNA library from P. falciparum strain 3D7 as a template. The derived fragment of PfSAHH of correct size was cloned into pHSG398 with EcoRI; the fragment of PfOAT was cloned into pDrive. Both fragments were sequenced and subcloned into pET28a using NcoI and XhoI.
Heterologous expression of PfSAHH and PfOAT and purification of the proteins
PfSAHH and PfOAT were expressed in the E. coli strain BL 21. Cells containing the plasmid of PfSAHH were grown in Terrific Broth medium supplemented with kanamycin (25 µg/ml). BL 21 cells containing PfOAT were cultivated in Luria Bertani medium in the presence of kanamycin (25 µg/ml). The cells were grown at 23°C to an optical density at 600 nm of 0.7, and expression was subsequently induced for 15 h with 0.2 mM (PfSAHH) or 0.5 mM (PfOAT) isopropyl-β-D-1-thiogalactopyranoside. Cells were harvested, resuspended in 50 mM sodium phosphate, 300 mM NaCl, pH 8.0, and sonicated in the presence of protease inhibitors. After centrifugation, the supernatant was applied to a Ni-NTA column, and recombinant proteins were eluted with sodium phosphate buffer containing 50 mM (PfSAHH) or 100 mM (PfOAT) imidazole.
BIAcore surface plasmon resonance detection
Surface plasmon resonance experiments were performed using a BIAcore X biosensor system (Biacore AB, Uppsala, Sweden). The carboxymethylated surface of the sensor chip CM5 was activated with a 1∶1 mixture of 0.1 M N-hydroxysuccinimide (NHS) and 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) (provided in the amine coupling kit; Biacore AB). Subsequently, 35 µl (60 µg/ml) of SAHH or OAT in 10 mM sodium acetate, pH 4.5 were injected to flow cell 2 (FC2) to be immobilized on the sensor surface via primary amine groups. No protein was injected into FC1. Residual unreacted active ester groups were blocked with 1 M ethanolamine-HCl, pH 8.5 (amine coupling kit). Experiments were performed at 25°C in HBS buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, 0.005% Nonidet P-40, pH 7.4) at a flow rate of 10 µl/min. To analyze the binding of any of the redox-active proteins (-mutants) with SAHH or OAT, the following cycles were conducted: Firstly, SAHH or OAT on the sensor surface was oxidized using 30 µl of 0.5 mM 5,5′-dithiobis(2-nitrobenzoate) (DTNB) before 30 µl of the analyte (10 µM in HBS buffer) were injected, followed by buffer flow over the chip and an elution step with 2 mM DTT in a volume of 30 µl. In order to analyze a non-covalent interaction of Grx with SAHH or OAT, 100 µM of the GrxC29S/C32S mutant were treated with 10 mM iodoacetamide for 2 h in the dark. Excess iodoacetamide was eliminated by gelfiltration chromatography using NAP-5 columns (Amersham Biosciences). Difference resonance spectra (FC2–FC1) were recorded. After equilibrating the surface with HBS buffer it was ready for the next cycle. Data were evaluated using the software BIAevaluation 3.0 (Biacore AB).
Ornithine aminotransferase assay
The conversion of ornithine to glutamate-5-semialdehyde was determined spectrophotometrically using a modified method of Kim et al. [65]. OAT was added to an assay mixture of 0.5 ml containing 100 mM phosphate buffer (pH 7.4), 50 mM L-ornithine, 20 mM α-ketoglutarate and 0.05 mM pyridoxal-5-phosphate and incubated for 30 min at 37°C. The reaction was stopped by adding 0.4 M HCl and 0.16% (w/v) ninhydrin. After heating for 5 min at 100°C 500 µl ethanol was added and the absorbance was measured spectrophotometrically at 512 nm.
Supporting Information
Oligonucleotide primers used for site-directed mutagenesis.
(0.03 MB DOC)
Comparison of the DTT-eluate fractions of the Trx, Grx, and Plrx pull-down experiments. The respective active site mutant of each of the three proteins was immobilized on CNBr-activated Sepharose 4B resin before incubating the column with Plasmodium falciparum cell lysate and washing extensively with NaCl-containing buffer. Putative target proteins were eluted with 10 mM DTT. The obtained protein samples were separated on a 12% polyacrylamide gel and stained with Coomassie blue.
