Abstract
Bacillus thuringiensis (Bt)1 insecticidal toxins bind to receptors on midgut epithelial cells of susceptible insects, and binding triggers biochemical events that lead to insect mortality. Recently, a 100-kDa aminopeptidase N (APN) was isolated from brush border membrane vesicles (BBMV) of Anopheles quadrimaculatus and shown to bind Cry11Ba toxin with surface plasmon resonance (SPR) detection [Abdullah et al. (2006) BMC Biochemistry 7, 16]. In our study, a 106-kDa APN, called AgAPN2, released by phosphatidylinositol-specific phospholipase C (PI-PLC) from Anopheles gambiae BBMV was extracted by Cry11Ba bound to beads. The AgAPN2 cDNA was cloned and analysis of the predicted AgAPN2 protein revealed a zinc-binding motif (HEIAH), three potential N-glycosylation sites and a predicted glycosylphosphatidylinositol (GPI) anchor site. Immunohistochemistry localized AgAPN2 to the microvilli of the posterior midgut. A 70-kDa fragment of the 106-kDa APN was expressed in Escherichia coli. When purified, it competitively displaced 125I-Cry11Ba binding to An. gambiae BBMV, and bound Cry11Ba on dot blot and microtiter plate binding assays with a calculated Kd of 6.4 nM. Notably, this truncated peptide inhibited Cry11Ba toxicity to An. gambiae larvae. These results are evidence that the 106-kDa GPI-anchored APN is a specific binding protein, and a putative midgut receptor, for Bt Cry11Ba toxin.
Keywords: Bacillus thuringiensis, Cry toxin, mosquitocidal protein, aminopeptidase
The mosquito, Anopheles gambiae, is the principal vector of Plasmodium falciparum that causes human malaria. Anopheles species are typically controlled at the adult stage by chemical pesticides. Depending on the larval habitat, treatment with Bacillus thuringiensis (Bt)1 biopesticides, however can have an important role in vector management programs (1, 2).
Insecticidal Cry toxins recognize receptor molecules located in the brush border of target insects. In lepidopteran larvae, the Cry1 toxins bind cadherins, aminopeptidases (APN), alkaline phosphatase (ALP), and glycolipids. According to proposed models for toxin action [reviewed in (3)], binding to multiple receptors is required to induce toxin processing, pre-pore formation and membrane insertion (4).
The Cry4 and Cry11 toxins of Bt israelensis bind to the apical microvilli of midgut cells in gastric caeca and posterior midgut of mosquito larvae (5-8). A 65-kDa protein binds Cry4Ba and Cry11Aa in Aedes larvae (9). The 65-kDa protein was identified as an ALP and confirmed as a Cry11Aa receptor (10). Recently, the AgCad1 cadherin from An. gambiae was identified as a binding protein and putative receptor for Cry4Ba toxins (11).
APNs are exoenzymes located in the brush border of insect larvae. Lepidopteran larvae have at least four paralogues of APN located on the surface of their midgut brush border membrane. Presumably, the different APNs have cleavage preferences for various classes of amino acids. These molecules also function as adventitious receptors for Bt Cry1 toxins. In the mosquito An. quadrimaculatus, a 100-kDa APN (APNAnq100) has tight binding to Cry11Ba, but not to Cry11Aa or Cry4Ba toxin according to binding analysis by SPR (12).
In the present study, we describe the cloning and identification of an APN-like protein from the gut of An. gambiae. Immunohistochemistry study shows that the APN localizes on the apical cell membrane in the posterior midgut of the mosquito. Binding studies using APN released from BBMV of the mosquito larvae and E. coli cell-expressed truncated APN peptide are evidence that the protein is a Cry11Ba binding protein. Bioassays using the truncated peptide display an inhibition effect with Cry11Ba, which provide further evidence that the APN is a Cry11Ba receptor. This is the first report of an APN type protein as a Cry toxin binding protein in An. gambiae.
EXPERIMENTAL PROCEDURES
Purification of Cry11Ba toxin
The Bt strain 407, harboring plasmid pJEG80.1 encoding cry11Ba (13), was kindly provided by Dr. D.H. Dean (Ohio Sate University). A complex sporulation medium supplemented with erythromycin antibiotic was used according to (14): 2 gm/L peptone (Difco™, BD), 5 gm/L yeast extract (Difco™, BD), 0.07 M K2HPO4, 0.02 M KH2PO4, 6 × 10-3 M glucose, 2 × 10-4 M MgSO4·7H2O, 5 × 10-4 M CaCl2·2H2O, 6 × 10-6 M MnSO4·7H2O, 1 × 10-6 M FeSO4·7H2O. The strain was grown in 1 L media at 30°C overnight, then 1 L of sodium phosphate solution (0.06 M Na2HPO4, 0.04 M NaH2PO4·H2O) was added into the growing culture. Upon completion of sporulation spores and crystals were harvested by centrifugation, re-suspended in 0.1 M NaCl, 2% Triton -X 100, 20 mM Bis-Tris, pH 6.5, sonicated on ice, and then washed in the same buffer three times. The spore-crystal mixture was then washed in 1 M NaCl (twice), and distilled H2O (twice). All centrifugation steps were 10, 000 × g for 10 min at 5°C. Crystals were separated from spores by centrifugation through a 30-60% (w/v) NaBr step gradient at 47,000 × g for 2 h at 5°C. Purified crystals were washed twice with distilled water, dissolved in 100 mM 3-(cyclohexylamino)propanesulfonic acid (CAPS), 0.05% β-mercaptoethanol, pH 10.6, for 2 h at 37°C, and insoluble material was removed by centrifugation 10, 000 × g for 10 min. Activation of Cry11Ba protoxin was performed by incubating the toxin with bovine pancreatic trypsin (Sigma) at a mass ratio of 50:1 (toxin:trypsin) for 2 h at 37°C.
Mosquito rearing
The colony of An. gambiae (CDC G3 strain) was maintained at 27°C with a photoperiod of 14 h light/ 10 h dark. Adults were given access to 10% sucrose and females were fed blood on restrained mice. Fresh eggs were washed with 0.1% bleach and hatched in distilled water. Larvae (ca. 200/pan) were fed with ground TetraMin® daily. Larvae were reared to 4th instar, collected and used immediately for midgut dissection, section preparation or bioassay.
Midgut dissection, RNA isolation and cDNA synthesis
The 4th instar larvae were placed in RNAlater™ (Sigma) for fixation and dissection. The whole intestine, observed under a dissecting microscope, was gently pulled out using a pair of dissecting forceps. The midgut was then isolated by carefully removing the foregut and hindgut, immediately frozen on dry ice, and aliquots of ~ 50 mg were stored at -80°C. Total RNA was extracted from 50 mg midgut tissue using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized from total RNA using Superscript II reverse transcriptase (Gibco-BRL) and oligo(dT)17 primer.
APN cDNA cloning
Three pairs of specific primers were designed according to the Ensembl gene ENSANGG00000018740 and used sequentially as shown in Table 1 and Figure 1A. PCR amplifications were performed using synthesized midgut cDNA with 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 2 or 3 min. The resultant PCR products were cloned into pGEM-T easy vector (Promega) and sequenced from both forward and reverse directions at the Molecular Genetics Instrumentation Facility (University of Georgia). The cloned 2.9 kb cDNA, designated AgAPN2, contains an open reading frame encoding a predicted 106-kDa APN.
