Abstract
Based on abscisic acid (ABA) inhibition of seed germination and seedling growth assays, we isolated an ABA overly sensitive mutant (abo4-1) caused by a mutation in the Arabidopsis thaliana POL2a/TILTED1(TIL1) gene encoding a catalytic subunit of DNA polymerase ε. The dominant, ABA-insensitive abi1-1 or abi2-1 mutations suppressed the ABA hypersensitivity of the abo4-1 mutant. The abo4/til1 mutation reactivated the expression of the silenced Athila retrotransposon transcriptional silent information (TSI) and the silenced 35S-NPTII in the ros1 mutant and increased the frequency of somatic homologous recombination (HR) ∼60-fold. ABA upregulated the expression of TSI and increased HR in both the wild type and abo4-1. MEIOTIC RECOMBINATION11 and GAMMA RESPONSE1, both of which are required for HR and double-strand DNA break repair, are expressed at higher levels in abo4-1 and are enhanced by ABA, while KU70 was suppressed by ABA. abo4-1 mutant plants are sensitive to UV-B and methyl methanesulfonate and show constitutive expression of the G2/M-specific cyclin CycB1;1 in meristems. The abo4-1 plants were early flowering with lower expression of FLOWER LOCUS C and higher expression of FLOWER LOCUS T and changed histone modifications in the two loci. Our results suggest that ABO4/POL2a/TIL1 is involved in maintaining epigenetic states, HR, and ABA signaling in Arabidopsis.
INTRODUCTION
Plants are sessile and unable to escape acute environmental stresses, but they have developed sophisticated responses to cope with and survive stress conditions. Plants are able to quickly counteract environmental stresses through actions, such as up- or downregulating gene expression and accumulating adaptive metabolic compounds. Abiotic stresses, such as drought, high salinity, and low temperature, induce the accumulation of the phytohormone abscisic acid (ABA) (Finkelstein et al., 2002; Koornneef et al., 2002). ABA modulates embryogenesis, maturation, and dormancy during seed development and plays vital roles in a wide range of biological processes throughout plant growth, development, and stress responses (Finkelstein et al., 2002; Koornneef et al., 2002). Through forward and reverse genetic studies, many genes involved in ABA responses have been isolated and characterized (Finkelstein et al., 2002; Koornneef et al., 2002; Xiong et al., 2002; Shinozaki and Yamaguchi-Shinozaki, 2007). Increased ABA in plant cells inhibits DNA replication and cell division, which results in retarded plant growth (Finkelstein et al., 2002; Swiatek et al., 2002; Chandrasekharan et al., 2003). However, the molecular mechanism for ABA inhibition of plant growth is largely unknown. One hint is that ABA induces expression of the cyclin-dependent kinase inhibitor KRP1/ICK1 (Wang et al., 1998). Cyclin-dependent kinases regulate the progression of the cell cycle in eukaryotes. Studies thus far have focused on ABA-mediated rapid physiological responses that are important for plant stress resistance in the short term. Whether ABA or an ABA-regulated signal can affect epigenetic states and genome stability, which might be important for the long-term adaptation of plants to environmental stresses and possibly contribute to plant evolution, is not known.
During the cell cycle, both DNA methylation patterns and specific chromatin structures must be duplicated with high fidelity throughout DNA replication. The DNA replication process involves DNA biosynthesis, transient disruption of parental nucleosomes, and nucleosome reassembly (Groth et al., 2007), and many different proteins participate in this complex process. During S-phase, the leading and lagging strand are replicated by different mechanisms in a continuous and discontinuous fashion, respectively. In the lagging strand synthesis, DNA polymerase α uses RNA primers initiated by the primase to synthesize a short DNA and form an RNA–DNA hybrid. Replication factor C binds to the RNA–DNA junction and recruits the homotrimers of PCNA (a ring-shaped replication factor) to encircle the DNA. Then, most possibly, the DNA polymerase δ replaces the DNA polymerase α, together with PCNA, to copy the DNA template (Garg and Burgers, 2005; Moldovan et al., 2007). Pol ε is a leading strand DNA polymerase during chromosomal DNA replication in yeast (Pursell et al., 2007). In yeast, DNA polymerase ε is a four-subunit complex (consisting of the catalytic subunit, POL2p, and three regulatory subunits, DPB2p, DBP3p, and DBP4p), and this structure is conserved in Homo sapiens (Pospiech and Syvaoja, 2003). In Arabidopsis thaliana, DNA polymerase ε subunits consist of DPB2 (encoded by At5g22110, corresponding to yeast DPB2p), DPB4 (encoded by At1g09030, corresponding to yeast DPB4p), and DBP3a (encoded by At5g50490, corresponding to yeast DBP3p) for the regulatory subunits, and POL2a and POL2b for the catalytic subunits (Jenik et al., 2005; Ronceret et al., 2005). A null mutation in either DPB2 or POL2a is lethal (Ronceret et al., 2005). Knockout mutants with T-DNA insertions in POL2b exhibit no morphological phenotypes (Ronceret et al., 2005). Studies of Pol ε in yeast and humans implicated it in DNA repair and recombination (Pospiech and Syvaoja, 2003). Pol ε, together with RPA (replication protein A), replication factor C, PCNA, and DNA ligase I, was found to refill the repaired DNA patch through a reconstituted protein system in mammals and yeast (Aboussekhra et al., 1995; Shivji et al., 1995). Genetic studies in yeast indicated the importance of both Pol ε and δ in filling the gaps of repair DNA (Pospiech and Syvaoja, 2003). Moreover, DNA polymerase ε binds double-stranded DNA and promotes epigenetic silencing at telomeres in budding yeast, but this activity is not associated with its polymerase activity (Tsubota et al., 2006). Establishment of transcriptional gene silencing (TGS) and histone deposition in yeast seems to be independent of DNA replication (Kirchmaier and Rine, 2001; Li et al., 2001; Green et al., 2005). In Arabidopsis, lesions in POL2a/TIL1 lead to longer cell cycles and developmental defects (Jenik et al., 2005; Ronceret et al., 2005). Other biological roles for POL2a in planta have not yet been characterized because of the lack of a suitable mutant. In this study, we characterized an ABA overly sensitive mutant (abo4-1), a novel weak allele of POL2a. We found that the mutation in ABO4/POL2a/TIL1 releases TGS in some genomic regions and greatly increases the frequency of somatic homologous recombination (HR); both effects were reinforced by ABA treatment. Our studies on the abo4 mutant reveal previously unknown functions of POL2a in epigenetic regulation, HR, and ABA signaling in plants.
RESULTS
Phenotypic Characterization of an ABA Overly Sensitive Mutant, abo4-1
An ABA overly sensitive mutant, abo4-1, was isolated from an ethyl methanesulfonate–mutagenized population of Arabidopsis (Columbia [Col] gl1 background) using a root bending assay on Murashige and Skoog (MS) medium containing 30 μM ABA. The abo4-1 mutant was backcrossed with the wild type four times before phenotypic analysis.
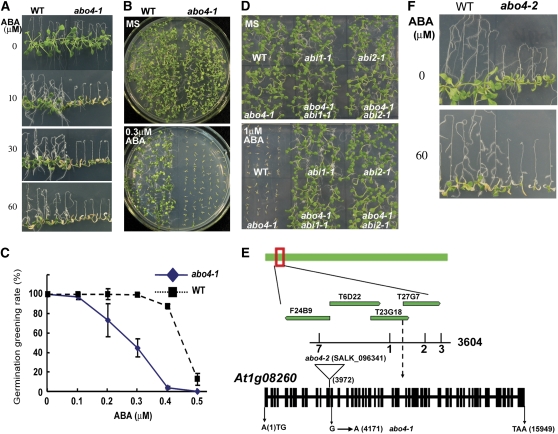
As shown in Figure 1A, different concentrations of ABA greatly inhibited root growth and made abo4-1 cotyledons become yellow, but not wild-type. We also tested the seed germination sensitivity of abo4-1 to ABA. In the absence of ABA, both abo4-1 and the wild type showed similar seed germination rates (Figure 1B, MS). In the presence of different concentrations of ABA, both germination and postgermination growth of abo4-1 were greatly inhibited compared with the wild type (Figure 1B, 0.3 μM ABA). Figure 1C shows the statistical analysis of seedling greening rate (percentage of greening seedlings over all germinated seedlings) after germination on day 7 at different concentrations of ABA. Supplementation of 0.3 μM ABA had a striking effect on abo4-1 seedling greening, whereas even 0.4 μM ABA did not significantly influence the greening rate of the wild type (Figure 1C).
Figure 1.
ABA Sensitivity of the abo4-1 Mutant.
(A) ABA sensitivity of abo4-1 seedlings grown on MS media containing different concentrations of ABA. Five-day-old seedlings grown on MS medium were transferred to MS medium containing different concentrations of ABA, respectively, and cultured for another 7 d before taking pictures.
(B) The seed germination sensitivity of abo4-1 to ABA. Wild-type and abo4-1 seeds were directly sowed on MS medium or MS medium containing 0.3 μM ABA, respectively. After being kept at 4°C for 3 d, the plates were removed to a growth chamber and cultured for 7 d before taking pictures.
(C) The statistic data of seed germination of wild-type and abo4-1 on MS medium containing different concentrations of ABA. The experimental conditions were the same as in (B). Values are means ± se, n = 3 independent experiments. At each time, >100 seeds were counted.
(D) The dominant mutations of the negative regulators ABI1 and ABI2 block the ABA sensitivity of abo4-1. The seeds of wild-type, abo4-1, abi1-1, abi2-1, abo4-1 abi1-1, and abo4-1 abi2-1 double mutants were germinated on MS medium containing no ABA or 1 μM ABA. The pictures were taken after seedlings were grown for 7 d in a growth chamber.
(E) Mapping-based cloning of ABO4. The ABO4 locus was mapped between two BAC clones F24B9 (recombinants, 7 of 1802) and T27G7 (recombinants, 3 of 1802). Further mapping delimited the ABO4 locus to a region within BAC T23G18. A point mutation (G4171A, in the 13th exon) was found in AT1G08260/POL2a/TIL1. A T-DNA insertion (abo4-2, SALK_096341) line was obtained with a T-DNA inserted at position 3972 (in the 12th exon).
(F) Comparison of ABA sensitivity of wild-type and abo4-2 seedlings. The seedlings were first grown on MS medium for 5 d. abo4-2 seedlings were picked up according to its grown phenotype and were, together with the wild type, transferred to MS medium containing different concentrations of ABA. Here, we only show the seedlings grown on MS medium containing 60 μM ABA for 7 d. The similar growth phenotypes were observed when abo4-2 seedlings were grown on other concentrations of ABA (10, 20, 30, and 50 μM, respectively).
