Abstract
Plasmodesmata (Pds) traverse the cell wall to establish a symplastic continuum through most of the plant. Rapid and reversible deposition of callose in the cell wall surrounding the Pd apertures is proposed to provide a regulatory process through physical constriction of the symplastic channel. We identified members within a larger family of X8 domain–containing proteins that targeted to Pds. This subgroup of proteins contains signal sequences for a glycosylphosphatidylinositol linkage to the extracellular face of the plasma membrane. We focused our attention on three closely related members of this family, two of which specifically bind to 1,3-β-glucans (callose) in vitro. We named this family of proteins Pd callose binding proteins (PDCBs). Yellow fluorescent protein-PDCB1 was found to localize to the neck region of Pds with potential to provide a structural anchor between the plasma membrane component of Pds and the cell wall. PDCB1, PDCB2, and PDCB3 had overlapping and widespread patterns of expression, but neither single nor combined insertional mutants for PDCB2 and PDCB3 showed any visible phenotype. However, increased expression of PDCB1 led to an increase in callose accumulation and a reduction of green fluorescent protein (GFP) movement in a GFP diffusion assay, identifying a potential association between PDCB-mediated callose deposition and plant cell-to-cell communication.
INTRODUCTION
In plants, cell-to-cell communication through plasmodesmata (Pds) is vital, with many diverse areas of plant biology being dependent upon non-cell-autonomous functions. Hence, numerous transcription factors have functions that are spatially distinct from their sites of synthesis (Lucas et al., 1995; Gallagher et al., 2004; Kim et al., 2005; Kurata et al., 2005), and small RNA species involved in RNA silencing move symplastically from cell to cell (Yoo et al., 2004; Gomez et al., 2005). Pds are channels that traverse the cell wall to establish a symplastic continuum through most of the plant. In doing so, they provide a path for the pathogenic invasion of tissues by viruses (Wolf et al., 1989; Escobar et al., 2003; Howard et al., 2004; Canto and Palukaitis, 2005) and potentially for biotrophic invasion by some fungi (Kankanala et al., 2007). Despite their importance, very little is known about either the constituent components of Pds or the mechanisms that regulate both their size and the specificity of molecules that traffic through them.
The most detailed description of Pds has come from high-resolution electron microscopy (Burgess, 1971; Olesen, 1979; Ding et al., 1992a; Turner et al., 1994; Overall and Blackman, 1996; Bayer et al., 2004). Pds are plasma membrane–lined channels with a central axial component derived from the endoplasmic reticulum, called the desmotubule. The space between these membranous components appears to be interrupted by spoke-like structures that leave microchannels through which molecular trafficking is proposed to occur (Burgess, 1971; Ding et al., 1992a; Cantrill et al., 1999). How this flow is regulated is not fully understood. However, a role for the cell wall at the apertures, or neck regions, through the deposition of the 1,3-β-glucan, callose, has been proposed (Hughes and Gunning, 1980; Levy et al., 2007a, 2007b).
Callose is deposited on the cell plate during division (Samuels et al., 1995), around the neck regions of Pds in mature and immature tissues (Turner et al., 1994; Bayer et al., 2004), as an integral process in the symplastic isolation during cotton fiber elongation (Ruan et al., 2004), and during pollen maturation (Meikle et al., 1991; Parre and Geitmann, 2005). Callose deposition at Pds is stimulated by physical and physiological stresses (Hughes and Gunning, 1980; Beffa and Meins, 1996; Iglesias and Meins, 2000; Sivaguru et al., 2000; Rinne et al., 2005; Ueki and Citovsky, 2005; Maeda et al., 2006) and is proposed to provide a mechanism for regulating Pd flux (Botha and Cross, 2000; Iglesias and Meins, 2000; Levy et al., 2007a). Hence, for example, plants with reduced accumulation of the glycolytic enzyme 1,3-β-glucanase had increased callose accumulation and a reduction in the experimental molecular size exclusion limit. This was demonstrated by a reduced ability of green fluorescent protein (GFP) to diffuse through Pds (Levy et al., 2007b) and reduced susceptibility to virus invasion (Iglesias and Meins, 2000).
One of the major hurdles to our increased understanding of Pd function has been our limited knowledge of its protein constituents. Various approaches have been taken to identify such constituents, with limited success (reviewed in Oparka, 2004; Maule, 2008). The proteins identified include cytoskeletal elements (e.g., actin and myosin VIII) (Ding et al., 1996; Blackman and Overall, 1998; Reichelt et al., 1999), proteins associated with the endoplasmic reticulum (e.g., calreticulin) (Baluska et al., 1999), a protein kinase 1 that phosphorylates Tobacco mosaic virus movement protein (Lee et al., 2005), a 1,3-β-glucanase (Levy et al., 2007b), a class 1 reversibly glycosylated protein (Sagi et al., 2005), and, recently, a receptor-like transmembrane protein located on the plasma membrane in Pds (Thomas et al., 2008). In this article, we report the identification of a family of small glycosylphosphatidylinositol (GPI)-linked proteins that exhibit specific callose binding activity in vitro and that localize to Pds as fluorescent protein fusions. Although insertional knockout mutants of some of the genes showed no adverse phenotypes, overexpression of some of the proteins in Arabidopsis thaliana resulted in an increase in callose concentration around Pds and a reduction in molecular diffusion from cell to cell. Hence, these proteins have the potential to provide an anchor between Pds and callose in the neighboring region of the cell wall and may identify an unexpected factor in the regulation of callose deposition and cell-to-cell communication.
