SUMMARY
Telomerase is a ribonucleoprotein complex that replicates the 3’ ends of linear chromosomes by successive additions of telomere repeat DNA. The telomerase holoenzyme contains two essential components for catalysis, a telomerase reverse transcriptase (TERT) and telomerase RNA (TER). The TER includes a template for telomere repeat synthesis as well as other domains required for function. We report the solution structure of the wild type minimal conserved human TER pseudoknot (PKWT) refined with an extensive set of RDCs, and a detailed analysis of the effect of the bulge U177 on pseudoknot structure, dynamics analyzed by RDC and 13C relaxation measurements, and base pair stability. The overall structure of PKWT is highly similar to the previously reported ΔU177 pseudoknot (PKDU) that has a deletion of a conserved bulge U important for catalytic activity. For direct comparison to PKWT, the structure of PKDU was re-refined with a comparable set of RDCs. Both pseudoknots contain a catalytically essential triple helix at the junction of the two stems, including two stem 1-loop 2 minor groove triples, a junction loop 1-loop 2 Hoogsteen base pair, and stem 2-loop 1 major groove U•A-U Watson-Crick-Hoogsteen triples located directly above the bulge U177. However there are significant differences in stabilities of base pairs near the bulge and the dynamics of some nucleotides. The stability of the base pairs in stem 2 surrounding the bulge U177 is greatly decreased, with the result that the Watson-Crick pairs in the triple helix begin to unfold before the Hoogsteen pairs, which may affect telomerase assembly and activity. The bulge U is positioned in the minor groove on the opposite face from the triple helical interactions, and sterically blocks the A176 2’OH which has recently been proposed to play a role in catalysis. The bulge U may serve as a hinge to provide backbone flexibility in this region.
Keywords: NMR, telomerase, RNA pseudoknot, structure, dynamics
Introduction
Telomerase is a ribonucleoprotein complex (RNP) that extends the 3’ ends of linear chromosomes by successive additions of telomere repeats, dTTAGGG in vertebrates.1 In most somatic cells, telomerase activity is low or undetectable, and telomeres shorten with accumulated cell divisions. Shortening of telomeres below a critical length leads to apoptosis and cell death, and there is a strong correlation between telomere length and aging.2–4 Telomerase is highly active in more than 90% of cancer cell lines and this activity appears to be required for the proliferation of cancer cells.5,6 Mutations in both the RNA and protein components of telomerase have been linked to diseases of the haemopoietic system such as dyskeratosis congenita and aplastic anemia.7–10 The correlations between telomerase activity, telomere length, aging, cancer, and disease have led to tremendous interest in understanding how telomerase functions.
The core components of telomerase for catalytic activity are telomerase reverse transcriptase (TERT) and telomerase RNA (TER).11 In addition to TERT, there are also other proteins associated with telomerase, which are important for ribonucleoprotein assembly, maturation, localization, processivity, and stability.11–16 The TER contains a template that is used by the unique reverse transcriptase TERT to copy the telomeric sequence.12,17,18
TERs have been identified and sequenced in many eukaryote species, and are divergent in both size and sequence among different organisms. However phylogenetic and mutational analyses revealed that highly conserved secondary structures exist in TER, which in addition to the template are essential for telomerase function.8,19–22 In vertebrates there are four conserved components: the pseudoknot/core, CR4-CR5, H/ACA, and CR7 domains (Figure 1).19 The H/ACA and CR7 domains are important for TER accumulation, localization, and 3’ end processing.14,23 The pseudoknot/core includes the template and template boundary element and together with the CR4-CR5 domain is required for TERT binding, nucleotide and telomere repeat processivity and fidelity, and catalytic activity.19,24–28 Telomerase catalytic activity can be reconstituted in vitro with TERT plus the TER pseudoknot/core and CR4-CR5 domains only.29,30 The pseudoknot in vertebrate TERs contains two helical regions, P2b and P3, a loop (J2b/3), and the last 4 nucleotides of loop J2a/3, with highly conserved sequence. The remainder of the pseudoknot, the helical regions P2a.1 and P2a as well as the single stranded J2a/3 region, is not highly conserved, and except for the last 4 nucleotides, J2a/3 is not required for catalytic activity in vitro.20,24,26,28
Figure 1. The pseudoknot domain of hTER.
Secondary structure of human telomerase RNA8,19 showing the pseudoknot/core, CR4-CR5, CR7, and H/ACA domains. The conserved region of the pseudoknot from which the minimal pseudoknot construct (PKWT) used in this study was derived is boxed. (Inset) The sequence and secondary structure of PKWT. PKWT is composed of stem1 (P2b), loop1 (J2b/3), stem 2 (P3), and loop 2 (last 8 nucleotides of J2a/3). The nucleotides shown in green are 100%, uppercase black ≥80%, and lowercase black <80% conserved in vertebrate TERs. The bulge U177 is colored in red. The first two G-C base pairs in stem 1 were added to optimize transcription yields.
The structural basis of the sequence conservation in vertebrate TER pseudoknots was revealed by the solution structure of a minimal hTER pseudoknot construct (PKDU), which showed it contains a unique triple helical region at the junction of the two stems.28 The triple helix was further shown to be critical for catalysis, since mutations that destabilized the tertiary interactions also decreased telomerase activity, and compensatory mutations at least partially restored activity. Recent mutational and modeling studies have led to the proposal that a similar triple helix is present in the pseudoknots in the much larger TERs in Kluyveromyces species of yeasts31 and S. cerevisiae,32 and provide further evidence that the tertiary structure of the pseudoknot plays an essential role in catalysis.
