Abstract
With the increasing wealth of structural information available for human pathogens, it is now becoming possible to leverage that information to aid in rational selection of targets for inhibitor discovery. We present a methodology for assessing the drugability of all small-molecule binding pockets in a pathogen. Our approach incorporates accurate pocket identification, sequence conservation with a similar organism, sequence conservation with the host, and structure resolution. This novel method is applied to 21 structures from the malarial parasite Plasmodium falciparum. Based on our survey of the structural genome, we selected enoyl-acyl carrier protein reductase (ENR) as a promising candidate for virtual screening based inhibitor discovery.
Introduction
Identification of novel drug-binding targets remains an insufficiently met need in drug discovery. Recently, structural genomics initiatives have begun to play a significant role in the rapid production of structural data. However, many of the structures determined by such initiatives remain uncharacterized (Todd et al. 2005), leading to a discrepancy between the number of proteins elucidated and the number of druggable targets available. It is clear that as the number of available protein structures in a particular genome increases, the need for understanding and annotation of these structures also increases. In turn, this important functional information can be exploited for structure-based drug design. One application of such information is the identification of small-molecule binding sites. Traditionally, these sites have been elucidated through co-crystallization with a known ligand or through site-directed mutagenesis of putative active site residues. However, if such data is not available, then other means must be explored. An alternative method of determining binding sites is through computational structural analysis. This type of approach has been shown recently to detect successfully greater than 95% of the ligand binding sites in a benchmark of over 5000 protein structures (An et al. 2005).
Once potential ligand binding sites are identified, the next question is whether small molecules bound to those sites will interfere with the function of the protein. One method for determining the functional significance of a protein region is through sequence conservation to related homologues. This method has been shown to be particularly useful in determining interaction sites on proteins (Chelliah et al. 2004).
Due to the unfulfilled requirement of target discovery, we decided to approach this problem by surveying all proteins in a genome-wide scale. Here we present a methodology for the comprehensive computational evaluation of all small-molecule binding sites across a genome, and the potential to target these sites for structure-based drug design.
We apply our method to malaria, a human parasitic disease that causes 300–500 million acute infections and over 1 million deaths per year (WHO 2005). The species Plasmodium falciparum is responsible for the majority of all malarial deaths. The drugs chloroquine and sulfadoxine currently used to treat malaria are rapidly becoming ineffective due to resistance. As such, it is important to identify novel targets in the malarial genome for rational drug design. The use of X-ray crystallography has led to structures of many P. falciparum proteins and has helped elucidate the structural basis for drug activity and resistance in this species. As a logical next step, our approach surveys the structural genome of P. falciparum for the identification of drug targets. Based on the results of this analysis, we selected enoyl-acyl carrier protein reductase, which we believe to be a prime candidate for virtual screening-based inhibitor discovery.
Materials and Methods
Protein model generation
Molecular models were based on crystallographic PDB entries with released coordinates. In cases where multiple entries existed for a given protein, those with highest resolution, sequence identity closest to wild-type, and bound ligands were chosen with higher priority.
For PDB entries representing more than one instance of the biologically functional quaternary structure, the molecule with the best overall similarity to the other molecules was used (Overall similarity was measured by summing the pair-wise RMSD with the other structures.) For PDB entries representing a subset of the functional protein, BIOMT transformations were applied to create the missing subunits. For structures without BIOMT annotation, the missing subunits were generated using crystallographic symmetry transformations and quaternary structure information from the literature. Finally, hydrogen atoms were added to the functionally relevant structure and optimized for the lowest energy orientation. Incorrectly formed histidine residues were also corrected.
Pocket identification
Pockets were identified using a recently published algorithm for identifying ligand-binding sites in proteins (An et al. 2005). Briefly, the algorithm is based on a van der Walls grid potential map using a carbon probe. Bracketing and successive smoothing of that map with two parameter sets produces two distinct maps. One indicates volume considered inside the molecule. The other indicates cavities of empty space. Applying a threshold to the product of those two maps creates solid geometrical objects, which represent the identified pockets. Pockets below a minimum volume are discarded. The algorithm and all pocket analysis routines made use of the ICM software package (MolSoft 2006).
Initially, only proteins were included in the calculation of the van der Walls potential map. For molecules with pockets containing prosthetic groups or cofactors, pocket identification was repeated with the inclusion of these bound molecules, and thus each complex was treated as a separate protein. Duplicate pockets within multimeric proteins were identified by clustering the pockets by residue interaction. Within a group of similar pockets, only the best scoring pocket was kept.
