Abstract
Gene silencing using small interfering RNA (siRNA) has several potential therapeutic applications. In the present study, we investigated nanoparticles formulated using the biodegradable polymer, poly(d,l-lactide-co-glycolide) (PLGA) for siRNA delivery. A cationic polymer, polyethylenimine (PEI), was incorporated in the PLGA matrix to improve siRNA encapsulation in PLGA nanoparticles. PLGA-PEI nanoparticles were formulated using double emulsion-solvent evaporation technique and characterized for siRNA encapsulation and in vitro release. The effectiveness of siRNA-loaded PLGA-PEI nanoparticles in silencing a model gene, fire fly luciferase, was investigated in cell culture. Presence of PEI in PLGA nanoparticle matrix increased siRNA encapsulation by about 2-fold and also improved the siRNA release profile. PLGA-PEI nanoparticles carrying luciferase-targeted siRNA enabled effective silencing of the gene in cells stably expressing luciferase as well as in cells that could be induced to overexpress the gene. Quantitative studies indicated that presence of PEI in PLGA nanoparticles resulted in 2-fold higher cellular uptake of nanoparticles while fluorescence microscopy studies showed that PLGA-PEI nanoparticles delivered the encapsulated siRNA in the cellular cytoplasm; both higher uptake and greater cytosolic delivery could have contributed to the gene silencing effectiveness of PLGA-PEI nanoparticles. Serum stability and lack of cytotoxicity further add to the potential of PLGA-PEI nanoparticles in gene silencing-based therapeutic applications.
Keywords: RNA interference; siRNA; poly(d,l-lactide-co-glycolide); luciferase; sustained release
1. Introduction
RNA interference (RNAi) has emerged as a powerful tool for targeted gene silencing. RNAi is triggered by using small interfering RNA (siRNA), that are about 20-25 base pairs (bp) long. Upon introduction into cells, siRNAs assemble into endoribonuclease-containing complexes known as RNA-induced silencing complexes (RISCs), unwinding in the process. The siRNA strands guide the RISCs to complementary RNA molecules, where they cleave and destroy the target RNA (Elbashir et al., 2001). Gene silencing using siRNA is more efficient and specific to the target gene than that with antisense oligoneucleotides. While some studies have raised concerns over the possibility of siRNAs eliciting immune reactions via interactions with Toll-like receptor 3 and consequent interferon responses (Bridge et al., 2003; Kim et al., 2004; Sledz et al., 2003), other studies have shown that it is possible to administer synthetic siRNAs in vivo and downregulate an endogenous target without inducing interferon response (Heidel et al., 2004). Gene silencing using siRNA has potential applications in the treatment of cancer (Devi, 2006), cardiovascular diseases (Quarck and Holvoet, 2004) and in wound healing (Cheema et al., 2007).
A major limitation in the therapeutic use of siRNA is its rapid degradation in plasma and cellular cytoplasm, resulting in short half-life (Sioud, 2005). Further, siRNA molecules cannot penetrate into the cell efficiently, necessitating the use of a carrier system for its delivery (Zhang et al., 2007). Viral and non-viral vectors have been investigated for gene silencing. Viral vectors enable stable inhibition of gene expression (Pichler et al., 2005; Xu et al., 2005); however, viral vectors are associated with concerns of toxicity and immunogenicity (Merdan et al., 2002; Schagen et al., 2004). A number of therapeutic applications require sustained gene inhibition with minimal toxicity following repeated administration of the vector.
Non-viral vectors are generally considered safer than viral vectors (Aigner, 2007). Among non-viral vectors, nanoparticles formulated using the biocompatible polymer poly(d,l-lactide-co-glycolide) (PLGA) have demonstrated potential for sustained nucleic acid delivery (Cohen et al., 2000; Khan et al., 2004; Langer and Tirrell, 2004; Luby et al., 2004; Wang et al., 1999). Previous studies have shown that PLGA nanoparticles are non-toxic and biocompatible (Panyam et al., 2002a; Panyam et al., 2003c), and are suitable for in vivo gene delivery applications (Perlstein et al., 2003). Following internalization into the cell by endocytosis, PLGA nanoparticles escape the endo-lysosomal compartment and slowly release the encapsulated payload in the cytoplasm (Panyam et al., 2002b). These observations suggest that PLGA nanoparticles could be suitable for siRNA-mediated gene silencing applications as well. While large molecular weight plasmid DNA (>3 Kbp) can be efficiently encapsulated in PLGA nanoparticles (Cohen et al., 2000), it is not clear, however, whether relatively smaller molecular weight (∼20 bp) siRNA molecules can be incorporated effectively. Our studies indicate that nanoparticles fabricated using PLGA alone result in poor encapsulation of siRNA. We rationalized that introduction of polyethylenimine (PEI), a cationic polymer commonly used in gene delivery applications, in the PLGA matrix could improve the retention of anionic siRNA molecules. Several studies have used PEI in PLGA particles to introduce cationic charges on the surface, which allows the complexation of nucleic acids on the surface of the particles (Kim et al., 2005; Oster et al., 2005; Pai Kasturi et al., 2006; Takashima et al., 2007; Zhang et al., 2008). In our studies, we incorporated PEI in the PLGA matrix to encapsulate siRNA molecules in nanoparticles rather than to adsorb on the surface.
2. Materials and Methods
2.1 Materials
PLGA and polylactic acid (PLA) polymers were purchased from Absorbable Polymers (Pelham, AL). For the initial optimization studies, a double stranded 20-bp oligonucleotide sequence was used as a model for siRNA. Oligonucleotide sense sequence 5′-CCATCCCGACCTCGCGCTCC-3′ and the antisense sequence 3′-GGTAGGGCTGGAGCGCGAGG-5′ were synthesized by Operon Biotechnologies, Inc. (Huntsville, AL). Annealing of the two sequences was performed as per manufacturer's instructions. Luciferase-targeted siRNA (21 bp), non-targeted siRNA having four mismatched base pairs (siCONTROL), and siRNA transfection reagent (DharmaFECT®) were obtained from Dharmacon (Lafayette, CO). Acetylated bovine serum albumin (BSA), polyvinyl alcohol (PVA, average Mw 30,000-70,000 Da), and PEI (average Mw 25,000 Da) were obtained from Sigma (St. Louis, MO). EMT-6 G/L (murine mammary carcinoma) cells were kindly provided by Dr. John Ohlfest, University of Minnesota. MDA-Kb2 cells and Leibovitz L-15 medium were obtained from American Type Culture Collection (Manassas, VA). Dulbecco's Modified Eagle Medium (DMEM) with high glucose, TOTO®-3 iodide and LysoTracker® Green DND-26, penicillin/streptomycin, fetal bovine serum, Quant-iT™ picogreen dsDNA reagent and Trypsin-EDTA (1X) solution were obtained from Invitrogen Corporation (Carlsbad, CA). Luciferase assay system, cell culture lysis reagent 5X, CellTiter 96® AQueous non-radioactive cell proliferation assay (MTS) reagent powder and Tris-EDTA (1X) buffer were purchased from Promega (Madison, WI).
2.2 Nanoparticle formulations
Oligonucleotide or siRNA loaded nanoparticles were prepared using the double emulsion solvent evaporation technique, with some modifications (Panyam et al., 2002b). Three different variations of the technique were followed to prepare PLGA nanoparticles, PLGA-PEI nanoparticles and PLGA nanoparticles loaded with preformed oligo-PEI complex. In order to determine the effect of polymer molecular weight and lactide to glycolide ratio on oligo encapsulation and release, polymers of different molecular weight and composition were used (Table 1).
Table 1.