RESULTS
At5g61130 Encodes a Plasmodesmal Protein
Previous work (Bayer et al., 2006) identified a series of membrane-associated proteins from cell wall preparations of Arabidopsis suspension cells. As part of a survey of the subcellular targeting of these proteins as fusions to fluorescent markers, we have identified At5g61130 as a gene encoding a novel Pd protein. Because of its associated callose binding activity, we have named this protein Pd callose binding protein 1 (PDCB1).
At5g61130 encodes a GPI-anchor protein (Elortza et al., 2003) with N- and C-terminal domains that are cleaved during processing from the preprotein. To establish the subcellular location of PDCB1, the yellow fluorescent protein (YFP) coding sequence (pCitrine; Tian et al., 2004) was cloned between sequences encoding the N-terminal secretion signal (after amino acid 19) and the region for the mature protein (Figure 1). Since GPI-anchor proteins are usually covalently attached to the GPI moiety in the endoplasmic reticulum and become located on the extracellular face of the plasma membrane (Lodish et al., 2000), YFP was selected owing to its greater stability in the more acidic environment of the apoplast (Griesbeck et al., 2001). Transgenic expression of YFP-PDCB1 from the cauliflower mosaic virus (CaMV) 35S promoter (35Spro:YFP-PDCB1) in Arabidopsis resulted in spots of fluorescence located on the walls of epidermal cells in expanded leaf tissues (Figure 2A) in a pattern widely interpreted as Pd labeling (Lucas, 2006). (Supplemental Figure 1 online shows a confocal stack through YFP-PDCB1-expressing Arabidopsis epidermal cells, confirming the wall location in multiple focal planes.) A similar pattern was seen after transient expression of 35Spro:YFP-PDCB1 in Nicotiana benthamiana plants (see Supplemental Figure 2 online). In addition, in spongy mesophyll cells, the fluorescent spots were restricted to wall-to-wall connections between neighboring cells (Figures 2D to 2F). (Supplemental Figure 3 online illustrates YFP-PDCB1 in spongy mesophyll cells viewed at varying confocal laser intensities and shows unique association with Pds.) A similar pattern of fluorescence was observed in transgenic Arabidopsis plants accumulating YFP-PDCB1 following expression from its native promoter (PDCB1pro:YFP-PDCB1; Figures 2B and 2C). Confirmation that these spots were wall associated was obtained following plasmolysis when the fluorescence was spatially distinct from the retracted protoplast (Figures 2D to 2F). Interestingly, plasmolysis often resulted in fluorescence being leached into the apoplastic space (see Supplemental Figure 4 online), a technical problem that was resolved by preincubation of the tissues with a phospholipase C chemical inhibitor. GPI anchors are highly susceptible to stress-induced phospholipase C (Svetek et al., 1999), providing independent evidence that PDCB1 is a GPI-anchor protein.
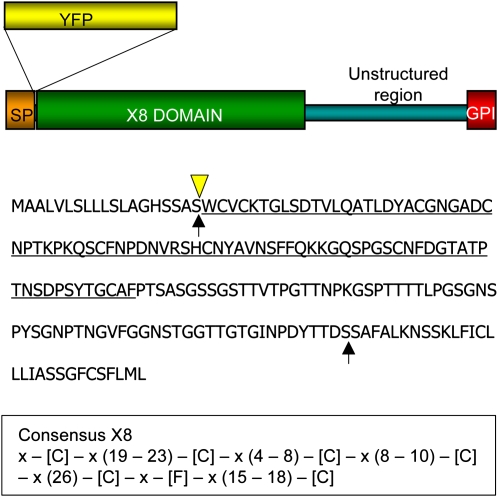
Figure 1.
Structural Organization of PDCB1.
The predicted locations of the N- and C-terminal signal cleavage sites (arrows), the conserved X8 domain (underlined), and the insertion point for YFP (triangle) are marked on the complete PDCB1 amino acid sequence. The consensus sequence for the X8 domain for all Arabidopsis X8 proteins is shown below.
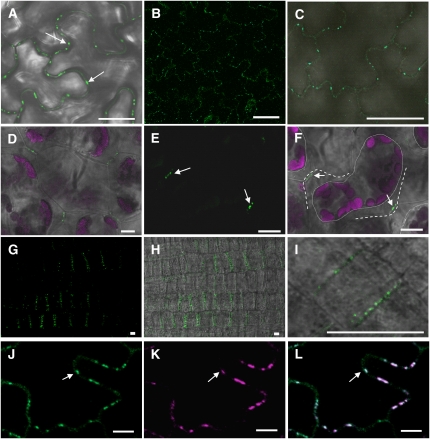
Figure 2.
