Abstract
H37Rv and H37Ra have been widely used as models of virulent and avirulent strains, respectively, of Mycobacterium tuberculosis. Since the sequencing of H37Rv, microarrays have been used to investigate gene expression of M. tuberculosis strains under various conditions, and to compare gene expression of specific isolates of the organism. Because differences in the virulence of these organisms could also be manifest via their differential induction of host genes, we used Affymetrix Human Genome Arrays U133A and U133B to evaluate human alveolar macrophage (AM) responses to infection with H37Rv and H37Ra. H37Rv altered expression of far more genes than did H37Ra. Moreover, the genes induced by H37Rv to a greater extent than by H37Ra were predominantly associated with the development of effective immunity. H37Rv markedly increased expression of IL-23 p19, whereas neither organism significantly induced IL-12 p35 expression. Quantitative PCR confirmed that H37Rv induced significantly more AM p19 expression than did H37Ra. After low-level infection of both AM and peripheral blood monocytes (MN) with H37Rv, neither cell type produced IL-12 (by ELISA). In contrast, AM displayed significant IL-23 production in response to H37Rv, whereas MN did not. Our findings thus suggest an important role for IL-23 in human host responses to pulmonary infection with M. tuberculosis, and are consistent with epidemiologic and genetic studies that imply that H37Rv may not have unusual capacity to cause human disease.
Keywords: tuberculosis, virulence, alveolar macrophage, microarray, IL-23
CLINICAL RELEVANCE
The findings of this study are relevant to ongoing efforts to characterize the virulence mechanisms of M. tuberculosis. More generally, our results provide insight into the development of protective Th1-like immune responses in the human lung.
Since Robert Koch's identification of Mycobacterium tuberculosis as the cause of tuberculosis in 1882, much speculation has been given to the relative roles of host susceptibility and strain virulence in the wide range of clinical outcomes that follow infection with M. tuberculosis. Early in the following century, a clinical isolate of the organism identified as strain H37 and first cultivated at Saranac laboratories became valued by a variety of researchers for its consistent virulent behavior in animal models of infection. The usefulness of this strain in studies of virulence was increased when new stocks of the organism displayed a sudden change to an avirulent phenotype that no longer caused progressive disease in animals. Although this change coincided with the use of a different culture medium for passage of the organism, reversion to use of the prior medium did not restore virulence, and this avirulent derivative was designated H37Ra. Alternative stocks that retained their initial virulence were renamed H37Rv (1). In the subsequent decades, investigations of the virulence of M. tuberculosis have frequently involved comparisons of the H37Rv and H37Ra. In animal models infection, H37Rv displays greater in vivo replication than does H37Ra, resulting in higher bacterial loads of M. tuberculosis in the lungs and other organs (2, 3).
The widespread acceptance of H37Rv as a standard virulent organism was reflected in the decision to make it the first M. tuberculosis strain to be fully sequenced (4). Subsequent comparison with H37Ra indicated several deletions in the genome of the avirulent strain, although complementation of H37Ra with these sequences did not restore virulence (5). The availability of the H37Rv genome also allowed for the design of microarrays suitable for the examination of global gene expression of M. tuberculosis. These have been used to assess gene expression of H37Rv under various conditions (6, 7), and to compare gene expression of H37Ra and H37Rv with those of selected clinical isolates of M. tuberculosis (8).
H37Rv and H37Ra also display differing capacities for intracellular growth within human mononuclear phagocytes (9), including alveolar macrophages (10). The relevance of intracellular growth to strain virulence is supported by recent studies indicating that so-called “hypervirulent strains,” identified by molecular fingerprinting as being responsible for outbreak “clusters” of active tuberculosis, grow more rapidly within human phagocytes than do nonclustered isolates (11, 12). Although differences in the interactions of M. tuberculosis strains of varying virulence with human phagocytes have been assessed using focused gene expression arrays (13, 14), these responses have not previously been evaluated using a genome-wide approach. We therefore assessed differences in gene expression of human alveolar macrophages infected with H37Ra and H37Rv using an Affymetrix human genome array system. We hypothesized that avirulent strain H37Ra would induce protective host responses, whereas the virulent H37Rv strain would suppress, or fail to induce, specific genes associated with protective immunity. Instead, however, we found that H37Ra had relatively little impact on macrophage gene expression, whereas H37Rv induced responses of genes associated with development of protective Th1-like immunity.
MATERIALS AND METHODS
Growth and Preparation of Organisms
M. tuberculosis H37Ra and H37Rv were obtained from American Type Tissue Collection (#25177 and #25618, respectively; ATCC, Rockville, MD). Processing of the organisms was performed as previously described in detail (15). Briefly, organisms were grown as broth cultures in Middlebrook 7H9 medium (#271310; Becton Dickinson, Sparks, MD) with 10% Middlebrook ADC enrichment (#212352; Becton Dickinson) and 0.2% glycerol within 1.7-L roller-bottles. Cultures were harvested during mid-log growth, aliquoted, and stored at −70°C. The initial aliquots were used to develop subsequent stocks for use in infections to limit the passaging of organisms in our hands. At the time of infection, bacterial clumps were removed using a series of mechanical disruptions and centrifugations as previously described (15). Quantification of the infecting inoculums was based on plating of serial dilutions of bacteria prepared in this manner on Middlebrook 7H10 agar with 10% OADC enrichment (#262710 and # 212240, respectively; Becton Dickinson), and subsequent manual counting of colony-forming units (CFU).
Cell Populations
All human subjects protocols were reviewed and approved by the Institutional Review Board of University Hospitals of Cleveland and the Case Western Reserve University School of Medicine.
Bronchoalveolar lavage (BAL) was performed on healthy nonsmoking volunteers with four 60-cc aliquots of normal saline. Resulting BAL samples were immediately placed on ice for transport to the laboratory. Cell pellets obtained by centrifugation were combined, resuspended in Iscove's Modified Dulbecco's Medium (IMDM) (#12–722F; Cambrex Bio Science, Walkersville, MD) with 5% fresh autologous serum (AS) and 1% penicillin G (PCN, #P-7749; Sigma, St. Louis, MO) and counted using a hemocytometer. Cytospin slides were prepared for each subject and, after modified Wright's staining (Diff-Qwik, #47733–150; VWR, West Chester, PA), differential cell count was performed on a minimum of 300 cells under light microscopy. Over 90% of BAL cells from each subject were identified as alveolar macrophages (AM); these cells are therefore subsequently referred to as AM.
Peripheral blood monocytes (MN) were isolated from venous blood by density sedimentation using Ficoll-Hypaque followed by adherence to plastic, as previously described (15).
Infections
After counting, all AM samples were resuspended in 4 ml of IMDM with 30% AS and 1% penicillin G and aliquoted into four sterile screw-topped microfuge tubes (#72.692.005; Sarstedt, Newton, NC). For some studies, MN from each subject were likewise re-suspended into 4 ml of medium, aliquoted in the same manner, and subsequently processed in parallel. For both the AM and MN aliquots, two tubes were used as uninfected controls at times 0 and 24 hours. The time 0 control tube was immediately centrifuged at 3,000 rpm (750 × g) for 10 minutes and processed as described below. Frozen stocks of H37Rv and H37Ra were thawed and prepared for infection by vortexing with sterile glass beads followed by centrifugation to remove clumped bacteria, as previously described (15). H37Rv and H37Ra then were added in a 3:1 bacteria-to-cell ratio to the third and fourth tubes, respectively. After 2 hours of incubation, microfuge tubes were centrifuged at 3,000 rpm (750 × g). Supernatants were discarded to remove non-phagocytosed organisms, and cell pellets were resuspended in 1 ml IMDM with 10% AS and 1% PCN. Cells were then incubated at 37°C in a 5% CO2 incubator. Twenty-four hours after the addition of bacteria, the remaining three tubes of each cell type were again centrifuged for further RNA processing.
Quantification of Intracellular Infection
To maximize the RNA yield for subsequent microarray analysis, the entire volume of infected BAL cells were lysed for RNA preparation as described below. To confirm that comparable infection of AM was achieved after incubation of H37Rv and H37Ra, BAL cells of an additional seven subjects were infected in the manner described above. Both immediately after completion of the 2-hour incubation with bacteria and 24 hours subsequently, supernatants were removed and quantification of M. tuberculosis performed as previously described (15). Briefly, cells were lysed by incubation with 0.067% SDS in Middlebrook 7H9 medium at 37°C for 10 minutes, which was followed by addition of an equal volume of 20% BSA. Serial dilutions of the resulting lysate were prepared and plated onto 7H10/OADC plates, which were placed in a 5%CO2 incubator at 37°C until colonies were visible. Colonies were counted under a microscope, and results expressed as CFU per 1 × 106 infected BAL cells.
Collection of RNA Samples
After centrifugation of infected and control cells as described, all cell pellets were resuspended in freshly prepared Qiagen RTL buffer (#79216; Qiagen, Valencia, CA) with 1% β-mercaptoethanol according to the manufacturer's recommendations for specific numbers of cells (i.e., 350 μl of buffer was added to samples containing < 5 × 106 cells for subsequent “mini-prep” processing, whereas 600 μl was added to samples containing 5 × 106 or more cells for “midi-prep” use). Samples were then stored at −70°C until all samples had been obtained, at which time they were shipped on dry ice to Wyeth Research for further processing.
Preparation of RNA from Cell Lysates
Frozen RNA lysates in RLT buffer were heated at 37°C for 10 minutes, then passed through QIAshredder spin columns, followed by RNA isolation using Qiagen RNA spin columns (as components of RNeasy Mini and Midi kits, #74104 and #75144, respectively; Qiagen) as per the manufacturer's instructions.
Preparation of First- and Second-Strand cDNA, and In Vitro Transcribed and Biotin-Labeled RNA for Gene Chip Hybridization
RNA samples from five subjects were prepared for microarray analysis. Two micrograms of total RNA, in 10 μl, was used to make first-strand cDNA by combining with 2 μl T7/T24 primer (100 pmol/μl, #900375; Affymetrix, Santa Clara, CA) for 10 minutes at 70°C. After addition of 4 μl 5× first-strand buffer (#Y02321; Invitrogen, Carlsbad, CA), 2 μl 100 mM DTT, and 1 μl 10 mM dNTPs (#18427; Invitrogen), samples were incubated at 50°C for 2 minutes. One microliter of Superscript II reverse transcriptase (200 U/μl, #18064; Invitrogen) was then added and the mixture incubated at 50°C for 1 hour. To generate the second-strand cDNA, 130 μl of the following mixture was added to the first-strand solution: 91 μl deionized water treated with the RNAse inhibitor diethyl pyrocarbonate (DEPC dH2O), 30 μl 5× second-strand buffer (#53972; Invitrogen), 3 μl 10 mM dNTPs, 1 μl Escherichia coli ligase (10 U/μl, #18052-019; Invitrogen,), 4 μl E. coli DNA polymerase (10 U/μl, #460634; Invitrogen), 1 μl E. coli RNase H (2 U/μl, #Y01425; Invitrogen). The second-strand solution was incubated for 2 hours at 16°C, after which 2 μl T4 DNA polymerase (6 U/μl, #18005-017; Invitrogen) was added and the reaction continued for another 5 minutes. The cDNA was purified by vortexing with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), followed by centrifugation through a Phase Lock Gel (#E0032; Fisher Scientific, Pittsburgh, PA) at 12,000 rpm for 20 seconds. The upper phase was precipitated with 10 μl of 7.5 M ammonium acetate and 450 μl of 100% ethanol. Samples were then placed at −80°C for at least 5 minutes, after which the precipitate was again centrifuged at maximum speed for 20 minutes. The pellet was washed with 70% ethanol, spun, air dried, and resuspended in 10 μl DEPC dH2O.