Encapsulation efficiency of different PLGA and PLGA-PEI nanoparticles
| Polymer used | Lactide to Glycolide Ratio | Mol. Wt (Daltons) | % Encapsulation efficiency | Oligo loading a (μg/30 mg nanoparticles) |
|---|---|---|---|---|
| PLGA-5K | 50:50 | ∼ 5000 | 36 ± 4 | 88 ± 10 |
| PLGA-40K | 50:50 | ∼ 40,000 | 43 ± 5 | 104 ± 14 |
| PLGA-100K | 50:50 | ∼100,000 | 17 ± 3 | 42 ± 8 |
| PLGA-75/25 | 75:25 | 76,000-120,000 | 17 ± 3 | 42 ± 5 |
| PLA-100 | 100:0 | 75,000-120,000 | 47 ± 4 | 115 ± 12 |
| PLGA-40K-PEI | 50:50 | ∼ 40,000 | 79 ± 4 | 194 ± 10 |
Initial amount of oligo added was 245 μg/30 mg polymer
2.2.1 PLGA nanoparticles containing oligo or siRNA
An aqueous solution of BSA (2 mg) and oligos/siRNA (245 μg) in 0.2 ml of RNase and DNase-free TE buffer was emulsified for 30 sec over an ice bath (Sonicator® XL, Misonix, NY) in 1 ml of chloroform containing 30 mg of polymer. The water-in-oil emulsion formed was further emulsified in 6 ml of 2.5% w/v PVA solution in TE buffer by sonication (24 W for 3 min) over an ice bath to form a multiple water-in-oil-in-water emulsion. This emulsion was stirred for 18 h at ambient conditions followed by for 2 h under vacuum to remove the residual chloroform. Nanoparticles were recovered by ultracentrifugation (35,000 rpm for 35 min at 4° C, Optima™ LE-80K, Beckman, Palo Alta, CA), washed two times with sterile distilled water to remove PVA, unentrapped BSA and oligos/siRNA, and then lyophilized (Labconco, FreeZone 4.5, Kansas City, MO). BSA was used in the nanoparticle formulation to facilitate the release of encapsulated siRNA from nanoparticles. Acetylated BSA was used because it is nuclease-free and thus does not contribute to the degradation of nucleic acids during processing. Previous studies have used BSA in PLGA nanoparticles to improve encapsulation and release of other proteins and plasmid DNA (Panyam et al., 2003a; Panyam et al., 2002b).
2.2.2 PLGA-PEI nanoparticles containing oligomer/siRNA
A similar procedure was used as described above, except the polymer solution in chloroform also contained varying concentrations of PEI (5 to 5000μg PEI/30 mg PLGA). For some studies, nanoparticles containing TOTO-labeled oligos were used; oligos (245 μg) were initially incubated with 100 μl of TOTO®-3 iodide dye solution (1 nM) for 1 h and then used for nanoparticle formulation.
2.2.3 PLGA nanoparticles containing oligo-PEI complex
A pre-formed complex of oligo and PEI was initially prepared by adding PEI (100 μg/ml) to 0.2 ml of an aqueous solution containing oligo (245 μg) and BSA (20 mg/ml) and incubating for 30 min at room temperature. This mixture was later emulsified in 1 ml of chloroform containing 30 mg of polymer as described above. The emulsion formed was processed as described in section 2.2.1 to prepare PLGA nanoparticles containing preformed oligo-PEI complexes.
2.3 Encapsulation efficiency
Oligo loading in nanoparticles was determined as described previously (Panyam et al., 2002b). The amount of unentrapped oligos recovered in the nanoparticle wash solutions obtained during the formulation step was quantified using the Quant-iT™ picogreen reagent. Fluorescence generated by the binding of picogreen to double stranded oligo or siRNA was measured using FLx-800 microplate reader (BIO-TEK Instruments, Winooski, VT). Loading in nanoparticles was determined by subtracting the amount of oligos recovered in the wash solutions from initial amount of oligos added. Encapsulation efficiency was defined as the percent of oligos initially added that was encapsulated in nanoparticles.
2.4 Particle size and zeta potential measurements
Hydrodynamic diameter and zeta potential of nanoparticles were determined using ZetaPlus particle size and zeta potential analyzer (Brookhaven Instruments, Holtsville, NY) equipped with a 35 mW solid state laser (658 nm). Mean hydrodynamic diameters were calculated for size distribution by weight, assuming a lognormal distribution, and the results were expressed as mean ± S.E.M. of five runs. Zeta potential values were calculated from measured velocities using Smoluchowski's equation, and results were expressed as mean ± S.E.M. of five runs. Particle size and zeta potential of nanoparticle formulations having different concentrations of PEI were determined in distilled water, serum-containing growth medium or in 0.001M HEPES buffer adjusted to pH 5 or pH 7.4 with either 0.1 N sodium hydroxide or 0.1 N hydrochloric acid as necessary.
2.5 In vitro release of oligo/siRNA from nanoparticles
About 1 mg of oligo or siRNA-loaded nanoparticles was incubated with 1 ml of release medium (TE buffer, pH 7.4) in a rotary shaker at 100 rpm and 37° C. Samples were taken in triplicate for each time point. At predetermined intervals, the nanoparticle suspension was centrifuged at 7,500 rpm and 4° C for 10 min. The amount of oligo or siRNA in the supernatant was determined by Quant-iT™ Picogreen assay.
2.6 Cellular uptake of PLGA and PLGA-PEI nanoparticles
MDA-kb2 cells were seeded in 24-well plates at a seeding density of 5×104 cells/well. Following attachment, cells were treated for 30 min with fluorescently-labeled PLGA and PLGA-PEI nanoparticle formulations (100 μg/ml/well). The treated cells were washed with PBS twice, and then lysed with Triton X solution in PBS (300 μl/well). Cell lysates were lyophilized and then extracted with methanol (1 ml/sample) for 4 hrs. The extracts were centrifuged at 5,000 rpm for 10 min and the supernatants were subjected to HPLC analysis. The fluorescent label (6-coumarin) was eluted on a C8 column (Phenomenex) by using acetonitrile and sodium heptane sulfonate buffer in isocratic mode and detected using a fluorescence detector (Laballiance, Jasco, PA) at excitation and emission wavelengths of 450 nm and 490 nm, respectively (Panyam et al., 2003b). Nanoparticle concentrations were determined based on 6-coumarin loading in nanoparticles, and then normalized to total cell protein content determined using Pierce protein assay kit.
2.7 Gene silencing with siRNA-loaded nanoparticles
Two different cell lines were used in the study. EMT-6 G/L cells used in the study were stably transfected with the fire-fly luciferase gene, and constitutively express high levels of the protein. These cells were cultured at 37° C, 5% CO2 in Dulbecco's Modified Eagle Medium with high glucose and 10% fetal bovine serum. The MDA-kb2 cell line was derived from the breast cancer cell line MDA-MB-453, and is stably transformed with the MMTV.luciferase.neo reporter gene construct. Compounds that act through the glucocorticoid receptor can activate the MMTV luciferase reporter in this cell line (Wilson et al., 2002). In our studies, luciferase expression was induced by treating the cells with dexamethasone, a glucocorticoid receptor agonist. MDA-Kb2 cells were cultured in Leibovitz L-15 medium with 10% FBS at 37° C without CO2.