YFP-PDCB1 Shows Targeting to Pds.
(A) Transgenic expression of 35SproYFP-PDCB1 in Arabidopsis showing fluorescence as punctate spots on the walls of epidermal pavement cells.
(B) and (C) Transgenic expression of PDCB1pro:YFP-PDCB1 in Arabidopsis showing similar patterns of fluorescence as in (A), in the absence of overexpression.
(D) to (F) Similar patterns of fluorescence for YFP-PDCB1 were seen in spongy mesophyll cells where the fluorescent punctae were restricted to regions of wall–wall contact between adjacent cells and were stably located following plasmolysis; (F) is the same as (E) except with combined differential interference contrast microscopy. Arrows indicate YFP-PDCB1 fluorescence on adjoining walls. Dotted lines indicate periphery of retracted protoplast and dashed lines the position of the cell wall. Fluorescence is shown for GFP in green and chlorophyll autofluorescence in magenta.
(G) to (I) YFP-PDCB1 labeling on new anticlinal division walls in newly divided root epidermal cells immediately behind the root meristem.
(J) to (L) Colocalization (e.g. arrows) of YFP-PDCB1 (J) and aniline blue staining for callose (K) supporting the identification of the fluorescent punctae as Pds; merged images are shown in (L). Aniline blue fluorescence is shown using magenta (K) false color.
Bars = 10 μm.
Unlike some viral movement proteins (e.g., Tobacco mosaic virus movement protein) that only associate with complex Pds late in tissue development (Ding et al., 1992b; Oparka et al., 1999), PDCB1 was also observed as punctate labeling on new division walls close to meristems. For example, microscopy of newly divided epidermal cells behind the root meristem of transgenic Arabidopsis expressing 35Spro:YFP-PDCB1 showed YFP-PDCB1 located on the anticlinal division walls (Figures 2G to 2I).
Absolute confirmation that these punctate patterns represented the expressed protein located at Pds was obtained using immunolocalization. Anti-PDCB1 antibodies were used to probe tissue sections prepared for electron microscopy. Since At5g61130 in wild-type plants was predicted to be expressed only weakly in leaves (www.genevestigator.ethz.ch), tissues for electron microscopy were prepared from 35Spro:YFP-PDCB1 transgenic Arabidopsis; immunogold labeling of wild-type tissues gave no significant labeling. Immunogold labeling of transgenic lines showed that YFP-PDCB1 was located at the outer neck region of Pds (Figure 3A) where callose becomes deposited (Figure 3C) and not through the central region. Quantification of the gold labeling showed a strong preponderance for labeling associated with Pds and significantly less background labeling elsewhere in the cell. Control preimmune serum showed no significant labeling at these positions (Figure 3B). Furthermore, using aniline blue staining (Figures 2J to 2L) and confocal microscopy, we demonstrated that YFP-PDCB1 precisely colocalized with Pd-associated callose.
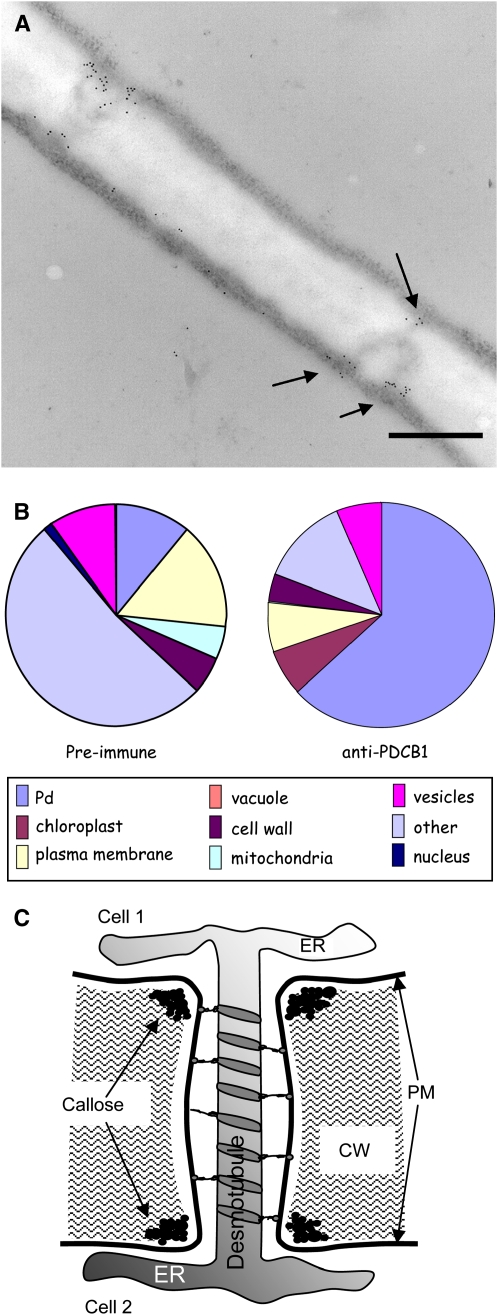
Figure 3.