In the PKDU minimal pseudoknot construct, the bulge U177 in stem 2 was removed in order to stabilize the pseudoknot conformation that is in equilibrium with a stem 1 hairpin folding intermediate.33 However, this mutation results in a 2-fold decrease in activity for TER plus TERT reconstituted in rabbit reticulocyte lysate and a 10-fold decrease for telomerase assembled in vivo and assayed in vitro.28,34 It has been suggested that the lower activity of telomerase hTERΔU177 might result from an alteration of the pseudoknot structure, e.g. a single nucleotide shift in the register of the triple helix.31 Molecular modeling studies also predict very different tertiary interactions for the wild-type vs the ΔU177 pseudoknot.35
We have determined the solution structure of the wild-type minimal conserved hTER pseudoknot (PKWT), refined with an extensive set of RDCs, and analyzed the effect of the bulge U177 on pseudoknot structure, dynamics, and stability. PKWT has the same overall fold and almost all the same tertiary interactions as PKDU, including the unusual major groove triples.28 However there is a significant difference in the dynamic behavior of some nucleotides, which has important implications for catalysis. The presence of the bulge U177 in PKWT greatly alters the relative stability of the surrounding base pairs, resulting in an altered unfolding pathway of the functionally critical triple helix. U177 is positioned in the minor groove directly over the A176 ribose, sterically blocking the 2’-OH that has recently been proposed to play a direct role in catalysis.32
RESULTS
Chemical shift mapping of wild-type hTER pseudoknot vs ΔU177
In order to investigate how the bulge U177 affects the structure of the pseudoknot and telomerase activity, we used a minimal wild-type pseudoknot construct (PKWT) with the same sequence as the previously solved ΔU177 pseudoknot (PKDU) except that it includes the bulge U177 (Figure 1 inset).28 Therefore, the wild type pseudoknot construct contains all of the highly conserved P3 (stem 2) and U-rich J2b/3 (loop 1) and the conserved P2b (stem 1) and J2a/3 (loop 2) nucleotides. A single nucleotide bulge at the same position in P3 is highly conserved in all vertebrate TERs (79%) sequenced to date36,37 and when present is always a pyrimidine. Only in some fish is the bulge either not present or in a different location in P3.38
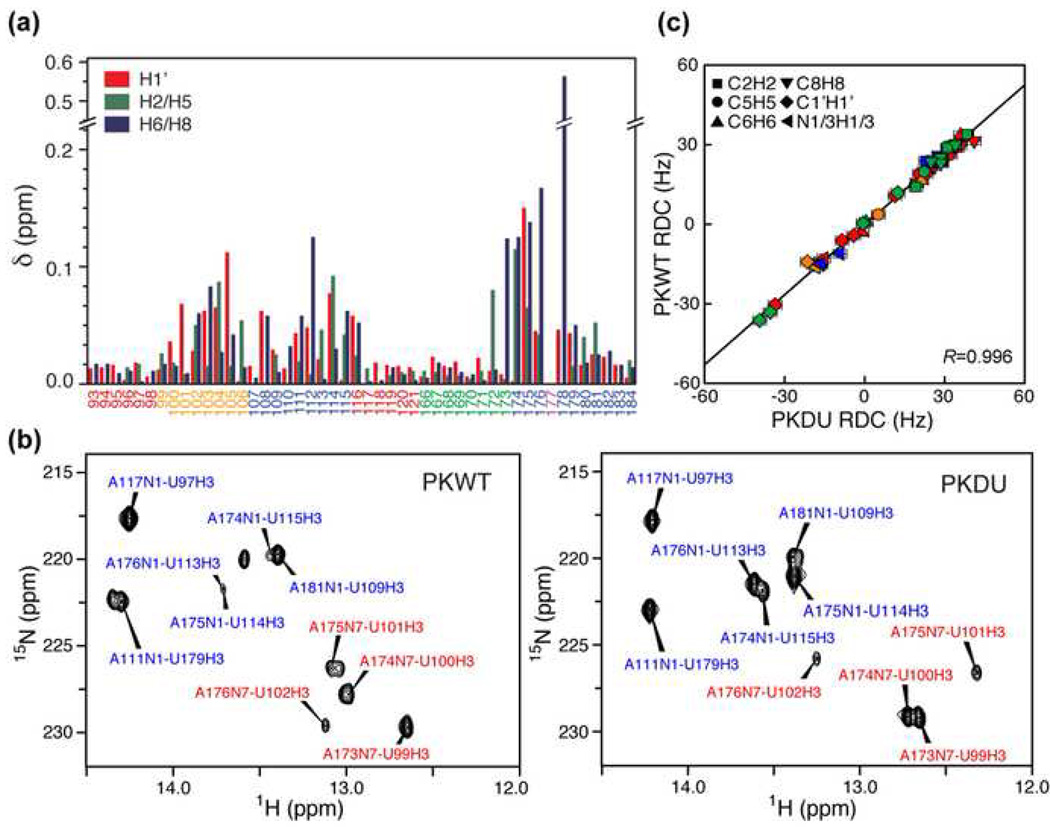
The NMR spectra of PKWT are complicated by the resonances from the presence of a small amount of the stem 1 hairpin conformation (formed by stem 1 and loop 1), a partially unfolded intermediate, which is in slow exchange with the pseudoknot conformation.33,34 The population of folded pseudoknot is ≥85% according to native gel and NMR spectral analysis. Despite the presence of resonances from the hairpin, it was possible to complete assignments of PKWT by initial comparison to the resonance assignments for PKDU28 and the isolated stem 1 hairpin.33 The chemical shift difference plot for the H1’ and aromatic proton resonances shows that the largest chemical shift differences between PKDU and PKWT are for the stem 2 residues around the bulge U177 and for loop 1 (Figure 2(a)). There are almost no differences in chemical shifts for the stem 1 and loop 2 region, except for small difference in loop 2 nucleotides near the junction of stem 1 and 2, and only minor differences are observed in the lower part of stem 2. Thus, chemical shift mapping indicates that the bulge nucleotide, U177, does not cause global changes in the PKWT structure relative to PKDU.
Figure 2. Chemical shift mapping, hydrogen bonding, and RDC analysis of PKWT vs PKDU.
(a) Plot of chemical shift differences between PKWT and PKDU for the H1’, H2/H5, and H8/H6 resonances. (b) JNN-HNN-COSY spectra for (left) PKWT and (right) PKDU showing region of NH crosspeaks from Watson-Crick and Hoogsteen A-U pairs. Crosspeaks are labeled blue for Watson-Crick base pairs and red for Hoogsteen base pairs. Spectra were taken at 293K on 600 MHz NMR. (c) Structural dynamics of PKDU and PKWT by RDCs. Shown is a correlation plot between RDCs of C2H2 (squares), C5H5 (circles), C6H6 (triangles), C8H8 (inverted triangles), C1’H1’ (diamonds), and N1/3-H1/3 (left triangles) measured for PKDU and PKWT.
Base pairs in the wild-type hTER pseudoknot
In the PKDU structure, all of the Watson-Crick and Hoogsteen base pairs were identified on the basis of imino-imino and imino-aromatic NOEs observed in NOESY and 1H-15N CPMG NOESY spectra, and most were confirmed by direct detection of hydrogen bonds using JNN-HNN-COSY experiments.39,40 A similar set of NOE connectivities is observed for PKWT (not shown). Figure 2(b) shows the AN1/N7-UH3 crosspeaks for Watson-Crick and Hoogsteen A-U base pairs in PKWT and PKDU observed at 20°C in the J NN-HNN-COSY spectra. JNN crosspeaks corresponding to all of the stem Watson-Crick, junction Hoogsteen, and stem-loop Hoogsteen base pairs including the three U-A-U triples observed for PKDU are also present in the PKWT spectra. Assignments to specific base pairs were confirmed in the 1H-15N CPMG-NOESY spectra (spectra not shown). These results indicate that PKWT forms the same secondary and tertiary base pairs as PKDU.
Residual dipolar coupling values reveal that PKWT and PKDU have similar conformations and dynamics
Residual dipolar couplings (RDCs) can provide long-range angular constraints for structure calculations, and can also provide characterization of motions faster than μs-ms timescale.41,42 An extensive set of 70 and 82 RDCs were measured on the bases and riboses of PKWT and PKDU, respectively (Supplementary Table 1). Figure 2(c) shows a correlation plot of the 51 RDCs shared in common between PKWT and PKDU. The high correlation factor R=0.996 indicates that both PKWT and PKDU have the same overall alignment, consistent with very similar overall structural and dynamical properties. Therefore the bulge U177 has at most a small effect on the overall conformation of the pseudoknot. In addition, during the structure refinements (discussed below), it was possible to fit the 63 and 74 RDCs for PKWT and PKDU, respectively, which do not display significant dynamics based on 13C spin relaxation measurements, to a single alignment tensor while simultaneously satisfying all other distance and dihedral angle restraints. This suggests that PKWT and PKDU have folded into stable overall conformations in which there are no observable inter-domain motions on a timescale faster than μs-ms. However, it should be noted that given the similar sizes of each domain, it is formally possible that inter-domain motions could evade detection based on RDCs if they contribute similarly to the overall alignment.