For transfection studies, EMT-6 G/L or MDA-Kb2 cells were plated in 24-well plates (30,000 cells/well/ml) 24 h prior to the experiment. Following attachment, the medium was removed and the cells were treated with nanoparticles containing luciferase-targeted siRNA (300 μg nanoparticles/ml/well), siRNA (dose equivalent to that in the nanoparticle treatment) along with the transfecting agent DharmaFECT®, nanoparticles containing non-targeted siRNA, or empty nanoparticles. In studies with MDA-Kb2 cells, each treatment group also received 100 nM dose of dexamethasone to induce luciferase expression. All the treatments were prepared in the respective growth medium containing serum. After 24 h incubation with the treatments, cells were washed with PBS and incubated with fresh medium. Luciferase expression was investigated at the end of 24 and 72 h. Cells were lysed by adding 100 μl/well of 1X cell culture lysis reagent. The luciferase activity in cell extracts was measured using a luciferase assay kit. Measurements were made in a single-tube luminometer (Zylux FB15/Sirius). The relative light units (RLUs) were normalized to the total cell protein of the extracts determined using Bio-Rad protein assay reagent. One-way ANOVA was used to test for the significance of differences between groups.
2.8 Intracellular trafficking of oligos
MDA-kb2 cells were used in this study. Cells were seeded in a two-chamber well slide at 100,000 cells/well seeding density and allowed to attach overnight. Cells were then treated for 24 h with TOTO-labeled oligos encapsulated in nanoparticles (300 μg nanoparticles/well) or transfected with equivalent amount of labeled oligos using DharmaFECT®. Cells were washed with PBS twice and then incubated with fresh growth medium. At 72 h post-treatment addition, cells were stained with LysoTracker® Green DND-26 (100 nM) for 2 h and then imaged using Fluoview 1000 confocal microscope (Center Valley, PA). Images captured using 568 nm (rhodamine) filter were overlaid with those captured using 488 nm (fluorescein) filter to determine the localization of oligos inside the cells.
2.9 Cytotoxicity evaluation
Cytotoxicity of selected formulations was determined using MTS assay in EMT-6 G/L cells. This assay is based on the ability of living cells to reduce a water-soluble yellow dye, MTS, to a purple colored formazan product by mitochondrial enzyme succinate dehydrogenase. About 10,000 cells were seeded per well in 96-well plates and allowed to attach for 24 h. Cells were then treated with empty PLGA-PEI nanoparticles or PLGA-PEI nanoparticles loaded with siRNA. Cells treated with PEI dissolved in growth medium was used as a positive control Cell viability was determined 72 h after treatment addition. The formazan formed was quantified by measuring the absorbance at 490 nm on a microplate reader.
3. Results
3.1 Encapsulation of oligos in PLGA nanoparticles
Custom synthesized double stranded oligomers (∼20 bp, similar in size to luciferase-targeted siRNA) were used in the initial formulation studies. The effect of lactide to glycolide ratio and the polymer molecular weight on the oligo encapsulation in nanoparticles is shown in Table 1. The efficiency of oligo encapsulation was observed to be generally low, with highest efficiencies of 43% and 47% observed for PLGA-40K and PLA-100 polymers, respectively. No specific trend was observed in the oligo encapsulation with change in either the polymer composition or the molecular weight. In vitro release studies indicated a 20-30% burst release during the first 24 hrs, followed by negligible release (less than 40-50% of the encapsulated oligos released over 14 days; not shown).
3.2 Effect of PEI on particle size and zeta potential of nanoparticles
Particle size of nanoparticles was found to be dependent on PEI concentration. In the absence of PEI, PLGA nanoparticles had an effective hydrodynamic diameter of 280 nm in deionized water and 230 nm in serum-containing growth medium. At low concentrations of PEI (<100 μg/30 mg PLGA), no significant effect on particle size was observed (Table 2). However, at concentrations higher than 500 μg of PEI, particle size increased significantly, suggesting aggregation of nanoparticles.
Table 2.
Effect of PEI on particle size of nanoparticles
| PEI Amount (μg/30 mg PLGA) | Particle Size (nm)a | |
|---|---|---|
| Water | Medium with serum | |
| 0.5 | 330 ± 6 | 214 ± 3 |
| 50 | 345 ± 5 | 263 ± 6 |
| 100 | 353 ± 4 | 244 ± 4 |
| 500 | 846 ± 20 | 454 ± 12 |
| 3000 | 815 ± 15 | 568 ± 26 |
| 5000 | 531 ± 17 | 537 ± 18 |
mean ± S.D. of five runs
Concentration of PEI in nanoparticles also influenced the surface charge of nanoparticles (Fig. 1). Increasing the concentration of PEI in PLGA nanoparticles increased the zeta potential of nanoparticles possibly because of the cationic charge of PEI. At concentrations above 500 μg PEI/30 mg PLGA, nanoparticles were cationic at both pH 7.4 and pH 5.0. At concentrations below 100 μg, nanoparticles were anionic at both pH 7.4 and pH 5.0. Nanoparticles containing 100 μg PEI/30 mg PLGA were anionic at pH 7.4 but were slightly cationic at pH 5.0. Nanoparticles loaded with preformed PEI-siRNA complex had a particle size in the range of 230 to 270 nm and anionic zeta potential (Table 3).
Figure 1.
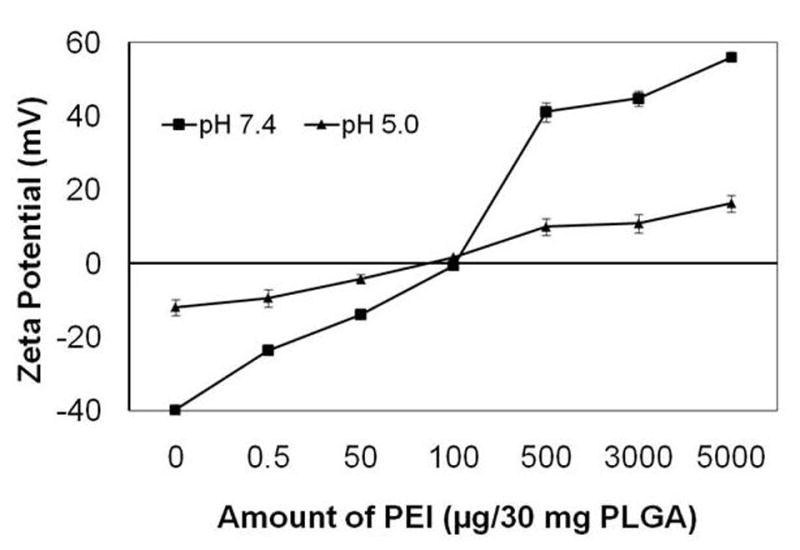
Effect of PEI incorporation on zeta potential of PLGA-40K nanoparticles measured in pH 5 and 7.4 buffers.
Table 3.
Particle size and zeta potential of nanoparticles encapsulating PEI-siRNA complex
| Formulation | Particle size (nm) | Zeta Potential (mV) | ||
|---|---|---|---|---|
| Water | Medium with serum | Water | Medium with serum | |
| PLGA-40K-siRNA-PEI complex | 270 ± 5 | 230 ± 8 | -18 ± 2 | -15 ± 4 |
3.3 Encapsulation of oligo and siRNA in the presence of PEI
PLGA-40K polymer was chosen for this study, as initial studies indicated that this polymer resulted in the highest encapsulation of model oligo among the polymers studied. PEI concentration (100 μg/30 mg PLGA) that did not significantly affect the particle size and retained the anionic charge of nanoparticles at pH 7.4 was chosen for this study. The oligo encapsulation efficiency in the presence of PEI increased by about 2-fold relative to that without PEI (Table 1). Similar to that observed with the oligo, luciferase-targeted siRNA was also encapsulated efficiently (86% ± 4 encapsulation efficiency). We also determined the encapsulation of pre-formed PEI-oligo complex in PLGA-40K nanoparticles. The efficiency of encapsulation of this complex was about 96 ± 2%.