Immunogold Localization of YFP-PDCB1 to Pds.
Thin sections of YFP-PDCB1-transgenic Arabidopsis leaves were subjected to immunogold labeling using anti-PDCB1 antiserum.
(A) A length of cell wall on which two branched Pds are labeled at the cytoplasmic ends of the structures (arrows). Note that there is little labeling with the antibody in the central cavity region of the Pds. Bar = 500 nm.
(B) A proportional quantification of the immunogold distribution where the majority of gold particles are associated with Pds and a minority scattered over other areas; total particle count from randomized electron micrographs is 1136. The latter labeling corresponded with the weak labeling observed with the preimmune serum; total particle count is 389.
(C) The general organization of Pds with the location of callose at the peripheral neck regions. ER, endoplasmic reticulum; PM, plasma membrane.
The PDCB Gene Family
PDCB1 encodes a 21-kD protein with additional N- and C-terminal signal sequences that direct the protein to the external face of the plasma membrane where the mature protein (with signal sequences removed) is secured through a covalent GPI anchor. The amino acid sequence of mature PDCB1 reveals two domains, a C-terminal unstructured domain rich in Pro residues and an N-proximal X8 domain (Figure 1). Homology searches of the Arabidopsis genome identified At5g08000 (PDCB2) and At1g18650 (PDCB3), which shared >50% amino acid sequence similarity with PDCB1. PDCB2 (At5g08000) also has been identified biochemically as a GPI-anchor protein (Elortza et al., 2003).
Phylogenetic analysis (Figure 4A; see Supplemental Figure 5 and Supplemental Data Set 1 online) showed that PDCB1, -2, and -3 fall within a tight cluster of seven proteins among a larger cluster of similarly structured proteins that lack the GPI-anchor signal sequence and within a larger superfamily of proteins, which includes proteins with X8 domains adjacent to 1,3-β-glucanase catalytic domains. The X8 domain is characterized by a conserved distribution of six Cys residues and a Phe residue (Figure 1) and likely plays a role in carbohydrate binding (Barral et al., 2005).
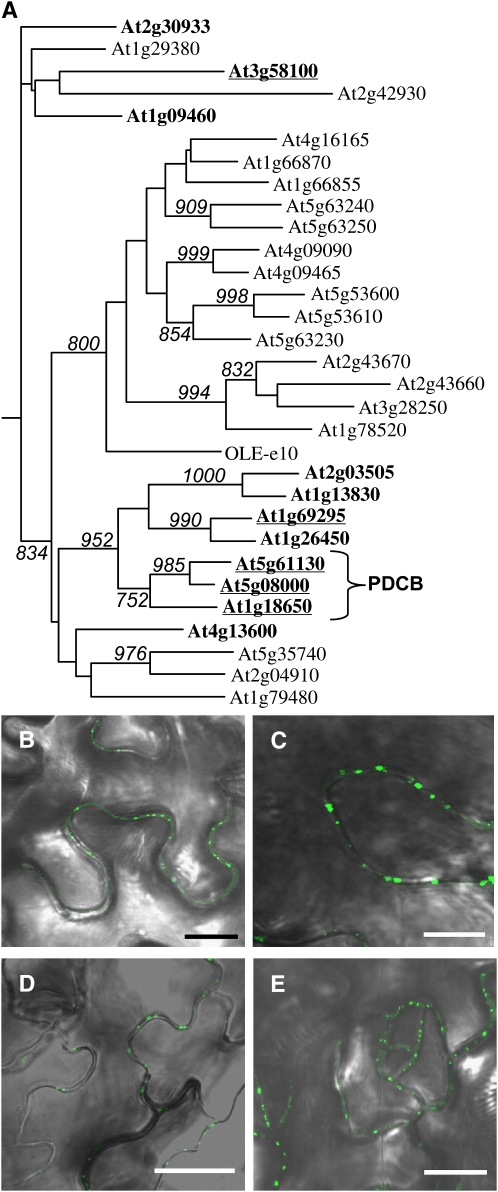
Figure 4.
The Larger PDCB Family of Pd Proteins.
(A) Phylogenetic tree of the X8 domains of Arabidopsis PDCB-related proteins. The analysis, which includes X8 domains from 30 PDCB structural homologs, is presented as a midpoint rooted phylogenetic tree of proteins containing X8 as their only identifiable functional domain; 1,3-β-glucanases are excluded from this analysis. X8 domains from proteins with (bold) and without GPI anchors are listed as Arabidopsis Genome Initiative numbers. Olive OLE-e10 is included and shows closer similarity to the clade of non-GPI X8 proteins than to the GPI-anchored proteins. Only bootstrap values >70% are shown. The PDCB subfamily is indicated. Proteins tested for their subcellular targeting as YFP fusion proteins are underlined. The sequences used to generate this phylogeny are presented in Supplemental Data Set 1 online.