We crossed two dominant ABA-insensitive mutants, abi1-1 and abi2-1, to abo4-1. ABI1 and ABI2, two protein phosphatases 2C, are key negative regulators in ABA signal transduction pathways (Leube et al., 1998; Rodriguez et al., 1998; Gosti et al., 1999; Merlot et al., 2001). The germination and postgermination growth of abo4-1 abi1-1 and abo4-1 abi2-1 double mutants were compared in the presence of 1 μM ABA. As shown in Figure 1D, the two double mutants showed similar ABA insensitivity as abi1-1 and abi2-1. However, the abi1-1 or abi2-1 mutations did not affect the growth phenotypes of abo4-1 under normal growth conditions. This result suggests that the enhanced ABA response in the abo4-1 mutant is mediated by ABA signal transduction pathway(s).
abo4-1 Is a Weak Allele of POL2a
We used map-based cloning to identify the ABO4 gene. ABO4 (At1g08260) encodes a catalytic subunit of DNA polymerase ε (POL2a) (Jenik et al., 2005; Ronceret et al., 2005). This locus had been previously identified as TILTED1 (TIL1) (Jenik et al., 2005). POL2a has 47 introns and 48 exons. A point mutation that changes Gly534 to Arg (G to A in position 4171 counting from the first putative ATG, in the 13th exon) was found in abo4-1 (Figure 1E). A Salk T-DNA insertion line (SALK_096341, abo4-2), in which the T-DNA was inserted at position 3972 (in the 12th exon), counting from the first putative ATG of the genomic coding sequence, was obtained. In yeast, the N-terminal catalytic domain of DNA polymerase ε is not essential for survival but is required for the DNA damage checkpoint control (Feng and D'Urso, 2001; Ohya et al., 2002). Because null mutations in Arabidopsis POL2a are embryonic lethal (Jenik et al., 2005; Ronceret et al., 2005), both abo4-1 and abo4-2 should be weak alleles of POL2a. abo4-2 showed similar ABA sensitivity of seedling growth as abo4-1 (Figure 1F). Because the homozygous abo4-2 mutants were sterile in both male and female, we selected heterozygous T-DNA mutant lines and crossed them with abo4-1. The F1 seedlings showed abo4-1 and wild-type growth phenotypes at a ratio of ∼1:1 (62:64) when grown on MS medium and also showed abo4-1 and wild-type ABA-sensitive phenotypes at a ratio of ∼1:1 (57:58) when grown on MS medium containing 30 μM ABA in the root bending assay. Seven sensitive seedlings were randomly picked up and checked by PCR, all were heterozygous plants of abo4-1 and abo4-2, which is consistent with the above genetic analysis. These results confirmed that ABO4 is At1g08260/POL2a/TIL1 (please also see the segregation analysis of F1 seedlings on methyl methanesulfonate [MMS] medium in Figure 2).
Figure 2.
abo4 Mutants Are Sensitive to DNA Damage.
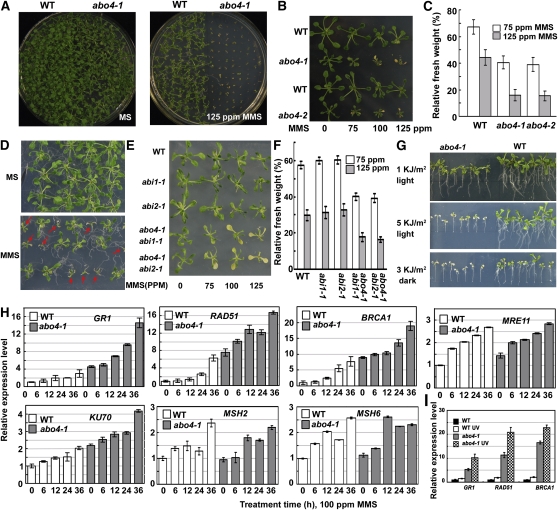
(A) Comparison of the wild type and abo4-1 seed germination on MS medium containing no (left) or 125 ppm (right) MMS. Seedlings were grown for 10 d on MS medium containing no MMS or 125 ppm MMS.
(B) Seedling growth sensitivity of wild-type gl1, abo4-1 (gl1), the wild type, and abo4-2 in MS liquid medium supplemented with different concentrations of MMS. Five-day-old seedlings were removed to the MS liquid medium containing different concentrations of MMS and cultured for 10 more days before taking pictures.
(C) Relative fresh weight of wild-type, abo4-1, and abo4-2 seedlings grown on 75 and 125 ppm MMS. The fresh weight of wild-type, abo4-1, and abo4-2 seedlings grown on MS agar medium for 10 d was used to compare with the fresh weight of seedlings grown on 75 and 125 ppm MMS, respectively. For each data point, 30 seedlings were collected and weighed. Data are the mean ± sd of three independent experiments.
(D) The segregation of F1 seedlings from heterozygous abo4-2 mutant lines crossed with abo4-1 grown on MS medium containing 100 ppm MMS (MMS sensitive to tolerant seedlings: 58/58). We randomly picked up 20 sensitive seedlings and checked by PCR, and all were heterozygous plants of abo4-1 and abo4-2. Arrows point to the segregated mutants.
(E) abi1 and abi2 mutations did not improve the DNA damage sensitivity caused by MMS in abo4-1. The similar experiments were done as in (B) for abo4-1 abi1-1 and abo4-1 abi2-1 double mutant seedlings.
(F) Relative fresh weight of wild-type, abi1-1, abi2-1, abo4-1 abi1-1, and abo4-1 abi2-1 seedlings grown on 75 and 125 ppm MMS. The fresh weight of wild type, abi1-1, abi2-1, abo4-1 abi1-1, and abo4-1 abi2-1 seedlings grown on MS agar medium for 10 d was used to compare with the fresh weight of seedlings grown on 75 and 125 ppm MMS, respectively. For each data point, 30 seedlings were collected and weighed. Data are the mean ± sd of three independent experiments.
(G) abo4-1 seedlings were more sensitive to UV-B light than the wild type. Five-day-old seedlings of the wild type and abo4-1 in the same plate were treated with different amounts of UV-B light and then grown in light (for 1 and 5 kJ/m2) or first in dark for 2 d and then in light (for 3 kJ/m2). The pictures were taken after seedlings were grown in light for another 5 d.
(H) Comparison of the gene expression in the wild type and abo4-1 under normal and MMS (100 ppm) treatment using real-time qRT-PCR. GR1, RAD51, BRCA1, MRE11, KU70, MSH2, and MSH6 were selected for comparison. The transcripts of the wild type in normal condition were used as a standard for normalization.
(I) Comparison of the transcripts of GR1, RAD51, and BRCA1 by qRT-PCR between the wild type and abo4-1 under normal and UV-B (3 kJ/m2, in dark for 6 h) treatment.
In both (H) and (I), three independent biological replications were performed for each experiment. The level of 18S rRNA was used as an internal control. Error bars indicate ±sd.
abo4-1 Mutants Were Hypersensitive to MMS and UV-B Light
In addition to bulk DNA synthesis, one of the important functions of DNA polymerase ε is nucleotide and base excision repair (Dianov et al., 2003; Pospiech and Syvaoja, 2003). To evaluate if ABO4/POL2a/TIL1 is involved in DNA repair, we examined the sensitivity of abo4-1 seedlings to the DNA-damaging reagent, MMS, at different concentrations and to UV-B light. Both MMS and UV-B light produce double-strand DNA breaks (DSBs) in cells. The postgermination growth of abo4-1 was completely inhibited by 125 ppm MMS, while the growth of wild-type plants was less affected (Figure 2A). Five-day-old seedlings grown on nutrient medium were transferred to nutrient medium supplemented with different concentrations of MMS and incubated for 10 more days. As shown in Figure 2B, at 75, 100, and 125 ppm MMS, the growth of shoots and roots in both abo4-1 and abo4-2 was more severely affected compared with the wild type. Growth measurement of fresh weight is shown in Figure 2C, indicating that the growth of abo4-1 and abo4-2 was more greatly inhibited by MMS than that of the wild type. To further confirm the effects of abo4-2 and abo4-1 on MMS sensitivity, the F1 seedlings from heterozygous T-DNA mutant lines crossed with abo4-1 were grown on MS medium containing 100 ppm MMS. As shown in Figure 2D, the ratio of MMS sensitive to tolerant seedlings was 1:1 (58:58). Twenty sensitive seedlings were randomly picked up and checked by PCR, and all were heterozygous for abo4-1 and abo4-2. These data indicate that the abo4 mutation cosegregated with MMS sensitivity. However, abi1-1 abo4-1 and abi2-1 abo4-1 double mutants showed similar MMS-sensitive growth phenotypes as the abo4-1 single mutant (Figures 2E and 2F), suggesting that the pathways leading to the sensitivity of abo4-1 to ABA and MMS are different. When treated with various dosages of UV-B light, the growth of abo4-1 seedlings was more profoundly affected than the wild type (Figure 2G). These results suggest that POL2a has functions in DNA repair.