Table 1.
Primers used in this study
| Primer | Primer Sequence (5’-3’) |
|---|---|
| Primers for cloning cDNA of AgAPN2 | |
| AgAPN2-F1 | 5’-ATGACATTGGCAGAAAAGCTAGCGCTG-3’ |
| AgAPN2-R1 | 5’-GCTAGACTTTATTCATCAATCGTGCTTG-3’ |
| AgAPN2-F2 | 5’-TTGGCCGCAGTTTAACAACGTTCGGA-3’ |
| AgAPN2-R2 | 5’-GCCACATTCGTGTAACTACCGTGCG-3’ |
| AgAPN2-F3 | 5’-TGGCCGCGGTACCTGATTTCTCTG-3’ |
| AgAPN2-R3 | 5’-TCAGGATCTGCTCTGTCGCCACGTT-3’ |
| Primers for cloning APN2t | |
| AgAPN2-F4 | 5’-CGCACATATGGGCCGCAGTTTAACAA-3’ |
| AgAPN2-R4 | 5’-AGGCCTCGAGCACATTCGTGTAACTA-3’ |
FIGURE 1.

(A) Diagram of the AgAPN2 cDNA, truncated APN2t clone and primer locations. A grey arrowhead locates the gluzincin aminopeptidase motif. (B) Deduced amino acid sequence of AgAPN2. The putative N-terminal cleavable peptide is underlined. The putative C-terminal GPI-anchor site is in boldface. The conserved gluzincin aminopeptidase motif is in boldface and underlined. The putative N-glycosylation sites are in bold-box. Peptides identified by MS analysis are in boldface and italics. The unique peptide of AgAPN2 is in boldface, italics and underlined. (C) Schematic representation of the APN gene cluster on chromosome 2R. Numbers indicate the position of the starting and the ending nucleotides of each gene. The gene cloned in this research is underlined.
Bioinformatic analyses of AgAPN2
The signal peptide of AgAPN2 was predicted by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/). Possible sites for posttranslational modification were analyzed using big-PI predictor (http://mendel.imp.ac.at/sat/gpi/gpi_server.html/) (15-18), Center for biological sequence prediction servers (http://www.cbs.dtu.dk/services/) and O-glycosylation prediction electronic tool (OGPET v1.0, http://129.108.112.23/OGPET).
Preparation of rabbit polyclonal serum against an E. coli expressed 70-kDa AgAPN2 peptide
A central region of the AgAPN2 cDNA encoding amino acid residues 228 to 843 was amplified by PCR using the pair primer AgAPN2-F4/R4 (Table 1, Figue 1A). The PCR fragment was then cloned into the protein expression vector pET-30a (+) (Novagen, Madison, WI) and transformed into E. coli strain BL21-CodonPlus (DE3)/pRIL. Protein expression and purification were performed according to the pET system manual 9th edition (Novagen). The recombinant truncated APN protein, APN2t, was purified by a HiTrap Ni2+-chelating HP column (GE Healthcare, Piscataway, NJ) according to manufacturer’s manual, dialyzed against 10 mM Tris-HCl (pH 8.0) and stored at -20°C until needed for experimentation. APN2t peptide was used to raise antiserum by immunization of a NZW (SPF) rabbit at the Animal Resources Facility of the University of Georgia.
Anti-AgAPN2 antibody was purified by Protein A Sepharose 6MB (GE Healthcare) beads using the low-salt method (19). The concentration of antibody was measured by Bio-Rad protein assay using bovine serum albumin (BSA) as standard (20).
Partial purification and preparation of APN from An. gambiae BBMV for mass spectrometry (MS) analysis
Frozen An. gambiae 4th instar larvae were kindly provided by the Malaria Research and Reference Reagent Resource Center (MR4) and stored in -80°C until use. BBMV were prepared from whole body of 4th instar larvae by MgCl2 precipitation according to (21) and stored in aliquots at -80°C. Protein was measured by the Bio-Rad protein assay. APN activity (22), a marker for brush border membranes, was enriched about 5-fold for the final BBMV preparation compared to the initial crude larval homogenate.
Sample preparation for MS analysis was as follows. Approximately 100 mg BBMV were solubilized overnight at 4°C in 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 2% (w/v) 3-[(cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) supplemented with 1 mM phelylmethylsulfonyl fluoride (PMSF). Subsequently, the solution was centrifuged at 39,000 × g at 4°C for 30 min. The resulting supernatant, diluted in buffer A [20 mM Tris-HCl, pH 8.0, 0.01% (w/v) CHAPS] was separated by anion exchange chromatography using an Acta Explorer system (GE Healthcare). Proteins were eluted with a continuous salt gradient using buffer A and buffer B [20 mM Tris-HCl, pH 8.0, 1 M NaCl, 0.01% (w/v) CHAPS]. Fractions with APN activity were pooled and resolved on SDS-12.5% PAGE. Separated proteins were detected by staining with Coomassie Blue G-250 and a duplicate gel was blotted to PVDF filter (Millipore) for probing with anti-AgAPN2 serum.
Protein bands/plugs were destained, reduced and alkylated, digested with trypsin, and the resulting tryptic peptides extracted for subsequent MS analysis. Specifically, plugs were washed twice with 50 mM ammonium bicarbonate/50% methanol for 20 min at room temperature. Plugs were washed with 75% acetonitrile for 20 min at room temperature and dried at 40°C for 10 min. Plugs were incubated in 10 mM DTT/20 mM ammonium bicarbonate at 37°C for 1 h. The DTT solution was removed and immediately replaced with 100 mM iodoacetamide/20 mM ammonium bicarbonate and incubated at room temperature in the dark for 30 min. Plugs were washed as above, and then incubated with 200 ng sequencing grade trypsin (Promega) at 37°C for 2 h. Peptides were extracted twice with 50% acetonitrile/0.1% TFA for 20 min at room temperature and concentrated by SpeedVac (Jouan). Approximately 25% of the resulting peptides were spotted on a matrix assisted laser / desorption ionization (MALDI) plate using 50% saturated a-cyano-4-hydroxy-cinnamic acid as a matrix (Sigma).
MS Analysis
MS and MS/MS data were acquired on a 4700 Proteomics Analyzer (Applied Biosystems) using standard acquisition methods. MS/MS data were acquired in a data dependent fashion, selecting the top 10 most intense peaks from the MS spectrum. MS spectra were calibrated using two trypsin autolysis peaks (1045.5 and 2211.1 m/z). MS/MS spectra were calibrated using the instrument default processing method. An An. gambiae protein sequence database described in (23) was incorporated into a licensed copy of Mascot 1.9.05 (http://www.matrixscience.com/). Mass lists were submitted to the database considering fixed cysteine carbamidomethylation and partial methionine oxidation modifications, 1 missed tryptic cleavage, and 25 ppm mass accuracy. Identifications were cross-examined using mass accuracy and molecular weight.