(B) Transient expression of 35SproYFP-PDCB2 in N. benthamiana leaves showing fluorescence as punctate spots on the walls of epidermal pavement cells.
(C) Transgenic expression of 35SproYFP-PDCB3 in Arabidopsis showing fluorescence as punctate spots on the walls of epidermal pavement cells.
(D) Transgenic expression of 35SproYFP-At1g69295 in Arabidopsis showing fluorescence as punctate spots on the walls of epidermal pavement cells.
(E) Transgenic expression of 35SproYFP-At3g58100 in Arabidopsis showing fluorescence as punctate spots on the walls of epidermal pavement cells. Bars in (B) to (E) = 10 μm.
To explore the potential for PDCB1-related proteins to also target Pds, YFP fusion proteins were created for the two closest homologs to PDCB1 and for two additional proteins that represented the diversity of the PDCB1-related X8-GPI family. Hence, YFP-PDCB expression constructs were made for PDCB2, PDCB3, At1g69295, and At3g58100 (Figure 4); At1g69295 resides in a neighboring subclade to PDCB1, and At3g58100 is more distantly related with an unclear phylogenetic association with PDCB1. All these constructs were expressed transiently in N. benthamiana. YFP-PDCB3, YFP-At1g69295, and YFP-A3g58100 were also expressed as stable transgenics in Arabidopsis; no such lines could be obtained for YFP-PDCB2. In tissues expressing these constructs, the proteins again were observed in a punctate pattern of fluorescence around the periphery of the cell (Figures 4B to 4E). As for YFP-PDCB1, these punctae were restricted to cell-to-cell contact zones in spongy mesophyll, colocalized with callose, and were retained on the cell wall after plasmolysis. These data suggest that many or all of the 11 PDCB1-related X8-GPI proteins could constitute this family of Pd proteins.
PDCB1 Binds to Callose through Its X8 Domain
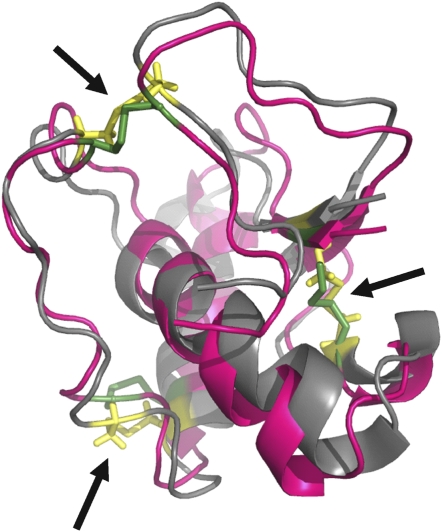
The X8 protein domain is present in a diverse range of Arabidopsis proteins, including as a nonenzymatic ancillary domain to some of the 1,3-β-glucanases. Barral et al. (2005) showed that an X8 protein, OLE-e10, from olive (Olea europaea) pollen had callose binding activity. Mature OLE-e10 comprises just the X8 domain and is phylogenetically distinct from the PDCB family, being more closely related to the X8-non-GPI proteins (Figure 4). Recently, the three-dimensional structure of the related OLE-e9 protein has been established by nuclear magnetic resonance (Trevino et al., 2008). Using a protein threading algorithm, the putative structure of the PDCB1 X8 domain was compared with the structure of OLE-e9. The X8 domain of PDCB1 showed 40% amino acid similarity with OLE-e9. The two structures were congruent, pointing to a common function for the two X8 domains (Figure 5).
Figure 5.
Congruent Tertiary Structures for PDCB1 and OLE-e9 X8 Domains.
The sequence of the PDCB1 X8 domain was analyzed by threading onto the tertiary structure of OLE-e9 determined by nuclear magnetic resonance (Trevino et al., 2008). The parent OLE-e9 structure is shown in magenta; the PDCB1 X8 domain is shown in gray. The disulphide bridges (arrows) are highlighted (OLE-e9, green; PDCB1 X8, yellow).