DSB-Inducible Genes Were Constitutively Expressed in abo4-1 Mutants
In some DNA damage–sensitive mutants, such as brushy1 (bru1) (Takeda et al., 2004), tebichi (teb) (Inagaki et al., 2006), the nucleosome assembly protein1-1 (nrp1-1) nrp2-1 (Zhu et al., 2006) and fasciata1 (fas1) fas2 double mutants (Endo et al., 2006; Kirik et al., 2006; Ramirez-Parra and Gutierrez, 2007), the expression of some DSB-inducible genes is enhanced. DNA damage caused by ionizing radiation, defects of nucleotide and base excision repair, or failure of mismatch repairing, can lead to DSBs which are repaired through the nonhomologous end joining (NHEJ) pathway (mainly in animals and plants) or through the HR pathway (mainly in yeast). We selected several marker genes functioning in DSB to check their expression. MEIOTIC RECOMBINATION11 (MRE11), one member of the MRN complex (composed of Mre11, Rad50, and Nbs1), plays a central role in early DSB repair (Puizina et al., 2004; Farah et al., 2005). KU70 functions in repairing DNA damage through NHEJ pathway (Tamura et al., 2002), while MSH2 and MSH6 proteins play critical roles in DNA mismatch, repairing and limiting HR (Emmanuel et al., 2006; Li et al., 2006; Lafleuriel et al., 2007). RAD51 is a homolog of the bacterial RecA recombinase in HR repair (Doutriaux et al., 1998). Arabidopsis BREAST CANCER SUSCEPTIBLITY1 (BRCA1) functions in concert with Rad51 and other genes to control double-strand break repair and HR (Scully et al., 2004; Reidt et al., 2006). GAMMA RESPONSE1 (GR1) is also suggested to play some roles in HR (Lafarge and Montane, 2003). We tested the transcript levels of these genes under normal and genotoxic stress conditions by real-time quantitative RT-PCR (qRT-PCR). Without stress, the transcripts of GR1, RAD51, BRCA1, MRE11, and KU70 were found to be higher in abo4-1 mutants compared with the wild type (Figure 2H). After MMS treatment, the transcripts of the five genes were upregulated in both abo4-1 and wild-type plants than under normal conditions at different time points (Figure 2H). It seems that abo4 mutation did not influence the expression of MSH2 and MSH6, but MMS treatment increased the transcripts of these two genes (Figure 2H). UV-B treatment also induced higher expression of GR1, RAD51, and BRCA1 in both the wild type and mutant (Figure 2I).
abo4-1 Affected Cell Cycle Regulation
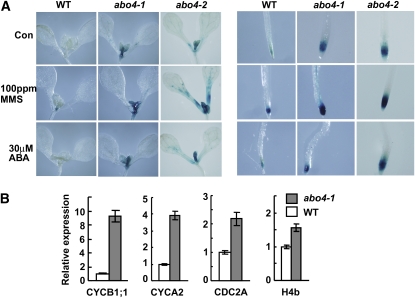
A previous study suggested important roles for POL2a in cell cycle regulation during embryo development (Jenik et al., 2005). Because abo4 affects meristem development, we investigated the role of ABO4/POL2a in the cell cycle in meristem cells. A transgenic line carrying the CYCB1;1 promoter fused with the β-glucuronidase (GUS) gene was crossed with abo4-1. In Arabidopsis, the CYCB1;1 promoter is expressed upon entry into the G2-phase, and GUS staining marks cells in G2- and early M-phase (Doerner et al., 1996). As illustrated in Figure 3A (top), much more GUS activity was detected in root tips and shoot meristems of abo4-1 and abo4-2 than the wild type, suggesting that G2/M progression in abo4-1 and abo4-2 was delayed. MMS treatment strongly induced GUS expression (Figure 3A, middle), suggesting a crucial role of DNA DSB repair in G2/M progression. However, ABA treatment did not apparently affect the expression of CYCB1;1 (Figure 3A, bottom), which is consistent with previous results showing that ABA only affects cell cycle progression in the G1 stage (Swiatek et al., 2002). We also tested the expression of CYCB1;1, CYCA2;1, CDC2A, and HISTONE H4b (S-specific Histone4) by qRT-PCR and found the transcripts of these genes were all more highly expressed in abo4-1 than wild-type plants (Figure 3B). The higher expression of H4b in abo4-1 suggests a prolonged S-phase, which is consistent with previous observations (Jenik et al., 2005).
Figure 3.
The abo4 Mutation Increases Cell Cycle–Related Gene Expression.
(A) The representative examples of GUS staining. CYCB1;1 promoter-GUS was expressed higher in shoot and root meristems of abo4-1 and abo4-2 than the wild type, and its expression was highly induced by MMS treatment but not affected by ABA.
(B) The relative expression levels of CYCB1;1, CYCA2;1, CDC2A, and H4b in the wild type and abo4-1 analyzed by qRT-PCR. The expression of each gene in the wild type was used as a standard. The representative results were from one of three independent experiments. Error bars indicate ±sd.
abo4 Mutation Greatly Elevated the Frequency of HR
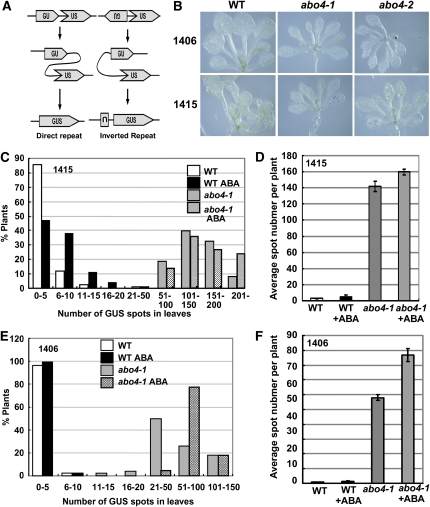
Because abo4-1 is sensitive to genotoxic stress and DSBs are repaired in part by HR, we examined whether or not the abo4 mutation influences the level of HR using a GUS reporter recombination substrate. Two transgenic lines, 1415 and 1406, with T-DNAs containing two inactive fragments of the GUS gene sharing an overlapping sequence in direct or inverted orientation (Lucht et al., 2002) (Figure 4A) were selected for these experiments. If an intrachromosomal recombination event occurs between the overlapping regions, a functional GUS gene will be reconstituted and expressed, which can be detected as a blue sector after GUS staining. The abo4-1 mutant was crossed with two recombination substrate lines 1415 and 1406, and we selected abo4-1 mutants or wild-type plants carrying the homozygous recombination substrate transgene from the F3 population and used them for GUS staining analysis. More than 100 seedlings were analyzed for each assay. The number of GUS staining spots was various among different lines. On average, we detected 2.9 spots per wild-type plant and 140.4 spots per abo4-1 plant in reporter line 1415, and one spot per wild-type plant and 59.3 spots per abo4-1 plant in reporter line 1406 with our conditions (Figures 4B to F). We obtained similar high GUS recombination rates for abo4-2 mutant in both 1415 and 1406 lines (Figure 4B, right). These results indicate that the abo4 mutations greatly increase HR.
Figure 4.
The Effects of abo4 Mutation and ABA on HR.
(A) The diagram of GUS reporter constructs in direct (1415, left) and inverted repeat (1406, right) used in this study (adapted from Lucht et al., 2002).
(B) Typical patterns of GUS staining in abo4-1, abo4-2, and the wild type carrying GUS reporter lines 1406 and 1415, respectively, under normal growth conditions.
(C) and (E) Quantitative analysis of HR in ∼100 plants of abo4-1 and the wild type carrying reporter line 1415 (C) and 1406 (E), respectively, under normal and ABA treatment conditions. The number of the GUS staining sectors was counted for each plant. The number of plants assayed was 115 for each assay. The proportions of plants showed frequency distribution of a given number of blue GUS spots in 1415 and 1406 populations.
(D) and (F) The average GUS spot number per seedling in ∼100 abo4-1 and wild-type seedlings under ABA and normal conditions. The number of plants assayed was 105 for each assay. Two independent experiments were performed. Error bars indicate ±sd. The abo4-1 showed a 48-fold increase in the number of blue sectors over the reporter line 1415 and 61-fold increase over the reporter line 1406 than the wild type under normal conditions.
ABA Treatment Tended to Increase HR in Both the Wild Type and abo4-1 Mutant
The influence of environment stresses, including pathogens, UV-B, UV-C, and x-rays, on somatic HR has been studied in plants (Schuermann et al., 2005; Boyko et al., 2006). However, the impact of key stress hormone ABA on HR has not yet been studied. Because of the sensitivity of abo4-1 to ABA, we examined HR after ABA treatment. Here, we used a relatively low concentration of ABA, as a slightly higher concentration of ABA greatly inhibited abo4-1 growth. Five-day-old seedlings were transferred to nutrient medium containing 5 μM ABA and grown for 7 d. Then, the seedlings were moved to nutrient medium and grown for 2 d. Because different plants have big variations in HR rate even if they are the same offspring, we used 100 to 120 seedlings and counted the GUS spots for each one and then obtained the average statistical number from these plants. Two independent experiments showed a similar HR increase in both the wild type and mutant. Figures 4C and 4E show the results of one typical experiment. The average number of blue sectors increased from 2.9 without ABA treatment to 7.1 with ABA treatment of the wild type, from 140.4 without ABA treatment to 159.4 with ABA treatment of abo4-1 in line 1415, from 1 without ABA treatment to 1.5 with ABA treatment of the wild type, and from 59.4 without ABA treatment to 85.5 with ABA treatment of abo4-1 in line 1406 (counting 100 to 120 seedlings for each line for each time). These results suggest that ABA treatment tended to increase HR frequency of both the wild type and the abo4-1 mutant.
ABA Treatment Increases DNA Breaks in abo4-1
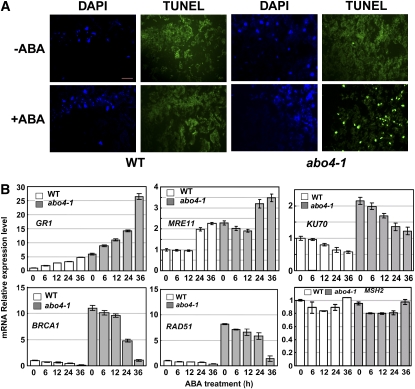
We tested whether the growth inhibition supersensitivity of abo4-1 to ABA is at least partially due to a lesion in DNA repair. DNA breaks were analyzed using the terminal deoxynucleotide transferase-mediated dUTP nick-end labeling (TUNEL) assay after ABA treatment. The TUNEL assay sensitively detects increased 3′-OH break ends (Gao and Showalter, 1999). Nuclei in leaves were simultaneously detected by 4′,6-diamidino-2-phenylindole staining. Under normal growth conditions, we did not detect any apparent TUNEL staining in both wild type and abo4-1 nuclei (Figure 5A, top). Here, we used 30 μM ABA to treat the seedlings, as this or higher concentrations of ABA apparently retarded the growth of mutant seedlings but does not greatly inhibit the growth of wild-type seedlings (the Col wild-type seedlings can tolerance as high as 150 μM ABA). Treatment with 30 μM ABA for 72 h greatly increased TUNEL staining in leaves of abo4-1 but did not have any measurable influence on TUNEL staining in the wild type (Figure 5A, bottom), suggesting that ABA treatment increases DNA breaks in the abo4-1 mutant.
Figure 5.
ABA Treatment Increases DNA Breaks and Upregulates GR1 and MRE11 Expression.
(A) The TUNEL assay under normal or ABA-treated conditions. Under the normal growth condition, TUNEL staining was negative in both the wild type and abo4-1 mutant (−ABA). Under ABA (30 μM for 3 d) treatment, only abo4-1 showed many TUNEL (bright green fluorescent signal) nuclei, but the wild type still showed negative TUNEL nuclei. The 4',6-diamidino-2-phenylindole (DAPI) staining (blue color) was used to indicate the nuclei.