To generate in vitro transcribed (IVT), biotin-labeled cRNA, the following solution was then mixed and added to the cDNA: 26.2 μl DEPC dH2O, 6 μl 10× Ambion IVT buffer (Ambion, Austin, TX), 6 μl rNTPs (G 30 mM, A 15 mM, C 12 mM, U 12 mM), 2.4 μl Biotin-11-CTP and 2.2 μl Biotin-11-UTP (#NEL542001 and NEL543001, respectively; PerkinElmer, Boston, MA), 2 μl RNase Inhibitor (#AM2682; Ambion), 3 μl 100 mM DTT, and 2 μl T7 RNA polymerase (2,500 U/μl, #TU950K; EPICENTRE Biotechnologies, Madison, WI). This was incubated for 16 hours at 37°C. The IVT was then purified by adding 40 μl DEPC-dH2O and 350 μl Buffer RLT (Qiagen). After mixing, 250 μl 100% ethanol was added to the sample, which was then passed through a Qiagen RNeasy spin column. The column was washed 1× with Buffer RPE (Qiagen), spun at maximum speed for 3 minutes. The filter was incubated for 5 minutes after the addition of 50 μl of RNase-free water, and the column was spun for 1 minute at 10,000 rpm. Another 5-minute incubation with 50 μl RNase-free water was then performed, after which the column was spun for 3 minutes at 10,000 rpm. The purified IVT was fragmented by adding 8 μl 5× fragmentation buffer to 32 μl cRNA (15 μg cRNA), and heating to 94°C for 35 minutes. The fragmented cRNA (40 μl) was then added to a hybridization cocktail composed of 30 μl Bio948 (2 μl/μg cRNA), 15 μl Wyeth cRNA standard (1 μl/μg cRNA), 3 μl herring sperm DNA (20 mg/ml), 3 μl acetylated BSA (50 mg/ml), 150 μl 2× hybridization buffer (final concentration 100 mM MES, 1 M NaCl, 20 mM EDTA, 0.01% Tween 20), and 59 μl RNase-free water. The hybridization mixture was heated to 99°C for 5 minutes, followed by heating at 45°C for 5 minutes.
Hybridization and Analysis of Gene Arrays
Two hundred microliters of the hybridization solution, together with Wyeth standards for quantitating the amount of each transcript, was then added to Human Genome U133A and Human Genome U133B arrays containing almost 45,000 probe sets (#900366 and #900368, respectively; Affymetrix) as previously described (16, 17). Hybridization was performed by incubation at 45°C for 16 to 18 hours with constant rotation on a rotisserie. Hybridized arrays were then washed and stained according to the manufacturer's protocol.
Arrays were scanned with a GeneArray scanner (Agilent, Santa Clara, CA). MAS 5.0 software (Gene Logic, Gaithersburg, MD) was used to generate expression measures including Signal values and Absent/Present, and a global scaling normalization was applied to the raw Signal intensity according to Affymetrix's algorithm (18). Briefly, a 2% trimmed-mean was calculated per chip, and was scaled to an arbitrary value of 100. A scaled Signal value was then computed for each gene by multiplying its original Signal intensity with the scale factor (100/trimmed-mean). A gene was selected for downstream analysis if it was identified as Present and its expression exceeded 50 (scaled) Signal units in at least one sample. Fold change was calculated on an Excel spreadsheet (Microsoft, Redmond, WA) by dividing the log2 transformation of the scaled Signal units for one treatment group by another's. Comparisons were made between AM gene expression at baseline and after 24 hours of incubation in medium alone, or after infection. The cutoff for differential gene expression was defined as fold-change of 1.5 and a P value ≤ 0.05 (as determined by performance of Student's t test based on log2 transformed scaled Signal units of each pairwise comparison).
Quantitative PCR
To confirm the expression (or lack thereof) of specific genes of interest, samples were examined further using quantitative PCR. For certain genes with low level of expression, contamination of RNA by genomic DNA was observed in initial experiments. Samples were therefore treated with DNase before performance of further studies, after which only three of the original five samples still had adequate amounts of RNA for performance of all the desired PCR studies. To generate a sufficient number of PCR results to allow for statistical analysis, BAL cells from another three subjects were prepared and infected. All three were found to have adequate RNA samples after DNase treatment. PCR studies were therefore performed on all six subjects for whom adequate RNA was available. The primers and probes used for quantitative PCR are listed in Table 1.
TABLE 1.
PRIMERS AND PROBES USED IN QUANTITATIVE PCR
| Oligos | Specificity | Sequence |
|---|---|---|
| KN0001 | hGAPDH forward | ACACCCACTCCTCCACCTTTG |
| KN0002 | hGAPDH reverse | CATACCAGGAAATGAGCTT GACAA |
| KN0005 | hIL23a forward | CAAATGATGTTCCCCATATCCA |
| KN0006 | hIL23a reverse | CAGAACTGACTGTTGTCCCTGAGT |
| KN0007 | p35 IL12 forward | CCTGGACCACCTCAGTTTGG |
| KN0008 | p35 IL12 reverse | GGTGAAGGCATGGGAACATT |
| KN0009 | p40 IL12 forward | CGGTCATCTGCCGCAAA |
| KN0010 | p40 IL12 reverse | TGCCCATTCGCT CCAAGA |
| KN0011 | hGMCSF forward | GAATGAAACAGTAGAAGTCATCTCAGAAA |
| KN0012 | hGMCSF reverse | GCTCCAGGCGGGTCTGTAG |
| KN0013 | hSLAMF1 forward | CCTCTTCAGAAGAAACTTGACTCCTT |
| KN0014 | hSLAMF1 reverse | AGGCTCTGTGGCAGCAACAT |
| Probes | Specificity | Sequence |
| KNP001 | hGAPDH #1 | CTGGCATTGCCCTCAACGACCA |
| KNP003 | hIL23a | TGGAGATGGCTGTGACCCCCAAG |
| KNP004 | hp35 IL12 | CAGAAACCTCCCCGTGGCCA |
| KNP005 | hp40 IL12 | CGGGCCCAGGACCGCTACTATAGCT |
| KNP006 | hGM-CSF | TTTGACCTCCAGGAGCCGACC |
| KNP007 | hSLAMF1 | AGCTCAGGACCCTTGCACCACC |
All data were normalized to GAPDH and then quantitated relative to standard curves of RNA from LPS-stimulated MN that were used as a positive control. Induction of specific genes 24 hours after infection with H37Ra or H37Rv was expressed as fold change relative to RNA induction in BAL samples incubated in medium alone for 24 hours. Statistical analysis was based on the Wilcoxon signed-rank test.
Comparison of Cytokine Production by Blood Monocytes and Alveolar Macrophages in Response to Infection with H37Rv
To determine whether the IL-23 and GM-CSF responses observed were of particular importance for local immunity to M. tuberculosis in the lung, we compared production of the two cytokines by paired AM and MN samples of ten healthy nonsmokers. Both cell types were infected with H37Rv using a 3:1 bacteria-to-cell ratio as described. Culture supernatants were collected from both infected and uninfected cells of these ten subjects at 24 and 48 hours. Concentrations of the IL-12 and IL-23 heterodimers as well as of GM-CSF in culture supernatants were measured using commercially available enzyme-linked immunosorbent assay (ELISA) kits (#88-7239-29 and #88-7126-22, respectively' eBioscience, San Diego, CA, and DGM00; R&D Systems, Minneapolis, MN). Statistical evaluation was performed using Wilcoxon's signed-rank test.
RESULTS
Comparison of Intracellular Infection of BAL Cells with M. tuberculosis Strains H37Rv and H37Ra
To evaluate whether our infection protocol resulted in comparable initial infectious burdens of H37Rv and H37Ra, quantification of intracellular bacilli (by CFU determination) was performed immediately following a 2-hour incubation of AM with the organisms. For H37Rv, initial CFU was 8.31 × 105 (± 4.47 × 105) per 1 × 106 BAL cells. Initial CFU of H37Ra in the same seven subjects was 3.24 × 105 (± 2.72 × 105). This represented a 2.56-fold difference (P = 0.068 by paired t test). As expected, H37Rv displayed greater intracellular growth after overnight incubation of infected cells than did H37Ra, as CFU of H37Rv increased 1.63-fold to 1.35 × 106 (± 7.67 × 105) 24 hours after infection, whereas CFU of H37Ra increased only 1.32-fold, to 4.29 × 105 (± 4.98 × 105). At this time point, corresponding to the time at which infected cell lysates were prepared for RNA extraction, intracellular CFU of H37Rv was 3.15-fold higher than that of H37Ra.
Because CFU studies were not performed on the same samples used for microarray analysis, we did not attempt to correct gene expression data for differences in growth of the organisms. Instead, our subsequent studies were focused on genes for which the variation in gene induction by H37Rv and H37Ra was most marked, so as to reduce the likelihood that these differences were due to disparities in infectious burden alone.
Microarray Evaluation
Microarray assessments of gene expression were performed on BAL samples of five subjects. One thousand nine hundred eighty-four genes displayed differential expression as defined by the cutoff criteria of fold-change (fc) of ≥ 1.5 and P value of ≤ 0.05. Because gene expression in uninfected AM after 24 hours of incubation was itself substantially different than that at baseline, our analysis focused primarily on differences in gene expression after the various overnight incubation conditions. The data tables noted below present results of gene expression comparisons for all five subjects studied.
Comparison of gene expression in uninfected AM and AM infected with H37Ra indicated significant changes in expression of only 17 genes. Of these, 5 were up-regulated in response to infection, whereas 12 were down-regulated. In contrast, infection with H37Rv resulted in altered expression of 439 genes. For 117 of these, infection resulted in upregulation of the gene, whereas 322 genes were down-regulated in response to H37Rv. (Complete lists of all of the genes that met criteria for significant alteration of expression are presented in the online supplemental materials.)
The comparison considered to be of greatest interest was that of AM infected with H37Ra and with H37Rv. Compared with H37Ra, infection with the virulent strain H37Rv resulted in up-regulation of 52 genes. These were grouped according to function, based on summaries obtained from the NCBI's Entrez Gene website (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene). The findings of this comparison are noted in Table 2. Compared with both uninfected cells and cells infected with avirulent strain H37Ra, infection with virulent strain H37Rv was most notable for its up-regulation of proinflammatory genes, particularly those associated with induction of Th1-like immunity (Table 2, Section 1). The gene for the IL-23 p19 subunit was the most highly up-regulated by infection with H37Rv, followed by genes for GM-CSF, the neutrophil chemotactic molecule CXCL1 (GRO α), IL-6, and TNF-α. Also up-regulated was signaling lymphocyte activation molecule (SLAM, also known as CD150), which contributes to CD28-independent activation of T cells to produce IFN-γ. An unexpected finding was the lack of up-regulation of IL-12, a heterodimer that shares a p40 subunit with IL-23, and is also composed of an IL-12–specific p35 subunit.
TABLE 2.