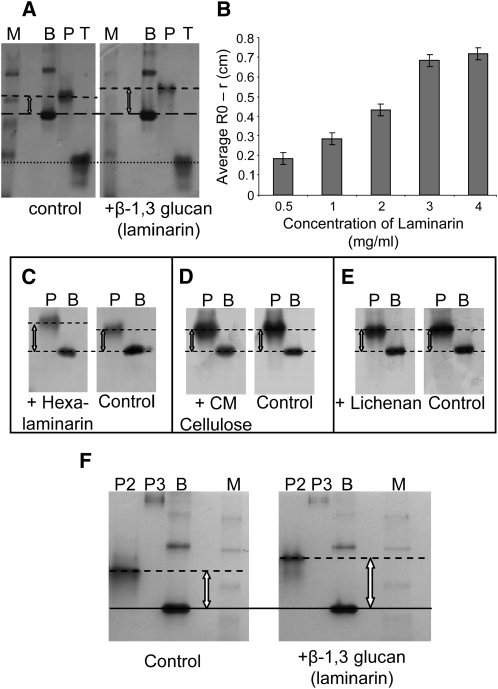
To assess biochemically whether, like OLE-e10, PDCB1 might show callose binding activity, the mature protein, lacking the N- and C-terminal signal sequences, was expressed in Escherichia coli and assessed for its interaction with 1,3-β-glucan in a gel retardation assay. The protein was expressed with thioredoxin fused to the N terminus to enhance its solubility, with an intervening 6x histidine (6xHis) tag to assist protein purification. Protein extracts were subjected to Ni2+-affinity chromatography followed by gel filtration. We were unsuccessful in our attempts to purify the PDCB1 X8 domain alone as a thioredoxin fusion. Purified thioredoxin.PDCB1 was electrophoresed through a nondenaturing gel into which selected polysaccharides had been incorporated. BSA and thioredoxin (also fused to a 6xHis tag), neither of which binds to polysaccharides, were run in parallel as internal negative controls. In the presence of laminarin (a long-chain 1,3-β-glucan), the migration of thioredoxin-PDCB1 was reduced relative to BSA and thioredoxin and relative to migration in the absence of laminarin (Figure 6A). Thioredoxin-PDCB1 retardation was proportional to the concentration of laminarin (Figure 6B). Retarded migration was also observed with hexalaminarin (short-chain 1,3-β-glucan) (Figure 6C). No change in migration was seen when laminarin was replaced with carboxymethylcellulose (derivatized 1,4-β-glucan) or lichenan (mixed 1,4;1,3-β-glucan) (Figures 6D and 6E).
Figure 6.
In Vitro Callose Binding Properties of PDCB Proteins.
Soluble purified thioredoxin-PDCB1, -2, or -3 fusion proteins were assessed for ability to bind to polysaccharides in a nondenaturing gel retardation assay.
(A) Callose binding was seen as retarded migration of the fusion protein (P) in the presence of the 1,3-β-glucan laminarin ligand when compared with the gel lacking laminarin (control) and to the unchanged migration of BSA (B) or thioredoxin alone (T). M, molecular weight markers.
(B) Bar chart showing a positive correlation between gel retardation and substrate concentration. Bars indicate ±sd from triplicate assays.
(C) to (E) PDCB1-poysaccharide binding specificity tested against hexalaminarin (1,3-β-glucan), carboxymethyl (CM) cellulose (derivatized 1,4-β-glucan), and lichenan (mixed 1,4;1,3-β-glucan), respectively. Binding was only observed with hexalaminarin.
(F) Assay for the binding of thioredoxin-PDCB2 and thioredoxin-PDCB3 to laminarin in a gel retardation assay. In comparison to the negative control, BSA (B), thioredoxin-PDCB2 (P2) shows retarded migration. It was not possible to assay binding for thioredoxin-PDCB3 (P3) due to aggregation and low migration in both test and control gels. M, molecular weight markers.
To determine if this activity was a unique property to PDCB1 or whether other PDCB homologs might also bind to callose, we also expressed PDCB2 and -3 similarly in E. coli and assayed the purified fusion proteins by nondenaturing gel retardation (Figure 6F). Relative to the negative control (BSA), thioredoxin-PDCB2 also showed retardation and therefore callose binding activity; binding activity for thioredoxin-PDCB3 could not be measured as it appeared to aggregate, resolving only as higher mass material in the gel.
PDCB Function in Vivo
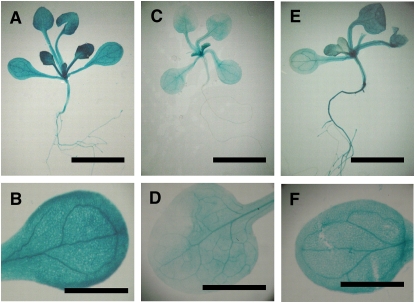
The similarity in structure between PDCB proteins and their common in vitro activity (at least for PDCB1 and PDCB2) suggested that they could cover redundant functions in vivo. For this to be the case, the genes should show overlapping patterns of tissue-specific expression. The public expression data (www.genevestigator.ethz.ch) point to strong tissue-specific expression of PDCB1, -2, and -3 in the vegetative/floral apex with weaker expression in the vegetative organs. To formally assess the expression patterns for PDCB1, -2, and -3, PDCBpro:β-glucuronidase (GUS) constructs were transformed into Arabidopsis, and transformed seedlings were stained histochemically for GUS activity (Figure 7). GUS staining of multiple independent lines showed consistent expression patterns that generally reflected the aggregated public data (www.genevestigator.ethz.ch). Activity for PDCB2pro was consistently lower than for PDCB1pro and PDCB3pro, although the overall patterns of activity were similar. GUS staining was most intense in the shoot apical region and in young leaves, but activity was still readily detected in the laminar and vasculature of mature leaves (Figure 7).
Figure 7.
Tissue-Specific Expression of PDCB1, -2, and -3.