(B) Comparison of GR1, MRE11, KU70, RAD51, BRCA1, and NSH2 expression by qRT-PCR under ABA treatment. The expression of GR1 and MRE11 was increased, and KU70, RAD51, and BRCA1 decreased by ABA treatment (30 μM) especially in the later time points in both abo4-1 and the wild type. The expression of MSH2 was not influenced by ABA. The transcripts of each gene under normal condition were used a comparison standard. The representative data were from one of three independent experiments showing similar results. Error bars indicate ±sd.
ABA Increases the Expression of Some DSB-Inducible Genes
Because ABA treatment led to increased DSBs in abo4-1 mutants, we analyzed the expression of GR1, RAD51, BRCA1, KU70, MRE11, and MSH2 after ABA treatment. ABA treatment increased the expression of GR1 and MRE11 but repressed the expression of KU70, BRCA1, and RAD51, especially at the later time points in both abo4-1 and the wild type (Figure 5B). However, we did not observe any clear expression changes of MSH2 (Figure 5B) and MSH6 (data not shown) by ABA, suggesting the ABA might not impose influence on the expression of the two genes. These results indicate that the molecular mechanisms regulating the expression of these genes by ABA and MMS are different.
abo4 Mutation Releases Transcriptional Gene Silencing of 35S-NPTII in the ros1 Mutant and TSI without Changing DNA Methylation
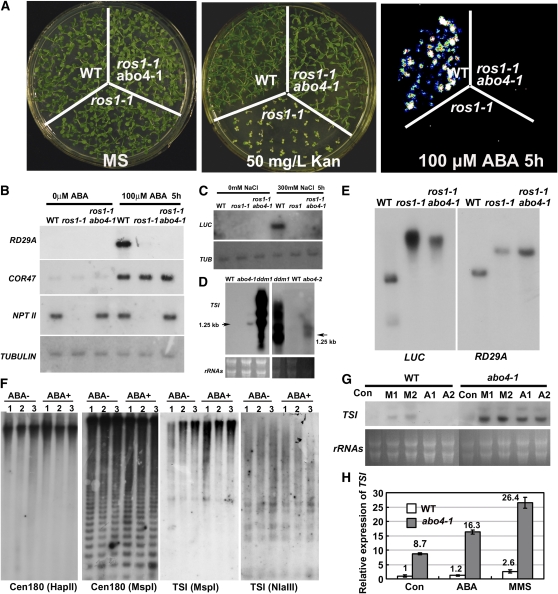
Mutation in the REPRESSOR OF SILENCING1 (ROS1) gene encoding a DNA demethylation enzyme led to the TGS of the originally activated 35S-NPTII, RD29A-LUC gene, and endogenous RD29A gene (Gong et al., 2002). Screening for ros1RD29A-LUC suppressors identified several genes that release the TGS of NPTII when mutated (Xia et al., 2006; Wang et al., 2007). To see whether the abo4/POL2a mutation is able to release the silenced 35S-NPT II in ros1RD29A-LUC, we crossed abo4-1 with ros1-1RD29A-LUC and made abo4-1 ros1-1RD29A-LUC double mutant carrying 35S-NPTII and RD29A-LUC transgenes. As shown in Figure 6A (middle), abo4-1 ros1-1RD29A-LUC seedlings were resistant to kanamycin, suggesting that the abo4 mutation reactivated the originally silenced 35S-NPTII in ros1-1RD29A-LUC mutant. RNA gel blot analysis confirmed the reexpression of NTPII gene in the abo4-1 ros1-1RD29A-LUC double mutant (Figure 6B). However, as in the ror1-1 ros1-1RD29A-LUC or tousled ros1RD29A-LUC mutant (Xia et al., 2006; Wang et al., 2007), the abo4-1 mutation did not release the TGS of RD29A-LUC and endogenous RD29A in ros1-1RD29A-LUC as analyzed by luminescence imaging assay (Figure 6A, left), RNA gel blot analysis of endogenous RD29A expression under ABA treatment (Figure 6B, RD29A), and LUC induction under NaCl treatment (Figure 6C). Under the same ABA and NaCl treatments, the expression of endogenous RD29A and RD29A-LUC in wild-typeRD29A-LUC was induced. As a positive control, a stress-inducible marker gene, COR47 (Gong et al., 2002), was also induced in both wild-typeRD29A-LUC and abo4-1. These results suggest the diversification of the TGS pathways at different loci (Gong et al., 2002; Xia et al., 2006; Wang et al., 2007).
Figure 6.
Releasing of TGS of 35S-NPTII in ros1 and TSI by abo4 Mutation Independent on DNA Methylation.
(A) abo4 mutation reactivated the silenced 35S-NPTII but did not affect the silenced RD29A-LUC in ros1-1 mutant. The seeds of wild-typeRD29A-LUC, ros1-1RD29A-LUC, and abo4-1 ros1-1RD29A-LUC carrying RD29A-LUC transgene were germinated and grown on MS medium (left plate) or MS medium supplemented with 50 mg/L kanamycin (middle plate) for 2 weeks. Both wild-typeRD29A-LUC and abo4-1 ros1-1RD29A-LUC seedlings showed the kanamycin-resistant phenotype from expression of the NPTII gene, while ros1-1RD29A-LUC seedlings were sensitive to kanamycin due to transgene silencing. Two-week-old seedlings were treated with 100 μM ABA for 5 h to induce the expression of RD29A-LUC. LUC expression was only detected in wild-typeRD29A-LUC but not in ros1-1RD29A-LUC and ros1-1 abo4-1RD29A-LUC double mutants (right plate), indicating that abo4 mutation did not reactivate the silenced RD29A-LUC in ros1-1.
(B) RNA gel blot analysis of endogenous RD29A and 35S-NPTII expression under ABA treatment. Total RNAs extracted from different seedlings treated or not treated by 100 μM ABA for 5 h were used for hybridization. A stress-inducible marker gene, COR47, was used as positive control to indicate that ABA treatment equally induced its expression in wild-typeRD29A-LUC, ros1-1RD29A-LUC, and ros1-1 abo4-1RD29A-LUC. RD29A expression was only detected in the wild type by ABA treatment, while NPTII transcripts were detected in both wild-typeRD29A-LUC and ros1-1 abo4-1RD29A-LUC. The endogenous RD29A and the transgene copy of 35S-NPTII were silenced in ros1-1RD29A-LUC. TUBULIN was used as a loading control.
(C) abo4 mutation did not release the TGS of RD29A-LUC. RNA gel blot analysis of RD29A-LUC expression in wild-typeRD29A-LUC, ros1-1RD29A-LUC, and ros1-1 abo4-1RD29A-LUC under 300 mM NaCl for 5 h. LUC transcripts were only detected in the wild type and not in ros1-1 and ros1-1 abo4-1.
(D) TSI expression was enhanced in the abo4-1 and abo4-2 mutants. RNA gel blot analysis of TSI expression in ddm1 was used as a control for comparison.
(E) abo4 mutation did not affect the DNA hypermethylation in the transgene and endogenous RD29A promoter in ros1-1RD29A-LUC. Genomic DNAs extracted from wild-typeRD29A-LUC, ros1-1RD29A-LUC, and ros1-1 abo4-1RD29A-LUC were digested with DNA methylation-sensitive enzymes MluI (AmCGmCGT) and BstUI (mCGmCG) as described (Gong et al., 2002) and hybridized with 32P-labeled LUC (for transgene RD29A promoter) or RD29A (for endogenous RD29A promoter).
(F) Methylation status of centromeric DNA (Cen180) or TSIs was neither influenced by abo4 mutations nor by ABA treatment. Genomic DNAs were extracted from (1) the wild type; (2) abo4-1; (3) abo4-2; ABA−, without ABA treatment; ABA+, seedlings were treated with 10 μM ABA for 7 d and digested by DNA methylation-sensitive restrictive enzymes HpaII (mCmCGG methylation), MspI (mCCGG methylation), and Nla III (mCATG methylation). The membranes were hybridized with 32P-labeled Cen 180 bp or TSI fragment, respectively.
(G) Both ABA and MMS treatment increased the TSI transcripts more in abo4-1 than the wild type. Seven-day-old seedlings were treated with MMS (M1, 50 ppm; M2, 100 ppm) for 7 d or treated with ABA (A1, 5 μM; A2, 10 μM) for 7 d. RNA gel blot analysis was performed using TSI as probe. RNAs were used as loading control.
(H) qRT-PCR analysis of TSI expression in the wild type and abo4-1 treated by 5 μM ABA or 50 ppm MMS for 7 d. Untreated wild type was used as a standard control. Three biological replications were performed for each experiment. Error bars indicate ±sd.
Several Arabidopsis mutants defective in DNA/chromatin assembly and replication showed instability of epigenetic states and release of TGS at the pericentromeric Athila retrotransposons, which are referred to as transcriptional silent information (TSI) (Steimer et al., 2000; Takeda et al., 2004; Elmayan et al., 2005; Kapoor et al., 2005; Inagaki et al., 2006; Wang and Liu, 2006; Xia et al., 2006; Zhu et al., 2006; Ramirez-Parra and Gutierrez, 2007; Wang et al., 2007). RNA gel blot analysis indicated that the abo4-1 and abo4-2 mutations released the TGS of TSIs, although the effect was much less than that of the ddm1 mutation (Figure 6D).
Because DNA methylation is the hallmark of heterochromatin and closely related to TGS, we measured the DNA methylation status in the TSI, centromeric DNA, and RD29A promoter regions by DNA gel blot analysis using DNA methylation-sensitive enzymes, as these regions are hypermethylated (Gong et al., 2002). DNA gel blot analysis indicated that abo4-1 did not affect DNA methylation in both foreign and endogenous RD29A promoters (Figure 6E) as well as in TSI and centromeric DNA regions (Figure 6F, ABA−). The DNA methylation status of TSI and centromeric DNA was not affected by ABA treatment (Figure 6F, ABA+). Like ror1/rpa2a and tsl1 mutations (Xia et al., 2006; Wang et al., 2007), abo4 mutation influences the epigenetic instability likely independent on DNA methylation.