HUMAN ALVEOLAR MACROPHAGE GENES UP-REGULATED TO A GREATER EXTENT BY H37Rv THAN BY H37Ra, AND/OR SIGNIFICANTLY UP-REGULATED BY H37Ra 24 h AFTER INFECTION
| Fold change (Fc)
|
P Value
|
Rank
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Rv vs Ra | Rv vs Uninf | Ra vs Uninf | (Rv vs Ra) | (Rv vs Ra) | Function | ||||||
| 1. Proinflammatory | ||||||||||||
| IL23, p19 | 8.34 | 13.13 | 1.57 | 0.0109 | 1 | Dimerizes with IL-12 p40 to form IL-23. Role in Th1 responses, particularly of memory cells. Activates tyk2, jak 2, STAT 1,3,4. Also contributes to development of Th17 cells. | ||||||
| CSF2 (GM-CSF) | 8.1 | 11.21 | 1.38 | 0.0174 | 2 | Differentiation of MN into M1 (Th1 promoting) macrophage phenotype. Recruitment of DC(CD11b+, CD8α−), stimulation of DC maturation (increased expression of B7-1 CD1d). | ||||||
| CXCL1 (Gro-α) | 4.52 | 6.85 | 3.11 | <0.0001 | 3 | Growth-related oncogene protein-alpha (Gro-alpha). Role in neutrophil chemotaxis via action on CXCR2. | ||||||
| IL-6 | 4.44 | 15.41 | 3.47 | 0.0040 | 4 | Promotes terminal differentiation of B-cells to plasma cells, stimulates Ab secretion, promotes myeloid stem cell differentiation, induces acute phase proteins. Inhibits TNF, IL-1. | ||||||
| TNFα | 3.14 | 9.65 | 3.07 | 0.0460 | 5 | Mainly secreted by macrophages, it binds to and functions through its receptors TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR. Involved in the regulation of cell proliferation, differentiation, apoptosis, lipid metabolism, and coagulation. Implicated in autoimmune diseases, insulin resistance, and cancer. | ||||||
| SLAM | 2.93 | 4.44 | 1.51 | 0.0217 | 8 | Signaling lymphocyte activation molecule (CD150). CD28-independent activation of T-cells roles in induction of IFN-γ (Th1) and cell proliferation. Role in IL-1–dependent DC activation. | ||||||
| DUSP5 | 2.35 | 2.43 | 1.08 | 0.0124 | 11 | Dual-specificity phosphatase 5. Regulates IL-2–dependent phosphorylation and catalytic activity of ERK 1/2. Up-regulated by IL-2, IL-7, and IL-15, and by TALL-1, but not by IL-4. | ||||||
| EHD1 | 2.33 | 2.28 | −1.02 | 0.0057 | 13 | EH domain containing 1. Functions as tubule-inducing factor in Arf6 pathway for recycling of plasma membranes after clathrin-independent endocytosis. Enhances MHC-I recycling. | ||||||
| RGS1 | 2.2 | 1.96 | −1.12 | 0.0238 | 16 | Regulator of G-protein signaling 1. Inhibits signal transduction by increasing GTPase activity of G protein alpha subunits, thereby driving them into their inactive GDP-bound form. May be involved in the regulation of B-cell activation and proliferation. | ||||||
| TNIP1 | 2.19 | 2.17 | −1.01 | 0.0057 | 17 | Interacts with zinc finger protein a20/TNFAIP3 and inhibits TNF-induced NF-κB–dependent gene expression by TRFA2-mediated transactivation signal. Increases cell surface expression of CD4, interacts with HIV-1 matrix protein, and can inhibit viral replication. | ||||||
| PTGS2 | 2.11 | 4.8 | 2.28 | 0.0030 | 18 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase), aka cyclooxygenase. Key enzyme in prostaglandin biosynthesis, and acts both as a dioxygenase and as a peroxidase. PTGS2 is the inducible isoenzyme of PTGS and is regulated by specific stimulatory events, suggesting that it is responsible for prostanoid synthesis in inflammation and mitogenesis. | ||||||
| TNFRSF5 | 2.07 | 2.01 | −1.03 | 0.0475 | 19 | aka CD40: Receptor for TNFSF5/CD40L. Essential in mediating T cell–dependent immunoglobulin class switching, memory B cell development, and germinal center formation. | ||||||
| IVNS1ABP | 1.95 | 2.09 | 1.07 | 0.0154 | 24 | Influenza virus NS1A binding protein. Role in mediating effects of virus on mRNA splicing. | ||||||
| (CXCL1, CXCL2) | 1.92 | 5.96 | 3.11 | 0.0222 | 25 | GRO-α and GRO-β. See GRO-α above. GRO-β also acts on CXCR2. Has a role in menstrual cycle by regulation of neutrophil trafficking into the endometrium. | ||||||
| HIVEP2 | 1.91 | 2.71 | 1.42 | 0.0239 | 26 | HIV1 enhancer binding protein 2. Implicated as playing a role in HIV production from latently infected T-cells. | ||||||
| RDC1 | 1.84 | 1.77 | −1.04 | 0.0043 | 30 | G-coupled protein receptor. Involved in increasing affinity of neutrophil integrin molecules for endothelial adhesion molecules. Transduces information provided by extracellular stimuli into intracellular messengers via coupling to G-protein–coupled receptors. | ||||||
| UREB1 | 1.84 | 1.58 | −1.16 | 0.0150 | 31 | May function as a DNA-binding transcriptional regulator. Encodes a protein that contains a HECT domain associated with ubiquitin-protein ligase activity. | ||||||
| XBP1 | 1.82 | 2.12 | 1.17 | 0.0170 | 32 | Basic region leucine zipper transcription. Essential for terminal B-cell differentiation. | ||||||
| IVNS1ABP | 1.8 | 2.22 | 1.23 | 0.0492 | 33 | Encoded by NS1A protein. Multifunctional, involved in protein–protein and protein–RNA interactions. Binds nonspecifically to dsDNA and specific protein targets and regulates several post-transcriptional processes. | ||||||
| CCL3 | 1.75 | 2.74 | 1.57 | 0.0209 | 37 | Macrophage inflammatory protein 1-alpha (MIP1α). Acts on CCR5 (present on MN, macrophages, Th1 cells, activated T-cells, NK cells). | ||||||
| PLA2G4A | 1.73 | 2.61 | 1.51 | 0.0420 | 41 | Phospholipase A2, group IVA. Cytosolic, Ca-dependent phospholipase that cleaves archadonic acid (AA) from membrane glycerophospolipids. Implicated in initiation of the inflammatory response. | ||||||
| TNFAIP2 | 1.7 | 2.21 | 1.3 | 0.0370 | 43 | TNF-α–induced protein 2. Implicated in immune and proinflammatory responses. Induced also by retinoic acid receptor alpha fusion protein—suggests possible retinoic acid target gene in promyelocytic leukemia. | ||||||
| CXCL3 (Gro-γ) | 1.66 | 4.63 | 2.8 | 0.0590 | (n/a) | Growth-related oncogene protein-γ (Gro-γ). Promotes neutrophil recruitment via effects on CXCR2. Promotes angiogenesis. | ||||||
| PBEF | 1.6 | 1.72 | 1.07 | 0.0451 | 48 | Pre–B-cell colony-enhancing factor. Growth factor for early stage B-cells. Up-regulated in neutrophils as an inhibitor of apoptosis in response to inflammatory stimuli (e.g., LPS, IL-1β, TNF, IL-6). Implicated in acute lung injury in setting of sepsis. | ||||||
| CD44 | 1.44 | 2.02 | 1.4 | 0.0465 | 52 | Encodes a cell-surface glycoprotein involved in cell–cell interactions, cell adhesion, and is a receptor for hyaluronic acid (HA) and can also interact with other ligands, such as osteopontin, collagens, and matrix metalloproteinases (MMPs). Contributes to lymphocyte activation, recirculation, and homing, as well as to hematopoesis and tumor metastasis. | ||||||
| BF | 1.36 | 3.39 | 2.49 | 0.0199 | (n/a) | B factor, properdin migration.. Encodes complement factor B, a component of the alternative pathway of complement activation. Factor B circulates in the blood as a single chain polypeptide. Upon activation of the alternative pathway, it is cleaved by complement factor D, yielding the noncatalytic chain Ba and the catalytic subunit Bb. Bb is a serine protease which associated with C3b to form the alternative pathway C3 convertase. Bb is also involved in the pre-activated B lymphocytes, whereas Ba inhibits their proliferation. | ||||||
| 2. Cytotoxicity/apoptosis-related | ||||||||||||
| IER3 | 2.05 | 3.14 | 1.53 | 0.0140 | 20 | Pro-apoptotic gene induced by TNF-α, depends on NF-κB activation. | ||||||
| SERPINB9 | 2.01 | 2.27 | 1.13 | 0.0080 | 21 | Anti-apoptotic, inhibitor of granzyme B. | ||||||
| TRAF1 | 1.97 | 2.64 | 1.34 | 0.0462 | 22 | Adapter protein and signal transducer, links members of TNFR family to different signaling pathways by association with receptor cytoplasmic domain and kinases. Mediates activation of NF-κB and jnk and is involved in apoptosis. TRAF1/TRAF2 complex recruits apoptotic suppressors birc2, birc3 to TNFRSF1b/TNFR2. | ||||||
| PHLDA2 | 1.75 | 1.98 | 1.13 | 0.0145 | 38 | Tumor-suppressing subchromosomal transferable DNA fragment (STF3). Similar to TDAG51, implicated in FAS (APT1) expression and apoptosis. Role in tumor development. | ||||||
| STK17A | 1.66 | 2.1 | 1.27 | 0.0474 | 45 | Serine/threonine kinase 17a (aka DRAK1 kinase). Located in nucleus, acts as positive regulator of apoptosis. Highly expressed in placenta. | ||||||
| PBEF | 1.6 | 1.72 | 1.07 | 0.0451 | 48 | Pre–B-cell colony-enhancing factor. Growth factor for early stage B-cells. Up-regulated in neutrophils as an inhibitor of apoptosis in response to inflammatory stimuli (e.g., LPS, IL-1β, TNF, IL-6). Implicated in acute lung injury due in setting of sepsis. | ||||||