3.4 In vitro release of oligos and siRNA from PLGA-PEI nanoparticles
Introduction of PEI in the organic phase of nanoparticle formulation improved the total amount and percent of siRNA released from nanoparticles. In the presence of PEI, a burst effect (∼30% over 24 hrs) was still observed but the release was continuous after that and the total percent of encapsulated siRNA released was ∼70% in 15 days (Fig. 2). When PEI was used to form a complex with oligo and the complex was encapsulated in nanoparticles, a very poor release profile was observed. Thus, despite 96 ± 2% encapsulation efficiency, only 9% of the encapsulated oligo was released over 15 days (Fig. 3).
Figure 2.
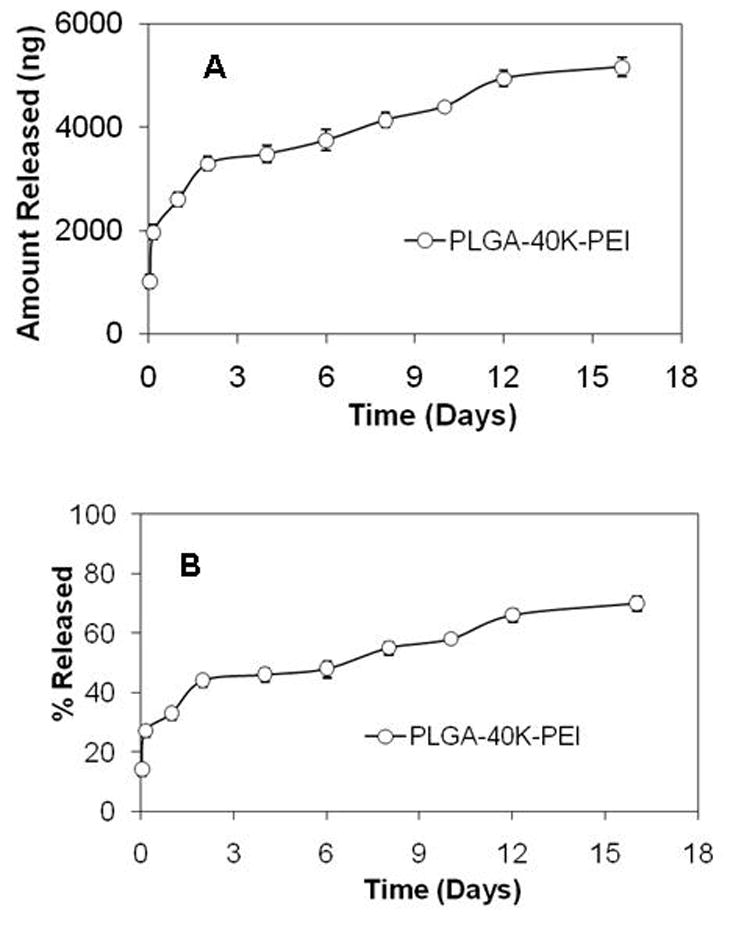
In vitro release of siRNA from PLGA-40K-PEI nanoparticles. (A) Amount of siRNA released from 1 mg nanoparticles and (B) Percent of encapsulated siRNA released.
Figure 3.
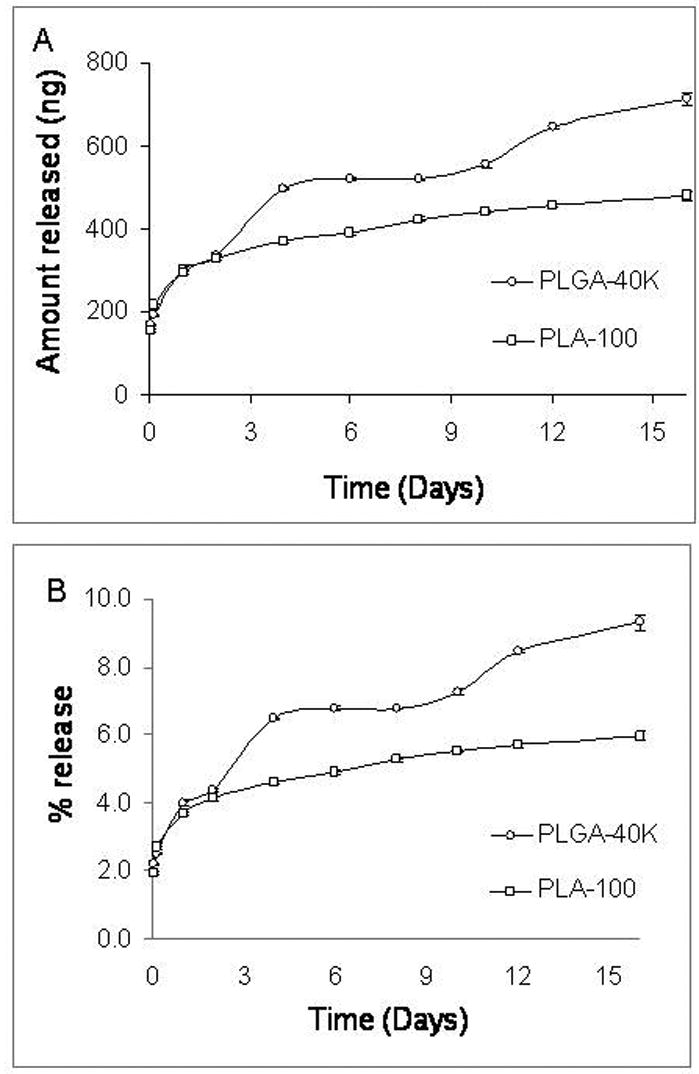
In vitro release of oligo from nanoparticles containing pre-formed oligo-PEI complex. (A) Amount of oligos released from 1 mg nanoparticles and (B) Percent of encapsulated oligos released.
3.5 Luciferase gene silencing study
Because nanoparticles fabricated using PLGA-40K-PEI demonstrated sustained release of siRNA, we used this formulation in the gene silencing studies. The ability of PLGA-PEI nanoparticles to decrease luciferase expression was determined in both a stably transfected cell line (EMT-6 G/L) as well as in an inducible cell line (MDA-Kb2). In both the cases, inhibition of luciferase expression was found to be greater and more sustained in cells treated with siRNA nanoparticles than in those transfected using a commercial transfecting agent (Figs. 4A and B). In the case of MDA-kb2 cells, treatment with dexamethasone increased luciferase expression by about 3-4 folds. PLGA-PEI nanoparticles reduced the luciferase expression in dexamethasone stimulated cells by about 70%, and this effect was sustained over three days.
Figure 4.