Solution structure of PKWT
The chemical shift difference mapping, JNN correlations, and RDC analysis all indicate that PKWT has the same global fold and tertiary interactions as PKDU, and thus the bulge U does not affect the overall fold. To further explore the role of the bulge U in telomerase RNA structure and function, we determined the solution structure of PKWT. The NOE distance restraints for the residues in stem 1 and loop 2 are almost identical between PKWT and PKDU, consistent with the small differences in chemical shifts. However, many of the NOE distance restraints obtained from analysis of 2D filtered/edited NOESY spectra43 of PKWT were different than PKDU for the junction, stem 2, and loop 1 regions, and new NOEs were observed for the residues around the bulge U. Resonance assignments and structure calculations are described in Methods. The solution structure of PKWT was determined using 756 NOE distance restraints and 242 dihedral restraints, and further refined using 63 RDCs (Table 1). The PKWT structure is well defined with a pairwise r.m.s.d. to the mean of 1.08 ± 0.27 Å for all heavy atoms for the 20 lowest energy structures (Figure 3 (a),(c) and Table 1).
Table 1.
Restraints and Structural Statistics for hTER Pseudoknots
| PKWT | PKDU | |
|---|---|---|
| NOE distance and dihedral constraints | ||
| Distance restraints | ||
| Total NOE | 756 | 744 |
| Intra-residue | 345 | 337 |
| Inter-residue | ||
| Sequential (|i−j| = 1) | 290 | 286 |
| Nonsequential (|i−j| > 1) | 121 | 121 |
| Hydrogen bond | 90 | 91 |
| Total dihedral angle restraints | 242 | 240 |
| Total RDCs (1DCH and 1DNH) | 63 | 74 |
| Qfree(%) | 7.53 | 7.61 |
| Structure statistics | ||
| Violations | ||
| Distance constraints (Å)a | 0.029 ± 0.001 | 0.038 ± 0.002 |
| Dihedral angle constraints (°)b | 0.163 ± 0.022 | 0.129 ± 0.018 |
| Dipolar couplings (Hz)c | 0.410 ± 0.035 | 0.376 ± 0.021 |
| Deviation from idealized geometry | ||
| Bond lengths (Å) | 0.004 ± 0.0003 | 0.005 ± 0.0001 |
| Bond angles (°) | 1.083 ± 0.003 | 1.100 ± 0.005 |
| Impropers (°) | 0.473 ± 0.009 | 0.505 ± 0.008 |
| Average r.m.s.d. (Å) from mean structure for 20 | 1.08 ± 0.27 | 1.02 ± 0.19 |
| lowest energy structures for all heavy atoms | (Residues 1–47) | (Residues 1–46) |
| a. Number of NOE violations > 0.2 (Å) | 0.65 ± 0.81 | 6.55 ± 2.10 |
| Number of NOE violations > 0.5 (Å) | 0 | 0 |
| b. Number of Dihedral violations > 5 (°) | 0 | 0 |
| c. Number of RDC violations > 2 (Hz) | 0 | 0 |
Figure 3. NMR solution structures of the hTER pseudoknots.
(a, b) Superpositions of the 20 lowest energy structures over all heavy atoms (a) PKWT and (b) PKDU. (c, d) Lowest energy structure of (c) PKWT and (d) PKDU. The phosphodiester backbone is outlined by a gray ribbon. Due to unwinding at the stem 1-stem 2 junction, the base interactions of loop 2 in the minor groove and loop 1 in the major groove are on the same face of the pseudoknot. (e) Schematic of the tertiary fold of PKWT. Nucleotides are colored red (stem 1), blue (stem 2), gold (loop 1), green (loop 2), and magenta (U177).
As in PKDU, the two phylogenetically predicted stems, stem 1 (P2b) and stem 2 (P3), are separated by an A-U Hoogsteen pair formed by the first nucleotide (U99) in loop 1 and the last nucleotide (A173) in loop 2. Above the junction A-U base pair, there are two minor groove base triple interactions (C116•G98-A172 and A117•U97-A171) between loop 2 and the last two base pairs of stem 1. Below the junction, three successive Hoogsteen triples are formed by base pairing of loop 1 nucleotides (U100, U101, U102) with the three A nucleotides in the Watson-Crick A•U pairs at the top of stem 2 (Figure 4 (a), (c), (e)). The junction Hoogsteen paired A173 stacks directly over the Hoogsteen base pairs in the stem 2-loop 1 triples and beneath the loop 2 A172, which forms the bottom base triple with stem 1 (C116•G98-A172).
Figure 4. Details of the tertiary structure of PKWT and PKDU.
(a, c) Lowest energy structure of (a) PKWT and (c) PKDU showing the triple helical region. (b, d) Schematic showing the base interactions in the triple helix and below in (b) PKWT and (d) PKDU. The nucleotides are colored as in Figure 3. (e) The U114-A175-U101 Hoogsteen base triple in PKWT. (f) The C112-G178-U103 base triple in PKDU. (g) Detailed view of the base pairs surrounding U177, showing the roll and tilt of the flanking base pairs and the position of U177 base over the A176 ribose. The U177 base (magenta) and A176 2’OH are shown as van der Waals surface. For A–F, the nucleotides are colored as in Figure 3, and in G the nucleotides are in CPK colors.
In PKWT, as in PKDU, the bulge U177 is located directly below the three Hoogsteen U•A-U triples in stem 2. U177 is flipped out of the helix and is situated in the minor groove of stem 2, on the opposite face of the helix from where the tertiary interactions that form the triple helix are located. Only a few inter-residue NOEs were observed for U177, and consistent with this as well as dynamics measurements (discussed below) its position in the minor groove is slightly less well defined than the base paired regions of the pseudoknot. The bulge U177 did not disrupt its neighboring C112•G178 and U113•A176 base pairs, as clearly indicated by the presence of their hydrogen bonded iminos (Figure 2 and 7). However, the presence of the bulge U creates a large roll (opening toward minor groove) and tilt (opening toward the bulge U) between the U113•A176 and C112•G178 base pairs, thus disrupting the base stacking between A176 and G178. This generates more space in the major groove for the loop 1 nucleotides below the triple helix, which is consistent with the changes in chemical shift and dynamics (discussed below) observed for the 3’ loop 1 nucleotides.
Figure 7. Comparison of base pair stabilities between PKWT and PKDU.
Imino proton spectra of (a) PKWT and (b) PKDU from 5 to 45 °C. Resi due numbers (labeled on the 20 °C and 45 °C spectra) are color coded as in Figure 3, with stem 2 Watson-Crick paired residues 113–115 shown in darker blue for emphasis. For PKWT, small resonances below 11.6 ppm are from the stem 1 hairpin iminos in U-U base pairs;33 other resonances from the hairpin are also visible in the spectra, as well as some small peaks from a fraction of N-1 molecules.