(0.92 MB TIF)
Pull-down controls with PfTrx1 (A), PfTrx1C30S/C33S (B), PfGrx1 (C), PfGrx1C29S/C32S (D), PfPlrx (E), and CNBr-activated Sepharose 4B resin without immobilized protein (F). These control experiments were carried out in analogy to the original pull-downs; hardly any bands can be detected indicating that the binding to the single mutants of Trx, Grx, and Plrx in our pull-down experiments is specific. According to mass spectrometric analysis of the comparable band in the pull-down assays (see Figure 1), band (1) corresponds to 1-Cys peroxiredoxin, the other strong bands in the lower MW range represent the bait redoxins.
(5.68 MB TIF)
Acknowledgments
The authors wish to thank Elisabeth Fischer for her excellent technical assistance in cell culture, as well as Proteome Sciences R&D GmbH & Co. KG (Frankfurt am Main, Germany) for collaborating on the peptide mass fingerprints.
Footnotes
The authors have declared that no competing interests exist.
The study was supported by the Deutsche Forschungsgemeinschaft (SFB 535, TPA12 to KB). Funds were obtained from the German Research Council and the German Academic Exchange Service. None of the funders played a role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript.
References
- 1.Becker K, Koncarevic S, Hunt NH. Oxidative stress and antioxidant defense in malarial parasites. In: Sherman I, editor. Molecular Approaches to Malaria. American Society of Microbiology Press; 2005. pp. 365–383. [Google Scholar]
- 2.Rahlfs S, Schirmer RH, Becker K. The thioredoxin system of Plasmodium falciparum and other parasites. Cell Mol Life Sci. 2002;59:1024–1041. doi: 10.1007/s00018-002-8484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanzok SM, Schirmer RH, Turbachova I, Iozef R, Becker K. The thioredoxin system of the malaria parasite Plasmodium falciparum. Glutathione reduction revisited. J Biol Chem. 2000;275:40180–40186. doi: 10.1074/jbc.M007633200. [DOI] [PubMed] [Google Scholar]
- 4.Kawazu S, Komaki K, Tsuji N, Kawai S, Ikenoue N, et al. Molecular characterization of a 2-Cys peroxiredoxin from the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2001;116:73–79. doi: 10.1016/s0166-6851(01)00308-5. [DOI] [PubMed] [Google Scholar]
- 5.Kawazu S, Tsuji N, Hatabu T, Kawai S, Matsumoto Y, et al. Molecular cloning and characterization of a peroxiredoxin from the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2000;109:165–169. doi: 10.1016/s0166-6851(00)00243-7. [DOI] [PubMed] [Google Scholar]
- 6.Krnajski Z, Walter RD, Muller S. Isolation and functional analysis of two thioredoxin peroxidases (peroxiredoxins) from Plasmodium falciparum. Mol Biochem Parasitol. 2001;113:303–308. doi: 10.1016/s0166-6851(01)00219-5. [DOI] [PubMed] [Google Scholar]
- 7.Rahlfs S, Becker K. Thioredoxin peroxidases of the malarial parasite Plasmodium falciparum. Eur J Biochem. 2001;268:1404–1409. doi: 10.1046/j.1432-1327.2001.02005.x. [DOI] [PubMed] [Google Scholar]
- 8.Sztajer H, Gamain B, Aumann KD, Slomianny C, Becker K, et al. The putative glutathione peroxidase gene of Plasmodium falciparum codes for a thioredoxin peroxidase. J Biol Chem. 2001;276:7397–7403. doi: 10.1074/jbc.M008631200. [DOI] [PubMed] [Google Scholar]
- 9.Farber PM, Arscott LD, Williams CH, Jr, Becker K, Schirmer RH. Recombinant Plasmodium falciparum glutathione reductase is inhibited by the antimalarial dye methylene blue. FEBS Lett. 1998;422:311–314. doi: 10.1016/s0014-5793(98)00031-3. [DOI] [PubMed] [Google Scholar]
- 10.Harwaldt P, Rahlfs S, Becker K. Glutathione S-transferase of the malarial parasite Plasmodium falciparum: characterization of a potential drug target. Biol Chem. 2002;383:821–830. doi: 10.1515/BC.2002.086. [DOI] [PubMed] [Google Scholar]