| GG2-1 | 1.49 | 2.14 | 1.44 | 0.0208 | 51 | aka, TNFAIP8 (TNF-α–induced protein 8), is an anti-apoptotic factor. Inhibition with siRNA(small interfering RNA) enhances apoptosis, decreases proliferation and production of MMP-1 in response to TNF-α. | ||||||
| 3. Cell-signaling related | ||||||||||||
| TM4SF1 | 2.58 | 2.68 | 1.04 | 0.0169 | 10 | Transmembrane 4 superfamily member 1. A member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most members are cell-surface proteins characterized by the presence of four hydrophobic domains. Mediates signal transduction events involved in regulation of cell development, activation, growth, and motility. Highly expressed in specific carcinomas. | ||||||
| TM4SF1 | 2.35 | 2.43 | 1.03 | 0.0482 | 12 | Transmembrane 4 superfamily member 1. | ||||||
| TM4SF1 | 2.3 | 2.21 | −1.04 | 0.0223 | 14 | Transmembrane 4 superfamily member 1. | ||||||
| RGS1 | 2.2 | 1.96 | −1.12 | 0.0238 | 16 | Regulator of G-protein signaling 1. Inhibits signal transduction by increasing GTPase activity involved in the regulation of B-cell activation and proliferation. High constitutive expression in B-cell malignancies (NHL, hairy-cell leukemia). | ||||||
| RDC1 | 1.84 | 1.77 | −1.04 | 0.0043 | 30 | G-coupled protein receptor. Involved in inducing a conformation change in integrin molecules of the neutrophil membrane, increasing their affinity for the endothelia Ig-superfamily adhesion molecules. Transduces information provided by extracellular stimuli into intracellular messengers via coupling to G-protein–coupled receptors. | ||||||
| PALM2 | 1.79 | 2.49 | 1.39 | 0.0466 | 34 | Paralemmin 2. Scaffold protein coordinates cAMP-mediated signaling complexes by binding to type I and II protein kinase A. | ||||||
| TM7SF1 | 1.62 | 1.63 | 1.01 | 0.0421 | 47 | Transmembrane 7 superfamily member 1—part of family of G-protein–coupled receptors. Up-regulated in the course of kidney development. | ||||||
| 4. Cell-cycle/tumor related | ||||||||||||
| TFPI2 | 3.11 | 5.03 | 1.62 | 0.0385 | 7 | Tissue factor pathway inhibitor 2. TFPI2 is down-regulated on aggressive tumors. TFP12 treatment inhibits tumor growth, metastases, suggesting role in anti-tumor immunity. | ||||||
| NBS1 | 1.96 | 3.01 | 1.53 | 0.0397 | 23 | Encodes protein Nijmegen breakage syndrome 1 (nibrin), involved in processing/repair of DNA double-strand breaks and in cell cycle checkpoints. | ||||||
| G0S2 | 1.86 | 2.55 | 1.38 | 0.0338 | 29 | Putative lymphocyte G0/G1 switch gene. Potential oncogene and regulator of latent HIV. | ||||||
| GADD45A | 1.78 | 4.7 | 2.64 | 0.0327 | 35 | Growth arrest and DNA-damage inducible, alpha. Encodes protein products with antiproliferative activities. Induction regulated in p53 manner after ionizing radiation. Transcription target for cancer. | ||||||
| ARHE | 1.76 | 2.03 | 1.15 | 0.0341 | 36 | Ras homolog gene family, member E. Regulator of growth, differentiation, and malignant transformation. | ||||||
| EREG | 1.74 | 3.52 | 2.02 | 0.0204 | 40 | Epiregulin. Epidermal growth factor (EGF) family growth factor. May be a mediator of localized cell proliferation. Involved in cell growth of epithelial malignancies (bladder, lung, kidney, colon). May stimulate cell proliferation and/or angiogenesis. | ||||||
| NBS1 | 1.7 | 2.83 | 1.66 | 0.0266 | 42 | Nijmegen breakage syndrome 1 (nibrin, see #23 above). | ||||||
| OPTN | 1.66 | 1.79 | 1.08 | 0.0310 | 44 | Optinuerin (?hypothetical). Involved with signal transduction, associated with TNF pathways. | ||||||
| 5. Cytoskeleton | ||||||||||||
| DMN | 3.13 | 3.93 | 1.26 | 0.0075 | 6 | Desmuslin. Is a member of the intermediate filament (IF) family of cytoskeletal proteins that confer resistance to mechanical stress and are encoded by a dispersed multigene family. Has been found to form a linkage between desmin (a subunit of the IF network) and the extracellular matrix. Provides an important structural support in muscle. | ||||||
| LIMK2 | 1.75 | 1.72 | −1.02 | 0.0483 | 39 | LIM domain kinase 2. Phosphorylates (inactivates) cofilin, an actin depolymerizing factor, and induces actin cytoskeleton reorganization. | ||||||
| 6. Lysosome/organelle-associated | ||||||||||||
| LAMP3 | 2.89 | 2.1 | −1.39 | 0.0474 | 9 | Lysosomal-associated membrane protein 3 (CD63). | ||||||
| EHD1 | 2.33 | 2.28 | −1.02 | 0.0057 | 13 | EH domain containing 1. Function as tubule-inducing factor in Arf6 pathway for recycling of plasma membranes internalized by clathrin-independent endocytosis. Overexpression of EHD1 enhances MHC-I recycling. | ||||||
| EHD1 | 1.9 | 2.29 | 1.16 | 0.0290 | 27 | EH-domain containing 1. | ||||||
| NUPL1 | 1.86 | 2.14 | 1.15 | 0.0040 | 28 | Nucleoporin type 1. Novel basic helix-loop-helix expressed broadly during early embryonic organogenesis and prominently in developing dorsal root ganglia. The protein is localized to the nuclear rim and is a component of the nuclear pore complex (NPC). All molecules entering or leaving the nucleus either diffuse through or are actively transported by the NPC. | ||||||
| FLJ90005 | 1.66 | 1.68 | 1.01 | 0.0295 | 46 | (?hypothetical) Mediates ubiquination of cellular proteins. Expressed throughout nervous system. | ||||||
| HSPA1A | 1.58 | 1.34 | −1.19 | 0.0278 | 49 | HSP-70 protein 1A. Molecular chaperonins that assist other proteins in their folding, transport, and assembly into complexes. Protect cells from environmental hazards. Involved in signal transduction in cooperation with HSP-90. In all functions, recognize non-native conformations of other proteins, bind extended hydrophobic segments exposed during translation, membrane translocation, or after stress-induced damage. | ||||||
| SEC24A | 1.56 | 1.78 | 1.14 | 0.0414 | 50 | SEC-related gene family, member A (S. cervisiae). Component of COPII coat that covers ER-derived vesicles involved in transport from ER to golgi apparatus. Act to promote transport of secretory, plasma membrane, and vacuolar proteins from ER to golgi. Expressed in fibroblasts, hepatocytes, and lymphocytes. | ||||||
| 7. Unknown | ||||||||||||
| FLJ23231 | 2.28 | 2.9 | 1.27 | 0.0103 | 15 | Hypothetical protein. | ||||||
Genes are grouped according to function. Fold change is listed for the comparisons of H37Rv-infected vs. H37Ra-infected cells, H37Rv-infected vs. uninfected cells incubated in medium alone for 24 hours, and H37Ra-infected vs. uninfected cells. Italicized values in fold-change columns indicate genes that met the study's significance criteria of fold change of ≥ 1.5 and P value of ≤ 0.05 for samples obtained from five healthy subjects.
H37Rv infection also affected the expression several genes associated with apoptosis and cytotoxicity (Table 2, Section 2). Several pro-apoptotic genes were up-regulated by H37Rv (IER3, TRAF1, PHLDA2, STK17A); however, anti-apoptotiwc genes (PBEF, GG2-1) were up-regulated as well. H37Rv also induced increased expression of SERPINB9, an inhibitor of the granzyme B. Infection with H37Rv also resulted in up-regulation of genes associated with cell signaling (most notably transmembrane 4 superfamily member 1; Table 2, section 3), cell cycle and tumor regulation (Table 2, Section 4), cytoskeleton (Table 2, Section 5), and lysosomes as well as other organelles (Table 2, Section 6). Up-regulation of several genes of unknown function was observed as well (Table 2, Section 7).
The five genes significantly up-regulated by H37Ra were all associated with immune response, and are also indicated in Table 2. Of note, only one gene was found to be significantly up-regulated by infection with H37Ra, but not with H37Rv. This gene, BF, encodes complement factor B, a component of the alternative complement pathway that plays a role in B-cell activation.
No genes were down-regulated by infection with H37Rv to a significantly greater degree than by H37Ra. A partial list of genes down-regulated by H37Rv is presented in Table 3, which indicates the 41 genes that were down-regulated by 2.5- or greater fold-change. (The complete list of down-regulated genes is contained in the online supplement.) Infection with H37Rv resulted in significant down-regulation of relatively few genes associated with inflammation and immunity (Table 3, Section 1). As indicated in Table 3, Section 4, H37Rv down-regulated Rab-coupling protein (RCP), which regulates fusion of intracellular vesicles. The majority of down-regulated genes were associated with lipid metabolism and with general metabolic functions (Table 3, Sections 5 and 6), suggesting that H37Rv infection generally resulted in a switch from expression of genes associated with resting cell metabolism to those associated with an immune response.
TABLE 3.