Sequence alignment and conservation
Homologous sequences were identified using the online NCBI protein-protein BLAST tool (Altschul et al. 1990). PDB sequence data was queried, using default parameters, against the non-redundant compilation of GenBank CDS translations, RefSeq proteins, PDB, SwissProt, PIR, and PRF protein sequences. Homologous sequences were selected from each Plasmodium species. If multiple sequences were returned per species, the sequence representing the greatest overall consensus was selected. The single highest scoring sequence from Homo sapiens was selected.
Sequence alignment used the Needleman and Wunsch algorithm (Needleman and Wunsch 1970) with zero end gap penalties. For each pocket, sequence conservation statistics were gathered for nearby residues whose side chains were within 2.5 Å of the pocket. Residue conservation was defined as (number of identical residues)/(total number of nearby residues). Relative conservation was defined as (residue conservation)/(solvent accessible residue conservation). Solvent accessible residues were those with at least 25% of their surface accessible to a water probe molecule.
Pocket scoring
Pockets were ranked by a scoring function with six terms, with lower scores indicating a better pocket. The general equation of the scoring function is
where CA is the absolute residue conservation and CR is the relative residue conservation. The absolute and relative conservation measures were calculated from pair-wise alignments with Plasmodium yoelii. CH is the absolute conservation with Homo sapiens. V and A are the pocket volume and surface area, respectively. Both V0 and A0 were set to 450, an ideal value for these variables (unpublished data). R is the resolution of the crystallographic structure. The relative contributions of the terms were balanced by scaling each so that the interquartile range (IQR) was 1 and the mean was 0. Resolution was scaled to half the weight of the other terms with an IQR of 0.5.
Results
Pocket Identification
Of the 59 crystal structures of Plasmodium falciparum obtained from the PDB, 21 were distinct proteins (Table 1). These proteins encompass a wide variety of functions, including glycolysis, gluconeogenesis, purine salvage, vesicular traffic, and hemoglobin degradation activity.
Table 1.
List of 21 P. falciparum proteins whose structures were analyzed for the presence of druggable pockets. The resolution of the structures ranged from 1.1 to 3.0 Å.
| Protein | Abbreviated Name | PDB ID | Resolution (Å) |
|---|---|---|---|
| acyl-CoA binding protein | ACBP | 1hbk | 2.00 |
| fructose-1,6-bisphosphate aldolase | ALDO | 1a5c | 3.00 |
| adenylosuccinate synthetase | AdSS | 1p9b | 2.00 |
| cyclophilin | CyP | 1qng | 2.10 |
| dihydrofolate reductase-thymidylate synthase | DHFR-TS | 1j3i | 2.33 |
| enoyl-acyl carrier protein reductase | ENR | 1vrw | 2.35 |