Phosphatidylinositol-specific phopholipase C (PI-PLC) treatment and GPI-anchor detection
BBMV (56 mg) suspended in 3 ml of MET buffer (0.3 M mannitol, 5 mM EGTA, 20 mM Tris-HCl, pH 7.5) were incubated with 2 U of PI-PLC (Sigma) supplemented with Complete™ protease inhibitor cocktail (Roche, EDTA free) overnight at room temperature. The suspension was centrifuged at 16,000 × g for 10 min at 5°C and the released proteins recovered in the supernatant. The presence of a cleaved GPI group was detected by using polyclonal antibody against the cross-reacting determinant (CRD) of GPI-anchored proteins (kindly provided by Dr. K. Mensa-Wilmot, University of Georgia).
Cry11Ba toxin binding to GPI-anchored proteins
About 187 mg of cyanogen bromide (CNBr) activated sepharose 6MB (Sigma) was mixed with 2 mg of Cry11Ba toxin in 2 ml of 0.1 M NaHCO3, 0.5 M NaCl, pH 9.0 at room temperature for 4 h according to the manufacturer’s manual. After precipitation, the toxin-bead complex was removed from the coupling buffer and re-suspended in 2 ml of 2 M ethanolamine, pH 8.0. The mixture was incubated overnight at 4°C to saturate all remaining activated groups. After three washes with phosphate buffered saline solution (PBS), the toxin-bead complex was mixed with the supernatant described in the section above. The mixture was incubated overnight at 4°C with gentle rotation. After six washes with PBS buffer, the toxin-bead complex was boiled in SDS-PAGE sample buffer for 10 min and proteins were separated by SDS-12.5% PAGE and transferred to PVDF filter for blotting experiments. The membrane was blocked with 5% skim milk (Difco™, BD) in PBST (PBS + 0.1% Tween 20) for 1 h at room temperature. Then the filter was incubated with anti-AgAPN2 or anti-CRD antibody (1:2000 and 1:10000 dilution, respectively) in blocking buffer overnight at 4°C, washed four times and followed by anti-rabbit IgG horseradish peroxidase conjugate (Sigma) (1:25,000 dilution) in the same buffer for 1 h at room temperature. The bound antibody was visualized by chemiluminescent substrate (ECL kit, GE Healthcare) and X-ray film.
Immunohistochemistry
An. gambiae 4th instar larvae were punctured in the thorax and end of the abdomen by a dissecting needle and immediately fixed with 4% paraformaldehyde in PBS for 2 h on ice, then dehydrated, embedded in plastic mixture and sectioned. Immuno-detection was according to (24). The localization of AgAPN2 was detected by anti-AgAPN2 serum diluted 1:500 in blocking solution [5% (w/v) BSA, 0.2% (v/v) Tween-20 in PBS], whereas a control section was probed by pre-immune serum diluted in the same solution.
Dot blot binding assays
APN2t inclusion bodies were solubilized in 50 mM NaOH overnight at 37°C and dialyzed against 50 mM Na2CO3, pH 10.5. CR11-MPED peptide from An. gambiae cadherin1 was purified as described in (11). One μg of each peptide was dotted onto a PVDF filter directly and probed with 0.125 nM of 125I-Cry11Ba. The toxin (1 μg) was labeled with 0.5 mCi of Na125I (PerkinElmer) using the chloramine-T method (25) and specific activity was 7.4 μCi/μg of input toxin. The filters were exposed to X-ray film at −80 °C for autoradiography.
Membrane vesicles binding assays
Binding assays were performed as described in (25). Duplicate samples of 8 μg BBMV were incubated with increasing amounts of 125I-Cry11Ba toxin in 100 μl of binding buffer (20 mM Na2CO3, pH 10.5, 0.15 M NaCl, 0.1% Tween-20, 1.5% BSA) for 18 h at 4 °C with shaking. Binding reactions were stopped by centrifugation and pellets washed twice with 1 ml of ice-cold binding buffer. Radioactivity of the final pellets was measured with a Beckman model Gamma 4000 detector. For homologous and heterologous competition assays, 10 μM of unlabeled Cry11Ba toxin or peptides were used to compete the binding of 125I-Cry11Ba toxins to BBMV. The experiments were repeated twice.
Microtiter plate binding assays
A protein-protein binding assay using coated microtitre plates and enzyme-linked immunosorbent assay (ELISA) for detection was developed from the protocol of Braitbard et al. (26). The purified APN2t peptide was biotinylated using 50-fold molar excess N-hydroxysulfosuccinimide ester-PC-biotin (Pierce, Rockford, IL) according to the manufacturer’s instructions. The final reaction was dialyzed against 20 mM Na2CO3, pH 9.6. Microtiter plates (high binding 96-well, Immulon® 2HB, Thermo Fisher Scientific Inc., Waltham, MA) were coated overnight at 4°C with 0.5 μg Cry11Ba/well in 100 μl coating buffer (100 mM Na2CO3, pH 9.6), and blocked 1 h at room temperature with PBS /0.5% BSA /0.05% Tween 20. For the binding assays, biotinylated APN2t peptide was diluted to the desired concentrations (0.001 nM to 20 nM) in 100 μl wash buffer (PBS/0.05% Tween 20). For the competition assays, 1000-fold molar excess non-labeled APN2t peptide was added into solution containing biotinylated APN2t. A 100 μl aliquot of biotin-AgAPN2t solution was added to each well and the plates incubated at room temperature for 2 h. The plates were then washed three times and 100 μl of horse radish peroxidase conjugated streptavidin (SA-HRP; Pierce) diluted 1:10,000 in wash buffer were added in each well. After incubation at room temperature for 1 h and washing as above, 50 μl of HRP chromogenic substrate (1-Step™ Ultra TMB-ELISA, Thermo Fisher Scientific Inc.) were added to detect bound SA-HRP. The reaction was stopped by the addition of 50 μl 2 M sulfuric acid; after 5 min the absorbance at 450 nm was measured with a microplate reader (MDS Analytical Technologies, Sunnyvale, CA). Specific binding was determined by subtracting non-specific binding from total binding. All samples were in duplicate and the assay was repeated. Data were analyzed using SigmaPlot software (Version 11; Systat Software Inc., San Jose, CA) and the curves were fitted based on a best fit of the data to a one site saturation binding equation.
Toxicity bioassays
Soluble Cry11Ba was mixed with APN2t or CR11-MPED inclusion bodies at 1:100 toxin:peptide mass ratio. Cry11Ba alone or toxin-peptide mixtures was transferred to wells of a 6-well Costar culture plate (Corning). Ten early 4th instar larvae were added to each well and the plates stored at 27°C. Each treatment was at least duplicated and the bioassays were conducted three times, and there was a control of inclusion bodies alone. Larval mortality was recorded after 48 h.