HUMAN ALVEOLAR MACROPHAGE GENES DOWN-REGULATED BY INFECTION WITH H37Rv AND/OR BY INFECTION WITH H37Ra
| H37Rv vs Uninf
|
H37Ra vs Uninf
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Fc | P Value | Rank | Fc | P Value | Rank | Function | |||||||
| 1. Immune response/inflammation | ||||||||||||||
| SPP1 | −3.94 | 0.0046 | 6 | −1.85 | 0.1100 | (n/a) | Secreted phoshoprotein 1. Cytokine activity, growth factor activity, and integrin and protein binding. Involved in Th1 immune response, anti-apoptosis, cell adhesion, cell–cell signaling, chemotaxis, ossification, T-cell proliferation, myeloid, blood cell differentiation. | |||||||
| KAL1 | −2.74 | 0.0008 | 23 | −1.83 | 0.0775 | (n/a) | Kallmann syndrome 1 sequence. Functions in neural cell adhesion, axonal migration, cell adhesion, motility, chemotaxis. Involved in protein binding, extracellular matrix endopeptidase inhibitor activity. | |||||||
| MME | −2.71 | 0.0009 | 26 | −1.13 | 0.7300 | (n/a) | Membrane metallo-endopeptidase. Acute lymphocytic leukemia antigen. Present on all leukemic cells of pre-B phenotype. Glycoprotein abundant in kidney on brush border of proximal tubules and glomerular epithelium. AA sequence identical to enkephalinase, which inactivates glucagons, enkephalins, substrate P, neurotensin, oxytocin, and bradykinin when it cleaves peptides at amino side of hydrophobic residues. | |||||||
| FN1 | −2.70 | 0.0063 | 27 | −2.16 | 0.0167 | 1 | Fibronectin 1. Involved in cell adhesion and migration processes including embryogenesis, wound healing, blood coagulation, host defense, and metastasis. Aids in acute phase response, collagen, binding, heparin binding, and oxidoreductase activity. | |||||||
| ADAM 10 | −2.52 | 0.2580 | 38 | −1.51 | 0.3000 | (n/a) | A disintegrin and metalloproteinase domain 10. Cell surface proteins with unique structure possessing both potential adhesion and protease domains. SH2 domain binding, hydrolase, metalloendopeptidase, and protein homodimerization activity and protein receptor, zinc binding. Cell–cell signaling, negative regulation of cell adhesion, notch receptor processing, nucleocytoplasmic transport and protein amino acid phosphorylation. | |||||||
| ITGAM | −2.07 | 0.0121 | 85 | −1.97 | 0.0174 | 3 | Integrin, alpha M (complement component receptor 3, alpha), aka CD11b. Encodes the integrin alpha M chain. I domain containing alpha integrin combines with the beta 2 chain (ITGB2) to form a leukocyte-specific integrin, macrophage receptor 1 (MAC1) or inactivated C3b. Importance in adherence of neutrophils and monocytes to stimulated endothelium and phagocytosis of complement-coated particles. Aids in matrix adhesion, integrin-mediated signaling pathway, Mg binding receptor activity. | |||||||
| 2. Apoptosis-associated | ||||||||||||||
| SOX4 | −2.73 | 0.0197 | 24 | −1.46 | 0.3200 | (n/a) | SRY (sex-determining region Y) box 4. Intronless gene encodes a member of the SOX family of transcription factors. Involved in regulation of embryonic development and in determination of cell fate. May act in apoptosis pathway, cell death and forming complexes with other proteins. | |||||||
| 3. Signal transduction | ||||||||||||||
| NRP1 | −4.03 | 0.0171 | 3 | −1.46 | 0.5100 | (n/a) | Neuropilin 1. Membrane-bound co-receptor to a tyrosine kinase receptor for vascular endothelial signal transduction. | |||||||
| CD9 | −4.02 | 0.0034 | 4 | −1.69 | 0.1500 | (n/a) | CD9 antigen (p24). Member of transmembrane 4 superfamily of cell surface proteins with four hydrophobic domains. Mediates signal transduction regulating cell development, activation, growth and motility. Role in platelet activation and aggregation, also in myocyte fusion and myotubule maintenance. | |||||||
| FPRL2 | −2.80 | 0.0031 | 20 | −1.66 | 0.0900 | (n/a) | Formyl peptide receptor-like 2. N-1-formyl peptide receptor and rhodopsin-like receptor activity. Involved in G-protein–coupled protein signaling, cell motility, chemotaxis, and signal transduction. | |||||||
| 4. Lysosome, other intracellular compartments | ||||||||||||||
| ABCG2 | −3.95 | 0.0006 | 5 | −1.46 | 0.4300 | (n/a) | ATP binding cassette, subfamily G. Membrane-associated gene in superfamily of ATP-binding cassette transporters. ABCGs transport molecules across intra- and extra-cellular membrane. Part of white subfamily, which functions as a xenobiotic transporter, may play a role in multidrug resistance. Significant expression in placenta. | |||||||
| RCP | −2.75 | 0.0009 | 22 | −1.70 | 0.1100 | (n/a) | Rab coupling protein. Regulation of formation, targeting, and fusion of intracellular transport vesicles. | |||||||
| H2AFY | −2.59 | 0.0001 | 35 | −1.23 | 0.6700 | (n/a) | H2A histone family, member Y. Basic nuclear proteins responsible for the structure of the chromosomal nucleosome assembly. | |||||||
| 5. Lipid metabolism | ||||||||||||||
| CYP27A1 | −5.24 | 0.0045 | 1 | −3.19 | 0.3400 | (n/a) | Cytochrome P450, family 27, subfamily A, polypeptide 1. Encodes a member of cytochrome P450 superfamily of genes (catalyzing reactions involved in drug metabolism and synthesis of cholesterol, steroids, and other lipids). Roles in electron transport, steroid hydroxylase activity. | |||||||
| CYP1B1 | −3.21 | 0.0173 | 12 | −1.82 | 0.1500 | (n/a) | Cytochrome P-450, family 1, subfamily B, polypeptide 1. Located in ER, metabolizes pro-carcinogens(polycyclic aromatic hydrocarons and 17-beta estradiol). Also metabolizes signaling molecules involved in eye development. | |||||||
| FABP3 | −3.03 | 0.0281 | 15 | −1.58 | 0.3600 | (n/a) | Fatty-acid binding protein 3. Involved in uptake, intracellular metabolism, and/or transport of long chain fatty acids. Arrests growth of mammary epithelial cells. | |||||||
| ALOX5 | −2.87 | 0.0012 | 18 | −1.55 | 0.13 | (n/a) | Arachidonate 5-lipoxygenase. Iron ion binding, lipoxygenase and oxidoreductase activity. Involved in electron transport, inflammatory response, and leukotriene biosynthesis. | |||||||
| OSBPL11 | −2.72 | 0.0015 | 25 | −1.55 | 0.2000 | (n/a) | Oxysterol binding protein 11. Encodes a member of the OSBP family, a group of intracellular lipid receptors. Involved in lipid transport, steroid metabolism. | |||||||
| HPGD | −2.67 | 0.0114 | 29 | −1.48 | 0.2800 | (n/a) | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity. Electron transport activity, oxidoreductase activity, prostaglandin metabolism. | |||||||
| ABCG1 | −2.62 | 0.0008 | 32 | −1.37 | 0.3400 | (n/a) | ATP-binding cassette, sub-family G (white)—same as other ABCGs. Involved in macrophage cholesterol and phospholipd transport. Regulates lipid homeostasis in other cell types as well. L-tryptophan and purine nucleotide transport activity, lipid transport, and permease activity. | |||||||
| FABP5 | −2.54 | 0.0014 | 37 | −1.24 | 0/57 | (n/a) | Fatty-acid binding protein 5 (psoriasis-associated). Binds fatty acids in epidermal sites. | |||||||
| ALOX5 | −2.51 | 0.0034 | 40 | −1.35 | 0.3900 | (n/a) | Arachidonate 5-lipoxygenase (as above). | |||||||
| FACL1 | −1.78 | 0.7000 | (n/a) | −1.72 | 0.0306 | 5 | Fatty-acid Coenzyme A ligase, long-chain 2. An isoenzyme of the long-chain fatty acid co-enzyme A ligase. Converts free long-chain fatty acids into fatty acyl-CoA esters. Lipid biosynthesis and fatty acid degradation. Aids in digestion, fatty acid metabolism, ligase activity, Mg ion binding. | |||||||
| SC5DL | −1.49 | 0.1100 | (n/a) | −1.63 | 0.2770 | 7 | Sterol-C5-desaturase. Cholesterol biosynthesis, integral membrane protein. Aids in C-5 sterol desaturase activity, lathosterol oxidase and oxidoreductase activity. Lipid metabolism and sterol biosynthesis. | |||||||
| 6. General metabolism | ||||||||||||||
| FUCA1 | −3.67 | 0.0004 | 8 | −1.07 | 0.9200 | (n/a) | Fucosidase, alpha L-1, tissue. Associated with lysosomal storage, alpha-L-fucosidase activity, hydrolase activity. Also carbohydrate glycosaminoglycan metabolism, catabolism. | |||||||
| IDH1 | −3.36 | 0.0003 | 10 | −1.66 | 0.2300 | (n/a) | Isocitrate dehydrogenase 1. Catalyzes oxidative decarboxylation of isocitrate to 2-oxyoglutarate NADP-dependent, found in cytoplasm and peroxisomes. Important role in cytoplasmic NADPH production. | |||||||
| HK3 | −2.97 | 0.0456 | 16 | 1.45 | 0.4900 | (n/a) | Hexokinase 3 (white cell). Phosphorylates glucose to produce glucose-6-phosplate, committing glucose to the glycolytic pathway. Inhibited by glucose-6-phosphate. Also has ATP-binding, hexokinase, kinase, and transferase activity. | |||||||
| NPL | −2.87 | 0.0070 | 19 | −1.25 | 0.6300 | (n/a) | N-acetylneuraminate pyruvate lysate. Lysate activity, diaminopimelate and lysine biosynthesis. | |||||||
| GLUL | −2.79 | 0.0038 | 21 | −1.17 | 0.7900 | (n/a) | Glutamate-ammonia ligase (glutamine synthase). Glutamine synthases involving oxidatively modified proteins, glutamate-ammonia ligase activity. Involved in glutamine biosynthesis, nitrogen fixation, regulation of neurotransmitter levels. | |||||||
| HMOX1 | −2.69 | 0.0084 | 28 | −1.48 | 0.3300 | (n/a) | Heme oxygenase (decyling) 1. Essential enzyme in catabolism, cleaves heme to form biliverdin, which is converted to bilirubin. Positive regulation of I kappa B kinase. | |||||||
| SGK | −2.61 | 0.0005 | 33 | −1.37 | 0.5300 | (n/a) | Serum/glucocorticoid regulated kinase. Encodes serine/threonine protein kinase. Stimulates glycogenolysis and proteolysis, inhibits protein and glycogen synthesis. Activates potassium, sodium, and chloride channels. | |||||||
| MAN1A1 | −2.59 | 0.0001 | 34 | −1.68 | 0.1500 | (n/a) | Mannosidase, alpha, class 1A, member 1—Encodes a class I mammalian Golgi 1, 2 mannosidase, which is a type II transmembrane protein. Catalyzes the removal of three distinct mannose residues from peptide-bound Man (9)-GlcNAc (2) oligosaccharides. Calcium ion binding, hydrolase activity, N-linked glycosylation, carbohydrate metabolism. | |||||||
| IRS2 | −2.57 | 0.0182 | 36 | −1.32 | 0.5300 | (n/a) | Insulin receptor substrate 2. Encodes IRD2, a cytoplasmic signaling molecule that mediates effects of insulin, insulin-like growth factor 1 and other cytokines by acting as a molecular adaptor between receptor tyrosine kinase and downstream insulin receptor tyrosine kinase. Role in glucose metabolism, positive regulation of cell proliferation. | |||||||
| ADAM 10 | −2.52 | 0.0258 | 38 | −1.53 | 0.3000 | (n/a) | A disintegrin and metalloproteinase domain 10. Cell surface protein with unique structure possessing potential adhesion and protease domains. SH2 domain binding, hydrolase, metalloendopeptidase and protein homodimerization activity, protein receptor and zinc ion binding. Cell-cell signaling, cell adhesion, negative regulation of cell adhesion, notch receptor processing. Nucleocytoplasmic transport and protein amino acid phosporylation. | |||||||
| SDR1 | −2.51 | 0.0001 | 39 | −1.41 | 0.3700 | (n/a) | Short-chain dehydrogenase/reductase 1. | |||||||
| AKR1C3 | −2.50 | 0.0407 | 41 | −1.30 | 0.5200 | (n/a) | Aldo-keto reductase family 1, member C3. Encodes a member of the aldo/keto reductase superfamily that catalyzes the conversion of aldehydes and ketones to their corresponding alcohols. | |||||||
| ATP1B1 | −1.59 | 0.0061 | (n/a) | −1.43 | 0.0209 | 11 | ATPase, Na+/K+ transporting, beta 1 polypeptide. Na+/K+, H+/K+ ATPases beta chain proteins. Integral membrane proteins responsible for establishing and maintaining the electrochemical gradients of Na+ and K+ ions across the plasma membrane. Required for osmoregulation, sodium-coupled transport, electrical excitability of nerve and muscle. | |||||||
| NDUFB3 | −1.39 | 0.0480 | (n/a) | −1.73 | 0.0288 | 4 | NADH dehydrogenease (ubiquinone) 1 beta subcomplex, 3, 12 kD. First complex in the electron transport chain of mitochondria. The multi-subunit NADH dehydrogenase complex. | |||||||
| 7. Cell cycle–associated | ||||||||||||||
| H2AFZ | −1.88 | 0.0450 | (n/a) | −1.58 | 0.0450 | 9 | H2A histone family, member Z. Encodes a replication-independent member of the histone H2A family. Role in embryonic development. Histone proteins are responsible for nucleosome structure of the chromosomal fiber in eukaryotes. | |||||||
| CPR8 | −1.82 | 0.0178 | (n/a) | −2.05 | 0.0157 | 2 | Cell cycle progression 8 protein. aka CCPG1. | |||||||
| HDAC2 | −1.43 | 0.1500 | (n/a) | −1.62 | 0.0160 | 8 | Histone deacetylase 2, transcriptional regulator homologous to yeast transcription factor RPD3. Regulates chromatin structure during transcription. Associated with zinc finger transcription factor, which negatively regulates transcription by tethering RPD3. | |||||||
| U2AF1 | −1.22 | 0.4100 | (n/a) | −1.63 | 0.0088 | 6 | U2 (RNU2) small nuclear RNA auxiliary factor 1. Non-snRNP protein required for binding of U2snRNP to the pre-mRNA branch site. Constitutively enhances dependent RNA splicing by directly mediating interactions between the large subunit and proteins bound to the enhancers. | |||||||
| 8. Cell signaling–associated | ||||||||||||||
| AKAP11 | −1.72 | 0.0129 | (n/a) | −1.55 | 0.0286 | 10 | A kinase (PRKA) anchor protein 11. Binds to the regulatory subunit of protein kinase A. Expressed in mature sperm, binds to R1 and R11 subunits of PKA in testis. Functions in cell cycle control of semate cells and germ cells. Protein phosphatase 1 binding. | |||||||
| 9. Cytoskeleton-associated | ||||||||||||||
| GSN | −4.76 | 0.0140 | 2 | −1.74 | 0.2600 | (n/a) | Gelsolin. Functions in actin binding, calcium ion binding, actin filament polymerization, severing, capping. | |||||||
| SDC2 | −3.72 | 0.0129 | 7 | −1.58 | 0.3300 | (n/a) | Syndecan 2. Cytoskeletal protein binding. | |||||||
| 10. Unknown | ||||||||||||||
| GSN | −3.59 | 0.0201 | 9 | −1.67 | 0.2600 | (n/a) | Gelsolin. See #2 above. | |||||||
| SDC2 | −3.13 | 0.0182 | 13 | −1.43 | 0.4200 | (n/a) | Syndecan 2. See #7 above. | |||||||
| 10) Unknown | ||||||||||||||
| KIAA0930 | −3.28 | 0.0180 | 11 | −1.44 | (n/a) | |||||||||
| (unnamed) | −3.10 | 0.0033 | 14 | −1.64 | (n/a) | |||||||||
| DKFZP564 | −2.95 | 0.0039 | 17 | −1.50 | (n/a) | |||||||||
| LOC345780 | −2.64 | 0.0190 | 30 | −1.68 | (n/a) | |||||||||
| FLJ20152 | −2.63 | 0.0023 | 31 | −1.34 | (n/a) | |||||||||
| TPT1, na | −1.15 | 0.3000 | (n/a) | −1.4 | 0.0378 | 12 | Tumor protein, translationally controlled 1. Little known. | |||||||
Genes are again grouped according to function. For comparisons of both H37Rv-infected vs. uninfected AM, and of H37Ra vs. uninfected AM, fold-change, P value, and rank for the five subjects studied are indicated. Italicized values indicate genes that met the study's significance criteria of fold change of ≥ 1.5 and P value of ≤ 0.05. (Listings of rank as “n/a”—“not applicable”—indicate genes that did not meet criteria for a significant change in expression for that comparison).