Due to the sensitivity of abo4-1 to ABA, we examined TSI expression under ABA treatment. Interestingly, TSI transcripts were upregulated by ABA only in abo4-1 but were not detected in the wild type under our RNA gel blot conditions (Figure 6G). qRT-PCR analysis indicated that TSI expression was induced two times in abo4-1 by ABA, but only 1.2 times in the wild type (Figure 6H). However, MMS treatment induced TSI transcripts in both the wild type and abo4-1 but at a little higher level in abo4-1 (2.6 times in wild type and three times in abo4-1) (Figures 6G and 6H).
abo4 Mutants Show Early Flowering and Reduced Expression of FLOWERING LOCUS C and Increased Expression of FLOWERING LOCUS T with Changing Histone H3 Modifications
abo4-2 plants grown on MS medium were a little smaller than abo4-1 plants, and the two mutants were both noticeably smaller than the wild type when grown on agar plates or soil (Figures 7A and 7B). The F1 seedlings obtained from the crossing of abo4-1 with heterozygous abo4-2 showed abo4-1 and wild-type growth phenotype at a ratio of ∼1:1 (54:56) when grown in soil (Figure 7C). Under long-day conditions (16 h light/8 h dark), wild-type plants flowered at ∼24 ± 1 d after germination with a leaf number of ∼11 ± 1, while abo4-1 plants flowered at ∼17 ± 1 d after germination and had a leaf number of 8 ± 1 (counting 50 plants each time, three times, Figures 7B and 7D). The F1 heterozygous abo4-1 and abo4-2 plants flowered earlier than abo4-1 or abo4-2 mutants (Figure 7B). abo4-1 produced fewer seeds than the wild type (on an average 27 seeds per silique in abo4-1 and 40 seeds per silique in the wild type, counting 50 siliques, Figure 7E). Occasionally, some plants were found to have fasciated stems and abnormal flowers (Figure 7F, wild-type stem; Figure 7G, abo4-1 stem; Figure 7H, abo4-2 stem; Figure 7I, wild-type flower; Figure 7J, abo4-1 flower), suggesting stochastic incidents might have happened during development. The root meristem of abo4-1 is much shorter than that of the wild type (Figure 7K, wild type; Figure 7L, abo4-1). The shoot apical meristem (SAM) in the wild type exhibits three classical zones: the central zone in the top, the peripheral zone in the middle, and the ribzone in the bottom (Xia et al., 2006). However, it is hard to clearly differentiate these three zones in the SAM of abo4-1 (Figure 7M, wild type; Figure 7N, abo4-1).
Figure 7.
Growth phenotypes of abo4 mutants.
(A) Seedling comparison of wild-type, abo4-1, abo4-2, and abo4-1 abo4-2 heterozygous plants on MS agar plates. Please notice that abo4-2 shows a smaller phenotype, and abo4-1 abo4-2 heterozygous plants show abo4-1 phenotype.
(B) Comparison of wild-type, abo4-1, abo4-2, and abo4-1 abo4-2 heterozygous plants grown in soil.
(C) The F1 seedlings of heterozygous abo4-2 crossed with abo4-1. The ratio of abo4-1 to wild-type phenotype is ∼1:1 (54:56). abo4-2/abo4-1 mutants showing abo4-1 phenotypes are indicated with arrows.
(D) Comparison of the rosette leaves of wild-type (gl1), abo4-1(gl1), wild-type (Col), and abo4-2 (Col) before flowering. abo4-1 is in Col gl1 background, and abo4-2 is in Col background. Because there is no growth difference in Col gl1 and Col except for Col gl1 with no trichomes, we only used Col gl1 as control in later analysis.
(E) Comparison of siliques in wild-type, abo4-1, and abo4-2. Please notice that no seeds were set in abo4-2 siliques, and fewer seeds were set in abo4-1 than the wild type.
(F) to (H) Comparison of stems of the wild type (F), abo4-1 (G), and abo4-2 (H). Here, we only show abo4-1 and abo4-2 stems with fasciation phenotype. However, most of the abo4-1 and abo4-2 stems grew normally.
(I) and (J) Comparison of flowers of the wild type (I) and abo4-1 (J).
(K) and (L) Comparison of roots of the wild type (K) and abo4-1 (L). The root elongation zone is shorter in abo4-1 than the wild type. Bars = 5 mm.
(M) and (N) Comparison of SAMs of the wild type (M) and abo4-1 (N). Three classical zones were clearly observed in the SAM of the wild type but not in the SAM of abo4-1. Bar = 100 μm.
(O) qRT-PCR analysis of FLC, FT, and AP1 expression. In each case, RNAs from the wild type were used as a standard control for normalization for each RNA. The representative data were from one of three independent experiments showing similar results. The level of 18S rRNA was used as an internal control. Error bars indicate ±sd.
(P) ChIP analysis of histone H3K27me3 and H3K4me2 modifications in the first intron of FLC (the same primers are used as in Cao et al., 2008) and in the FT promoter region by qRT-PCR. Three independent immunoprecipitations were performed for each experiment. The immunoprecipitated DNA was quantified by real-time qRT-PCR. We used ACTIN as internal controls for the H3K4me2 level and TA3 for the H3K27me3 level. Error bars indicate ±sd.
The floral repressor FLOWERING LOCUS C (FLC) is a central player that quantitatively represses the floral pathway integrators FLOWERING LOCUS T (FT), SOC1, and LFY for delaying flowering (Boss et al., 2004; Amasino, 2005; Dennis and Peacock, 2007). We used qRT-PCR to check the expression of FLC, FT, and AP1, a downstream target of FT. Two-week-old seedlings grown on MS plates under long-day conditions (16 h light/8 h dark) were used to extract total RNAs. As shown in Figure 7O, the expression of FLC was lower in abo4-1 and abo4-2 compared to that in the wild type. However, the expression of FT and AP1 was much higher in abo4-1 and abo4-2 than that in the wild type. Because the expression of FLC and FT is mediated by histone modifications (Amasino, 2005), and ABO4 suggests to be involved in epigenetic inheritance, we performed a chromatin immunoprecipitation (ChIP) analysis using histone H3K4me2 and H3K27me3 antibodies to test whether the abo4 mutation affects epigenetic marks on FLC and FT. The dimethylation of Lys-4 of histone H3 (H3K4me2) is a hallmark of active genes in euchromatin, while the trimethylation of Lys-27 of histone H3 (H3K27me3) functions in the epigenetic silencing of FLC (Finnegan and Dennis, 2007; Greb et al., 2007). We found a clear increase of H3K27me3 in the first intron regions of FLC (FLC-P3 and FLC-P5, two fragments amplified by PCR; please see Cao et al., 2008), but only a little decrease in the promoter region of FT in abo4-1 compared to the wild type (Figure 7P, top). However, a noticeable enrichment of H3K4me2 was found in the promoter of FT but no change (FLC-P5) or a little decrease (FLC-P3) of H3K4me2 in the first intron regions of FLC (Figure 7P, bottom). These results suggest that abo4 mutation influences the histone H3 modifications around FLC gene body and FT promoter region.
DISCUSSION
Plant hormones act as important regulators of cell division by controlling the expression of cell cycle–related genes (Dewitte and Murray, 2003). Previous studies suggested that ABA inhibits cell division through an increase in the expression of the cyclin-dependent kinase inhibitor KRP1/ICK1 (Wang et al., 1998, 2000). However, inhibition of DNA synthesis by ABA treatment is considered to occur earlier than inhibition of either RNA or protein synthesis (Stewart and Smith, 1972), suggesting that ABA or ABA signaling might directly target existing DNA synthesis or cell cycle proteins to arrest cell division. Here, we showed that a mutation in ABO4/POL2a/TIL1 leads to Arabidopsis plants that are overly sensitive to ABA. However, the ABA sensitivity of abo4-1 was blocked by two of the ABA-insensitive mutations, abi1 and abi2, suggesting that ABA itself is not directly targeting the POL2a protein. We also found that the growth of fas1-101 (a mutation in chromatin assembly factor CAF-1) and ror1-3 (a T-DNA insertion mutation in replication protein A2) (Xia et al., 2006) was also sensitive to ABA, but a polymerase α mutant (isolated as a ros1-1 suppressor) (see Supplemental Figure 1 online) did not show ABA-sensitive phenotypes. ABA did not change the expression level of ICK between abo4-1 and the wild type (data not shown). It is conceivable that ABA signal is transduced to downstream target(s), which then affect the DNA synthesis and cell cycles through POL2a and other proteins.
abo4-1 mutants are supersensitive to DNA-damaging agents MMS and UV-B light, suggesting that in Arabidopsis, POL2a plays a crucial role in DNA repair. We found that abo4-1 constitutively expresses higher levels than the wild type of GR1, BRCA1, RAD51, MER11, and KU70, all of which are induced in response to the accumulation of DNA damage by MMS. These DNA damage–induced genes are thought to function in DNA repair, cell cycle checkpoints, or HR (Lafarge and Montane, 2003; Puizina et al., 2004; Inagaki et al., 2006). Interestingly, ABA treatment also induced more DNA breaks in abo4-1 than in the wild type and increased the expression of GR1 and MRE11, but suppressed the transcript levels of RAD51, BRCA1, and KU70. These data suggest that the plant responses to DNA damage caused by ABA are different from those caused by MMS because MMS highly induced the expression of all these genes.
In the abo4-1 and abo4-2 mutant, cell cycle progression is arrested at the G2/M-phase with a hallmark of high CYCB1;1 expression. Recent studies indicated that defective cell cycle progression was also observed in mutants, such as teb, bru1/tsk/mgo3, fas1, fas2, nrp1/nrp2, tsl, ror1/rap2a, and tso2-1 rnr2a-1, all of which are defective in DNA repair and replication (Ehsan et al., 2004; Endo et al., 2006; Inagaki et al., 2006; Wang and Liu, 2006; Zhu et al., 2006). The Arabidopsis POL2a mutation leads to the lengthening of the S-phase of the cell cycle with a high expression of H4b in Arabidopsis (this study; Jenik et al., 2005). It is possible that a POL2a lesion impairs DNA and chromatin replication, leading to more stalled replication forks during S-phase, which activates the S-phase checkpoint response. DNA synthesis and chromatin assembly must be precisely duplicated before cells enter into the mitotic stage. Retardation in the G2/M-phase with the accumulation of CYCB1;1-GUS expression could be important for cells to repair S-phase accumulated DNA damage. However, MMS, but not ABA treatment, increased the accumulation of CYCB1;1-GUS, suggesting that the DNA damages caused by MMS delay the cell cycle in G2/M-phase, which was not influenced by ABA. These results imply that the delayed cell cycle by ABA treatment did not happen in the G2/M but might be in the G1/S, which is supported by the previous results that ABA negatively influences the cell cycle progression at the G1/S transition but not in the G2/M in tobacco BY-2 cells or during maize (Zea mays) germination (Swiatek et al., 2002; Sanchez Mde et al., 2005). However, how ABA inhibits G1/S transition remains an enigma. Because ABA did not influence the cell cycle in G2/M in the abo4 mutants, this means that ABA does not directly restrain the ABO4/POL2a activity, but instead, ABA might target an unidentified protein(s) that might mainly function in the G1/S and manipulate the activities of ABO4/POL2a and other proteins, such as FAS1 and ROR1/RPA2A. However, this protein would not affect the polymerase α, as its mutation did not show an ABA phenotype. These results suggest that POL2a, FAS1, and ROR1/RPA2A are in a different pathway from polymerase α in the cell cycle.