- 11.Hiller N, Fritz-Wolf K, Deponte M, Wende W, Zimmermann H, et al. Plasmodium falciparum glutathione S-transferase—structural and mechanistic studies on ligand binding and enzyme inhibition. Protein Sci. 2006;15:281–289. doi: 10.1110/ps.051891106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahlfs S, Fischer M, Becker K. Plasmodium falciparum possesses a classical glutaredoxin and a second, glutaredoxin-like protein with a PICOT homology domain. J Biol Chem. 2001;276:37133–37140. doi: 10.1074/jbc.M105524200. [DOI] [PubMed] [Google Scholar]
- 13.Akoachere M, Iozef R, Rahlfs S, Deponte M, Mannervik B, et al. Characterization of the glyoxalases of the malarial parasite Plasmodium falciparum and comparison with their human counterparts. Biol Chem. 2005;386:41–52. doi: 10.1515/BC.2005.006. [DOI] [PubMed] [Google Scholar]
- 14.Deponte M, Sturm N, Mittler S, Harner M, Mack H, et al. Allosteric coupling of two different functional active sites in monomeric Plasmodium falciparum glyoxalase I. J Biol Chem. 2007;282:28419–28430. doi: 10.1074/jbc.M703271200. [DOI] [PubMed] [Google Scholar]
- 15.Martin JL. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 16.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal. 2000;2:811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 18.Becker K, Kanzok SM, Iozef R, Fischer M, Schirmer RH, et al. Plasmoredoxin, a novel redox-active protein unique for malarial parasites. Eur J Biochem. 2003;270:1057–1064. doi: 10.1046/j.1432-1033.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- 19.Buchholz K, Rahlfs S, Schirmer RH, Becker K, Matuschewski K. Depletion of Plasmodium berghei plasmoredoxin reveals a non-essential role for life cycle progression of the malaria parasite. PLoS ONE. 2008;3:e2474. doi: 10.1371/journal.pone.0002474. doi:10.1371/journal.pone.0002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 21.Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, et al. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci U S A. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaire SD, Guillon B, Le Marechal P, Keryer E, Miginiac-Maslow M, et al. New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2004;101:7475–7480. doi: 10.1073/pnas.0402221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindahl M, Florencio FJ. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc Natl Acad Sci U S A. 2003;100:16107–16112. doi: 10.1073/pnas.2534397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci U S A. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouhier N, Villarejo A, Srivastava M, Gelhaye E, Keech O, et al. Identification of plant glutaredoxin targets. Antioxid Redox Signal. 2005;7:919–929. doi: 10.1089/ars.2005.7.919. [DOI] [PubMed] [Google Scholar]
- 26.Wong JH, Cai N, Balmer Y, Tanaka CK, Vensel WH, et al. Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry. 2004;65:1629–1640. doi: 10.1016/j.phytochem.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Becker K, Tilley L, Vennerstrom JL, Roberts D, Rogerson S, et al. Oxidative stress in malaria parasite-infected erythrocytes: host-parasite interactions. Int J Parasitol. 2004;34:163–189. doi: 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Nickel C, Rahlfs S, Deponte M, Koncarevic S, Becker K. Thioredoxin networks in the malarial parasite Plasmodium falciparum. Antioxid Redox Signal. 2006;8:1227–1239. doi: 10.1089/ars.2006.8.1227. [DOI] [PubMed] [Google Scholar]
- 29.Nickel C, Trujillo M, Rahlfs S, Deponte M, Radi R, et al. Plasmodium falciparum 2-Cys peroxiredoxin reacts with plasmoredoxin and peroxynitrite. Biol Chem. 2005;386:1129–1136. doi: 10.1515/BC.2005.129. [DOI] [PubMed] [Google Scholar]
- 30.Holmgren A. Thioredoxin structure and mechanism: conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure. 1995;3:239–243. doi: 10.1016/s0969-2126(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 31.Hisabori T, Hara S, Fujii T, Yamazaki D, Hosoya-Matsuda N, et al. Thioredoxin affinity chromatography: a useful method for further understanding the thioredoxin network. J Exp Bot. 2005;56:1463–1468. doi: 10.1093/jxb/eri170. [DOI] [PubMed] [Google Scholar]