| ferredoxin | Fd | 1iue | 1.70 |
| glutathione reductase | GR | 1onf | 2.60 |
| glutathione S-transferase | GST | 1q4j | 2.20 |
| hypoxanthine-guanine-xanthine phosphoribosyltransferase | HGXPRT | 1cjb | 2.00 |
| kinesin | KinI | 1ry6 | 1.60 |
| L-lactate dehydrogenase | LDH | 1t24 | 1.70 |
| peptide deformylase | 1rl4 | 2.18 | |
| gamete antigen 27 | Pfg27 | 1n81 | 2.10 |
| phosphoglcerate kinase | PGK | 1ltk | 3.00 |
| protein kinase 5 | PK5 | 1v0o | 1.90 |
| plasmepsin II | PM2 | 1lf2 | 1.80 |
| purine nucleoside phosphorylase | PNP | 1nw4 | 2.20 |
| Rab6 | Rab6 | 1d5c | 2.30 |
| triosephosphate isomerase | TIM | 1o5x | 1.10 |
| thioredoxin | Trx | 1syr | 2.95 |
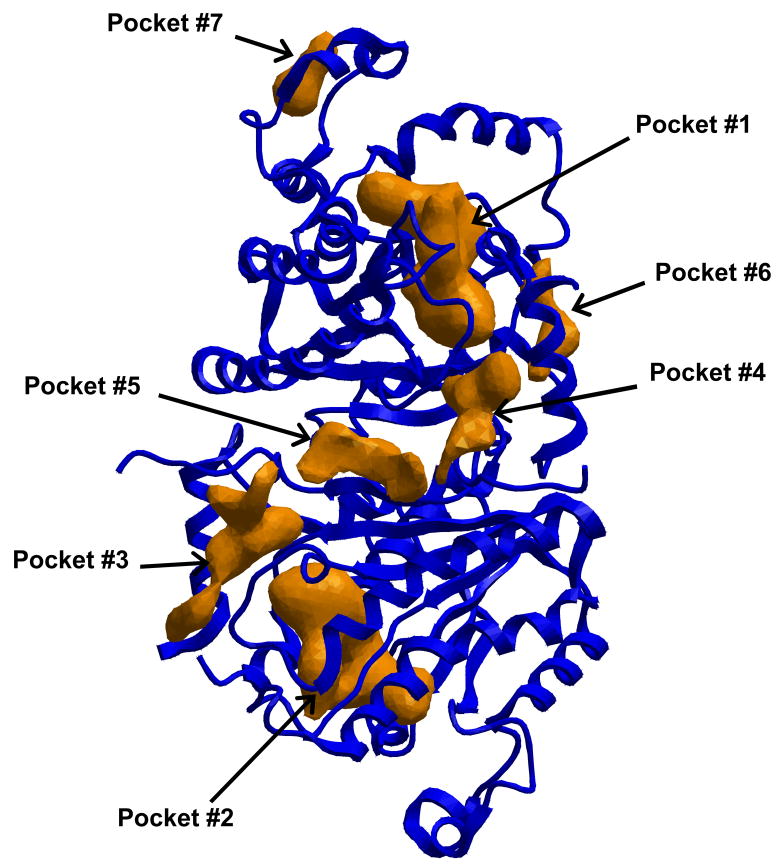
The PocketFinder module in ICM was executed on all 21 proteins (see Material and Methods). This method identifies all cavities on the surface of a protein and numbers them based on volume, as depicted in Figure 1. However, because several proteins contained repeated subunits in the quaternary structure, many pockets were replicated 2–6 times. As such, the number of pockets per protein varied greatly, from 1 to 24. When duplicates were found, only the pockets with the best score were considered, described below.
Figure 1.
Structure of ENR, one of 21 proteins targeted in this study. All drug-like binding sites are identified with the ICM pocketFinder module. Each of these pockets are numbered by volume, and combined with all the pockets from the other proteins for final scoring.
Structures for three proteins yielded large pockets (volume > 600 Ǻ3) that also contained prosthetic groups or cofactors. These included enoyl-acyl carrier protein reductase (ENR), dihydrofolate reductase-thymidylate synthase (DHFR-TS), and glutathione reductase (GR). Small molecule inhibitors do not necessarily need to compete out all cofactors and in some cases may have difficulty displacing these tightly bound molecules. As such, we created a second set of proteins excluding their cofactors and performed PocketFinder analysis on both the apo- and cofactor-bound protein set. Scoring and elimination of redundant pockets produced a list of 133.
Sequence Conservation
In order to determine which of the pockets identified with the PocketFinder module are functionally relevant, we compared the same regions with another important Plasmodium species, the rodent malarial parasite P. yoleii. The recent whole genome sequencing of P. yoleii (Carlton et al. 2002) made this task possible for all 21 P. falciparum proteins. Sequence conservation between P. falciparum and P. yoleii (Table 2) was performed only on the pocket residues, as identified with PocketFinder. The mean pair-wise sequence identity to P. yoleii for all pockets was 0.92.
Table 2.
Top 20 pockets across the P. falciparum proteome ranked by drugability score.