The 12 genes that displayed down-regulation after infection with H37Ra (as compared with uninfected AM after 24 hours of incubation) are also included in Table 3. As indicated, many of these were not significantly down-regulated by H37Rv, but only the fibronectin1 and cell cycle progression 8 protein displayed fc of greater than 2.0 in response to H37Ra.
Quantitative PCR
To confirm the most striking microarray findings, quantitative PCR for IL-23 p19, IL-12p35, and the shared subunit p40, as well as for GM-CSF, was performed.
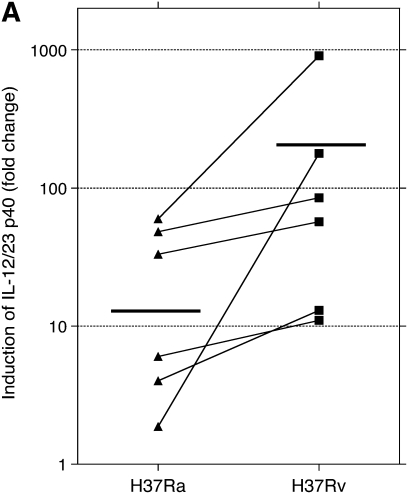
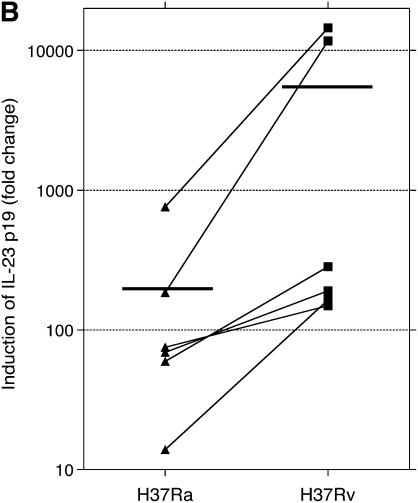
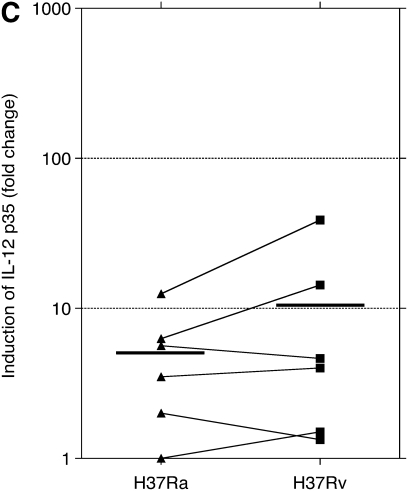
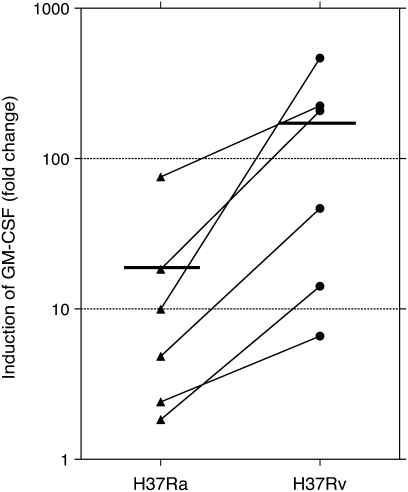
PCR studies were originally performed using the same RNA samples evaluated by microarray. In preliminary studies, contamination by genomic DNA was found to interfere with PCR assessments, so samples were subsequently treated with DNase. After treatment, only three of the original five sets of RNA samples were found to be adequate for PCR evaluation. BAL samples were therefore collected from an additional three subjects and were infected and otherwise prepared in the same manner to have an adequate number of samples to allow for statistical evaluation. The findings for PCR studies for these six subjects are presented in Figures 1 and 2 as fold-change in mRNA compared with that of uninfected cells at 24 hours of culture. Expression of mRNA for mRNA p40, p35, and p19 are displayed in Figure 1. Compared with uninfected cells, both H37Ra and H37Rv induced significant expression of IL-12/IL-23 p40, IL-23 p19, and IL-12 p35. Induction of gene expression by H37Rv was significantly greater than by H37Ra for both p40 and IL-23 p19 subunits (P = 0.031 for both p40 and p19). However, induction of IL-12 p35 by H37Rv was not significantly different from that observed with H37Ra. Expression of GM-CSF is displayed in Figure 2. Compared with uninfected cells, GM-CSF expression was also significantly up-regulated by both H37Ra and H37Rv. Induction of GM-CSF by H37Rv was significantly greater than by H37Ra, however (P = 0.031).
Figure 1.
Assessment of alveolar macrophage IL-23 and IL-12 gene expression in response to infection with Mycobacterium tuberculosis H37Rv and H37Ra. Panels indicate fold increases in expression (y axis) of RNA for (A) IL-12/IL-23 p40, (B) IL-23 p19, and (C) IL-12 p35, as normalized to that of uninfected cells at 24 hours of incubation. As illustrated, mean fold-change of p40 was 23.9 in response to H37Ra and 208.1 in response to H37Rv. H37Ra induced a 193.3 fold-change increase in p19 expression, compared with 4,495.5 increase induced by H37Rv. For both components of the IL-23 heterodimer, H37Rv induced significantly more gene expression than did H37Ra (P = 0.031 for both p40 and p19). In contrast, H37Ra and H37Rv both induced relatively modest changes in p35 induction (5.16-fold and 10.7 fold, respectively), and the differences between responses to the two strains were not significantly different for the IL-12 specific subunit (P = 0.440). Figure indicates individual data for each of six subjects studied. Statistics were determined using the Wilcoxon signed-rank test.
Figure 2.
QT-PCR confirms greater induction of GM-CSF by H37Rv than by H37Ra. Compared with uninfected cells at 24 hours of incubation, H37Ra induced a mean 18.8 fold-change in expression of GM-CSF, whereas H37Rv induced a 160.7 mean fold-change. This difference was statistically significant (P = 0.031) for the six subjects studied (using the Wilcoxon signed-rank test).
AM Display Greater Production of GM-CSF and IL-23 and in Response to Infection with H37Rv than Do Peripheral Blood Monocytes
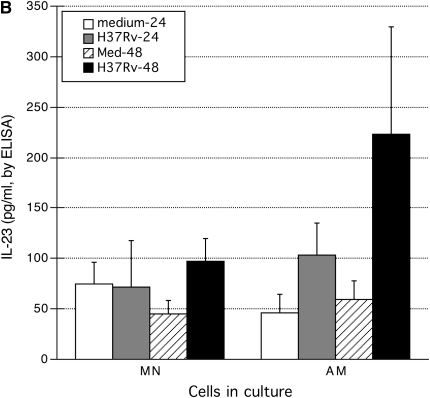
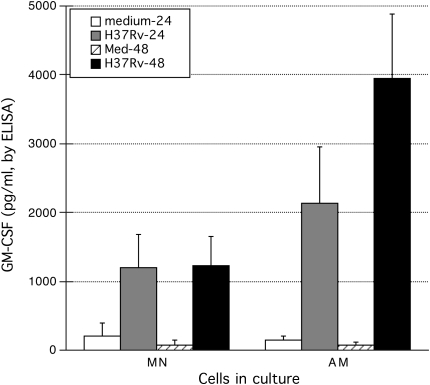
To evaluate the possibility that IL-23 and GM-CSF were prominent components of the early immune response to M. tuberculosis within the lung in particular, we assessed cytokine responses of paired AM and MN from 10 individuals after in vitro infection with H37Rv. IL-12 responses were assessed as well. For both IL-12 and IL-23, ELISA kits were selected specifically for detection of the functional dimer of the common p40 subunit associated with either p35 or p19, respectively. As illustrated in Figure 3A, low-level infection with H37Rv induced relatively little IL-12 production by either MN or AM, and production of IL-12 was not statistically significant for either cell type at 24 or 48 hours. MN likewise did not produce significant amounts of IL-23 in response to infection with H37Rv (Figure 3B). In contrast, production of IL-23 by AM was significant at 48 hours (P = 0.010, by Wilcoxon's signed-rank test). However, the difference in IL-23 production by AM and MN in response to H37Rv was not significant at either time point. In contrast, both MN and AM produced substantial (and statistically significant) amounts of GM-CSF protein after infection with H37Rv (Figure 4). Although production of GM-CSF by AM was somewhat higher than that of MN, these differences were not significant at either 24 or 48 hours after infection.
Figure 3.