- 32.Hosoya-Matsuda N, Motohashi K, Yoshimura H, Nozaki A, Inoue K, et al. Anti-oxidative stress system in cyanobacteria. Significance of type II peroxiredoxin and the role of 1-Cys peroxiredoxin in Synechocystis sp. strain PCC 6803. J Biol Chem. 2005;280:840–846. doi: 10.1074/jbc.M411493200. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:18–27. doi: 10.1093/pcp/pch019. [DOI] [PubMed] [Google Scholar]
- 34.Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:5. (E-pub 2003 Aug 18). doi: 10.1371/journal.pbio.0000005. doi:10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deponte M, Rahlfs S, Becker K. Peroxiredoxin systems of protozoal parasites. Subcell Biochem. 2007;44:219–229. doi: 10.1007/978-1-4020-6051-9_10. [DOI] [PubMed] [Google Scholar]
- 36.Kawazu S, Komaki-Yasuda K, Oku H, Kano S. Peroxiredoxins in malaria parasites: parasitologic aspects. Parasitol Int. 2008;57:1–7. doi: 10.1016/j.parint.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Boucher IW, McMillan PJ, Gabrielsen M, Akerman SE, Brannigan JA, et al. Structural and biochemical characterization of a mitochondrial peroxiredoxin from Plasmodium falciparum. Mol Microbiol. 2006;61:948–959. doi: 10.1111/j.1365-2958.2006.05303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayala A, Parrado J, Bougria M, Machado A. Effect of oxidative stress, produced by cumene hydroperoxide, on the various steps of protein synthesis. Modifications of elongation factor-2. J Biol Chem. 1996;271:23105–23110. doi: 10.1074/jbc.271.38.23105. [DOI] [PubMed] [Google Scholar]
- 39.Vignols F, Mouaheb N, Thomas D, Meyer Y. Redox control of Hsp70-Co-chaperone interaction revealed by expression of a thioredoxin-like Arabidopsis protein. J Biol Chem. 2003;278:4516–4523. doi: 10.1074/jbc.M210080200. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. Embo J. 2004;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trebitsh T, Levitan A, Sofer A, Danon A. Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol Cell Biol. 2000;20:1116–1123. doi: 10.1128/mcb.20.4.1116-1123.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson LE, Li D, Prakash N, Stevens FJ. Identification of potential redox-sensitive cysteines in cytosolic forms of fructosebisphosphatase and glyceraldehyde-3-phosphate dehydrogenase. Planta. 1995;196:118–124. doi: 10.1007/BF00193225. [DOI] [PubMed] [Google Scholar]
- 43.Li D, Stevens FJ, Schiffer M, Anderson LE. Mechanism of light modulation: identification of potential redox-sensitive cysteines distal to catalytic site in light-activated chloroplast enzymes. Biophys J. 1994;67:29–35. doi: 10.1016/S0006-3495(94)80484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robien MA, Bosch J, Buckner FS, Van Voorhis WC, Worthey EA, et al. Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site. Proteins. 2006;62:570–577. doi: 10.1002/prot.20801. [DOI] [PubMed] [Google Scholar]
- 45.Souza JM, Radi R. Glyceraldehyde-3-phosphate dehydrogenase inactivation by peroxynitrite. Arch Biochem Biophys. 1998;360:187–194. doi: 10.1006/abbi.1998.0932. [DOI] [PubMed] [Google Scholar]
- 46.Brodie AE, Reed DJ. Reversible oxidation of glyceraldehyde 3-phosphate dehydrogenase thiols in human lung carcinoma cells by hydrogen peroxide. Biochem Biophys Res Commun. 1987;148:120–125. doi: 10.1016/0006-291x(87)91084-9. [DOI] [PubMed] [Google Scholar]
- 47.Brodie AE, Reed DJ. Cellular recovery of glyceraldehyde-3-phosphate dehydrogenase activity and thiol status after exposure to hydroperoxides. Arch Biochem Biophys. 1990;276:212–218. doi: 10.1016/0003-9861(90)90028-w. [DOI] [PubMed] [Google Scholar]
- 48.Lind C, Gerdes R, Schuppe-Koistinen I, Cotgreave IA. Studies on the mechanism of oxidative modification of human glyceraldehyde-3-phosphate dehydrogenase by glutathione: catalysis by glutaredoxin. Biochem Biophys Res Commun. 1998;247:481–486. doi: 10.1006/bbrc.1998.8695. [DOI] [PubMed] [Google Scholar]
- 49.Buchanan BB. Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol. 1980;31:341–374. [Google Scholar]