RDC-refined structure of PKDU
The solution structure of PKDU (PDB ID 1YMO) was previously solved using a limited set of RDCs (20) in the final stages of refinement. To obtain a structure of comparable precision to the solution structure of PKWT presented here, the structure of PKDU was re-refined using the more extensive set of RDCs acquired in this study (Supplementary Table 1). In this refinement calculation (hereafter referred to as RDC-refined), all NOE and dihedral angle restraints remained the same, and the 74 newly measured RDCs that did not show significant dynamics were used. Additionally, in the initial calculations for the RDC-refined structures, U103 N3H was consistently within hydrogen bond distance of G178 N7. Re-examination of the spectra revealed a previously unassigned weak imino proton resonance that we identified as U103, which shows crosspeaks consistent with a hydrogen bond to G178 N7. This additional hydrogen bond, which was not observed in PKWT spectra, was included in the final PKDU structure calculations. The RDC-refined PKDU structures have a pairwise r.m.s.d. to the mean of 1.02 ± 0.19 Å for all heavy atoms for the 20 lowest energy structures (Figure 3 (b), (d) and Table 1) vs 1.28 ± 0.29 Å in the earlier refinement. The RDC-refined PKDU structure is slightly more bent than the earlier refinement, with most of the difference due to an increased bend toward the major groove of the lower part of stem 2. U103 is slightly repositioned to form the C112•G178-U103 triple (Figure 4(f)) at the bottom of the stem 2-loop 1 triplex. The major groove of stem 2 is a little wider and as a consequence the nucleotides at the bottom (turn) of loop 1 are less conformationally restrained.
Comparison of PKWT and PKDU solution structures
Despite their similar global fold and tertiary structure as indicated by the high correlation between RDCs shared in common, direct comparison of the RDC-refined structures of PKDU with PKWT does reveal some important differences between them (Figure 4). Computationally, these differences arise from some differences in the NOEs and the restraints from the bulge U, and a subset of the RDCs not shared in common. Back calculation of the set of PKDU RDCs shared in common with PKWT using the structures of PKWT and the original order tensors in the PKWT structure refinement gave an averaged Q value of 11.0%. A similar Q value of 9.9% was obtained for the equivalent calculations of PKWT RDCs. These low Q values are consistent with the high correlation of the shared RDCs (Figure 2 (c)). While same back-calculation of the RDCs not shared in common gave averaged Q values of 64.5% and 38.6% for PKWT and PKDU RDCs, respectively, a subset of them (8 out of 17 PKWT RDCs and 15 out of 27 PKDU RDCs) are within 2 sigma error (<5–6 Hz) from the back-calculated values. These subsets of the RDCs not shared in common are consistent with the similar overall global structures between PKWT and PKDU, while the rest reflect the differences in the structures of PKWT and PKDU.
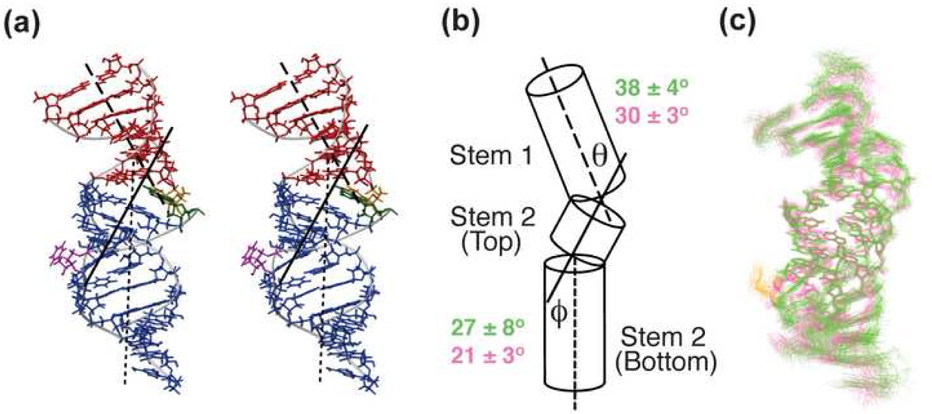
In both pseudoknots, two helical bends are found, one at the junction region between stems 1 and 2 where a significant unwinding of two stems is observed, and the other in stem 2 between the bottom of the U•A-U triplex and the C112•G178 below, where the bulge U is located in PKWT (Figure 5). The accommodation of the bulge U177 in PKWT introduces small helical bending differences in these two regions compared to PKDU. At the junction between stem 1 and stem 2, insertion of the Hoogsteen U99-A173 base pair prevents coaxial stacking of stem 1 and stem 2 and results in a bend of 30±3° in PKDU. In PKWT, this bend increases to 38±4°. In the PKDU structure, there is a small bend of 21 ±3° in stem 2 between the Watson-Crick partners of the last Hoogsteen triple (A176-U113) and the following Watson-Crick base pair (C112•G178), but the base pairs remain stacked. In the presence of the bulge U, the bases of A176 and G178 are partially unstacked as described above, and there is a slightly larger bend toward the major groove in the stem of PKWT at this position (27±8° vs 21 ±3°). Despite the small differences between the average inter-helical angles, we note that the positions of the two bends in PKWT and PKDU are such that the overall alignment of the two molecules is the same within experimental error.
Figure 5. Helical bends in PKWT and PKDU.
(a) Stereoview of the lowest energy structure of PKWT showing stem 1, stem 2, and the junction Hoogsteen base pair, with the helical axes shown as dashed (stem 1), solid (stem 2 top) and dotted (stem 2 bottom) lines, respectively. (b) Schematic showing the bends in the stems of the pseudoknots, with the stems shown as cylinders. Helical axes are defined for stem 1, region of stem 2 containing the three U-A base pairs in the triples (stem 2 top), and region of stem 2 below the three A-U base pairs (stem 2 bottom), and bends between them are defined as θ and ϕ, respectively. Helical angles were measured for the 20 lowest energy structures of PKWT (green) and PKDU (pink) and averaged (Curves 5.3 software). (c) Superpositions of the 20 lowest energy structures of PKWT (green) and PKDU (pink), with both families superimposed on the three Hoogsteen base triples (U100-A174-U115→U102-A176-U113).
Deletion of U177 does allow loop 1 to interact more tightly with stem 2 than in PKWT, resulting in formation of the C112•G178-U103 base triple in PKDU (Figure 4 (f)). The position of the bulge U177 in the minor groove and helical bending of stem 2 at U177 in PKWT generate a wider stem 2 major groove compared to PKDU. As a result, the loop 1 nucleotides, especially U103 and C104, interact more loosely with stem 2 than is the case for PKDU. In PKWT structures, the U103 N3H is not within hydrogen bond distance of G178 N7. Consistent with this, no imino proton resonance is observed for U103 in PKWT. The bulge U177 thus generates a slight repositioning of loop 1 that propagates up to the junction, and affects the local dynamics of this region of the pseudoknot as discussed below.