To our surprise, we observed a higher number of HR events in abo4-1 than the wild type, suggesting that POL2a is involved in maintaining low HRs in the wild-type plants. This phenomenon contrasts with lower MAT switching with the yeast POL2 mutation (Holmes and Haber, 1999). In eukaryotes, DSBs are repaired by both NHEJ and the HR pathway. In the HR pathway, the undamaged sister chromatid is used as a DNA template to accurately repair the lesion strand in the late S-G2 phase. NHEJ mainly reconnects the broken DNA ends with no sequence similarity especially during the G1-phase when sister chromatids are not available (Kobayashi et al., 2008; Shrivastav et al., 2008). Yeast preferentially uses HR in DNA repair and possesses a high HR frequency, whereas plants and mammals prefer NHEJ repair. A model was proposed in which stalled replication forks were broken and repaired by HR before replication resumes when DNA replication encounters DNA damages (Rothstein et al., 2000). Studies in Arabidopsis indicate that greatly increased HR frequency was observed in fas1, fas2 (defective in genes encoding chromatin assembly factor 1 subunits p150 and p60, respectively) (Endo et al., 2006; Kirik et al., 2006), or cen2 (centrin2, defective in a calcium binding EF-hand protein) (Molinier et al., 2004); while mutation in BRU1 (a novel protein involved in DNA repair and replication) (Takeda et al., 2004), RAD50 (Gherbi et al., 2001), RAD17, or RAD9 (Heitzeberg et al., 2004) only slightly increases HR, whereas teb (a homolog of human DNA polymerase θ) (Inagaki et al., 2006), rad51c, or rad51d (a bacterial RecA recombinase homolog) (Durrant et al., 2007), mim (hypersensitive to MMS, irradiation, and MMC, a structural maintenance of chromosome protein) (Mengiste et al., 1999; Hanin et al., 2000), and ino80 (an ortholog of the SWI/SNF ATPase family) (Fritsch et al., 2004) mutations apparently decreased HR. We also observed that mutations in ROR1/RPA2A elevated the HR to a similar level as in abo4 mutants (see Supplemental Figure 2 online). It is possible that the POL2a works together with ROR1/RPA2A and FAS1 to regulate HR as these three proteins also coparticipate the DNA synthesis and DNA nucleotide excision repair (Gaillard et al., 1996; Lindahl et al., 1997; Pospiech and Syvaoja, 2003). It seems that defects of these protein activities lead to lengthening of S-phase, which accumulates DSBs and in turn activates cell cycle checkpoints and induces HR. We found that ABA treatment increased the HR frequency in both abo4-1 and wild-type plants. Interestingly, ABA downregulates the DNA end binding protein KU70, a key protein in NHEJ, which is consistent with a recent study on KU70 gene expression (Liu et al., 2008). It is possible that ABA-induced DNA breaks may partly result from the lower expression of KU70 because reduction of KU70 might lead to lower efficiently rejoining repair of DNA breaks in NHEJ. As NHEJ and HR pathways compete with each other during the cell cycle, the reduced NHEJ might increase the DNA breaks and activate the expression of MRE11 and GR1. GR1 has some homology with Ct IP, a mammalian protein phosphorylated by ATM (ataxia-telangiectasia mutated). MRE11 plays a crucial role in preventing the accumulation of DSBs resulting from stalled replication forks during DNA replication (Pichierri and Franchitto, 2004). Ct IP physically and functionally interacts with the MRE11 complex, and both are required for efficient HR (Sartori et al., 2007).
However, the high HR frequency observed in abo4-1 could have a more complex cause. A previous study indicates that MMS or other DNA breakage reagents are only able to increase HR severalfold (Lindhout et al., 2006) but not as high as in abo4-1 mutants. Recent studies of fas1 and fas2 suggest that the delayed chromatin assembly at S-phase would be another important factor for increasing HR in Arabidopsis (Endo et al., 2006; Kirik et al., 2006). However, mutations in ddm1, a chromatin remodeling SWI2/SNF2 greatly affecting the DNA methylation and chromatin structures in the whole genome in Arabidopsis (Gendrel et al., 2002), do not apparently affect HR (Shaked et al., 2006), and mutations in another SWI/SNF ATPase INO80 specifically reduce the HR (Fritsch et al., 2004). Considering that high HR mutants, including cen2, fas1, fas2, abo4, and ror1, are all involved in the NER pathway (Molinier et al., 2004; Endo et al., 2006; Kirik et al., 2006; this study), it is possible that another unidentified mechanism that might depend on the NER pathway, but be unique in plants, could regulate the HR. In any case, POL2a should be the core player because it directly synthesizes DNA, and defects of POL2a may lead to insufficient repair of damaged DNA and in turn increase HR.
The release of TGS in abo4-1 indicates that POL2a plays an essential role in maintaining epigenetic states in Arabidopsis. In budding yeast, a genetic screen for rDNA silencing defects identified many genes, including Pol ε (Ehrenhofer-Murray et al., 1999; Smith et al., 1999). It was also found that the mouse Pol ε B subunit interacts with SAP18, a protein associated with the transcriptional corepressor Sin3, which possesses histone deacetylase activity and functions in TGS (Wada et al., 2002). These results suggest the functional conservation of Pol ε in maintaining epigenetic states in different organisms. We found that abo4 mutation activated two originally silenced loci, including TSI and 35S-NPTII in the ros1-1RD29A-LUC mutant. However, the epigenetic regulation by ABO4/POL2a seems independent of DNA methylation as checked in several hypermethylation loci by DNA gel blot analysis. Previous studies on fas1, fas2, ror1, bru1, tousled, and mom1 (Takeda et al., 2004; Elmayan et al., 2005; Kapoor et al., 2005; Vaillant et al., 2006; Xia et al., 2006; Wang et al., 2007) indicate that the TGS regulated by these genes are independent of DNA methylation, which is different from the TGS pathway mediated by RNA-direct DNA methylation and heterochromatin formation (Matzke et al., 2007). However, MOM1 is a chromatin remodeling factor not participating in DNA repair pathway (Shaked et al., 2006) and should be in the different pathway from FAS1, FAS2, ROR1/RPA2, BRU1, TOUSLED, and ABO4/POL2a in mediating TGS. These results suggest that several different pathways mediate the TGS in plants. Studies on ROR1/RPA2A and TOUSLED indicate that the released TGS of 35S-NPTII in ror1 and tousled mutants might be due to a change in histone modifications (Xia et al., 2006; Wang et al., 2007). Here, we found that abo4 mutations influenced flowering time through manipulating the histone modifications in FLC and FT loci. In the icu2-1 study, the authors find that ICU2 DNA polymerase α interacts with TFL2/LHP1 to regulate chromatin-mediated gene expression (Barrero et al., 2007). Together, these studies indicate that DNA replication machinery is involved in chromatin-mediated gene expression in plants.
Our findings that ABA increases HR and activates the expression of retrotransposons are biologically significant because somatic recombination and transposon activation may be important mechanisms for restructuring the genome as an adaptive response to environmental challenge (Grandbastien, 1998). Changes to the genome of somatic cells can be transmitted to offspring and may produce new resistance traits during the evolutionary process (Molinier et al., 2006). Because ABA accumulation is induced by environmental stresses, we suggest that ABA might affect genome stability through the DNA replisome and allow plants to effectively adapt to environmental stresses over the course of evolution. However, further studies are needed to dissect the molecular mechanisms of this process.
METHODS
Plant Materials and Growth Conditions
We used Arabidopsis thaliana Col accession with a gl1 mutation resulting in no trichomes on the plants. The F2 ethyl methanesulfonate–mutagenized seeds were used for screening ABA-sensitive mutants. Root growth was analyzed by the root bending assay as described previously (Wu et al., 1996). Briefly, 5-d-old seedlings with 1- to 1.5-cm-long roots were transferred from the vertical agar plates, one by one, onto a second agar medium that contained 20 to 30 μM ABA. The seedlings were arranged in rows, and the plates were oriented vertically with the roots pointing upward. After growth for another 5 to 7 d, the putative mutants that showed inhibited root growth were picked up. The root growth phenotypes of putative mutants were rechecked in the next generation, genetic analysis was performed by backcrossing with the wild type, and the ABA-sensitive phenotype of abo4-1 was determined. All seedlings were cultured on agar-solidified MS medium supplemented with 3% (w/v) sucrose at 21°C under 24 h of light. Plant growth conditions in soil are the same as described previously (Xia et al., 2006).
Mapping-Based Cloning of ABO4
abo4-1 was crossed with an Arabidopsis Landsberg accession, and the F2 population (1802 samples) was used for mapping-based cloning by analyzing recombination events using simple sequence length polymorphism markers. The abo4 mutation was first mapped to chromosome 1 between F12K11 and F14J9. Then, markers F24B9, T23G18, and T27G7 were used to narrow down the abo4-1 mutation to the BAC clone T23G18. Sequencing the candidate gene At1g08260 identified a G-to-A point mutation. A Salk T-DNA insertion line SALK_096341 (abo4-2) was requested from The Arabidopsis Information Resource. We isolated genomic DNA from the progeny of SALK lines and performed PCR analysis using primer pair LP 5′-ACCTTCTCCTTGATTTCGTCC-3′ and RP 5′-CTATGGCTCTTTATGGGTTGC-3′, which covered the T-DNA insertion region, to find the homozygous lines. Because abo4-2 is sterile in both male and female, we used the heterozygous T-DNA plants (confirmed by PCR using primer pair RP 5′-CTATGGCTCTTTATGGGTTGC-3′ and TF 5′-ATTTTGCCGATTTCGGAAC-3′) to cross with abo4-1 (confirmed by Derived Cleaved Amplified Polymorphic Sequences using primer pair 5′-GGTCATGTAGAGTGCCTTGAAGGTG-3′ and 5′-GAATCATGCACACAAGAATGGGAAGC-3′; cut with HphI) to make heterozygous mutants that contained both the abo4-1 and abo4-2 mutations. The heterozygous plants of abo4-1 and abo4-2 showed abo4-1 phenotypes.