Predicted promoter sequences for PDCB1, -2, and -3 were isolated from the Arabidopsis genome and fused to a GUS reporter gene. Transgenic lines were stained for GUS activity as 11-d-old seedlings. Bars = 0.5 cm in (A), (C), and (E) and 0.2 cm in (B), (D), and (F).
(A) and (B) Views of a whole seedling (A) and an isolated leaf (B) from a PDCB1pro:GUS transgenic line after staining for GUS activity to reveal the distribution of gene-specific expression.
(C) and (D) As in (A) and (B) except for PDCB2pro:GUS.
(E) and (F) As in (A) and (B) except for PDCB3pro:GUS.
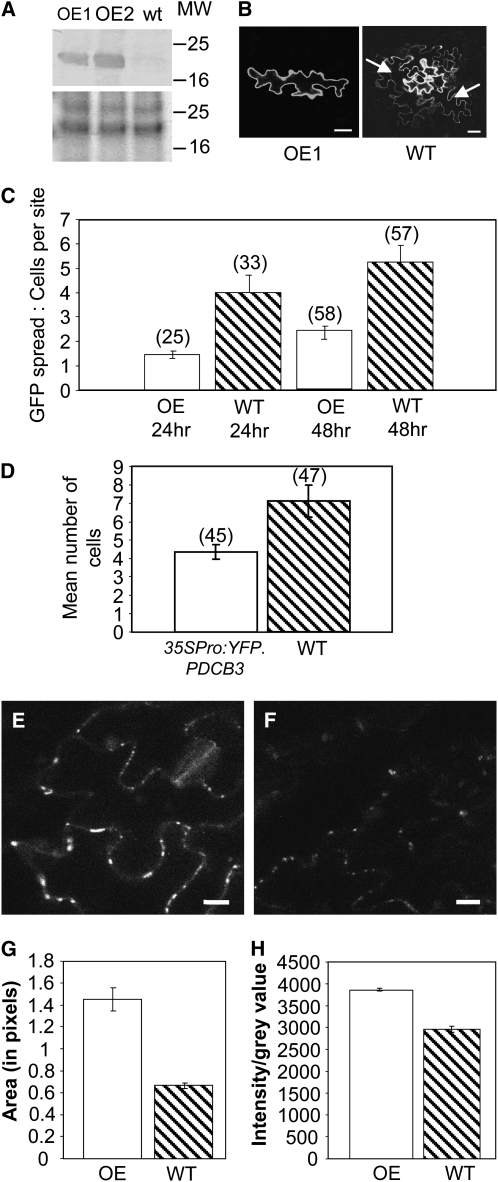
To address the consequences of reduced expression of PDCB1, -2, and -3, we obtained T-DNA insertional mutant lines for At5g08000 (PDCB2; Salk_034932) and At1g18650 (PDCB3; Salk_043611.56.00.x) (see Supplemental Figure 6 online). Because of potential problems with functional redundancy, crosses between mutants for At1g18650 and At5g08000 were also made. Insertional mutant lines for PDCB1 listed for the Gabi-Kat and Riken collections could not be confirmed. Hence, we also used artificial microRNAs (Schwab et al., 2006) in an attempt to generate transgenic lines with reduced PDCB1 expression. Constructs designed to target two independent sites within the PDCB1 gene, inserted into the pdcb2 pdcb3 double mutant, failed to give rise to any lines with significantly reduced PDCB1 expression. In a complementary strategy, we transformed wild-type plants with constructs expressing untagged PDCB1 constitutively from the CaMV 35S promoter; Arabidopsis plants similarly expressing YFP-PDCB1 or YFP-PDCB3 had been generated previously. Immunoblot analysis of leaf tissues from transgenic plants expressing the unfused PDCB1 showed increased accumulation of PDCB1 (Figure 8A). Accumulation of tagged YFP-PDCB1 and YFP-PDCB3 had been visualized previously as YFP fluorescence located at Pds (Figures 2 and 4).
Figure 8.
PDCB1 Transgenic Plants Show Increased Callose Accumulation and Decreased Plasmodesmal Molecular Diffusion.
(A) Crude protein extracts from two independent 35Spro:PDCB1 overexpressing hemizygous transgenic Arabidopsis Col-0 lines (OE1 and OE2) were assessed for PDCB1 accumulation by immunoblot analysis using anti-PDCB1 antibody compared with wild type (wt) Columbia-0 (Col-0) (top). Equal protein loading is shown from the equivalent section of the protein gel stained with Coomassie blue (bottom). MW, protein markers in kilodaltons.
(B) Cell-to-cell movement potential in the OE1 plants was assessed in a GFP diffusion assay where diffusion of soluble GFP from a bombarded epidermal cell to surrounding cells was measured. Representative examples of restricted and permitted GFP diffusion following bombardment of 35Spro:GFP into the leaves of a PDCB1 overxpressing (OE1) or a wild-type plant. GFP recipient epidermal cells are indicated (arrows). Bars = 10 μm.