| Protein | Pocket # | Ligands | Seq. Ident. | Rel. Ident. | Hs Ident. | Vol. (Å3) | Area (Å2) | Res. (Å) | Score |
|---|---|---|---|---|---|---|---|---|---|
| PNP | 1 | inhibitor (IMH) | 1.00 | 1.25 | 0.56 | 590 | 513 | 2.20 | −2.95 |
| PNP | 2 | 1.00 | 1.25 | 0.50 | 274 | 290 | 2.20 | −2.56 | |
| LDH | 1 | 1.00 | 1.07 | 0.56 | 466 | 406 | 1.70 | −2.49 | |
| ENR* | 1 | inhibitor (TCC) | 1.00 | 1.17 | 0.57 | 410 | 362 | 2.35 | −2.47 |
| TIM | 1 | 0.94 | 1.17 | 0.81 | 448 | 407 | 1.10 | −2.45 | |
| DHFR-TS* | 3 | inhibitor (WRA) | 1.00 | 1.25 | 0.79 | 432 | 375 | 2.33 | −2.35 |
| HGXPRT | 3 | 1.00 | 1.25 | 0.75 | 269 | 323 | 2.00 | −2.29 | |
| PNP | 3 | 1.00 | 1.25 | 0.50 | 174 | 323 | 2.20 | −2.18 | |
| PM2 | 1 | inhibitor (R37) | 0.88 | 1.28 | 0.80 | 518 | 485 | 1.80 | −2.15 |
| AdSS | 3 | product (GDP) | 0.95 | 1.23 | 0.82 | 396 | 442 | 2.00 | −2.06 |
| LDH | 2 | inhibitor (OXQ), cofactor | 1.00 | 1.07 | 0.76 | 420 | 390 | 1.70 | −1.88 |
| KinI | 2 | 1.00 | 1.10 | 0.65 | 263 | 297 | 1.60 | −1.83 | |
| AdSS | 2 | 0.94 | 1.22 | 0.83 | 469 | 348 | 2.00 | −1.65 | |
| GR | 4 | 1.00 | 1.16 | 0.71 | 269 | 416 | 2.60 | −1.57 | |
| ENR* | 2 | 0.94 | 1.10 | 0.63 | 381 | 412 | 2.35 | −1.57 | |
| ENR* | 3 | 1.00 | 1.17 | 0.64 | 232 | 298 | 2.35 | −1.56 | |
| KinI | 1 | 1.00 | 1.10 | 0.85 | 442 | 464 | 1.60 | −1.54 | |
| ACBP | 1 | payload (COA), payload (MYR) | 0.85 | 1.22 | 0.77 | 318 | 367 | 2.00 | −1.41 |
| DHFR-TS* | 7 | 1.00 | 1.25 | 0.69 | 210 | 210 | 2.33 | −1.36 | |
| ENR | 2 | 0.94 | 1.10 | 0.63 | 295 | 355 | 2.35 | −1.23 |
Starred protein names indicate pockets in which a cofactor was included in the template structure. The columns indicate abbreviated protein name, pocket number of the original protein, presence of cofactor or ligand, sequence identity between P. falciparum and P. yoelii, relative sequence identity between pocket residues and other surface residues, sequence identity to H. sapiens, volume and area of each pocket, and resolution of original protein. See supplementary material for full table.
We next sought a measure to identify pockets in the Plasmodium genome that when targeted, would avoid cross-reactivity with the human host. We thus calculated sequence conservation between each of the Plasmodium pockets and their equivalent pockets in the most homologous human proteins. To avoid using homologues similar in sequence but not function, we verified that the published annotation indicated a similar function and classification in both P. falciparum protein and H. sapiens homologue. The mean human protein sequence identity for all pockets was 0.73. It is important to note that this measurement does not express all types of cross-reactivity with the human host. In particular, it does not address non-specific reactivity or the toxicity of potential drugs designed for these pockets.
Pocket Scoring and Selection
We have developed a new ‘drugability’ formula for the scoring of pockets on a genome-wide scale (See Materials and Methods). The formula uses the following six terms: 1) absolute residue sequence identity 2) sequence identity relative to the protein surface 3) absolute sequence identity with the closest human homologue 4) pocket volume 5) pocket surface area, and 6) crystallographic resolution of the protein. All 133 pockets in the P. falciparum genome were ranked according to these criteria and are listed in the Supplementary Table.
We then rationalized the top 20 pockets of the malarial structural genome for their availability to drug targeting. Five of these pockets were binding sites of existing inhibitors. These included enoyl-acyl carrier protein reductase (ENR), dihydrofolate reductase-thymidylate synthase, plasmepsin II, as well as several pockets of L-lactate dehydrogenase and purine nucleoside phosphorylase. It is important to note that the scoring function specifically excludes any information about known binding sites or co-crystallized molecules. Another top-ranked pocket included adenylosuccinate synthetase bound to GDP. Ligand-bound pockets did not show up in the top 20 ranked pockets, primarily because of the human homologue similarity and the unfavorable volume/surface area. All such pockets showed either human conservation greater than 0.88 or volume greater than 800 Ǻ3. The “Ligands” column of Table 2 indicates ligands that, had they been left in the template, would have intersected with the pocket.
Our selection narrowed the results to two pocket candidates: purine nucleoside phosphorylase (PNP) pocket #1, whose inhibition has been shown to prevent P. falciparum growth (Kicska et al. 2002), and ENR pocket #1, which is involved in the critical process of fatty acid biosynthesis. Due to its important role in the life cycle of malaria, and a lack of suitable therapeutics targeting it, we decided ENR is most amenable for virtual screening based drug discovery.