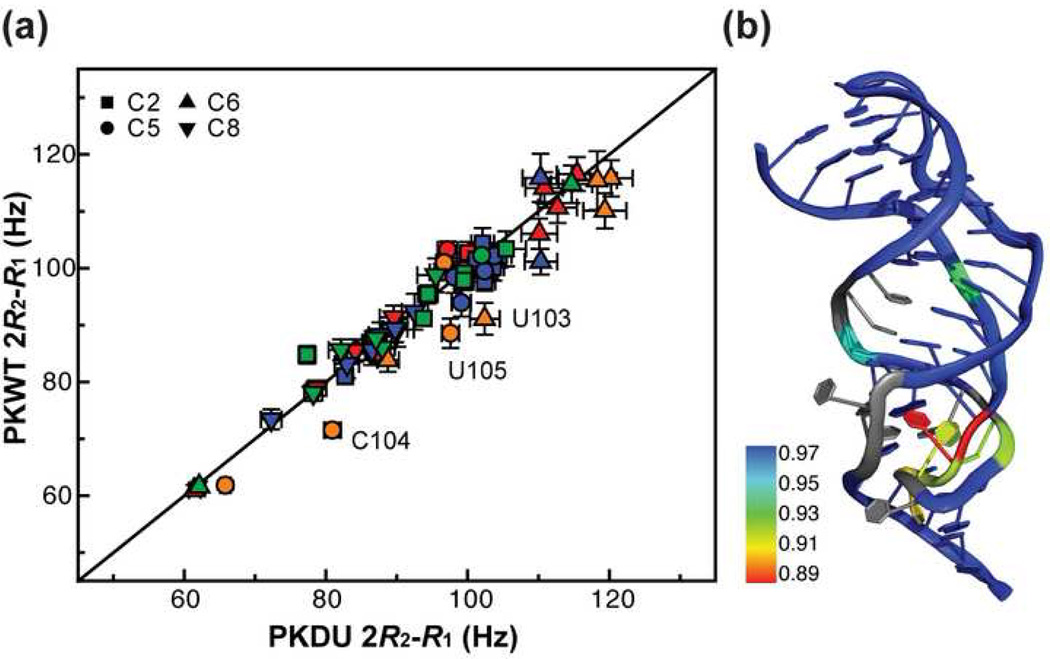
Dynamics of PKWT vs PKDU based on 13C spin relaxation
The RDCs showed that PKWT and PKDU have similar overall structural and dynamical properties, indicating that the bulge U177 does not result in a static bend large enough to affect the overall alignment of the pseudoknot (Figure 2(c)). To investigate whether U177 causes structural flexibility on the picosecond to nanosecond timescale, we measured the longitudinal (R1) and rotating-frame (R1ρ) relaxation rates of base carbons (Supplementary Table 2) in PKWT and PKDU. Since PKWT and PKDU have similar overall structures and thus similar hydrodynamics, the ratio between their 2R2-R1 values reflects the difference between the amplitudes of the internal motions (Figure 6(a)).44 The bases in stem 1, stem 2 and loop 2 have PKWT:PKDU ratios of 2R2-R1 of 1.01±0.02, 0.99±0.03, and 1.01±0.03, respectively. This indicates that the local dynamics of all three of these regions are similar in PKWT and PKDU. However, for loop 1, the 2R2-R1 ratio between PKWT and PKDU is 0.94±0.05, with U103, C104, U105 displaying the biggest differences (Figure 6(b)). Thus, loop 1 is in general more flexible in PKWT than PKDU. U103 is right below the three U•A-U triples in PKWT, and displays one of the biggest differences in flexibility between PKDU and PKWT aromatic resonances. This increase in flexibility and the large change in G178 H8 chemical shift are consistent with absence of the U103 pairing interaction with C112•G178 in PKWT. A few other residues are also more flexible in PKWT than PKDU; U113, which is located in the base pair above the bulge U177, U109, which is affected by the increased dynamics in loop 1, and the junction U99 (discussed below).
Figure 6. Comparison of dynamics of PKWT and PKDU by 13C spin relaxation measurements.
(a) Correlation plot of 2R2-R1 values for PKWT vs PKDU for C2 (squares), C5 (circles), C6 (triangles), and C8 (inverted triangles). Values for carbon atoms in residues in stem 1, loop 1, stem 2, and loop 2 are shown in red, orange, blue, and green, respectively. (b) Cartoon of PKWT structure showing mapping of the relative dynamics of nucleotides in PKWT vs PKDU. Differences are shown as a color scale from yellow to red, no difference is blue, and gray are residues for which there is no data for comparison. Increased dynamics are seen in the loop 1 nucleotides below the triple helix (U103, C104,U105), U109, U113, and U99.
While comparison between PKWT and PKDU provides a characterization of the effects due to the presence of the bulge U177, the 2R2-R1 values within PKWT and PKDU directly provide information on the local dynamics of bases in the pseudoknot (Supplementary Figure 1). For both pseudoknots, similar 2R2-R1 values are observed among each type of base carbon, consistent with overall stable local conformations on the ps-ns timescale. The bulge U177 is experiencing significant motions as indicated by the low 2R2-R1 value of C6. Interestingly, U99, which forms the junction Hoogsteen base pair, also displays significant local dynamics, and is more flexible in PKWT than in PKDU. In both structures, the stem 1-stem 2 junction is significantly underwound, and the U99 base is not well stacked on the base triple below. The only other nucleotides with low 2R2-R1 values are found at the ends of the pseudoknot (C121, C166 and A184).
Effect of bulge U177 on stability of PKWT vs PKDU
While the structures of PKWT and PKDU are highly similar, there is a 4.2 kcal difference in their thermodynamic stabilities, which is comparable with what would be expected for deletion of a bulge nucleotide in an A-form RNA helix. UV melting studies indicate that PKWT and PKDU melt in three transitions, which were attributed to sequential melting of tertiary structure, stem 2, and stem 1, with the tm of the first two transitions ~10°C lower for PKWT th an PKDU.28,33 The bulge U177 might be expected to decrease the stability of the base pairs in stem 2 surrounding the bulge and/or it could affect the stability of the tertiary (Hoogsteen) interactions by affecting the positions of nucleotides in loop 1.
Examination of the cross peaks in the JNN-HNN-COSY spectra revealed that AN1-UH3 cross peak intensities for all three of the stem 2 Watson-Crick A•U base pairs above the bulge U177 are much less intense for PKWT than for PKDU at 20°C (Figure 2(b)). In contrast, AN7-UH3 cross peak intensities for the Hoogsteen U-A base pairs are comparable for PKWT vs PKDU. In order to investigate whether the differences in crosspeak intensities correspond to differences in hydrogen bond strength, the JNN coupling constants were quantified (Table 2). Surprisingly, there are almost no significant differences in the JNN coupling constants for any of the base pairs except for the two Watson-Crick pairs below the bulge, which are smaller in PKWT than PKDU. The weak cross peaks for the three A•U base pairs above the bulge, involved in the triples, all had correspondingly small diagonal peaks. We attribute the weak cross and diagonal peaks in the JNN-HNN-COSY spectra from the stem 2 base pairs above the bulge U177 in PKWT to a small population of A•U base pairs opening and exchanging with water at 20 °C. Consistent with this, all of the weak Watson-Crick A•U imino cross peaks in the triples of PKWT are much stronger in JNN-HNN-COSY spectra acquired at 10°C (spectra not shown).
Table 2.
Imino proton chemical shifts and hydrogen bond JNN coupling constants of PKWT and PKDU at 20°C
| Residues | JNNa | JNN,PKWT/ JNN,PKDU |
||
|---|---|---|---|---|
| PKWT | PKDU | |||
| Stem 1 Watson-Crick |
G94-C120 | 5.5 ± 0.1 | 5.8 ± 0.1 | 0.96 |
| G95-C119 | 5.6 ± 0.1 | 5.7 ± 0.1 | 0.99 | |
| C96-G118 | 5.9 ± 0.1 | 6.0 ± 0.1 | 0.99 | |
| U97-A117 | 6.3 ± 0.1 | 6.5 ± 0.1 | 0.96 | |
| G98-C116 | 5.5 ± 0.1 | 5.5 ± 0.1 | 1.01 | |
| Stem 2 Watson-Crick |
U115-A174 | 5.4 ± 0.3 | 5.1 ± 0.1 | 1.05 |
| U114-A175 | 5.5 ± 0.3 | 5.9 ± 0.1 | 0.93 | |
| U113-A176 | 5.5 ± 0.3 | 5.6 ± 0.1 | 0.99 | |
| C112-G178 | 5.3 ± 0.1 | 6.4 ± 0.1 | 0.82 | |
| A111-U179 | 5.5 ± 0.1 | 6.1 ± 0.1 | 0.90 | |
| G110-C180 | 5.8 ± 0.1 | 5.6 ± 0.1 | 1.03 | |
| U109-A181 | 5.8 ± 0.1 | 5.5 ± 0.1 | 1.04 | |
| C108-G182 | 6.0 ± 0.1 | 5.7 ± 0.1 | 1.04 | |
| G107-C183 | 5.5 ± 0.1 | 5.6 ± 0.2 | 0.99 | |
| Loop 1 Hoogsteen |
U99-A173 | 4.9 ± 0.1 | 5.0 ± 0.1 | 0.99 |
| U100-A174 | 5.2 ± 0.7 | 5.3 ± 0.1 | 0.98 | |
| U101-A175 | 5.1 ± 0.1 | 4.9 ± 0.1 | 1.05 | |
| U102-A176 | 5.1 ± 0.2 | 5.6± 0.2 | 1.08 | |
Errors are calculated from the noise of the experiments.