RESULTS
Cloning AgAPN2 cDNA from the midgut of An. gambiae larvae
The APNAnq100 protein purified from An. quadrimaculatus larvae binds Cry11Ba with high affinity and specificity (12). Using the reported 20 amino acid residues of N-terminal APN sequence, we searched the An. gambiae whole genome sequence [Ensembl An. gambiae genome assembly AgamP3, February 2006 (http://www.ensembl.org/Anopheles_gambiae/index.html)] using BLAST. The search results yielded a single predicted protein (ENSANGP00000025222) as having 85% identity to the 20 residues of APNAnq100, and two other proteins (ENSANGP00000021245, ENSANGP00000025086) having a 80% match. These three predicted proteins matched to the three An. gambiae proteins specified as candidate APNAnq100 homologues by Abdullah et al. (12).
We selected ENSANGP00000025222 for further analysis. Using the RT-PCR strategy shown in Figure 1A, we cloned a cDNA encoding the predicted APN from midgut tissue of An. gambiae larvae. The cDNA was designated AgAPN2 (GenBank accession number EU827528), since Dinglasan et al. (27) had previously assigned AgAPN1 to another APN that is not one of these three and shows 20% identity to the 20-amino-acid N-terminal sequence of APNAnq100. The cloned AgAPN2 cDNA is 2904 bp long and contains a 2808 bp open reading frame. The predicted start codon is embedded in an ‘adequate’ Kozak consensus (AGAATGACA) (28). A polyadenylation signal sequence (AATAAA) is located 83 bp downstream of the stop codon TAG. AgAPN2 encodes a 935-amino acid protein with a theoretical molecular size of 106-kDa (Figure 1B). AgAPN2 has a 20-amino-acid sequence (residue 40-59) with 85% identity to the N-terminus of mature APNAnq100 identified previously. Analysis of the AgAPN2 protein predicts an N-terminal signal peptide, a C-terminal GPI site with 913Gly as the predicted ω-site, and 3 N-glycosylation sites. AgAPN2 contains a consensus gluzincin aminopeptidase motif, GAMENWG-X3-YRE-X23-HE-X2-H-X18-E (where X is any amino acid) (29), that is associated with Zn2+ binding (30).
AgAPN2 cloned from the G3 strain corresponds to a genomic region in the PEST strain that was used for the whole genome assembly. It is noteworthy that the region has not been computationally annotated to locate the APN gene in the Ensembl An. gambiae genome assembly AgamP3, thus the counterpart gene of AgAPN2 in the PEST strain was named AgAPN2PEST. The genes encoding the three APN proteins mentioned above, ENSANGP00000021245, ENSANGP00000025222 and ENSANGP00000025086, are located adjacent to AgAPN2PEST (Figure 1C, Table 2) and have been incorrectly combined and designated as AGAP003696 in Ensembl’s release 49 (March 2008). We tentatively designate them as AgAPN3, AgAPN4 and AgAPN5. AgAPN2 has one amino acid substitution (T884→S884) as compared to AgAPN2PEST. AgAPN2 shows 99% amino acid identity to AgAPN4, 95% to AgAPN3 and 56% to AgAPN5.
Table 2.
Annotation of four APN-like genes on chromosome 2R
| Genes | Ensembl identifiera | Positions | exons | ESTs Coverageb | Similarity to ESTs |
|---|---|---|---|---|---|
| AgAPN2 | N/Ac | 2R:42070199-42073368 | 6 | 85% | >99.37% |
| AgAPN3 | 18756 | 2R:42089971-42093136 | 6 | 84% | >98.99% |
| AgAPN4 | 18740 | 2R:42095012-42098180 | 6 | 85%d | >98.67%d |
| AgAPN5 | 18817 | 2R:42059966-42063174 | 6 | 93% | >99.35% |
ENSANGG000000, the prefix of Ensembl gene identifier is omitted.
Percentage of the gene squence that is supported by EST sequences deposited in GenBank ESTs (March 2008).
This gene has not been computationally annotated in the Ensembl database.
EST sequences support AgAPN2 and AgAPN4 are the same.
Peptide mass fingerprinting (PMF) identification of the 106-kDa APN prepared from BBMV as AgAPN2
The predicted protein encoded by AgAPN2 cDNA was correlated with an actual brush border membrane protein. An aminopeptidase-enriched protein fraction was isolated by anion exchange chromatography from solubilized BBMV (Figure 2A). As shown in Figure 2B, anti-AgAPN2 serum specifically recognized a 106-kDa protein(s) in the enriched fraction (Figure 2B, lane 2). The 106-kDa band (Figure 2B, lane 1) was excised, digested with trypsin and the resulting fragments analyzed by MALDI-TOF MS. Mascot software was used to search the database (23) of predicted Anopheles proteins with the 104 peptide masses for matched proteins having significant Mascot scores greater than 58 (p<0.05). The results yielded three highly similar APNs (AgAPN2PEST, AgAPN3 and AgAPN4) that matched with high possibilities. To further determine the presence of AgAPN2 in the 106-kDa band, we examined the unique peptide(s) of AgAPN2. As shown in Figure 1B, AgAPN2 had one unique peptide matched to the observed mass, indicating that AgAPN2 was present in the 106-kDa band and recognized by anti-AgAPN2. The MALDI-TOF MS data also suggests the presence of AgAPN3 in the 106-kDa band, since three unique peptides of AgAPN3 were identified in the peptide mixture (Figure S1 in the Supporting Information).
FIGURE 2.

Peptide mass fingerprinting identification of a 106-kDa protein from BBMV as AgAPN2. (A) Solubilized proteins from BBMV were fractionated by 0.0-1.0 M NaCl gradient elution. The left side Y-axis indicates the UV absorbance at 280 nm (mAU), and the right side Y-axis indicates the percent conductivity of buffer B (%). Run volumes (ml) and collected fractions are indicated at the bottom. (B) Fractions 26-31 (hatched area) with APN activities were pooled and separated on SDS-PAGE. The Coomassie blue stained band at ~100 kDa (arrowhead, lane 1) corresponds to the band recognized by anti-AgAPN2 serum on a blot (lane 2). The band was excised for mass fingerprinting.
Localization of AgAPN2 in An. gambiae in the gut tissue
We used immunohistochemistry with anti-AgAPN2 serum and sectioned whole larvae of An. gambiae to confirm that AgAPN2 is a brush border protein. The results are shown in Figure 3; AgAPN2 was localized to the posterior region of midgut on the apical portion of the brush border (i.e. microvilli). A control section probed by pre-immune serum was not immunostained in the midgut region (Figure 3).
FIGURE 3.