Recombination Assays
The recombination reporter lines 1406 (direct repeat line) and 1415 (inverted repeat line) (Col accession) (Gherbi et al., 2001) were crossed with abo4-1 and homozygous plants for both GUS reporter and abo4-1 or for GUS reporter only (control plants), and samples were selected from F3 plants and used for GUS activity analysis. The GUS spot numbers, each of which indicates a recombination event, were then determined under a dissecting microscope. For ABA treatment, 5-d-old seedlings grown on MS medium were transferred to MS medium containing 5 μM ABA and grown for another 7 d. Then the seedlings were used for the GUS activity assay. GUS staining was done as described previously (Gong et al., 2002). Approximately 100 individual seedlings were analyzed for each sample.
DNA Damage Sensitivity Assays
For DNA damage assays, seeds were directly planted on MS medium containing different concentrations of MMS and grown for 2 weeks after germination, or 2-d-old seedlings were transferred to liquid MS medium supplemented with different concentrations of MMS and grown for another 10 d before taking pictures. For UV-B treatment, 5-d-old seedlings were exposed to different levels of UV-B light (UV 256 nm in a Stratalinker) and then were kept in the dark for 2 d before being removed to light for growth for 3 kJ/m2 or directly grown under light (for 1 kJ/m2 or 5 kJ/m2). After 5 d in light, the pictures were taken. Each experiment was repeated at least three times independently. The representative results from one experiment are shown. The TUNEL assay was conducted as described previously (Wang and Liu, 2006) using the In Situ Cell Death Detection Kit-Fluorescein (Roche Diagnostics). Five-day-old seedlings grown on MS medium were transferred to MS medium containing 30 μM ABA for 3 d before performing TUNEL staining. Images were captured with an Olympus BX51 microscope equipped with fluorescein isothiocyanate and UV light filters and a COOL SNAP HQ photometrics CCD camera.
RNA Gel Blot Analysis
Shoots of 2-week-old seedlings were dipped into solution containing 30 μM ABA for different times. For TSI expression analysis, 7-d-old seedlings were treated with MMS (50 and 100 ppm) for 7 d or treated with ABA (5 and 10 μM) for 7 d. For gene expression analysis in abo4-1 ros1-1, 2-week-old seedlings were treated with 100 μM ABA or 300 mM NaCl for 5 h. Total RNAs was extracted and hybridized with different probes as described previously (Xia et al., 2006). Twenty micrograms of total RNA from each sample was separated on 1.2% (w/v) agarose formaldehyde gels. Tubulin was used as a loading control. Probes were amplified with primer pairs provided in Supplemental Table 1 online.
DNA Gel Blot Analysis
DNA methylation analysis in RD29A promoter, TSI, and centromere DNA region was performed as described previously (Gong et al., 2002).
CYCB1;1∷GUS Assay
The Arabidopsis Col plants carrying CYCB1;1∷GUS were crossed with abo4-1. The homozygous plants for both CYCB1;1∷GUS and abo4-1 or only for CYCB1;1∷GUS were selected from F3 and used for GUS staining (Colon-Carmona et al., 1999; Endo et al., 2006). For ABA or MMS treatment, 5-d-old seedlings were transferred to MS medium containing 30 μM ABA or 100 ppm MMS and grown for another 4 d before GUS staining.
Real-Time qRT-PCR
Approximately 2 μg of total RNA, extracted from seedlings under different conditions and digested with RNase-free DNase I, was used for RT-PCR, employing oligo(dT) primers with M-MLV (Invitrogen) in a 50-μL reaction. One microliter of the RT reaction was used as template to determine transcript levels of the tested genes on a PTC-200 DNA Engine Cycler (MJ Research) with Chromo4 Detector in 25-μL reactions. The levels of 18S rRNA were used as a reference, and the values given are expressed as a ratio of the values in the wild type. Three biological replications were performed for each experiment. The values shown represent averages of triplicate assays for each RT sample. PCR conditions were as follows: 5 min at 95°C (one cycle), 30 s at 95°C, 30 s at 58 to 61°C, and 60 s at 72°C (30 cycles). The primers and detail annealing temperatures used for real-time PCR are provided in Supplemental Table 1 online.
ChIP
ChIP was performed as described previously (Xia et al., 2006) using 2-week-old seedlings grown on MS plates under long-day conditions (16 h light/8 h dark). Anti-H3K4me2 and anti-H3K27me3 antibodies were purchased from Upstate Biotechnology. We used ACTIN as internal controls for the H3K4me2 level and TA3 for the H3K27me3 level. PCR conditions are same as above using 58°C as annealing temperature. The primers used for ChIP are provided in Supplemental Table 1 online.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number NM_001123775 (ABO4/POL2A/TIL1/At1g08260).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of ABA-Sensitive Phenotypes in ror1-3, fas1-101, and pol α Mutants.
Supplemental Figure 2. Homologous Recombination Patterns in ror1-3 (1415) and ror1-3 (1406).
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the ABRC (Columbus, OH) for providing the T-DNA lines, David Schuermann and Barbara Hohn for providing GUS recombination reporter lines, Peter Doerner for CYCB1;1∷GUS, and Jian-Kang Zhu for critically reading the manuscript and giving valuable comments for improving the manuscript. This work was supported by the National Nature Science Foundation of China (30630004, 90717004, and 30421002) and 111 project to Z.G.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhizhong Gong (gongzz@cau.edu.cn).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Aboussekhra, A., Biggerstaff, M., Shivji, M.K., Vilpo, J.A., Moncollin, V., Podust, V.N., Protic, M., Hubscher, U., Egly, J.M., and Wood, R.D. (1995). Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80 859–868. [DOI] [PubMed] [Google Scholar]
- Amasino, R.M. (2005). Vernalization and flowering time. Curr. Opin. Biotechnol. 16 154–158. [DOI] [PubMed] [Google Scholar]
- Barrero, J.M., Gonzalez-Bayon, R., del Pozo, J.C., Ponce, M.R., and Micol, J.L. (2007). INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19 2822–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J.S., and Dean, C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16 (suppl.): S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko, A., Hudson, D., Bhomkar, P., Kathiria, P., and Kovalchuk, I. (2006). Increase of homologous recombination frequency in vascular tissue of Arabidopsis plants exposed to salt stress. Plant Cell Physiol. 47 736–742. [DOI] [PubMed] [Google Scholar]
- Cao, Y., Dai, Y., Cui, S., and Ma, L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20 2586–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan, M.B., Li, G., Bishop, K.J., and Hall, T.C. (2003). S phase progression is required for transcriptional activation of the beta-phaseolin promoter. J. Biol. Chem. 278 45397–45405. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Dennis, E.S., and Peacock, W.J. (2007). Epigenetic regulation of flowering. Curr. Opin. Plant Biol. 10 520–527. [DOI] [PubMed] [Google Scholar]
- Dewitte, W., and Murray, J.A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54 235–264. [DOI] [PubMed] [Google Scholar]
- Dianov, G.L., Sleeth, K.M., Dianova, I.I., and Allinson, S.L. (2003). Repair of abasic sites in DNA. Mutat. Res. 531 157–163. [DOI] [PubMed] [Google Scholar]
- Doerner, P., Jorgensen, J.E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380 520–523. [DOI] [PubMed] [Google Scholar]
- Doutriaux, M.P., Couteau, F., Bergounioux, C., and White, C. (1998). Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol. Gen. Genet. 257 283–291. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., Wang, S., and Dong, X. (2007). Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA 104 4223–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray, A.E., Kamakaka, R.T., and Rine, J. (1999). A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics 153 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsan, H., Reichheld, J.P., Durfee, T., and Roe, J.L. (2004). TOUSLED kinase activity oscillates during the cell cycle and interacts with chromatin regulators. Plant Physiol. 134 1488–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan, T., Proux, F., and Vaucheret, H. (2005). Arabidopsis RPA2: A genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. 15 1919–1925. [DOI] [PubMed] [Google Scholar]
- Emmanuel, E., Yehuda, E., Melamed-Bessudo, C., Avivi-Ragolsky, N., and Levy, A.A. (2006). The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep. 7 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, M., Ishikawa, Y., Osakabe, K., Nakayama, S., Kaya, H., Araki, T., Shibahara, K., Abe, K., Ichikawa, H., Valentine, L., Hohn, B., and Toki, S. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25 5579–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah, J.A., Cromie, G., Steiner, W.W., and Smith, G.R. (2005). A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics 169 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W., and D'Urso, G. (2001). Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 21 4495–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., and Dennis, E.S. (2007). Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17 1978–1983. [DOI] [PubMed] [Google Scholar]
- Fritsch, O., Benvenuto, G., Bowler, C., Molinier, J., and Hohn, B. (2004). The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell 16 479–485. [DOI] [PubMed] [Google Scholar]
- Gaillard, P.H., Martini, E.M., Kaufman, P.D., Stillman, B., Moustacchi, E., and Almouzni, G. (1996). Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor I. Cell 86 887–896. [DOI] [PubMed] [Google Scholar]
- Gao, M., and Showalter, A.M. (1999). Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J. 19 321–331. [DOI] [PubMed] [Google Scholar]
- Garg, P., and Burgers, P.M. (2005). DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40 115–128. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Gherbi, H., Gallego, M.E., Jalut, N., Lucht, J.M., Hohn, B., and White, C.I. (2001). Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Z., Morales-Ruiz, T., Ariza, R.R., Roldan-Arjona, T., David, L., and Zhu, J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111 803–814. [DOI] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien, M.A. (1998). Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3 181–187. [Google Scholar]
- Greb, T., Mylne, J.S., Crevillen, P., Geraldo, N., An, H., Gendall, A.R., and Dean, C. (2007). The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 17 73–78. [DOI] [PubMed] [Google Scholar]