2JNNHNN COSY NMR spectra were taken at 20°C on 600MHz wit h a cryoprobe.
Two-bond 15N-15N coupling constants (2JNN) are calculated by using 2JNN = atan [(−Ic/Id)1/2]/(2πτ), where Ic and Id are the intensities of cross and diagonal peaks, 2τ is the COSY transfer time.
To further investigate the stability of individual base pairs in the pseudoknot, imino proton spectra were acquired as a function of temperature (Figure 7). Qualitative analysis of the chemical shifts, peak intensities and line widths as a function of temperature revealed that for PKWT the top part of stem 2, especially the three Watson-Crick A•U base pairs above the bulge and to a lesser extent the two base pairs below the bulge, is much less stable than PKDU. For example, at 45°C, resonance intensities for stem 2 base p airs U113•A176 and U114•A175 are broadened to baseline in PKWT while these peaks are still relatively sharp for PKDU (Figure 7).
This is consistent with the observation from the structure of PKWT that the base pairs immediately above and below the bulge are partially unstacked with respect to each other, while there is good stacking between these base pairs in PKDU. Without the stabilizing stacking interactions between the bottom Watson-Crick A•U base pair of the triple helix and the more stable stem 2 base paired region below, the relatively weak stacking of sequential A•U base pairs alone dictates the stability of this region. In contrast, the stability of the Hoogsteen base pairs is comparable between PKWT and PKDU, and the refined structures show similar stacking interactions for the Hoogsteen-paired Us in the triples. As a consequence, the Watson-Crick pairs in top half of stem 2 are less stable than the Hoogsteen pairs in the triples for PKWT (Figure 7). The junction Hoogsteen A-U base pair is remarkably stable for both PKDU and PKWT, indicating its critical importance in the overall folding of the pseudoknot. Taken together, these data suggest a model in which PKWT unfolds by initially melting stem 2 Watson-Crick base pairs surrounding the bulge U177, while the stem 2-loop 1 Hoogsteen base pairs remain intact (Supplementary Figure 2), while for PKDU the Hoogsteen pairs in the triples begin to melt first, followed immediately by the tightly coupled stem 2 duplex transition. Thus, the bulge U177 significantly influences the later folding and earlier unfolding steps in the hTER pseudoknot folding pathway, which may have relevance to telomerase RNP assembly.
DISCUSSION
The pseudoknot/core domain of vertebrate TER is critical to telomerase activity, providing the essential template, a TERT binding site, and a conserved pseudoknot (Figure 1). While the roles of the template and TERT have been clearly established, much less is known about the role of tertiary structure of the RNA in telomerase function. We have determined the structure of a minimal wild-type hTER pseudoknot that includes the bulge U177 in the stem 2, which was eliminated in the previous structure study for pseudoknot stability.28 Comparison of PKWT and PKDU structures reveals that overall they are highly similar. The pseudoknot contains a unique triple helix composed of two stem 1-loop 2 minor groove triples, a junction loop 1-loop 2 base pair, and three major groove U•A-U Watson-Crick-Hoogsteen triples located directly above the bulge U. The importance of all of these elements of the tertiary structure to telomerase activity28 is validated by the structure of this minimal pseudoknot containing the wild-type sequence. The similarity between the PKWT and PKDU structures is consistent with the relatively small decrease (2–10 fold) in telomerase activity in hTERΔU177 telomerase, since deletion of U177 does not result in the predicted structural rearrangement of the triple helix.31,35 Thus, U177 plays a more subtle but important role in telomerase catalytic activity.
While the overall structures of PKWT and PKDU are very similar, the bulge U177 causes a small but significant rearrangement of the positions of the loop 1 nucleotides and an increase in dynamics of loop 1 nucleotides below the triple helix and in the junction. The bulge U effectively terminates the triple helix, which otherwise is extended by another triple in PKDU. The base pairs on either side of the bulge U are partially unstacked, which causes a small increase in the helical bend compared to PKDU at the junction between the bottom of the triple helical region and the rest of stem 2. Somewhat unexpectedly, due to the changes in the positions of the bases in loop 1, there is a larger increase in the size of the bend at the stem 1-stem 2 junction for PKWT vs PKDU (Figure 5). Since the template RNA, and therefore likely also TERT, is thought to be positioned near this region of the pseudoknot,31,32,45 these small changes in helical trajectory and dynamics may be relevant to activity.
The bulge U destabilizes the wild-type pseudoknot relative to PKDU by decreasing the stability of the region of stem 2 around the bulge, rather than by destabilizing the tertiary Hoogsteen base pairing interactions. In PKWT, the Hoogsteen base pairs are therefore more stable than the Watson-Crick base pairs. Relative to PKDU, the three stem 2 A•U base pairs above the bulge, which interact with loop 1 to form the Hoogsteen triples, all show a large increase in exchange rate with water, as evidenced by their broader lines, while the two base pairs in stem 2 below the bulge have longer average hydrogen bonds based on JNN values (Table 2), indicative of an increased opening rate. As a consequence of the difference in the stability of stem 2 in PKWT vs PKDU, their initial unfolding (and later folding) pathways are different. Thus, the bulge U may affect telomerase RNP assembly. It has been previously proposed that a “molecular switch” between the pseudoknot and an unusual stem 1 hairpin may be important for the catalytic cycle of telomerase, e.g. during the translocation step required for each successive addition of a telomere repeat.33,34 As discussed, the presence of the bulge U177 in the wild type pseudoknot results in a shift of the pseudoknot-hairpin equilibrium toward the hairpin form for the free hTER pseudoknot. These two conformations are in slow exchange (> seconds) on the NMR time scale. While some mutational data indicate that formation of the hairpin is not required for activity,24 the data is not unequivocal.8 The results here show that the pathway for conversion of hairpin to pseudoknot, as well as the previously analyzed thermodynamics,33 is different for PKWT vs PKDU. Whether this has relevance to telomerase activity remains to be determined.
The junction loop 1-loop 2 Hoogsteen base pair is remarkably stable for both PKWT and PKDU, and is critical to the tertiary structure and telomerase activity.28 Current structural models of the yeast pseudoknots proposed for K. lactis and S. cerevisiae include only Hoogsteen triples formed between loop 1 and stem 2, as loop 2 and stem 1 were not included in the analysis.31,32 It will be interesting to see if the actual structures of the yeast pseudoknots include additional major groove triples and a junction base pair, as seen in the human TER pseudoknot.