Discussion
There exists an unmet need to successfully identify novel targets in important pathogens. Other groups have attempted to decipher drugability of active sites by relating to sequence conservation and pocket surface area/size, with mixed results. In the present study, we describe a novel protocol that incorporates these variables in a unique formula for identifying drug-like pockets. Furthermore, we apply this method to an entire structural proteome, a database of targets larger than any group has attempted previously.
We used ICM PocketFinder (An et al. 2005), a recently developed method for detection of sites on a protein surface that are most amenable to small molecule binding. PocketFinder calculates van der Waals potentials in the vicinity of the protein surface, and identifies and sorts protein surface cavities based on their ability to bind a virtual ligand. The method is independent from any physical ligand-related information and thus allows for pocket identification in apo- as well as bound structures. The efficiency of the method makes it applicable for identification of potential drug targets by screening every structurally-determined protein in a genome.
To validate our protocol on a large scale, we applied the method to the P. falciparum structural proteome. In order to select the most biologically significant targets and sites, we introduced a novel binding pocket scoring function, which included terms for the pocket volume, size, and shape, as well as evolutionary conservation of residues lining each pocket. A high conservation between proteins in the P. falciparum genome and homologous proteins from a related species, P. yoelii, was considered an advantage in the process of pocket scoring. We reasoned that this would ensure their functional relevance as well as increase the chances that drugs developed to target these pockets will cross-react with different members of the Plasmodium genus. However, it is important to mention that this is a very limited attempt to select for the most functionally relevant proteins in a genome. For optimal evaluation, this approach needs to be coupled with other biological and functional aspects of each protein.
Another sequence analysis used in the final pocket scoring function included the comparison between P. falciparum and H. sapiens. Conservation of the residues between each putative target and homologous proteins in human could potentially lead to toxicity due to cross-reactivity, and thus considered unfavorable. While this measure does not account for all types of potential cross-reactivity in human, it enriches the set of top-scoring targets with those that are more biologically significant.
The application of our novel method led to the identification of ENR pocket #1 as the best candidate for structure-based drug design. ENR has not been previously targeted by structure-based drug design; moreover, this is a very important enzyme in the Plasmodium life cycle. We have validated our results in a separate study by targeting ENR with virtual screening and successfully discovering novel small molecule inhibitors (Nicola et al. 2007). Based on the success of these experiments, we have confidence that the same approach can be used to identify druggable targets in other structural genomes.
Supplementary Material
Acknowledgments
The authors thank Irina Kufareva for critical analysis of this manuscript. This work was funded in part by NIH research grant 5R01GM071872-03 to RA.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, Peterson JD, Pop M, Kosack DS, Shumway MF, Bidwell SL, Shallom SJ, van Aken SE, Riedmuller SB, Feldblyum TV, Cho JK, Quackenbush J, Sedegah M, Shoaibi A, Cummings LM, Florens L, Yates JR, Raine JD, Sinden RE, Harris MA, Cunningham DA, Preiser PR, Bergman LW, Vaidya AB, van Lin LH, Janse CJ, Waters AP, Smith HO, White OR, Salzberg SL, Venter JC, Fraser CM, Hoffman SL, Gardner MJ, Carucci DJ. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- Chelliah V, Chen L, Blundell TL, Lovell SC. Distinguishing structural and functional restraints in evolution in order to identify interaction sites. Journal of molecular biology. 2004;342:1487–1504. doi: 10.1016/j.jmb.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Kicska GA, Tyler PC, Evans GB, Furneaux RH, Kim K, Schramm VL. Transition state analogue inhibitors of purine nucleoside phosphorylase from Plasmodium falciparum. J Biol Chem. 2002;277:3219–3225. doi: 10.1074/jbc.M105905200. [DOI] [PubMed] [Google Scholar]
- MolSoft. Program Manual. ICM; San Diego: 2006. [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of molecular biology. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nicola G, Smith CA, Lucumi E, Kuo MR, Karagyozov L, Fidock DA, Sacchettini JC, Abagyan R. Discovery of novel inhibitors targeting enoyl-acyl carrier protein reductase in Plasmodium falciparum by structure-based virtual screening. Biochem Biophys Res Commun. 2007;358:686–691. doi: 10.1016/j.bbrc.2007.04.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AE, Marsden RL, Thornton JM, Orengo CA. Progress of structural genomics initiatives: an analysis of solved target structures. Journal of molecular biology. 2005;348:1235–1260. doi: 10.1016/j.jmb.2005.03.037. [DOI] [PubMed] [Google Scholar]
- WHO; World Health Organization, U. Roll Back Malaria. World Health Organization; Geneva, Switzerland: 2005. World Malaria Report 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.