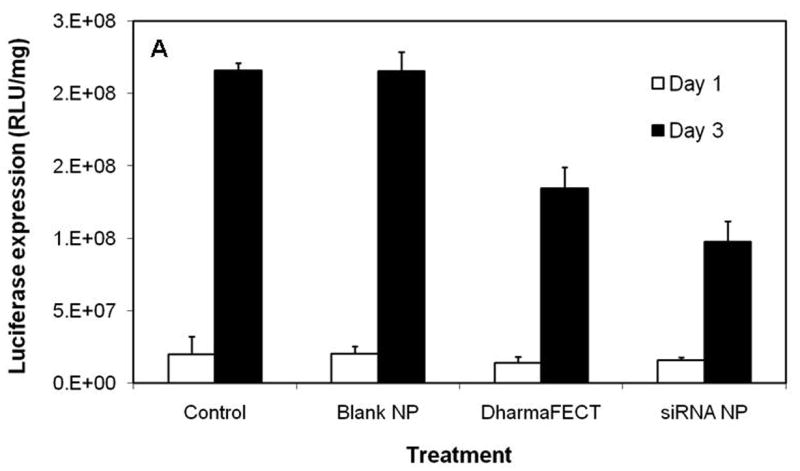
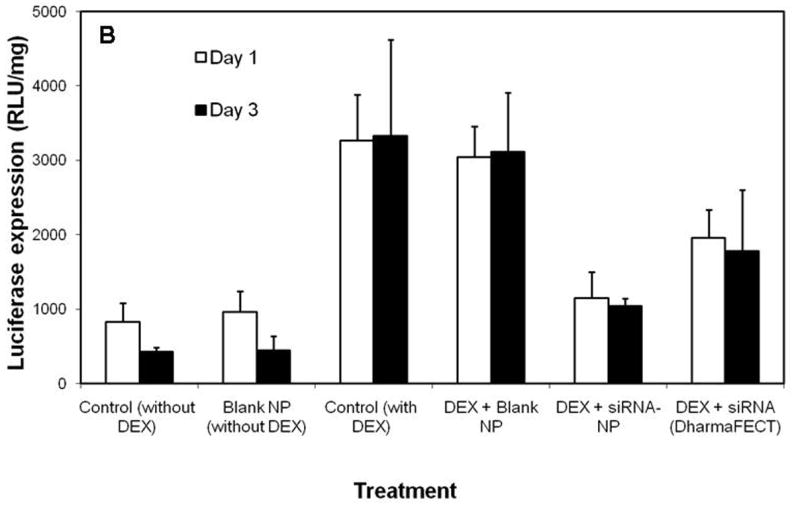
(A) Inhibition of luciferase expression in EMT-6 G/L cells. Luciferase expression was measured at the end of 24 and 72 h post siRNA addition. Control – cells treated with nanoparticles containing non-targeted siRNA; Blank NP – cells treated with blank nanoparticles; DharmaFECT - cells treated with DharmaFECT-siRNA combination; siRNA NP – cells treated with nanoparticles containing luciferase-targeted siRNA (B) Inhibition of luciferase expression in MDA-Kb2 cells. DEX refers to treatment with dexamethasone. Data as mean ± S.D., n =6, * P<0.05 (ANOVA).
3.6 Cellular uptake of PLGA and PLGA-PEI nanoparticles
Effect of PEI incorporation on the cellular uptake of PLGA nanoparticles was investigated in MDA-kb2 cells. Interestingly, the cellular uptake of PLGA-PEI nanoparticles was two-fold higher than PLGA nanoparticles (Fig. 5).
Figure 5.
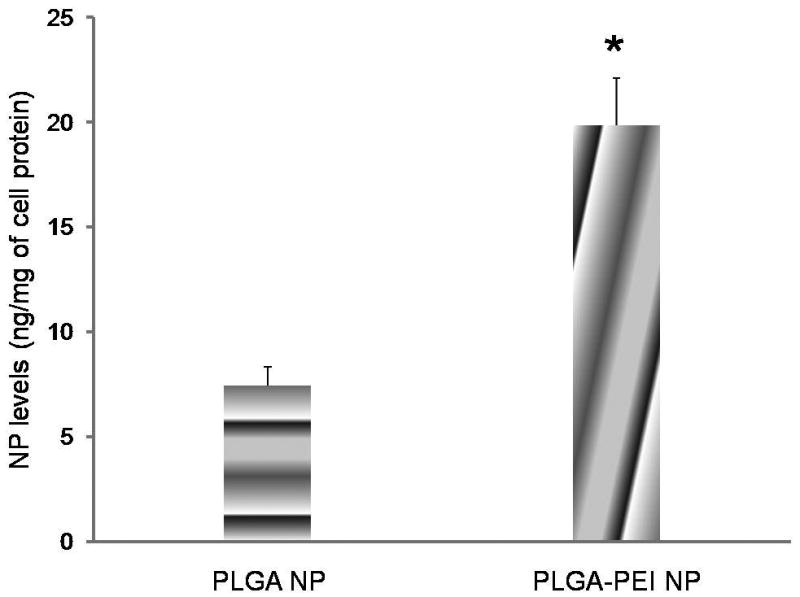
Effect of PEI on cellular uptake of PLGA nanoparticles (NP) in MBA-kb2 cells. Cells were incubated with nanoparticles for 30 min at 37° C. Data as mean ± S.D., n =6, * P<0.05, t-test.
3.7 Intracellular delivery of oligos
Intracellular delivery and distribution of labeled oligos was studied in MDA-kb2 cells counterstained for lysosomes. As can be seen in figure 6, incubation of cells with oligos encapsulated in nanoparticles resulted in both green and red fluorescence inside the cells with very little colocalization while treatment with free oligos resulted in predominantly yellow fluorescence, with the appearance of red fluorescence in some cells. This suggests that in the case of nanoparticle-treated cells, most of the released oligos were present outside the lysosomal vesicles, possibly in the cytoplasm. On the other hand, in the cells treated with free oligos, oligos appeared to be present in the lysosomal vesicles in most cells. Neither untreated nor blank nanoparticle treated cells showed autofluorescence (not shown).
Figure 6.
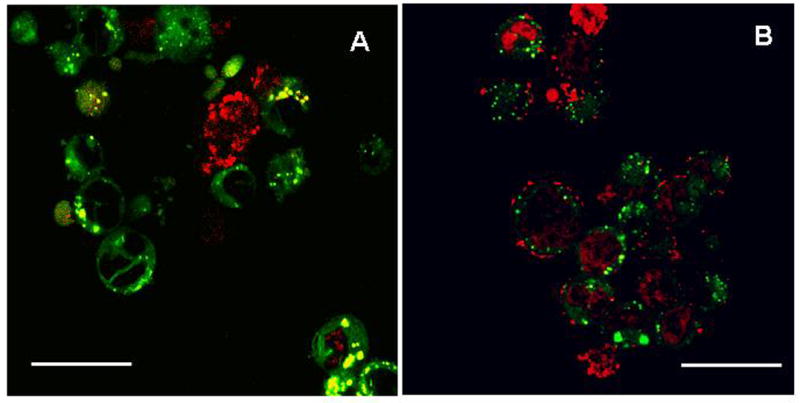
Intracellular trafficking of TOTO-3 labeled oligos in MDA-kb2 cells. Cells were treated with TOTO-3 labeled oligos either in the presence of DharmaFECT (A) or encapsulated in nanoparticles (B). Cells were imaged at the end of day 3 after counterstaining for lysosomes using LysoTracker® Green. Images obtained using rhodamine and fluorescein filters were overlaid to determine the oligo localization.
3.8 Cytotoxicity of PLGA-PEI nanoparticles
Effect of PEI and siRNA incorporation in PLGA nanoparticles on cytotoxicity was evaluated in the dose range that was used in the gene silencing studies. None of the nanoparticle formulations were found to be toxic to the cells in the dose range tested after incubation for 3 days (Fig. 7). Free PEI was found to be highly toxic to the cells at high concentrations.
Figure 7.
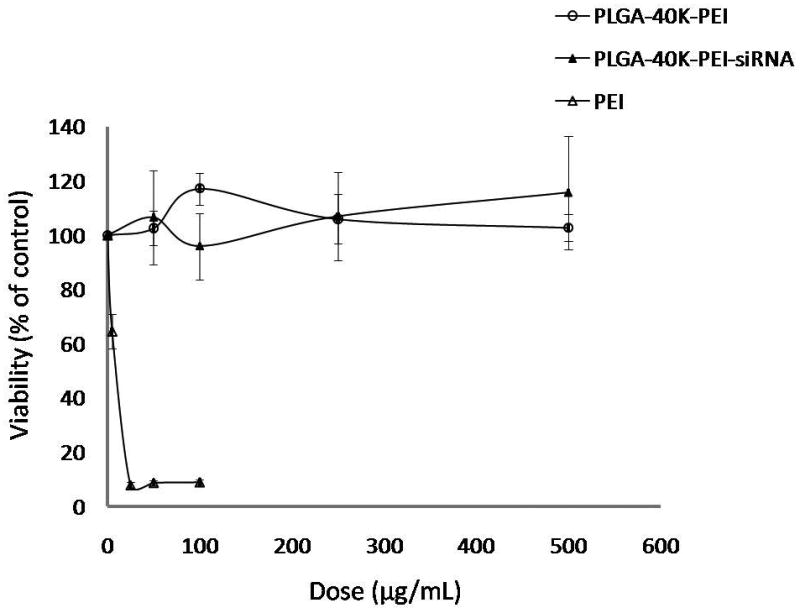
Cytotoxicity of nanoparticle formulations and PEI solution in EMT-6 G/L cells. PLGA-40K-PEI formulation did not have siRNA encapsulated. Cytotoxicity was determined using MTS assay at 72 h following treatment addition.