Substitution of the A176 ribose 2’-OH with 2’-H or 2’-OMe decreases the activity of in vitro reconstituted TERT + TER telomerase by 2-fold and 3-fold respectively.32 It has been proposed that this 2’-OH contributes directly to catalysis. In the PKWT structure the bulge U177 sits over the A176 2’-OH, sterically blocking it, (Figure 4(g)), and would therefore have to change position if the 2’-OH needs to be accessible. Thus, U177 could serve as a hinge to provide additional backbone flexibility for the A176 above. Alternatively, the decrease in telomerase activity with the A176 2’-H and 2’-OMe substituted TER could be due to a change in conformation at the bulge U177 due to the substitution. The 2’-OMe group would sterically contact the bulge U, while the 2’-H group may change the sugar pucker of A176 from C3’-endo to C2’-endo, both of which would alter the position of U177.
The decrease in telomerase activity for the A176 2’-OH substitutions is comparable to the decrease from deletion of the bulge U177. FRET studies of human TER45 and cross-linking experiments in the S. cerevisiae TER32 locate the TER template region, where the telomeric DNA being replicated binds, near the triple-helical region of the pseudoknot. The 3’ end of a DNA primer was photoaffinity cross-linked to two As in the A-rich strand of stem 2 in S. cerevisiae TER which form part of the triplex. In the equivalent region of the human TER pseudoknot, this would place the template-primer near the bulge U as well. Since the triple helix is essential for catalysis, the bulge U could contribute to whatever role it plays in the catalytic cycle. In S. cerevisiae, mutations of residues in the region of stem 2 that would be equivalent to stem 2 below the bulge U in human TER pseudoknot, cause a large decrease in TERT binding.22,32,46 Thus, the bulge U may be located at the heart of the telomerase catalytic center, between the catalytically essential triple helix and a site of TERT-TER interaction.
Major groove triples are not common in RNA tertiary structures, and the extensive interactions identified in PKDU28 and modeled for the yeast pseudoknots31,32 appeared to be unique. In addition to the structure of PKWT reported here, at least two other examples of sequential major groove triples have now been reported, in the structures of a SAM II riboswitch around the SAM binding site and at the catalytic core of a group II intron.47,48 The presence of a ‘catalytic triplex’ in the group II intron provides further support for the idea that the hTER pseudoknot triplex may play a central role with the TERT in telomerase catalysis.
MATERIALS AND METHODS
RNA synthesis and purification
Unlabeled, uniformly 13C,15N-labeled, and 13C,15N-base-specifically labeled PKDU and PKWT RNA were prepared by in vitro transcription using His6-tagged P266L T7 polymerase mutant49 with synthetic DNA templates, and purified as described.28 All purified RNAs were desalted and exchanged extensively into 10 mM potassium phosphate, 200mM KCl, 50µM EDTA NMR buffer (pH 6.3) by using the Amicon filtration system. For NMR studies, the RNA samples were annealed at 95°C in NMR buffer under dilute (1–10µM) conditions and concentrated to ~1 mM.
NMR spectroscopy and structure calculations
NMR spectra were recorded on Bruker DRX or Avance 500, 600, and 800 MHz spectrometers equipped with HCN cryoprobes. For the structure calculations, exchangeable proton spectra were taken in 95% H2O / 5% D2O at 283K and non-exchangeable proton spectra were measured in 100% D2O at 293K. NMR spectra were processed and analyzed with Bruker XWINNMR 3.5 and Sparky 3.110. (University of California, San Francisco, CA). The assignments were obtained from analysis of 2D NOESY, 1H-15N HMQC, and 15N-correlated CPMG-NOESY, 1H-13C HSQC, 2D HCCH-COSY, 3D HCCH-TOCSY, 3D NOESY-HSQC,50,51 and a suite of 2D-filtered/edited NOESY (F2f, F1fF2e, F1fF2f, and F1eF2e) experiments43 on the unlabeled and 13C,15N-base-type-specific labeled RNA samples, as previously described.28,52 The initial assignments of PKWT were made by comparing the spectra of PKWT to those of PKDU. The NOEs for the tertiary interaction for loop1/stem2 and loop2/stem1 were identified as described.28 JNN-HNN-COSY experiments were used to measure the hydrogen bond coupling constants and to confirm the hydrogen bonds for Watson–Crick and non-Watson-Crick base pairs.39,53 RDCs were measured for 1DCH and 1DNH on uniformly 13C,15N-labeled PKWT and PKDU samples in 14 mg/ml Pf1 phage (ASLA).54
Inter-proton distances were generated from 2D-filtered/edited NOESY as well as 2D NOESY spectra acquired in D2O and H2O. NOE distance restraints were classified as strong (1.8–3.5 Å), medium (2.5–4.5 Å), weak (3.5–5.5 Å), and very weak (4.5–6.5 Å), and the lower boundary was defined as the van der Waals radius (1.8 Å). Dihedral angle restraints (α, β, γ, and δ) were incorporated in the structure calculation as described in Wu et al. (2001).55 Final structure calculations included hydrogen bond distance restraints for the 15 Watson–Crick base pairs, and other non-Watson-Crick base pairs as previously described.28 The structures of PKWT were calculated using XPLOR-NIH 2.9.8 starting from extended, unfolded RNA by using NOE distances and dihedral angle restraints following standard XPLOR protocols. Structures with no experimental restraint violation (distances >0.2 Å and dihedral angles >5°) from the initial 200 calculated structures were further refined with 63 RDCs. For structure calculations, RDC values were normalized to C-H bond with a bond length of 1.0 Å. The optimal values for the magnitude and asymmetry of the alignment tensor from the grid search are Da = −36.4 Hz and R = 0.12, respectively. The force constants for RDCs were gradually increased from 0.001 to 0.5 kcal·mol−1·Hz−2 while the system was cooled from 1000K to 100K in 36 cycles of molecular dynamics.