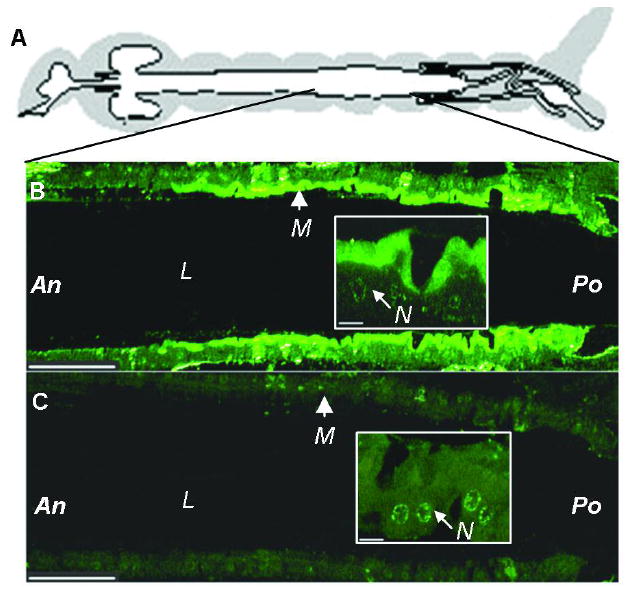
Immunolocalization of AgAPN2 on the microvilli of the posterior midgut of An. gambiae larvae. (A) Diagram of a mosquito gut [adapted from (40)]. (B) Sectioned midgut was probed by anti-AgAPN2 serum and detected by Alexa Fluor-488-conjugated goat-anti-rabbit IgG. (C) Control section was probed by pre-immune serum. (Insets) Higher-magnification views correspond to the area of apical membrane of epithelial cells in the lower-magnification image. M, microvilli. L, lumen. An, anterior. Po, posterior. N, nucleus. Scale bar: 100 and 50 μm for the low- and high- magnification images, respectively. All midgut sections were arranged anterior to the left.
The affinitive binding of Cry11Ba toxin to GPI-anchored AgAPN2 in BBMV
Since midgut APNs in insect larvae are typically attached to brush border membrane by GPI-anchors (22, 27, 31), we verified the predicted anchorage of AgAPN2. BBMV were treated with PI-PLC and strips from a blot of released proteins were probed with anti-CRD or anti-AgAPN2 antibodies. Anti-CRD antibody detects the glycan epitope on the protein’s C-terminus generated by PI-PLC cleavage. The anti-CRD antibody recognized proteins ranging in size from about 20- to 118-kDa indicating the presence and release of multiple GPI-anchored proteins from the BBMV (Figure 4A, lane 1). Anti-AgAPN2 and anti-CRD antibodies recognized a 106-kDa protein (Figure 4A, lanes 1 and 2). Since 100-kDa APNAnq from An. quadrimaculatus specifically binds Cry11Ba, we examined its homologue AgAPN2 for Cry11Ba binding. The supernatant from PI-PLC treated BBMV was applied to the Cry11Ba toxin-beads complex and bound proteins were examined by anti-CRD or anti-AgAPN2 antibodies. Cry11Ba precipitated three proteins including 106-kDa AgAPN2 (Figure 4B, lanes 4 and 6). The 116-kDa protein that was precipitated by Cry11Ba was tentatively identified as AgAPN1 based on cross-reactivity with anti-AgAPN1 serum (serum provided by R. Dinglasan, Johns Hopkins University; data not shown).
FIGURE 4.

Solubilization of AgAPN2 from An. gambiae BBMV proteins by PI-PLC digestion and extraction with Cry11Ba-beads. (A) PI-PLC-solubilized proteins were resolved by SDS-PAGE, transferred to PVDF filters and were probed with anti-CRD serum (lane 1) or anti-AgAPN2 antibody (lane 2). (B) Affinity extraction of GPI-anchored proteins with Cry11Ba-bead complex. After being washed, the beads with bound proteins were boiled in SDS-sample buffer, separated by SDS-PAGE, transferred to a PVDF filter and then probed with anti-CRD (lanes 3 and 4) or anti-AgAPN2 antibodies (lanes 5 and 6). A GPI-anchored protein migrating at 106-kDa was extracted by Cry11Ba-beads and detected by anti-AgAPN2 antibodies (arrow). Beads alone extracted no proteins (lanes 3 and 5).
The interaction of a truncated 70-kDa AgAPN2 peptide to Cry11Ba
We hypothesized that AgAPN2 was a specific binding protein of Cry11Ba, and performed assays to investigate in vitro binding of Cry11Ba to AgAPN2. To circumvent difficulties in purifying AgAPN2 from larval BBMV, we attempted to produce recombinant 106-kDa AgAPN2 in E. coli. Since this approach was not successful, we analyzed Cry11Ba binding on a blot of the truncated 70-kDa peptide used to raise antiserum. One μg of purified APN2t peptide was dotted onto a PVDF membrane, and the membrane was probed with 125I-labeled Cry11Ba toxin. Since the CR11-MPED peptide that originates from An. gambiae midgut cadherin protein was reported as a binding protein of a mosquitocidal toxin (Cry4Ba) in An. gambiae (11), purified CR11-MPED peptide was also dotted onto the membrane to test for Cry11Ba binding. The blots of each protein were probed separately. As shown in Figure 5B, Cry11Ba bound to purified 70-kDa APN2t peptide but weakly to CR11-MPED on blots.
FIGURE 5.

Analysis of the interaction between Cry11Ba and AgAPN2 protein. (A) Partially purified recombinant APN2t or AgCad1 CR11-MPED fragment (1 μg), was resolved by SDS-PAGE and stained with Coomassie blue. (B) Increasing amounts of 125I-Cry11Ba were incubated with An. gambiae BBMV (8 μg) with or without 10 μM of each competitor in the binding buffer. (Inset) APN2t or AgCad1 CR11-MPED (1 μg) was spotted in duplicate on a PVDF membrane and probed with 0.125 nM 125I-Cry11Ba. (C) Binding affinity of Cry11Ba to APN2t. Ninety-six-well microtiter plates coated with 0.5 μg trypsinized Cry11Ba were incubated with increasing nM concentrations of biotinylated APN2t peptide alone or with 1000-fold molar excess of unlabeled APN2t peptide to determine specific binding. (D) The 70-kDa APN2t inclusions were used for toxicity inhibition assay. Cry11Ba toxin or a mixture of Cry11Ba and APN2t or CR11-MPED (1:100 w/w) were diluted in plastic plates containing 2 ml of deionized water and tested against ten early 4th instar larvae of An. gambiae. Each treatment was at least in duplicate and the bioassays were conducted three times. Larval mortality was recorded after 48 h. P < 0.01.
To determine Cry11Ba specific binding, we performed homologous competition assays with increasing amounts of labeled toxin using An. gambiae BBMV. Under the highest concentration of toxin addition, 10 μM of unlabeled Cry11Ba displaced 42% of the 125I-Cry11Ba binding, reducing bound toxin from 16.9 fmole to 9.8 fmole (Figure 5B). Thus 0.625 nM of labeled Cry11Ba showed the specific binding of 0.90 pmole per mg of BBMV. To evaluate the capacity of toxin binding to AgAPN2, we performed heterologous competition assays using 10 μM of APN2t or CR11-MPED peptides as competitors. For each experiment, the binding of 125I-toxin was reduced from 16.9 fmole to 11.3 fmole or 15.0 fmole, respectively. The addition of APN2t displaced 33% of the 125I-Cry11Ba binding whereas CR11-MPED only competed 11% of the binding (Figure 5B). We could not calculate Kd or Kcom because no saturated binding was obtained for this experiment with up to 18.75 nM of input 125I-Cry11Ba.
The binding affinity of Cry11Ba to APN2t peptide was further analyzed by a protein-protein binding assay based on ELISA. As shown in Figure 5C, biotinylated APN2t peptide bound Cry11Ba and the addition of 1000-fold of the unlabeled peptide competed binding. The binding curves were fit to a one-site saturation binding model, yielding a Kd = 6.4 ± 1.4 nM dissociation constant for APN2t peptide binding to Cry11Ba. This indicated a strong binding of AgAPN2t to Cry11Ba.