Comparison of cytokine responses of alveolar macrophages (AM) and peripheral blood monocytes (MN) to infection with H37Rv. Culture supernatants were collected 24 and 48 hours after infection with M. tuberculosis H37Rv or after incubation in medium alone (n = 10 for all studies, with results are displayed as mean and SEM). As illustrated in A, neither AM nor MN produced significant quantities of the IL-12 heterodimer (as measured by ELISA) after infection with H37Rv. In contrast (B), AM did produce significant amounts of IL-23 after infection with H37Rv at 48 hours (P = 0.010), whereas MN did not. Although higher levels of IL-23 were observed after infection of AM than MN with H37Rv, these differences were not statistically significant for this number of subjects.
Figure 4.
Both MN and AM display significant production of GM-CSF in response to infection with M. tuberculosis H37Rv. GM-CSF responses of AM were somewhat higher than those of MN at both 24 and 48 hours after infection. Neither difference was statistically significant, however. Results again indicate means and SEM of responses of 10 subjects studied.
DISCUSSION
In this study, we used a genome-wide microarray system to assess differences in responses of human AM to infection with M. tuberculosis strains H37Rv and H37Ra. These organisms have been widely evaluated as representative virulent and avirulent laboratory strains, respectively, of M. tuberculosis. We had anticipated that infection with the avirulent strain would induce immune responses relevant to containment of the organism and that, in contrast, infection with the virulent strain would result in suppression of specific protective responses. Instead, we found that H37Rv induced significant changes in the expression of a much larger number of genes than did H37Ra. Moreover, the largest proportion of the genes up-regulated by H37Rv to a greater extent than by H37Ra represented proinflammatory responses associated with the development of effective cell-mediated immunity. Genes down-regulated by H37Rv were predominantly associated with general metabolic functions of the host cells.
IL-23 p19 was the gene most strongly induced by infection with H37Rv, whereas IL-12 was not significantly up-regulated by H37Rv or H37Ra. IL-23 and IL-12 are related heterodimeric cytokines associated with the development of Th1-mediated immunity. In addition, IL-23 has the unique ability to induce development of IL-17–producing CD4+ T cells, known as Th-17 cells (19). IL-12 and IL-23 share a common p40 subunit that joins either with p35 to form IL-12, or with p19 to form IL-23 (20). Quantitative PCR analysis confirmed that H37Rv induced greater AM expression of IL-23 p19 (as well as the shared p40 subunit) than did H37Ra, whereas the two strains did not demonstrate differential induction of IL-12 p35. Additional studies of responses of peripheral blood monocytes (MN) and AM confirmed that low-level infection with H37Rv failed to induce significant production of IL-12 by either cell type, and demonstrated that AM produced significant amounts of IL-23 in response to H37Rv, whereas MN did not. These observations raise the possibility that IL-23 rather than IL-12 may provide the predominant initial signal for the development of M. tuberculosis–specific cell-mediated immunity within the human lung. More generally, the finding that H37Rv induced strong responses of potentially protective cytokines also calls into question the proper designations of H37Ra and H37Rv within the expanding framework of epidemiologic and laboratory studies of mycobacterial virulence.
The use of a genome-wide microarray system in this study allowed for identification of genes that have not previously been evaluated for their potential role in the host response to M. tuberculosis. For example, our findings regarding apoptosis-related genes were different from those previously reported in a similar study by Spira and coworkers (13). Details of the designs of our respective studies may have led to some of the differences observed. However, our findings regarding M. tuberculosis–induced changes in the expression of PHLDA2, PBEF, IER3, and SERPINB9 all provide novel observations because these apoptosis-associated genes are not included in the Operon Human Array 1.0 array of selected apoptosis-associated genes used in the prior study. Likewise, a previous study of MN gene expression profiles in response to a range of bacterial pathogens noted that H37Rv was relatively ineffective in inducing IL-12, but the Affymetrix Hu6800 microarrays used in that assessment did not include the gene for IL-23 p19 (21). A comparison of gene responses to M. tuberculosis strains suggested by molecular epidemiology to be of varying virulence made use of Superarray Common Cytokine and Cytokine Receptor arrays that lacked p19 as well (14). Even comprehensive microarray-based studies have clear limitations, however. Because of post-transcriptional regulatory mechanisms, relative amounts of RNA induction may not correspond to differences in levels of functional protein. Further, the readout of gene expression is inherently descriptive, making conclusions regarding functional outcomes speculative.
In the case of our study, the differential intracellular burden of H37Rv and H37Ra provide an additional reason to view our findings with caution. In practice, many similar studies comparing infection with H37Rv and H37Ra have made no attempt to quantify burden of bacteria (other than performing infection using identical multiplicity of infection) (14, 22, 23). Given that clumping of bacteria occurs to varying degrees with different M. tuberculosis strains, it is difficult to achieve initial levels of infection as comparable as the 2.56-fold difference we observed currently (24), and the greater capacity of H37Rv for intracellular growth than H37Ra further adds to the difficulty in making quantitative comparisons regarding the effects of the two strains on host cells (10). In this regard, it is notable that our initial hypothesis was that H37Rv would down-regulate or suppress expression of “protective” genes induced by H37Ra. The fact that our infection protocol resulted in greater intracellular burden of H37Rv could have hindered our ability to identify genes up-regulated to a lesser extent by this strain than by H37Ra. Even in the face of higher infection with H37Rv, however, we did not identify any genes as being down-regulated by H37Rv to a greater extent than by H37Ra. Moreover, rather than down-regulating potentially protective responses, H37Rv infection primarily resulted in decreased expression of genes associated with the metabolic functions of resting host cells. Of the genes up-regulated by H37Rv to a greater extent than H37Ra, those we selected for further study (IL-23 and GM-CSF) were differentially expressed to substantially greater degrees than the 3.15-fold difference in intracellular CFU of the organisms 24 hours after infection, reducing the likelihood that this observation was an artifact of the differences in infectious burden alone.
The prominent attention given to the defects of IL-12 production and IL-12 receptor function in susceptibility to mycobacterial diseases has required reconsideration, as the findings suggesting these associations were based on the induction (or elimination) of the p40 subunit (25, 26), before the characterization of IL-23, and on studies of the IL-12Rβ1 subunit, which is a component of the IL-23 receptor as well (27, 28). Despite the known variability of human cytokine responses and the use of small numbers of subjects, we were able to demonstrate the predominance of induction of IL-23 rather than IL-12 in H37Rv-infected AM using three different methods: microarray analysis, quantitative PCR, and ELISA. An underlying assumption of our study is that these early responses to M. tuberculosis serve to prime subsequent development of antigen-specific cell-mediated immunity, rather than themselves mediating containment of the organism. Accordingly, it is difficult to confirm in humans the significance of the M. tuberculosis–induced IL-23 responses we observed. Using a murine model of infection, however, Cooper and colleagues have performed a series of studies that demonstrate a significant role for IL-23 in protective immunity to M. tuberculosis. These investigators first showed that disruption of the shared p40 subunit increases susceptibility to M. tuberculosis to a greater extent than disruption of the IL-12–specific subunit, p35 (29). Subsequent studies using p19 gene-disrupted mice suggest that, although IL-23 is not necessary for protection against M. tuberculosis in the presence of IL-12, it can serve to induce protective Th1-responses in the absence of IL-12 (30). The investigators have further shown that p19 gene–disrupted mice cannot develop protective recall responses to M. tuberculosis after experimental vaccination, although mice lacking p35 can do so. Treatment of p19-deficient mice with exogenous IL-17, a cytokine normally induced in response to IL-23, restored the capacity of these animals to develop protective responses after vaccination (31).
Our observation of significant production of IL-23 by AM, but not by MN, after infection with H37Rv also support the interpretation that IL-23 is of particular importance in the priming of protective immunity within the human lung. IL-23 may be uniquely well suited for promotion of Th1-responses in this local environment, as it acts predominantly on T cells that express the memory marker CD45RO (20). In marked contrast to peripheral blood lymphocytes, resting T cells of the lung overwhelmingly express CD45RO (32, 33). Robust local production of GM-CSF by AM may also contribute to the development of protective responses to M. tuberculosis. In murine studies, GM-CSF contributes to leukocyte recruitment to the lung in response to M. tuberculosis (34). Treatment of human peripheral blood monocytes with GM-CSF has been shown to induce development of a phagocyte population that produces IL-23 in response to infection with M. tuberculosis. This cell population is more effective than other phagocytes in inducing organism-specific Th1-like responses (35). Brisk production of GM-CSF by AM in response to M. tuberculosis infection thus may serve both to recruit additional phagocytes to the lung, and to promote differentiation of these recruited cells into a protective IL-23–producing phenotype.
The availability of molecular methods for strain identification of M. tuberculosis has indicated a need for reconsideration of the terms “virulent” and “avirulent” as they apply to laboratory strains and clinical isolates of M. tuberculosis. Molecular epidemiology has identified isolates of M. tuberculosis that are associated with outbreaks of active disease (11, 12, 36–38), as well as entire families of M. tuberculosis organisms, such as the Beijing strains, that are becoming increasingly prevalent throughout the world (39). Molecular analysis has, in addition, provided new tools for distinguishing H37Rv and H37Ra, which are frequently used as positive controls in clinical mycobacterial laboratories, from actual clinical isolates (40). In this context, the lack of identification of tuberculosis cases attributable to H37Rv throughout the era of molecular epidemiology supports the interpretation that this strain is not unusually virulent to humans.
The ability of widespread isolates of M. tuberculosis to rapidly induce disease without first being contained in the form of latent infection has led to their being termed “hypervirulent.” The basis for the hypervirulent phenotype has been investigated through comparison of immune responses induced by the Beijing-family outbreak strain H878 (36) with those induced by CDC1551, an isolate for which an unusually high incidence of tuberculosis skin-test conversion (but not of active disease) was observed in contacts of an infected individual (41). Manca and colleagues found that in contrast to CDC1551, HN878 was ineffective at inducing protective proinflammatory immune responses (14). Subsequent studies have attributed this difference to the capacity of a specific lipid, polyketide synthase–derived glycolipid (PGL), to suppress phagocyte cytokine production. The immune-inhibiting PGL is characteristic of HN878 as well as additional Beijing family isolates, but not of CDC1551. Like CDC1151, H37Rv lacks PGL, further suggesting that H37Rv may more closely resemble immunogenic, rather than hypervirulent, M. tuberculosis isolates (42). In contrast, the limited capacity of H37Ra to induce AM expression suggests that the “avirulence” of this strain may reflect an inherent inability of the organism to thrive in the intracellular environment (10, 43), rather than its capacity to stimulate effective immunity. This possibility is supported by in vivo studies, as H37Ra infection has been shown to be adequately controlled even in animals with significant impairments of cell-mediated immunity, such as SCID mice (44) and malnourished guinea pigs (45).
As molecular epidemiology continues to be more widely applied to the study of tuberculosis, additional isolates responsible for unusual clinical scenarios suggesting “hypervirulence” will be identified. Because of the world-wide availability of H37Rv and H37Ra, as well as the extensive existing literature comparing the two, these strains are likely to continue to serve as standards of reference for investigating the biology of clinical isolates of M. tuberculosis. Our findings suggest that the further investigation of mechanisms by which “hypervirulent” M. tuberculosis isolates evade host immunity may most appropriately compare these organisms to immunogenic strains such as H37Rv, rather than to poorly growing isolates whose containment may not require full activation of effective host immunity.