- Green, E.M., Antczak, A.J., Bailey, A.O., Franco, A.A., Wu, K.J., Yates III, J.R., and Kaufman, P.D. (2005). Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth, A., Rocha, W., Verreault, A., and Almouzni, G. (2007). Chromatin challenges during DNA replication and repair. Cell 128 721–733. [DOI] [PubMed] [Google Scholar]
- Hanin, M., Mengiste, T., Bogucki, A., and Paszkowski, J. (2000). Elevated levels of intrachromosomal homologous recombination in Arabidopsis overexpressing the MIM gene. Plant J. 24 183–189. [DOI] [PubMed] [Google Scholar]
- Heitzeberg, F., Chen, I.P., Hartung, F., Orel, N., Angelis, K.J., and Puchta, H. (2004). The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 38 954–968. [DOI] [PubMed] [Google Scholar]
- Holmes, A.M., and Haber, J.E. (1999). Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96 415–424. [DOI] [PubMed] [Google Scholar]
- Inagaki, S., Suzuki, T., Ohto, M.A., Urawa, H., Horiuchi, T., Nakamura, K., and Morikami, A. (2006). Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 18 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P.D., Jurkuta, R.E., and Barton, M.K. (2005). Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell 17 3362–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, A., Agarwal, M., Andreucci, A., Zheng, X., Gong, Z., Hasegawa, P.M., Bressan, R.A., and Zhu, J.K. (2005). Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr. Biol. 15 1912–1918. [DOI] [PubMed] [Google Scholar]
- Kirchmaier, A.L., and Rine, J. (2001). DNA replication-independent silencing in S. cerevisiae. Science 291 646–650. [DOI] [PubMed] [Google Scholar]
- Kirik, A., Pecinka, A., Wendeler, E., and Reiss, B. (2006). The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18 2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, J., Iwabuchi, K., Miyagawa, K., Sonoda, E., Suzuki, K., Takata, M., and Tauchi, H. (2008). Current topics in DNA double-strand break repair. J. Radiat. Res. (Tokyo) 49 93–103. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5 33–36. [DOI] [PubMed] [Google Scholar]
- Lafarge, S., and Montane, M.H. (2003). Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res. 31 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleuriel, J., Degroote, F., Depeiges, A., and Picard, G. (2007). Impact of the loss of AtMSH2 on double-strand break-induced recombination between highly diverged homeologous sequences in Arabidopsis thaliana germinal tissues. Plant Mol. Biol. 63 833–846. [DOI] [PubMed] [Google Scholar]
- Leube, M.P., Grill, E., and Amrhein, N. (1998). ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Lett. 424 100–104. [DOI] [PubMed] [Google Scholar]
- Li, L., Jean, M., and Belzile, F. (2006). The impact of sequence divergence and DNA mismatch repair on homeologous recombination in Arabidopsis. Plant J. 45 908–916. [DOI] [PubMed] [Google Scholar]
- Li, Y.C., Cheng, T.H., and Gartenberg, M.R. (2001). Establishment of transcriptional silencing in the absence of DNA replication. Science 291 650–653. [DOI] [PubMed] [Google Scholar]
- Lindahl, T., Karran, P., and Wood, R.D. (1997). DNA excision repair pathways. Curr. Opin. Genet. Dev. 7 158–169. [DOI] [PubMed] [Google Scholar]
- Lindhout, B.I., Pinas, J.E., Hooykaas, P.J., and van der Zaal, B.J. (2006). Employing libraries of zinc finger artificial transcription factors to screen for homologous recombination mutants in Arabidopsis. Plant J. 48 475–483. [DOI] [PubMed] [Google Scholar]
- Liu, P.F., Chang, W.C., Wang, Y.K., Chang, H.Y., and Pan, R.L. (2008). Signaling pathways mediating the suppression of Arabidopsis thaliana Ku gene expression by abscisic acid. Biochim. Biophys. Acta 1779 164–174. [DOI] [PubMed] [Google Scholar]
- Lucht, J.M., Mauch-Mani, B., Steiner, H.Y., Metraux, J.P., Ryals, J., and Hohn, B. (2002). Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 30 311–314. [DOI] [PubMed] [Google Scholar]
- Matzke, M., Kanno, T., Huettel, B., Daxinger, L., and Matzke, A.J. (2007). Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 10 512–519. [DOI] [PubMed] [Google Scholar]
- Mengiste, T., Revenkova, E., Bechtold, N., and Paszkowski, J. (1999). An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 18 4505–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. [DOI] [PubMed] [Google Scholar]
- Moldovan, G.L., Pfander, B., and Jentsch, S. (2007). PCNA, the maestro of the replication fork. Cell 129 665–679. [DOI] [PubMed] [Google Scholar]
- Molinier, J., Ramos, C., Fritsch, O., and Hohn, B. (2004). CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell 16 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier, J., Ries, G., Zipfel, C., and Hohn, B. (2006). Transgeneration memory of stress in plants. Nature 442 1046–1049. [DOI] [PubMed] [Google Scholar]
- Ohya, T., Kawasaki, Y., Hiraga, S., Kanbara, S., Nakajo, K., Nakashima, N., Suzuki, A., and Sugino, A. (2002). The DNA polymerase domain of pol(epsilon) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 277 28099–28108. [DOI] [PubMed] [Google Scholar]
- Pichierri, P., and Franchitto, A. (2004). Werner syndrome protein, the MRE11 complex and ATR: Menage-a-trois in guarding genome stability during DNA replication? Bioessays 26 306–313. [DOI] [PubMed] [Google Scholar]
- Pospiech, H., and Syvaoja, J.E. (2003). DNA polymerase epsilon - More than a polymerase. ScientificWorldJournal 3 87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina, J., Siroky, J., Mokros, P., Schweizer, D., and Riha, K. (2004). Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell, Z.F., Isoz, I., Lundstrom, E.B., Johansson, E., and Kunkel, T.A. (2007). Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra, E., and Gutierrez, C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidt, W., Wurz, R., Wanieck, K., Chu, H.H., and Puchta, H. (2006). A homologue of the breast cancer-associated gene BARD1 is involved in DNA repair in plants. EMBO J. 25 4326–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, P.L., Benning, G., and Grill, E. (1998). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421 185–190. [DOI] [PubMed] [Google Scholar]
- Ronceret, A., Guilleminot, J., Lincker, F., Gadea-Vacas, J., Delorme, V., Bechtold, N., Pelletier, G., Delseny, M., Chaboute, M.E., and Devic, M. (2005). Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 44 223–236. [DOI] [PubMed] [Google Scholar]
- Rothstein, R., Michel, B., and Gangloff, S. (2000). Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14 1–10. [PubMed] [Google Scholar]
- Sanchez Mde, L., Gurusinghe, S.H., Bradford, K.J., and Vazquez-Ramos, J.M. (2005). Differential response of PCNA and Cdk-A proteins and associated kinase activities to benzyladenine and abscisic acid during maize seed germination. J. Exp. Bot. 56 515–523. [DOI] [PubMed] [Google Scholar]
- Sartori, A.A., Lukas, C., Coates, J., Mistrik, M., Fu, S., Bartek, J., Baer, R., Lukas, J., and Jackson, S.P. (2007). Human CtIP promotes DNA end resection. Nature 450: 509–514. [DOI] [PMC free article] [PubMed]
- Schuermann, D., Molinier, J., Fritsch, O., and Hohn, B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21 172–181. [DOI] [PubMed] [Google Scholar]
- Scully, R., Xie, A., and Nagaraju, G. (2004). Molecular functions of BRCA1 in the DNA damage response. Cancer Biol. Ther. 3 521–527. [DOI] [PubMed] [Google Scholar]
- Shaked, H., Avivi-Ragolsky, N., and Levy, A.A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 221–227. [DOI] [PubMed] [Google Scholar]
- Shivji, M.K., Podust, V.N., Hubscher, U., and Wood, R.D. (1995). Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 34 5011–5017. [DOI] [PubMed] [Google Scholar]
- Shrivastav, M., De Haro, L.P., and Nickoloff, J.A. (2008). Regulation of DNA double-strand break repair pathway choice. Cell Res. 18 134–147. [DOI] [PubMed] [Google Scholar]
- Smith, J.S., Caputo, E., and Boeke, J.D. (1999). A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, G.R., and Smith, H. (1972). Effects of abscisic acid on nucleic acid synthesis and the induction of nitrate reductase in Lemna polyrhiza. J. Exp. Bot. 23 875–885. [Google Scholar]
- Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Mittelsten Scheid, O., and Paszkowski, J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek, A., Lenjou, M., Van Bockstaele, D., Inze, D., and Van Onckelen, H. (2002). Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol. 128 201–211. [PMC free article] [PubMed] [Google Scholar]
- Takeda, S., Tadele, Z., Hofmann, I., Probst, A.V., Angelis, K.J., Kaya, H., Araki, T., Mengiste, T., Mittelsten Scheid, O., Shibahara, K., Scheel, D., and Paszkowski, J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18 782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Adachi, Y., Chiba, K., Oguchi, K., and Takahashi, H. (2002). Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: Evidence for a role in the repair of DNA double-strand breaks. Plant J. 29 771–781. [DOI] [PubMed] [Google Scholar]
- Tsubota, T., Tajima, R., Ode, K., Kubota, H., Fukuhara, N., Kawabata, T., Maki, S., and Maki, H. (2006). Double-stranded DNA binding, an unusual property of DNA polymerase epsilon, promotes epigenetic silencing in Saccharomyces cerevisiae. J. Biol. Chem. 281 32898–32908. [DOI] [PubMed] [Google Scholar]
- Vaillant, I., Schubert, I., Tourmente, S., and Mathieu, O. (2006). MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 7 1273–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, M., Miyazawa, H., Wang, R.S., Mizuno, T., Sato, A., Asashima, M., and Hanaoka, F. (2002). The second largest subunit of mouse DNA polymerase epsilon, DPE2, interacts with SAP18 and recruits the Sin3 co-repressor protein to DNA. J. Biochem. (Tokyo) 131 307–311. [DOI] [PubMed] [Google Scholar]
- Wang, C., and Liu, Z. (2006). Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15 501–510. [DOI] [PubMed] [Google Scholar]
- Wang, H., Zhou, Y., Gilmer, S., Whitwill, S., and Fowke, L.C. (2000). Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24 613–623. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Liu, J., Xia, R., Wang, J., Shen, J., Cao, R., Hong, X., Zhu, J.K., and Gong, Z. (2007). The protein kinase TOUSLED is required for maintenance of transcriptional gene silencing in Arabidopsis. EMBO Rep. 8 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.J., Ding, L., and Zhu, J.K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R., Wang, J., Liu, C., Wang, Y., Wang, Y., Zhai, J., Liu, J., Hong, X., Cao, X., Zhu, J.K., and Gong, Z. (2006). ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 18 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Schumaker, K.S., and Zhu, J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Dong, A., Meyer, D., Pichon, O., Renou, J.P., Cao, K., and Shen, W.H. (2006). Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.