Because the 70-kDa APN2t peptide competed the binding of Cry11Ba to larval BBMV, we predicted that this peptide would interfere with the potency of Cry11Ba against An. gambiae. Bioassays were used to test the effect of APN2t peptide on Cry11Ba toxicity against An. gambiae. Fourth instar larvae of An. gambiae were fed with Cry11Ba toxin alone or in combination with APN2t or CR11-MPED inclusion body. The toxicity of Cry11Ba was reduced by 98% when larvae were fed with toxin and APN2t peptide (Figure 5D). The APN2t inclusion body alone showed no toxicity to An. gambiae larvae (data not shown). The addition of CR11-MPED peptide reduced Cry11Ba toxicity by 29%.
DISCUSSION
In this study, we cloned cDNA of AgAPN2 encoding a homologue of APNAnq100, from An. gambiae midgut tissue and characterized AgAPN2 with respect to its localization in larval midgut and Cry11Ba interactions. Aminopeptidases function as Cry1 toxin binding proteins in a number of lepidopteran species [reviewed in (3)]. Functional assays have demonstrated receptor function for combinations of Cry1 toxins and specific APNs in several species including tobacco hornworm (Manduca sexta), silkworm (Bombyx mori) and cotton leafworm (Spodoptera litura) reviewed in (3). Recently, the 100-kDa APN (APNAnq100) from An. quadrimaculatus was identified as a Cry11Ba toxin binding protein (12).
A phylogenetic tree of mature APN proteins was constructed by the maximum likelihood method based on 52 APN proteins from insects of Hemiptera, Coleoptera, Lepidoptera and Diptera (Figure 6). The putative mature APN sequences were predicted by the SignalP 3.0 to remove the signal peptides. Nine distinct classes of APNs were identified, of which five classes correspond to the lepidopteran clades reported previously (3, 32, 33, 34, 35) (Figure 6). Classes 6, 7, 8 and 9 are noteworthy because they are comprised of all the non-lepidopteran APNs that have not been phylogenetically characterized. AgAPN2 and its paralogues belong to the novel class 6. This class is also supported by LcAPN1 from an Australian sheep blowfly (Lucilia cuprina) (Figure 6). Class 7 includes AgAPN1 and an Ae. aegypti APN (AaAPNRc2) that shows 37% identity to AgAPN1. Class 8 is represented by two coleopteran APNs whereas class 9 is represented by a single hemipteran APN. Interestingly, Angelucci et al. (34) have recently extended the lepidopteran APN proteins to nine phylogenetic clusters with the addition of APNs including HaAPN6, HaAPN7 and their orthologues from B. mori. Although a different neighbor-joining method was used, the relationship between these two phylogenetic classifications needs to be further examined.
FIGURE 6.

Phylogenetic tree derived from ClustalX alignment of insect APNs. The tree was constructed by the maximum likelihood method. Classifications are according to (3, 31, 32, 33, 34). GenBank accession numbers are as follows: AaAPNRc2 (AAL85580), AgAPN1(Dinglasan, personal communication), AgAPN2, AgAPN3, AgAPN4, AgAPN5 (this study), AjFbAPN (ABE02186), AjMgAPN (ABH07377), ApAPN (ABD96614), BmAPN1 (AAC33301), BmAPN2 (BAA32140), BmAPN3 (AAL83943) BmAPN4 (BAA33715), CsAPN (ABC69855), EpAPN (AAF99701), HaAPN1 (AAK85538), HaAPN2 (AAK85539), HaAPN3 (AAN04900), HaAPN4 (AAM44056), HaAPN5 (AAW72993), HpAPN1 (AAF37558), HpAPN2 (AAF37559), HpAPN3 (AAF37560), HvAPN110 (AAK58066), HvAMPM (AAC46929), HvAPN170 (AAF08254), LcAPN1 (AAM77681), LdAPN2 (AAD31183), LdAPN2x (AAD31184), LdAPN3 (AAL26894), LdAPN4 (AAL26895), MsAPN1 (CAA61452), MsAPN2 (CAA66466), MsAPN3 (AAM18718), MsAPN4 (AAM13691), OfAPN1 (ABV01346), PiAPN (AAC36148), PxAPN (AAB70755), PxAMPN (CAA66467), PxAPN2 (CAA10950), PxAPN3f (AAF01259), SeAPN1 (AAP44964), SeAPN2 (AAP44965), SeAPN3 (AAP44966), SeAPN4 (AAP44967), SlAPN (AAK69605), TcAPN (XP_972951), TmAPN (AAP94045), TnAPN1 (AAX39863), TnAPN2 (AAX39864), TnAPN3 (AAX39865), TnAPN4 (AAX39866). Species names abbreviations are as follows: Aa, Aedes aegypti; Ag, Anopheles gambiae; Aj, Achaea janata; Ap, Acyrthosiphon pisum; Bm, Bombyx mori; Cs, Chilo suppressalis; Ep, Epiphyas postvittana; Ha, Helicoverpa armigera; Hp, Helicoverpa punctigera; Hv, Heliothis virescens; Lc, Lucilia cuprina; Ld, Lymantria dispar; Ms, Manduca sexta; Of, Ostrinia furnacalis; Pi, Plodia interpunctella; Px, Plutella xylostella; Se, Spodoptera exigua; Sl, Spodoptera litura; Tc, Tribolium castaneum; Tm, Tenebrio molitor; Tn, Trichoplusia ni. Bootstrap values represent the percentage frequency of which sequences resample in 100 replicates.
In this study AgAPN2 was detected as a 106-kDa GPI-anchored protein in BBMV prepared from An. gambiae larvae (Figure 2). Initial identification was by cross-reactivity with anti-AgAPN2 serum on a blot, and confirmation was by peptide mass fingerprinting. Although AgAPN2 was resolved as a single band on one-dimensional SDS-PAGE, other proteins, possibly the related AgAPNs listed in Table 2, may have been present in the 106-kDa gel band. Since AgAPN3 tryptic-digest peptides were detected in the 106-kDa band (Figure S1 in the Supporting Information), that paralogue was apparently present in the APN preparation. AgAPN3, also a predicted GPI-anchored protein, shares 95% amino acid identity to AgAPN2 and is probably recognized by polyclonal anti-AgAPN2 since the APN2t peptide used as an immunogen is 94% identical to the predicted AgAPN3 peptide. We lacked the information to support the presence of AgAPN4 in the 106-kDa band. Note the predicted AgAPN4 protein differs from AgAPN2 by only 5 amino acid residues making identification by peptide mass fingerprinting problematical (Figure S1 in the Supporting Information). The presence of multiple APN paralogues in a gene cluster may suggest that massive synthesis of APNs is needed for a rapid digestion of peptides over of a brief period of time.