Supplementary Material
Acknowledgments
The authors thank Wilma Mackay, M.S. of the Division of Infectious Diseases at Case Western Reserve University School of Medicine for helpful discussions regarding study design and statistical analysis.
These studies were supported by NIH RO1 HL-59859 (to R.F.S), ALA Career Investigator Award CI-024N, and a Merit Review Award of the US Department of Veterans Affairs Office of Research and Development. Research bronchoscopies were performed in the Clinical Research Unit (CRU) of the Clinical and Translational Science Collaborative at Case Western Reserve University located at University Hospitals Case Medical Center. The CRU is supported by NIH UL1 RR024989 (to Pamela B. Davis and Richard Rudick) from the National Center for Research Resources (NCRR).
A preliminary report of the findings of this article was presented at the 2006 International Conference of the American Thoracic Society and were published in abstract form, entitled “Differential induction of human alveolar macrophage gene expression by virulent and avirulent strains of Mycobacterium tuberculosis,” in the Proceedings of the American Thoracic Society (2006;3:A218).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2008-0219OC on September 11, 2008
Conflict of Interest Statement: B.A.J. is a Wyeth Employee. The company has no apparent interest in the results of the study. M.R.B. is employed by Wyeth Research and owns stock and stock options in Wyeth Corperations. K.N. is an employee of Wyeth Research. J.P.S. is a Wyeth Employee. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Steenken JWJ, Gardner LU. History of H37 strain of tubercle bacillus. Am Rev Tuberc 1946;54:62–66. [DOI] [PubMed] [Google Scholar]
- 2.Pierce CH, Dubos RJ, Schaefer WB. Multiplication and survival of tubercle bacilli in the organs of mice. J Exp Med 1953;92:189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins FM, Smith MJ. A comparative study of the virulence of Mycobacterium tuberculosis measured in mice and guinea pigs. Am Rev Respir Dis 1969;100:631–639. [DOI] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998;393:537–544. [DOI] [PubMed] [Google Scholar]
- 5.Brosch R, Phillip WJ, Stravropoulos E, Colston MJ, Cole ST, Gordon SV. Genomic analysis reveals variation between Mycobacterium H37Rv and the attenuated M. tuberculosis H37Ra strain. Infect Immun 1999;67:5768–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of M. tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 2002;43:717–731. [DOI] [PubMed] [Google Scholar]
- 7.Fisher MA, Plikaytis BB, Shinnick TM. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol 2002;184:4025–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Q, Kripke KE, Saldanha AJ, Holmes S, Small PM. Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology 2005;151:5–14. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M, Gong J, Lin Y, Barnes PF. Growth of virulent and avirulent Mycobacterium tuberculosis strains in human macrophages. Infect Immun 1998;66:794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silver RF, Li Q, Ellner JJ. Expression of virulence of Mycobacterium tuberculosis within human monocytes: virulence correlates with intracellular growth and induction of tumor necrosis factor alpha, but not with evasion of lymphocyte-dependent monocyte effector functions. Infect Immun 1998;66:1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M, Gong J, Yang Z, Samten B, Cave MD, Barnes PF. Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J Infect Dis 1999;179:1213–1217. [DOI] [PubMed] [Google Scholar]
- 12.Theus SA, Cave MD, Eisenach KD. Intracellular macrophage growth rates and cytokine profiles of Mycobacterium tuberculosis strains with different transmission dynamics. J Infect Dis 2005;191:453–460. [DOI] [PubMed] [Google Scholar]
- 13.Spira A, Carroll JD, Liu G, Aziz Z, Shah V, Kornfeld H, Keane J. Apoptosis genes in human alveolar macrophages infected with virulent or attenuated Mycobacterium tuberculosis: a pivotal role for tumor necrosis factor. Am J Respir Cell Mol Biol 2003;29:545–551. [DOI] [PubMed] [Google Scholar]
- 14.Manca C, Reed MB, Freeman S, Mathema B, Kreiswirth B, Barry CE III, Kaplan G. Differential monocyte activation underlies strain-specific Mycobacterium tuberculosis pathogenesis. Infect Immun 2004;72:5511–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silver RF, Li Q, Boom WH, Ellner JJ. Lymphocyte-dependent inhibition of growth of virulent Mycobacterium tuberculosis H37Rv within human monocytes: requirement for CD4+ T cells in purified protein derivative-positive, but not in purified protein derivative-negative subjects. J Immunol 1998;160:2408–2417. [PubMed] [Google Scholar]
- 16.Byrne MC, Whiteley MZ, Follettie MT. Preparation of mRNA for expression monitoring. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Hoboken, NJ: John Wiley and Sons; 2000.
- 17.Hill AA, Brown EL, Whitely MZ, Tucker-Kellogg G, Hunter CP, Slonim DK. Evaluation of normalization procedures for oligonucleotide array data based on spiked cRNA controls. Genome Biology 2001;2:0055.1–0055.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Affymetrix. Statistical algorithms description document. [Accessed 2002.] Available from: http://www.affymetrix.com/support/technical/whitepapers/sadd_whitepaper.pdf
- 19.Aggarwal S, Ghilardi N, Xie M-H, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of Interleukin-17. J Biol Chem 2003;278:1910–1914. [DOI] [PubMed] [Google Scholar]
- 20.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K,et al. Novel p19 protein engages IL-12p40 to form a new cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 2000;13:715–725. [DOI] [PubMed] [Google Scholar]
- 21.Nau GJ, Richmond JFL, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA 2002;99:1503–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadek MI, Sada E, Toossi Z, Schwander SK, Rich EA. Chemokines induced by infection of mononuclear phagocytes with mycobacteria and present in lung alveoli during active pulmonary tuberculosis. Am J Respir Cell Mol Biol 1998;19:513–521. [DOI] [PubMed] [Google Scholar]
- 23.Keane J, Remold HG, Kornfeld H. Virulent mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J Immunol 2000;164:2016–2020. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Whalen CC, Larkin R, Zukowski L, Albert J, Cave MD, Silver RF. Differences in rate and variability of intracellular growth of a panel of clinical isolates of Mycobacterium tuberculosis within a human monocyte model. Infect Immun 2002;70:6489–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fulton SA, Johnsen JM, Wolf SF, Sieburth DS, Boom WH. Interleukin-12 production by human monocytes infected with Mycobacterium tuberculosis: role of phagocytosis. Infect Immun 1996;64:2523–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper AM, Magram J, Ferrante J, Orme IM. Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with Mycobacterium tuberculosis. J Exp Med 1997;186:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altare F, Durandy A, Lammas D, Emile JF, Lamhamedi S, Le Deist F, Drysdale P, Jouanguy E, Doffinger R, Bernaudin F, et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science 1998;280:1432–1435. [DOI] [PubMed] [Google Scholar]
- 28.de Jong R, Altare F, Haagen IA, Elferink DG, Boer T, van Breda Vriesman PJ, Kabel PJ, Draaisma JM, van Dissel JT, Kroon FP, et al. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 1998;280:1435–1438. [DOI] [PubMed] [Google Scholar]
- 29.Cooper AM, Kipnis A, Turner J, Magram J, Ferrante J, Orme IM. Mice lacking bioactive IL-12 can generate protective, antigen-specific cellular responses to mycobacterial infection only if the IL-12 p40 subunit is present. J Immunol 2002;168:1322–1327. [DOI] [PubMed] [Google Scholar]
- 30.Khader SA, Pearl JE, Sakamoto K, Gilmartin L, Bell GK, Jelley-Gibbs DM, Ghilardi N, deSauvage F, Cooper AM. IL-23 compensates for the absence of IL-12 p70 and is essential for the IL-17 response during tuberculosis but is dispensable for protection and antigen-specific IFNγ responses if IL-12 p70 is available. J Immunol 2005;175:788–795. [DOI] [PubMed] [Google Scholar]
- 31.Khader S, Bell G, Pearl J, Fountain J, Rangel-Moreno J, Cilley G, Shen F, Eaton S, Gaffen S, Swain S, et al. IL-23 and IL-17 in the establishment of preective pulmonary CD4+ T-cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 2007;8:369–377. [DOI] [PubMed] [Google Scholar]
- 32.Robinson DS, Bentley AM, Hartnell A, Kay AB, Durham SR. Activated memory T helper cells in bronchoalveolar lavage fluid from patients with atopic asthma: relation to asthma symptoms, lung function, and bronchial responsiveness. Thorax 1993;48:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walrath J, Zukowski L, Krywiak A, Silver RF. Resident Th1-like effector memory cells in pulmonary recall responses to Mycobacterium tuberculosis. Am J Respir Cell Mol Biol 2005;33:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Juarrero M, Hattle JM, Izzo A, Junqueira-Kipnis AP, Shim TS, Trapnell BC, Cooper AM, Orme IM. Disruption of granulocyte macrophage-colony stimulating factor production in the lungs severely affects the ability of mice to control Mycobacterium tuberculosis infection. J Leukoc Biol 2005;77:914–922. [DOI] [PubMed] [Google Scholar]
- 35.Verreck FAW, de Boer T, Langenberg DML, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, de Waal-Malefyt R, Ottenhoff THM. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA 2004;101:4560–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kresiwirth BN, Whittam TS, Musser JM. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA 1997;94:9869–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agerton TB, Valway SE, Blinkhorn RJ, Shilkret KL, Reves R, Schluter WW, Gore B, Pozsik CJ, Plikaytis BB, Woodley C, et al. Spread of strain W, a highly drug-resistant strain of Mycobacterium tuberculosis, across the United States. Clin Infect Dis 1999;29:85–92. [DOI] [PubMed] [Google Scholar]
- 38.Milan SJ, Haugue KA, Kurepina NE, Lofy KH, Goldberg SV, Narita M, Nolan CM, McElroy PD, Kresiwirth BN, Cangelosi GA. Expanded geographical distribution of the N family of Mycobacterium tuberculosis strains within the United States. J Clin Microbiol 2004;42:1064–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glynn J, Witely J, Bifani P, Kremer K, van Soolingen D. Worldwide occurrence of Beijing/W strans of Mycobacterium tuberculosis: a systemic review. Emerg Infect Dis 2002;8:843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bifani P, Moghazeh S, Shopsin B, Driscoll J, Ravikovitch A, Kreiswirth B. Molecular characterization of Mycobacterium tuberculosis H37Rv/Ra variants: sintinguishing the mycobacterial laboratory strain. J Clin Microbiol 2000;38:3200–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valway S, Sanchez M, Shinnick T, Orme I, Argerton T, Hoy D, Jones J, Westermoreland H, Onorato I. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med 1998;10:633–639. [DOI] [PubMed] [Google Scholar]
- 42.Reed M, Domenech P, Manca C, Su H, Barczak A, Kreiswirth B, Kaplan G, Barry CEI. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 2004;431:84–87. [DOI] [PubMed] [Google Scholar]
- 43.McDonough KA, Kress Y, Bloom BR. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect Immun 1993;61:2763–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.North RJ, Izzo AA. Mycobacterial virulence: virulent strains of Mycobacteria tuberculosis have faster in vivo doubling times and are better equipped to resist growth-inhibiting functions of macrophages in the presence and absence of specific immunity. J Exp Med 1993;177:1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMurray D, Carlomango M, Cumberland P. Respiratory infection with attenuated Mycobacterium tuberculosis H37Ra in malnourished guinea pigs. Infect Immun 1982;39:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.