The structures of PKDU (1YMO) were re-refined. All the NOE and dihedral angle restraints were not changed except for the addition of distance restraints for U103 imino, but new 74 RDCs were used for structural calculations. The grid search on PKDU produced the optimal Da= −45.0 Hz and R = 0.07, respectively. The refinement steps were the same as for PKWT. Experimental restraints and structural statistics for the 20 lowest-energy structures for PKWT and PKDU are in Table 1. All structures were viewed and analyzed with MOLMOL and PYMOL. Helical parameters (Figure 5) were analyzed with Curves 5.3.56
RDC measurement and relaxation studies
All RDC and 13C spin relaxation experiments were conducted at 293 K at 600 and 800 MHz. NMR spectra were processed using NMRPipe/NMRDraw, and analyzed using NMRView. A total of 82 and 70 one bond C-H and N-H RDCs (1DC1’H1’, 1DC2H2, 1DC5H5, 1DC6H6, 1DC8H8, 1DN1H1, and 1DN3H3) were measured in duplicate from the difference in splittings along 1H and 13C/15N dimensions for PKDU and PKWT, respectively, in the absence and presence of Pf1 phage (~14mg/ml) (Supplementary Table 1). RDCs were averaged whenever possible for structure refinements. Longitudinal (R1) and rotating-frame (R1ρ) relaxation rates were measured for base carbons C2, C5, C6, and C8 using TROSY detected pulse sequences.57 The transverse relaxation rates (R2) were obtained from R1ρ rates according to R1ρ = R1 cos2θ + R2 sin2θ, where θ= arctan(vSL/Ω) is the effective tilt angle in the spin lock field, vSL is the spin lock field strength in Hz, and Ω is the resonance offset from the spin lock carrier frequency in Hz. A high power off-resonance spin lock was carefully calibrated as previously described (vSL=4154.0±42.0Hz), and was used in all R1ρ experiments. The offsets of spin lock were 3750 Hz for C2, C6, and C8, and −3000 Hz for C5 spin relaxation measurements. Relaxation delays were 20, 400 (x2), 800 (x2) ms for all R1 experiments; for R1ρ experiments, relaxation delays were 4, 32 (x2), 64 (x2) ms for C2, C6, and C8, and 4, 24 (x2), 48 (x2) ms for C5. Duplicated measurements were indicated as x2. A recycling delay of 1.5 s was used for all spin relaxation experiments. Relaxation rates and errors were determined by fitting intensities to a mono-exponential decay using NMRView and in-house software. Due to the asymmetric chemical shift anisotropy (CSA) tensor of base carbons and non-co-linearity of CC, CH and CSA interactions, R2/R1 values can not be easily interpreted as a function of overall dynamics, in contrast to the case for amide and imino nitrogen spin relaxation measurements. However, the 2R2-R1 value is mainly proportional to S2xJ(0), where S2 is the Lipari-Szabo order parameter ranging between 0 and 1 for maximum and minimum motion that is faster than the overall tumbling rate, and J(0) is the spectral density function at zero magnetic field.44 Thus, to a reasonable approximation, 2R2-R1 is independent of the timescale of internal dynamics, providing a good characterization of the amplitude of local dynamics (S2).
Coordinate deposition
Coordinates and restraints for the 20 lowest energy structures of PKWT and the RDC-refined PKDU have been deposited in the Protein Data Bank, with accession codes 2K95 and 2K96, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Hashim Al-Hashimi for providing NMR pulse sequences used in the 13C spin relaxation measurements. This work was supported by NIH and NSF grants to J.F. Q.Z. is a Baltimore Family Fellow of the Life Sciences Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 3.Aubert G, Lansdorp PM. Telomeres and aging. Physiol. Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 4.Blasco MA. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- 5.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 6.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat. Rev. Drug. Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 7.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin. Genet. 2008;73:103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 8.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Calado RT, Young NS. Telomere maintenance and human bone marrow failure. Blood. 2008;111:4446–4455. doi: 10.1182/blood-2007-08-019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends. Biochem. Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell. Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 13.Collins K. Physiological assembly and activity of human telomerase complexes. Mech. Ageing. Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theimer CA, Jady BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J. Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol. Cell. 2007;27:869–881. doi: 10.1016/j.molcel.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Venteicher AS, Meng ZJ, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Podell ER, Zaug AJ, Yang YT, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 17.Greider CW, Blackburn EH. A Telomeric Sequence in the RNA of Tetrahymena Telomerase Required for Telomere Repeat Synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 18.Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3' overhang. Proc. Natl. Acad. Sci. USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 20.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proc. Natl. Acad. Sci. USA. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dandjinou AT, Levesque N, Larose S, Lucier JF, Elela SA, Wellinger RJ, Grp R. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 22.Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. USA. 2004;101:14713–14718. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu D, Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell. 2003;11:1361–1372. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 24.Chen JL, Greider CW. Functional analysis of the pseudoknot structure in human telomerase RNA. Proc. Natl. Acad. Sci. USA. 2005;102:8080–8085. doi: 10.1073/pnas.0502259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucl. Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ly H, Blackburn EH, Parslow TG. Comprehensive structure-function analysis of the core domain of human telomerase RNA. Mol. Cell. Biol. 2003;23:6849–6856. doi: 10.1128/MCB.23.19.6849-6856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 28.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 30.Tesmer VM, Ford LP, Holt SE, Frank BC, Yi X, Aisner DL, Ouellette M, Shay JW, Wright WE. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell. Biol. 1999;19:6207–6216. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theimer CA, Finger LD, Trantirek L, Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. USA. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. A molecular switch underlies a human telomerase disease. Proc. Natl. Acad. Sci. USA. 2002;99:16998–17003. doi: 10.1073/pnas.262663599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yingling YG, Shapiro BA. The prediction of the wild-type telomerase RNA pseudoknot structure and the pivotal role of the bulge in its formation. J. Mol. Graph. Model. 2006;25:261–274. doi: 10.1016/j.jmgm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Podlevsky JD, Bley CJ, Omana RV, Qi XD, Chen JJL. The telomerase database. Nucl. Acids Res. 2008;36:D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones SM, M S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: Annotating Non-Coding RNAs in Complete Genomes. Nucl. Acids Res. 2005;33:D121–D141. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen JJ. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J. Biol. Chem. 2008;283:2049–2059. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- 39.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 40.Dingley AJ, Masse JE, Peterson RD, Barfield M, Feigon J, Grzesiek S. Internucleotide scalar couplings across hydrogen bonds in Watson-Crickand Hoogsteen base pairs of a DNA triplex. J. Am. Chem. Soc. 1999;121:6019–6027. [Google Scholar]
- 41.Prestegard JH, Al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q. Rev. Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 42.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 43.Peterson RD, Theimer CA, Wu H, Feigon J. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J. Biomol. NMR. 2004;28:59–67. doi: 10.1023/B:JNMR.0000012861.95939.05. [DOI] [PubMed] [Google Scholar]
- 44.Dethoff EA, Hansen AL, Musselman C, Watt ED, Andricioaei I, Al-Hashimi HM. Characterizing Complex Dynamics in the TAR Apical Loop and Motional Correlations with the Bulge by NMR, MD and Mutagenesis. Biophys. J. 2008 doi: 10.1529/biophysj.108.140285. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gavory G, Symmons MF, Krishnan Ghosh Y, Klenerman D, Balasubramanian S. Structural analysis of the catalytic core of human telomerase RNA by FRET and molecular modeling. Biochemistry. 2006;45:13304–13311. doi: 10.1021/bi061150a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chappell AS, Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol. Cell. Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 48.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gullerez J, Lopez PJ, Proux F, Launay H, Dreyfus M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl. Acad. Sci. USA. 2005;102:5958–5963. doi: 10.1073/pnas.0407141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001;338:371–399. doi: 10.1016/s0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- 51.Dieckmann T, Feigon J. Assignment methodology for larger RNA oligonucleotides: application to an ATP-binding RNA aptamer. J. Biomol. NMR. 1997;9:259–572. doi: 10.1023/a:1018622708674. [DOI] [PubMed] [Google Scholar]
- 52.Wu H, Henras A, Chanfreau G, Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. USA. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dingley AJ, Nisius L, Cordier F, Grzesiek S. Direct detection of N-H[…]N hydrogen bonds in biomolecules by NMR spectroscopy. Nat. Protoc. 2008;3:242–248. doi: 10.1038/nprot.2007.497. [DOI] [PubMed] [Google Scholar]
- 54.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 55.Wu H, Yang PK, Butcher SE, Kang S, Chanfreau G, Feigon J. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 2001;20:7240–7249. doi: 10.1093/emboj/20.24.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavery R, Sklenar H. Defining the Structure of Irregular Nucleic-Acids - Conventions and Principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 57.Hansen AL, Al-Hashimi HM. Dynamics of large elongated RNA by NMR carbon relaxation. J. Am. Chem. Soc. 2007;129:16072–16082. doi: 10.1021/ja0757982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.