4. Discussion
Successful gene silencing using RNA interference depends on effective cellular delivery and cytoplasmic entry of siRNA (Kim and Rossi, 2007). Once in the cytoplasm, siRNA molecules bind with RISCs to start the process of degrading the target mRNA molecules. However, like most other macromolecules, siRNA is degraded in the lysosomes following cellular uptake. Thus, the carrier used for RNA interference has to enable the lysosomal escape of siRNA. In addition, to be useful in vivo, the carrier has to be stable in the presence of serum (Yi et al., 2006).
Previous studies have shown that PLGA nanoparticles enter the cell efficiently and rapidly through a combination of specific and non-specific endocytic mechanisms (Panyam and Labhasetwar, 2003). Following uptake, a fraction of nanoparticles escapes the lysosomal compartment and reaches the cytosol. Reversal of the surface charge of PLGA nanoparticles from anionic to cationic in the acidic pH of the lysosomes is considered an important feature that enables PLGA nanoparticles to escape the lysosomal compartment (Panyam and Labhasetwar, 2003; Panyam et al., 2002b). Following lysosomal escape, PLGA nanoparticles are retained in the cellular cytoplasm for more than 2 weeks, slowly releasing their payload (Panyam and Labhasetwar, 2004).
Efficient cellular uptake, rapid lysosomal escape and sustained intracellular drug release characteristics make PLGA nanoparticles an attractive vehicle for gene silencing applications. However, our initial studies suggest that the encapsulation of siRNA in PLGA nanoparticles is not efficient. Because previous studies have suggested that encapsulation of nucleic acids in PLGA nanoparticles can be significantly improved by optimizing the polymer molecular weight and lactide composition (Prabha and Labhasetwar, 2004a), we evaluated the effect of polymer molecular weight and composition on siRNA encapsulation. Irrespective of the polymer used, the encapsulation efficiency was less than 50%. It is possible that the small size of siRNA (20 bp; compared to >3 kbp for plasmid DNA) and high water solubility enables rapid diffusion of siRNA molecules out of the polymer phase during nanoparticle fabrication. Such low encapsulation efficiencies in PLGA/PLA nanoparticles have been reported for low molecular weight, water-soluble drugs (Eroglu et al., 2001; Ubrich et al., 2004). In addition to poor loading, the release of siRNA from PLGA nanoparticles was incomplete. Poor encapsulation efficiency and incomplete release also resulted in poor gene silencing (not shown).
Because siRNA is highly anionic and polar, we rationalized that introduction of cationic charges in the polymer matrix could help retain siRNA molecules in nanoparticles. We used PEI, a popular cationic polymer used in gene delivery (Aigner, 2006; Jiang et al., 2008; Malek et al., 2008; Mao et al., 2006), to introduce cationic charges and increase siRNA encapsulation in nanoparticles. Also, because PEI is more hydrophilic than PLGA, it can be expected that PEI will enable a greater influx of water into the nanoparticle matrix, promoting greater degradation of PLGA and improved siRNA release from nanoparticles. A primary objective while designing the composite PLGA-PEI nanoparticles was to ensure that addition of PEI does not induce aggregation of nanoparticles and maintains the surface charge reversal of nanoparticles at the acidic pH (∼5) found in the lysosomes. Our studies indicated that a PEI concentration of 100 μg/30 mg PLGA resulted in nanoparticles that were anionic at physiologic pH, mildly cationic at pH 5 and were stable in the presence of serum. This concentration of PEI was, therefore, chosen for further studies. A previous study that investigated the use of PEI in PLGA nanoparticles reported a reduction in particle size with increase in the amount of PEI added (Takashima et al., 2007). This is in contrast to our results which show an increase in particle size with increasing PEI concentration. There are two possible reasons why these two PLGA/PEI formulations show different trends in particle size. First, the formulation processes used in the two reports are different. We used emulsion solvent evaporation technique for nanoparticle formulation while Takashima et al. used spray drying for the preparation of PLGA/PEI nanoparticles. Second, in the previous study, PLGA nanoparticle emulsion was dried with mannitol, and mannitol microparticles containing PLGA nanospheres were obtained. It is possible that combination of spray drying and the use of mannitol prevented aggregation of nanoparticles at high concentrations of PEI.
As hypothesized, introduction of PEI in PLGA nanoparticles significantly improved the encapsulation of oligos and siRNA and improved the siRNA release profile. Unlike nanoparticles fabricated using just PLGA, which showed a big burst followed by negligible release, oligo release was more continuous in the case of PLGA-PEI nanoparticles. As discussed earlier, there are several potential reasons for this observed enhancement in release. Being a more hydrophilic polymer than PLGA, PEI would dissolve more readily in aqueous buffers, possibly creating channels in the nanoparticle matrix. This will accelerate polymer degradation and siRNA release from nanoparticles. Similar enhanced drug release has been observed from PLGA/PLA particles containing hydrophilic additives like polyethylene glycol (Naraharisetti et al., 2005). Further studies are needed to confirm possible mechanisms of accelerated siRNA release in the presence of PEI.
We also determined whether introduction of PEI along with the oligos (pre-formed complex of PEI-oligo) rather than in the polymer phase would improve the oligo loading in nanoparticles. Several previous studies have investigated this approach, where PEI-DNA complex is encapsulated in microparticles for sustained release of PEI-DNA complex (De Rosa et al., 2002). This approach resulted in high encapsulation efficiency. However, in vitro release studies indicated that the loaded oligos were released very slowly. Less than 10% of the encapsulated oligos was released at the end of 15 days. This is possibly due to the poor diffusion of the large size (several nanometers) PEI-oligo complex out of the nanoparticle polymer matrix (Oster et al., 2005).
Further, we investigated whether incorporation of PEI in PLGA nanoparticles affected the cellular uptake of nanoparticles. We found that inclusion of PEI in the nanoparticle matrix, in fact, resulted in greater cellular accumulation of nanoparticles. While it is not clear why PLGA-PEI nanoparticles were taken up more than PLGA nanoparticles, it is possible that the mild cationic charge of PLGA-PEI nanoparticles in the acidic pH of the endo-lysosomal compartment may enable a greater fraction of the nanoparticles to escape the lysosomes. This, in turn, could result in reduced recycling and improved retention of nanoparticles within the cells, leading greater overall cellular accumulation of nanoparticles. Fluorescence microscopic studies examining the intracellular release and distribution of nanoparticle-encapsulated siRNA suggested that at least a fraction of the siRNA released was outside the lysosomes, providing support to the above possibility. Additionally, a fraction of the PEI that is either released or is present on the surface of the nanoparticles could enable nanoparticles to escape the endo-lysosomes through the proton sponge effect. Upon acidification of the endo-lysosome, PEI can cause osmotic swelling and subsequent endosomal rupture, followed by the escape of PEI/siRNA or oligo complexes into the cytosol (De Smedt et al., 2000). Our results are similar to previous studies (Bivas-Benita et al., 2004) that reported greater cellular uptake of PLGA nanoparticles with PEI adsorbed on the surface.