- 50.Scheibe R. Redox-Modulation of Chloroplast Enzymes: A Common Principle for Individual Control. Plant Physiol. 1991;96:1–3. doi: 10.1104/pp.96.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magnani M, Stocchi V, Ninfali P, Dacha M, Fornaini G. Action of oxidized and reduced glutathione on rabbit red blood cell hexokinase. Biochim Biophys Acta. 1980;615:113–120. doi: 10.1016/0005-2744(80)90014-5. [DOI] [PubMed] [Google Scholar]
- 52.Ziegler DM. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu Rev Biochem. 1985;54:305–329. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]
- 53.Anderson LE, Li AD, Stevens FJ. The enolases of ice plant and Arabidopsis contain a potential disulphide and are redox sensitive. Phytochemistry. 1998;47:707–713. doi: 10.1016/s0031-9422(97)00659-6. [DOI] [PubMed] [Google Scholar]
- 54.Bjelic S, Aqvist J. Computational prediction of structure, substrate binding mode, mechanism, and rate for a malaria protease with a novel type of active site. Biochemistry. 2004;43:14521–14528. doi: 10.1021/bi048252q. [DOI] [PubMed] [Google Scholar]
- 55.Clemente JC, Govindasamy L, Madabushi A, Fisher SZ, Moose RE, et al. Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor. Acta Crystallogr D Biol Crystallogr. 2006;62:246–252. doi: 10.1107/S0907444905041260. [DOI] [PubMed] [Google Scholar]
- 56.Bernstein NK, Cherney MM, Yowell CA, Dame JB, James MN. Structural insights into the activation of P. vivax plasmepsin. J Mol Biol. 2003;329:505–524. doi: 10.1016/s0022-2836(03)00444-3. [DOI] [PubMed] [Google Scholar]
- 57.Asojo OA, Gulnik SV, Afonina E, Yu B, Ellman JA, et al. Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparum. J Mol Biol. 2003;327:173–181. doi: 10.1016/s0022-2836(03)00036-6. [DOI] [PubMed] [Google Scholar]
- 58.Meek SE, Lane WS, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 59.Aitken A, Collinge DB, van Heusden BP, Isobe T, Roseboom PH, et al. 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem Sci. 1992;17:498–501. doi: 10.1016/0968-0004(92)90339-b. [DOI] [PubMed] [Google Scholar]
- 60.Aachmann FL, Fomenko DE, Soragni A, Gladyshev VN, Dikiy A. Solution structure of selenoprotein W and NMR analysis of its interaction with 14-3-3 proteins. J Biol Chem. 2007;282:37036–37044. doi: 10.1074/jbc.M705410200. [DOI] [PubMed] [Google Scholar]
- 61.Tamarit J, Belli G, Cabiscol E, Herrero E, Ros J. Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem. 2003;278:25745–25751. doi: 10.1074/jbc.M303477200. [DOI] [PubMed] [Google Scholar]
- 62.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 63.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 64.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 65.Kim HR, Rho HW, Park JW, Park BH, Kim JS, et al. Assay of ornithine aminotransferase with ninhydrin. Anal Biochem. 1994;223:205–207. doi: 10.1006/abio.1994.1574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers used for site-directed mutagenesis.
(0.03 MB DOC)
Comparison of the DTT-eluate fractions of the Trx, Grx, and Plrx pull-down experiments. The respective active site mutant of each of the three proteins was immobilized on CNBr-activated Sepharose 4B resin before incubating the column with Plasmodium falciparum cell lysate and washing extensively with NaCl-containing buffer. Putative target proteins were eluted with 10 mM DTT. The obtained protein samples were separated on a 12% polyacrylamide gel and stained with Coomassie blue.
(0.92 MB TIF)
Pull-down controls with PfTrx1 (A), PfTrx1C30S/C33S (B), PfGrx1 (C), PfGrx1C29S/C32S (D), PfPlrx (E), and CNBr-activated Sepharose 4B resin without immobilized protein (F). These control experiments were carried out in analogy to the original pull-downs; hardly any bands can be detected indicating that the binding to the single mutants of Trx, Grx, and Plrx in our pull-down experiments is specific. According to mass spectrometric analysis of the comparable band in the pull-down assays (see Figure 1), band (1) corresponds to 1-Cys peroxiredoxin, the other strong bands in the lower MW range represent the bait redoxins.
(5.68 MB TIF)