In agreement with bioinformatic analyses, AgAPN2 is tethered to the membrane by a GPI-anchor, as are other insect APNs (36). The AgAPN2 protein was localized to the microvilli in the posterior part of the midgut (Figure 3). The distribution of AgAPN2 agrees with the histochemical localization of cells secreting aminopeptidase activity reported in (37, 38), and microarray-based analyses of transcripts associated with nutrient metabolism (39). The localization of AgAPN2 to the posterior midgut is consistent with the role of APN as an exoenzyme that cleaves amino acids from the N-terminus facilitating uptake by amino acid transporters located in the intestinal epithelium.
Cry11Ba bound PI-PLC-released AgAPN2 in a native condition. There was specificity in the interaction as Cry11Ba affinitively precipitated AgAPN2 among the multiple PI-PLC-released proteins (Figure 4). As noted in Results the 116-kDa protein visible in Figure 4 is probably AgAPN1. The precipitated 116-kDa protein seems to be more abundant than AgAPN2 or alternatively has higher affinity to Cry11Ba. We further present evidence that the peptide region (residue 228-843) of AgAPN2 binds Cry11Ba toxin. The 70-kDa truncated APN2t peptide bound the toxin on dot blot and microtiter plates (Figure 5). The binding affinity of Cry11Ba to the peptide region of AgAPN2 was determined as ca. 6.4 nM. This is comparable to a homologue of AgAPN2 (APNAnq100) in An. quadrimaculatus that tightly bound Cry11Ba in the native state with a ca. 1 nM KD (12). An in vitro competition binding assay using unlabeled Cry11Ba toxin or peptides as competitors also illustrated Cry11Ba specifically bound to An. gambiae BBMV. In the experiment, the 70-kDa truncated APN2t peptide competed Cry11Ba binding to BBMV, eliminating 79% of the specific binding of 125I-Cry11Ba, whereas CR11-MPED from AgCad1 only competed 26% of the Cry11Ba specific binding. The ability of AgAPN2 to compete more Cry11Ba binding on BBMV agrees with the dot blot assay, for which CR11-MPED binding to Cry11Ba was considerably less intense than that of AgAPN2. These results demonstrated AgAPN2 is a specific binding protein for Cry11Ba. The Cry11Ba specificity for a peptide region of AgAPN2 is similar to the recognition of a conserved region shared on APNs from B. mori that is recognized by Cry1Aa and Cry1Ab (32).
An in vivo approach was taken to correlate toxin binding to AgAPN2 with insecticidal activity. In agreement with the in vitro assay, when truncated AgAPN2 peptide was fed to 4th instar larvae with solubilized Cry11Ba, toxicity was reduced by 98% (Figure 5C). This inhibition is explained by Cry11Ba binding to AgAPN2 70-kDa peptide, thereby reducing toxin binding to native AgAPN2 located on the brush border membrane. The ability of AgAPN2 to inhibit Cry11Ba toxin is evidence that Cry11Ba binding to AgAPN2 is relevant to in vivo toxin action.
In the Cry1A toxin – lepidopteran systems, APNs play a role with alkaline phosphatase in inserting toxin oligomers into the cell membrane (4). In Ae. aegypti, alkaline phosphatase is a functional receptor for Cry11Aa toxin (10). In this research, we showed that Cry11Ba bound on beads selectively extracted three proteins from BBMV proteins, including AgAPN2. Although additional research is needed to determine the relative roles of AgAPN2 and the other AgAPNs in Cry11Ba toxicity, the recognition of AgAPN2 by Cry11Ba toxin suggests that mosquitocidal Cry toxins also have a complex mode of intoxication.
Supplementary Material
Multiple sequence alignment of AgAPN2, AgAPN3 and AgAPN4. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
The authors thank Dr. Mark R. Brown for assistance in immunohistochemistry, and Paul I. Howell from Malaria Research and Reference Reagent Resource Center (MR4) for providing the An. gambiae frozen sample. We extend our gratitude to Dr. Mohd Amir Abdullah and Reben Kalladuri for development of the ELISA-based toxin binding assay. We thank Dr. Mark R. Brown and Dr. Judith H. Willis for critically reading the manuscript.
Footnotes
This research was supported by National Institutes of Health Grant R01 AI 29092 to D. H. Dean (The Ohio State University) and M.J.A.
Abbreviations: alkaline phosphatase (ALP), aminopeptidase N (APN), Bacillus thuringiensis (Bt), brush border membrane vesicles (BBMV), 3-(cyclohexylamino)propanesulfonic acid (CAPS), 3-[(cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), glycosylphosphatidylinositol (GPI), phelylmethylsulfonyl fluoride (PMSF), phosphatidylinositol-specific phospholipase C (PI-PLC), polymerase chain reaction (PCR), surface plasmon resonance (SPR).
References
- 1.Walker K, Lynch M. Contributions of Anopheles larval control to malaria suppression in tropical Africa: review of achievements and potential. Med Vet Entomol. 2007;21:2–21. doi: 10.1111/j.1365-2915.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- 2.Federici BA, Park HW, Bideshi DK, Wirth MC, Johnson JJ, Sakano Y, Tang M. Developing recombinant bacteria for control of mosquito larvae. J Am Mosq Control Assoc. 2007;23:164–175. doi: 10.2987/8756-971X(2007)23[164:DRBFCO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71:255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bravo A, Gomez I, Conde J, Munoz-Garay C, Sanchez J, Miranda R, Zhuang M, Gill SS, Soberon M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophys Acta. 2004;1667:38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Ravoahangimalala O, Charles JF. In vitro binding of Bacillus thuringiensis var. israelensis individual toxins to midgut cells of Anopheles gambiae larvae (Diptera: Culicidae) FEBS Lett. 1995;362:111–115. doi: 10.1016/0014-5793(95)00220-4. [DOI] [PubMed] [Google Scholar]
- 6.Yamagiwa M, Kamauchi S, Okegawa T, Esaki M, Otake K, Amachi T, Komano T, Sakai H. Binding properties of Bacillus thuringiensis Cry4A toxin to the apical microvilli of larval midgut of Culex pipiens. Biosci Biotechnol Biochem. 2001;65:2419–2427. doi: 10.1271/bbb.65.2419. [DOI] [PubMed] [Google Scholar]
- 7.Kamauchi S, Yamagiwa M, Esaki M, Otake K, Sakai H. Binding properties of Bacillus thuringiensis Cry1C delta-endotoxin to the midgut epithelial membranes of Culex pipiens. Biosci Biotechnol Biochem. 2003;67:94–99. doi: 10.1271/bbb.67.94. [DOI] [PubMed] [Google Scholar]