In vivo, some proteins like structural proteins are constitutively expressed while others are induced following exposure of cells to specific extracellular or intracellular signals (Ruffini et al., 2007). To determine the ability of siRNA-loaded nanoparticles to inhibit the expression of either type of protein, we used two different cell culture models. EMT-6 cells used here constitutively overexpress luciferase and were used as a model to study siRNA-mediated inhibition of a constitutively expressed gene. MDA-kb2 cells express increased levels of luciferase upon exposure to dexamethasone and were used to study the inhibition of inducible gene expression. PLGA-PEI nanoparticles were effective in reducing the luciferase expression in both models, suggesting the broad usefulness of this system. In vitro cell culture conditions do not permit chronic gene silencing studies, and therefore, in this report we studied gene silencing only for 3 days. We plan to examine sustained gene silencing in vivo in our future studies.
In order to demonstrate intracellular delivery of siRNA as a key mechanism in nanoparticle-mediated gene silencing, we investigated the intracellular distribution of labeled oligos following treatment with nanoparticles or with commercial transfecting reagent. Our studies indicate that nanoparticles slowly release the encapsulated oligos within the cells. Treatment with commercial reagent also resulted in sustained oligo levels within the cells; however, qualitative colocalization studies suggest that a greater fraction of the oligo was present in cytosol following nanoparticle treatment than that following treatment with the commercial reagent. A similar sustained cytosolic release has been observed for PLGA nanoparticles loaded with plasmid DNA (Prabha and Labhasetwar, 2004b). It was interesting to note that with nanoparticle treatment, oligo-associated red fluorescence was present in almost all the cells, indicating efficient delivery.
One concern with using PEI to improve siRNA delivery is the potential for cytotoxicity. Several studies have demonstrated the lack of cytotoxicity following treatment with PLGA nanoparticles (Basarkar et al., 2007). However, PEI has been shown to be toxic to cells, depending on the concentration used (Gwak and Kim, 2008). Our studies indicate that, at the concentrations used, PLGA-PEI nanoparticles do not induce significant cytotoxicity. This is possibly due to the low concentration of PEI used in the formulation (3 ng per μg of PLGA). While free PEI resulted in significant cytotoxicity at high doses, at doses equivalent to that used in nanoparticles (∼1 μg/ml), no significant toxicity was seen
5. Conclusion
Introduction of PEI in PLGA nanoparticles significantly improved the encapsulation of siRNA and its release. Encapsulation of siRNA in PLGA-PEI nanoparticles resulted in greater inhibition of the target gene expression in vitro. Intracellular siRNA release is the proposed mechanism of gene silencing observed with PLGA-PEI nanoparticles. Stability in the presence of serum and lack of cytotoxicity further add to the potential of PLGA-PEI nanoparticles for gene silencing applications. Future studies will focus on investigating PLGA-PEI nanoparticles for in vivo gene silencing applications.
Acknowledgments
Funding from NIH (7R21CA116641-02) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aigner A. Delivery Systems for the Direct Application of siRNAs to Induce RNA Interference (RNAi) In Vivo. J Biomed Biotechnol. 2006;2006:71659. doi: 10.1155/JBB/2006/71659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner A. Nonviral in vivo delivery of therapeutic small interfering RNAs. Curr Opin Mol Ther. 2007;9:345–352. [PubMed] [Google Scholar]
- Basarkar A, Devineni D, Palaniappan R, Singh J. Preparation, characterization, cytotoxicity and transfection efficiency of poly(DL-lactide-co-glycolide) and poly(DL-lactic acid) cationic nanoparticles for controlled delivery of plasmid DNA. Int J Pharm. 2007;343:247–254. doi: 10.1016/j.ijpharm.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivas-Benita M, Romeijn S, Junginger HE, Borchard G. PLGA-PEI nanoparticles for gene delivery to pulmonary epithelium. Eur J Pharm Biopharm. 2004;58:1–6. doi: 10.1016/j.ejpb.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Cheema SK, Chen E, Shea LD, Mathur AB. Regulation and guidance of cell behavior for tissue regeneration via the siRNA mechanism. Wound Repair Regen. 2007;15:286–295. doi: 10.1111/j.1524-475X.2007.00228.x. [DOI] [PubMed] [Google Scholar]
- Cohen H, Levy RJ, Gao J, Fishbein I, Kousaev V, Sosnowski S, Slomkowski S, Golomb G. Sustained delivery and expression of DNA encapsulated in polymeric nanoparticles. Gene Ther. 2000;7:1896–1905. doi: 10.1038/sj.gt.3301318. [DOI] [PubMed] [Google Scholar]
- De Rosa G, Quaglia F, La Rotonda MI, Appel M, Alphandary H, Fattal E. Poly(lactide-co-glycolide) microspheres for the controlled release of oligonucleotide/polyethylenimine complexes. J Pharm Sci. 2002;91:790–799. doi: 10.1002/jps.10063. [DOI] [PubMed] [Google Scholar]
- De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm Res. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- Devi GR. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 2006;13:819–829. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Eroglu H, Kas HS, Oner L, Turkoglu OF, Akalan N, Sargon MF, Ozer N. The in-vitro and in-vivo characterization of PLGA:L-PLA microspheres containing dexamethasone sodium phosphate. J Microencapsul. 2001;18:603–612. doi: 10.1080/02652040010019587. [DOI] [PubMed] [Google Scholar]
- Gwak SJ, Kim BS. Poly(lactic-co-glycolic acid) nanosphere as a vehicle for gene delivery to human cord blood-derived mesenchymal stem cells: comparison with polyethylenimine. Biotechnol Lett. 2008 doi: 10.1007/s10529-008-9676-7. [DOI] [PubMed] [Google Scholar]
- Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Lack of interferon response in animals to naked siRNAs. Nat Biotechnol. 2004;22:1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- Jiang G, Park K, Kim J, Kim KS, Oh EJ, Kang H, Park TG, Hahn SK. Hyaluronic acid - polyethylenimine conjugate for target specific intracellular delivery of siRNA. Biopolymers. 2008 doi: 10.1002/bip.20978. [DOI] [PubMed] [Google Scholar]
- Khan A, Benboubetra M, Sayyed PZ, Ng KW, Fox S, Beck G, Benter IF, Akhtar S. Sustained polymeric delivery of gene silencing antisense ODNs, siRNA, DNAzymes and ribozymes: in vitro and in vivo studies. J Drug Target. 2004;12:393–404. doi: 10.1080/10611860400003858. [DOI] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- Kim IS, Lee SK, Park YM, Lee YB, Shin SC, Lee KC, Oh IJ. Physicochemical characterization of poly(L-lactic acid) and poly(D,L-lactide-co-glycolide) nanoparticles with polyethylenimine as gene delivery carrier. Int J Pharm. 2005;298:255–262. doi: 10.1016/j.ijpharm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Langer R, Tirrell DA. Designing materials for biology and medicine. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- Luby TM, Cole G, Baker L, Kornher JS, Ramstedt U, Hedley ML. Repeated immunization with plasmid DNA formulated in poly(lactide-co-glycolide) microparticles is well tolerated and stimulates durable T cell responses to the tumor-associated antigen cytochrome P450 1B1. Clin Immunol. 2004;112:45–53. doi: 10.1016/j.clim.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Malek A, Czubayko F, Aigner A. PEG grafting of polyethylenimine (PEI) exerts different effects on DNA transfection and siRNA-induced gene targeting efficacy. J Drug Target. 2008;16:124–139. doi: 10.1080/10611860701849058. [DOI] [PubMed] [Google Scholar]
- Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjug Chem. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Deliv Rev. 2002;54:715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- Naraharisetti PK, Lew MD, Fu YC, Lee DJ, Wang CH. Gentamicin-loaded discs and microspheres and their modifications: characterization and in vitro release. J Control Release. 2005;102:345–359. doi: 10.1016/j.jconrel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Oster CG, Kim N, Grode L, Barbu-Tudoran L, Schaper AK, Kaufmann SH, Kissel T. Cationic microparticles consisting of poly(lactide-co-glycolide) and polyethylenimine as carriers systems for parental DNA vaccination. J Control Release. 2005;104:359–377. doi: 10.1016/j.jconrel.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Pai Kasturi S, Qin H, Thomson KS, El-Bereir S, Cha SC, Neelapu S, Kwak LW, Roy K. Prophylactic anti-tumor effects in a B cell lymphoma model with DNA vaccines delivered on polyethylenimine (PEI) functionalized PLGA microparticles. J Control Release. 2006;113:261–270. doi: 10.1016/j.jconrel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Panyam J, Dali MM, Sahoo SK, Ma W, Chakravarthi SS, Amidon GL, Levy RJ, Labhasetwar V. Polymer degradation and in vitro release of a model protein from poly(D,L-lactide-co-glycolide) nano- and microparticles. J Control Release. 2003a;92:173–187. doi: 10.1016/s0168-3659(03)00328-6. [DOI] [PubMed] [Google Scholar]
- Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- Panyam J, Labhasetwar V. Sustained cytoplasmic delivery of drugs with intracellular receptors using biodegradable nanoparticles. Mol Pharm. 2004;1:77–84. doi: 10.1021/mp034002c. [DOI] [PubMed] [Google Scholar]
- Panyam J, Lof J, O'Leary E, Labhasetwar V. Efficiency of Dispatch and Infiltrator cardiac infusion catheters in arterial localization of nanoparticles in a porcine coronary model of restenosis. J Drug Target. 2002a;10:515–523. doi: 10.1080/1061186021000038391. [DOI] [PubMed] [Google Scholar]
- Panyam J, Sahoo SK, Prabha S, Bargar T, Labhasetwar V. Fluorescence and electron microscopy probes for cellular and tissue uptake of poly(D,L-lactide-co-glycolide) nanoparticles. Int J Pharm. 2003b;262:1–11. doi: 10.1016/s0378-5173(03)00295-3. [DOI] [PubMed] [Google Scholar]
- Panyam J, Sahoo SK, Prabha S, Bargar T, Labhasetwar V. Fluorescence and electron microscopy probes for cellular and tissue uptake of poly(D,L-lactide-co-glycolide) nanoparticles. Int J Pharm. 2003c;262:1–11. doi: 10.1016/s0378-5173(03)00295-3. [DOI] [PubMed] [Google Scholar]
- Panyam J, Zhou WZ, Prabha S, Sahoo SK, Labhasetwar V. Rapid endo-lysosomal escape of poly(DL-lactide-co-glycolide) nanoparticles: implications for drug and gene delivery. FASEB J. 2002b;16:1217–1226. doi: 10.1096/fj.02-0088com. [DOI] [PubMed] [Google Scholar]
- Perlstein I, Connolly JM, Cui X, Song C, Li Q, Jones PL, Lu Z, DeFelice S, Klugherz B, Wilensky R, Levy RJ. DNA delivery from an intravascular stent with a denatured collagen-polylactic-polyglycolic acid-controlled release coating: mechanisms of enhanced transfection. Gene Ther. 2003;10:1420–1428. doi: 10.1038/sj.gt.3302043. [DOI] [PubMed] [Google Scholar]
- Pichler A, Zelcer N, Prior JL, Kuil AJ, Piwnica-Worms D. In vivo RNA interference-mediated ablation of MDR1 P-glycoprotein. Clin Cancer Res. 2005;11:4487–4494. doi: 10.1158/1078-0432.CCR-05-0038. [DOI] [PubMed] [Google Scholar]
- Prabha S, Labhasetwar V. Critical determinants in PLGA/PLA nanoparticle-mediated gene expression. Pharm Res. 2004a;21:354–364. doi: 10.1023/b:pham.0000016250.56402.99. [DOI] [PubMed] [Google Scholar]
- Prabha S, Labhasetwar V. Nanoparticle-mediated wild-type p53 gene delivery results in sustained antiproliferative activity in breast cancer cells. Mol Pharm. 2004b;1:211–219. doi: 10.1021/mp049970+. [DOI] [PubMed] [Google Scholar]
- Quarck R, Holvoet P. Gene therapy approaches for cardiovascular diseases. Curr Gene Ther. 2004;4:207–223. doi: 10.2174/1566523043346499. [DOI] [PubMed] [Google Scholar]
- Ruffini PA, Morandi P, Cabioglu N, Altundag K, Cristofanilli M. Manipulating the chemokine-chemokine receptor network to treat cancer. Cancer. 2007;109:2392–2404. doi: 10.1002/cncr.22706. [DOI] [PubMed] [Google Scholar]
- Schagen FH, Ossevoort M, Toes RE, Hoeben RC. Immune responses against adenoviral vectors and their transgene products: a review of strategies for evasion. Crit Rev Oncol Hematol. 2004;50:51–70. doi: 10.1016/S1040-8428(03)00172-0. [DOI] [PubMed] [Google Scholar]
- Sioud M. On the delivery of small interfering RNAs into mammalian cells. Expert Opin Drug Deliv. 2005;2:639–651. doi: 10.1517/17425247.2.4.639. [DOI] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Saito R, Nakajima A, Oda M, Kimura A, Kanazawa T, Okada H. Spray-drying preparation of microparticles containing cationic PLGA nanospheres as gene carriers for avoiding aggregation of nanospheres. Int J Pharm. 2007;343:262–269. doi: 10.1016/j.ijpharm.2007.05.042. [DOI] [PubMed] [Google Scholar]
- Ubrich N, Bouillot P, Pellerin C, Hoffman M, Maincent P. Preparation and characterization of propranolol hydrochloride nanoparticles: a comparative study. J Control Release. 2004;97:291–300. doi: 10.1016/j.jconrel.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Wang D, Robinson DR, Kwon GS, Samuel J. Encapsulation of plasmid DNA in biodegradable poly(D, L-lactic-co-glycolic acid) microspheres as a novel approach for immunogene delivery. J Control Release. 1999;57:9–18. doi: 10.1016/s0168-3659(98)00099-6. [DOI] [PubMed] [Google Scholar]
- Wilson VS, Bobseine K, Lambright CR, Gray LE., Jr A novel cell line, MDA-kb2, that stably expresses an androgen- and glucocorticoid-responsive reporter for the detection of hormone receptor agonists and antagonists. Toxicol Sci. 2002;66:69–81. doi: 10.1093/toxsci/66.1.69. [DOI] [PubMed] [Google Scholar]
- Xu D, McCarty D, Fernandes A, Fisher M, Samulski RJ, Juliano RL. Delivery of MDR1 small interfering RNA by self-complementary recombinant adeno-associated virus vector. Mol Ther. 2005;11:523–530. doi: 10.1016/j.ymthe.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Wu H, Jia GL. Formulation and characterization of poly (D,L-lactide-co-glycolide) nanoparticle containing vascular endothelial growth factor for gene delivery. J Clin Pharm Ther. 2006;31:43–48. doi: 10.1111/j.1365-2710.2006.00702.x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhao B, Jiang H, Wang B, Ma B. Cationic lipids and polymers mediated vectors for delivery of siRNA. J Control Release. 2007;123:1–10. doi: 10.1016/j.jconrel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Zhang XQ, Intra J, Salem AK. Comparative study of poly (lactic-co-glycolic acid)-poly ethyleneimine-plasmid DNA microparticles prepared using double emulsion methods. J Microencapsul. 2008;25:1–12. doi: 10.1080/02652040701659347. [DOI] [PubMed] [Google Scholar]