- 8.Clark TM, Hutchinson MJ, Huegel KL, Moffett SB, Moffett DF. Additional morphological and physiological heterogeneity within the midgut of larval Aedes aegypti (Diptera: Culicidae) revealed by histology, electrophysiology, and effects of Bacillus thuringiensis endotoxin. Tissue Cell. 2005;37:457–468. doi: 10.1016/j.tice.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Buzdin AA, Revina LP, Kostina LI, Zalunin IA, Chestukhina GG. Interaction of 65- and 62-kD proteins from the apical membranes of the Aedes aegypti larvae midgut epithelium with Cry4B and Cry11A endotoxins of Bacillus thuringiensis. Biochemistry (Mosc.) 2002;67:540–546. doi: 10.1023/a:1015594127636. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez LE, Aimanova KG, Gill SS, Bravo A, Soberon M. A GPI-anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem J. 2006;394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua G, Zhang R, Abdullah MA, Adang MJ. Anopheles gambiae cadherin AgCad1 binds the Cry4Ba toxin of Bacillus thuringiensis israelensis and a fragment of AgCad1 synergizes toxicity. Biochemistry. 2008;47:5101–5110. doi: 10.1021/bi7023578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdullah MA, Valaitis AP, Dean DH. Identification of a Bacillus thuringiensis Cry11Ba toxin-binding aminopeptidase from the mosquito, Anopheles quadrimaculatus. BMC Biochem. 2006;7:16. doi: 10.1186/1471-2091-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delecluse A, Rosso ML, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microbiol. 1995;61:4230–4235. doi: 10.1128/aem.61.12.4230-4235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhaber B, Bork P, Eisenhaber F. Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 1998;11:1155–1161. doi: 10.1093/protein/11.12.1155. [DOI] [PubMed] [Google Scholar]
- 16.Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 17.Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- 18.Eisenhaber B, Bork P, Yuan Y, Loffler G, Eisenhaber F. Automated annotation of GPI anchor sites: case study C. elegans. Trends Biochem Sci. 2000;25:340–341. doi: 10.1016/s0968-0004(00)01601-7. [DOI] [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. Antibodies: A Laboratory Manual. 1. Cold Spring Harbor Laboratory Press; New York: 1988. Storing and purifying antibodies; p. 310. [Google Scholar]
- 20.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Silva-Filha MH, Nielsen-Leroux C, Charles J-F. Binding kinetics of Bacillus sphaericus binary toxin to midgut brush-border membranes of Anopheles and Culex sp. mosquito larvae. Eur J Biochem. 1997;247:754–761. doi: 10.1111/j.1432-1033.1997.00754.x. [DOI] [PubMed] [Google Scholar]
- 22.Garczynski SF, Adang MJ. Bacillus thuringiensis CryIA(c) δ-endotoxin binding aminopeptidase in the Manduca sexta midgut has a glycosylphosphatidylinositol anchor. Insect Biochem Mol Biol. 1995;25:409–415. [Google Scholar]
- 23.He N, Botelho JMC, McNall RJ, Belozerov V, Dunn WA, Mize T, Orlando R, Willis JH. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect Biochem Mol Biol. 2007;37:135–146. doi: 10.1016/j.ibmb.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Brown MR, Hua G, Adang MJ. Comparison of the localization of Bacillus thuringiensis Cry1A delta-endotoxins and their binding proteins in larval midgut of tobacco hornworm, Manduca sexta. Cell Tissue Res. 2005;321:123–129. doi: 10.1007/s00441-005-1124-6. [DOI] [PubMed] [Google Scholar]
- 25.Garczynski SF, Crim JW, Adang MJ. Identification of putative insect brush border membrane-binding molecules specific to Bacillus thuringiensis delta-endotoxin by protein blot analysis. Appl Environ Microbiol. 1991;57:2816–2820. doi: 10.1128/aem.57.10.2816-2820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braitbard O, Glickstein H, Bishara-Shieban J, Pace U, Stein WD. Competition between bound and free peptides in an ELISA-based procedure that assays peptides derived from protein digests. Proteome Sci. 2006;4:12. doi: 10.1186/1477-5956-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinglasan RR, Kalume DE, Kanzok SM, Ghosh AK, Muratova O, Pandey A, Jacobs-Lorena M. Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito midgut antigen. Proc Natl Acad Sci U S A. 2007;104:13461–13466. doi: 10.1073/pnas.0702239104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laustsen PG, Rasmussen TE, Petersen K, Pedraza-Diaz S, Moestrup SK, Gliemann J, Sottrup-Jensen L, Kristensen T. The complete amino acid sequence of human placental oxytocinase. Biochim Biophys Acta. 1997;1352:1–7. doi: 10.1016/s0167-4781(97)00036-5. [DOI] [PubMed] [Google Scholar]
- 30.Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 31.Takesue S, Yokota K, Miyajima s, Taguchi R, Ikezawa H, Takesue Y. Partial release of aminopeptidase N from larval midgut cell membranes of the silkworm, Bombyx mori, by phosphatidylinositol-specific phospholipase C. Comp Biochem Physiol. 1992;102B:7–11. doi: 10.1016/0305-0491(92)90263-q. [DOI] [PubMed] [Google Scholar]
- 32.Nakanishi K, Yaoi K, Nagino Y, Hara H, Kitami M, Atsumi S, Miura N, Sato R. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella -- their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 2002;519:215–220. doi: 10.1016/s0014-5793(02)02708-4. [DOI] [PubMed] [Google Scholar]
- 33.Herrero S, Gechev T, Bakker PL, Moar WJ, de Maagd RA. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four Aminopeptiase N genes. BMC Genomics. 2005;6:96. doi: 10.1186/1471-2164-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Zhang X, Zhang J. Molecular characterization of four midgut aminopeptidase N isozymes from the cabbage looper, Trichoplusia ni. Insect Biochem Mol Biol. 2005;6:611–620. doi: 10.1016/j.ibmb.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Angelucci C, Barrett-Wilt GA, Hunt DF, Akhurst RJ, East PD, Gordon KHJ, Campbell PM. Diversity of aminopeptidases, derived from four lepidopteran gene duplications, and polycalins expressed in the midgut of Helicoverpa armigera: Identification of proteins binding the δ-endotoxin, Cry1Ac of Bacillus thuringiensis. Insect Biochem Mol Biol. 2008;38:685–696. doi: 10.1016/j.ibmb.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adang MJ. Insect aminopeptidase N. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of proteolytic enzymes. 2. Elsevier; Amsterdam: 2004. [Google Scholar]
- 37.Volkmann A, Peters W. Investigations on the midgut caeca of mosquito larvae--I. Fine structure. Tissue Cell. 1989;21:243–251. doi: 10.1016/0040-8166(89)90069-4. [DOI] [PubMed] [Google Scholar]
- 38.Volkmann A, Peters W. Investigations on the midgut caeca of mosquito larvae--II. Functional aspects. Tissue Cell. 1989;21:253–261. doi: 10.1016/0040-8166(89)90070-0. [DOI] [PubMed] [Google Scholar]
- 39.Neira Oviedo M, Vanekeris L, Corena-McLeod MD, Linser PJ. A microarray-based analysis of transcriptional compartmentalization in the alimentary canal of Anopheles gambiae (Diptera: Culicidae) larvae. Insect Mol Biol. 2008;17:61–72. doi: 10.1111/j.1365-2583.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- 40.Boudko DY, Moroz LL, Linser PJ, Trimarchi JR, Smith PJ, Harvey WR. In situ analysis of pH gradients in mosquito larvae using non-invasive, self-referencing, pH-sensitive microelectrodes. J Exp Biol. 2001;204:691–699. doi: 10.1242/jeb.204.4.691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of AgAPN2, AgAPN3 and AgAPN4. This material is available free of charge via the Internet at http://pubs.acs.org.