(C) Quantification of the GFP diffusion was determined after 24 and 48 h, at which times the overexpressing line showed significantly less (P < 0.05 and P < 0.0001, respectively) plasmodesmal flux; numbers in parentheses record the numbers of bombardment foci counted for each treatment. The experiment was repeated twice with similar results. Bars indicate ±se.
(D) GFP diffusion assay applied to leaves of a 35Spro:YFP-PDCB3 transgenic plant. Data from the 48 h time point are presented. GFP diffusion in the transgenic line was significantly (P < 0.05) less than in wild-type plants. Bars indicate ±se.
(E) and (F) Confocal micrographs of leaf tissue from transgenic PDCB1 overexpressing (OE1) and wild-type Col-0 plants, respectively, stained with aniline blue to reveal callose accumulation. Images were collected using identical confocal microscope settings. Bars = 20 μm.
(G) and (H) Quantification of the callose fluorescence on representative optical sections achieved using image intensity analysis with IMAGEJ software. Either the size of individual aniline blue–stained foci (in pixels) were compared (G), or gray values of individual foci were compared (H). For each treatment, 100 data sets were collected. As the data did not show a normal distribution, statistical analysis was done using a Mann-Whitney Hugh test. In either case, the data sets between the treatments are statistically different at P < 0001. Bars indicate ±se.
Neither the single nor double insertional mutant lines for pdcb2 and pdcb3 showed any morphological or developmental phenotypes, probably reflecting functional redundancy with PDCB1. For the overexpression lines, hemizygous 35Spro:PDCB1 lines showed normal growth and development. By contrast, homozygous lines produced stunted floral spikes, which were frequently dead shortly after the production of an influoresence under normal plant culture conditions and were generally infertile.
A key tool for assessing potential roles for proteins in Pd function is the use of monomeric soluble GFP as a reporter of molecular flux through Pds by passive diffusion (Liarzi and Epel, 2005; Levy et al., 2007b; Schönknecht et al., 2008). This is conveniently achieved by measuring the diffusion of GFP from a point of initial GFP expression following bombardment of 35Spro:GFP into the leaves of plants with altered expression of the genes in question. To assess any impact of altered PDCB expression on Pd function, insertional mutant and overexpression lines were subjected to the GFP diffusion assay following bombardment with gold particles cocoated with plasmids encoding GFP or RFPer (cell-autonomous endoplasmic reticulum–targeted red fluorescent protein to mark the bombardment site). The spread of GFP was monitored at 48 h after bombardment in comparison to that observed in wild-type plants. When single and double mutant lines of pdcb2 and pdcb3 were tested, no change in GFP diffusion was observed (see Supplemental Figure 7 online). By contrast, GFP diffusion in 35Spro:PDCB1 hemizygous plants measured at 24 and 48 h after bombardment was significantly less (P < 0.05 and < 0.0001, respectively) than in wild-type plants (Figure 8B). Equivalent data were obtained for plants expressing 35Spro:YFP-PDCB1 or 35Spro:YFP-PDCB3 (Figure 8C). Since overexpression of proteins can give rise to pleiotropic effects, as an additional control we assessed GFP diffusion in transgenic plants overexpressing the gene for an unrelated GPI-anchor protein, fasciclin-like protein 13 (FLA13; At5g44130), not targeted to Pds. These plants showed no change in GFP diffusion compared with wild-type plants (see Supplemental Figure 7 online).
Callose Accumulation in Response to PDCB1 Overexpression
Since bioinformatic analysis of the PDCB proteins failed to predict any associated catalytic activity, it was not immediately apparent why increased PDCB expression should lead to altered molecular flux through Pds. However, we had shown that thio-PDCB proteins, through their X8 domains, bind to callose at the Pd neck region, and callose deposition in this location had been shown to regulate trafficking of molecules through Pds (Iglesias and Meins, 2000; Levy et al., 2007a). In addition, for some 1,3-β-glucanases, the X8 domain exists as a C-terminal ancillary domain, and changes in 1,3-β-glucanase activity have been shown (Iglesias and Meins, 2000; Levy et al., 2007b) to affect both active and passive cell-to-cell trafficking. Hence, we asked whether plants overexpressing PDCB proteins might show altered callose accumulation. When leaf tissue from 35Spro:PDCB1 plants was stained with aniline blue and visualized by confocal microscopy, these lines were seen to contain more callose than nontransgenic segregant lines. Visually more intense aniline blue staining indicated increased callose accumulation (Figures 8E and 8F). Following computational quantification of representative images (Figures 8G and 8H), it was confirmed that lines overexpressing the unfused PDCB1 contained significantly (P ≤ 0.001) more callose than the wild type.
Callose deposition at Pds and at phloem sieve plates is frequently associated with applied chemical and physiological stresses. Since changes in PDCB expression also led to increased callose deposition, we investigated whether expression of PDCB1, -2, or -3 was responsive to applied stresses. Public array expression data (https://www.genevestigator.ethz.ch) did not, however, indicate any consistent changes for these genes across a wide range of treatments (see Supplemental Figure 8 online). To validate these public data, the expression patterns of PDCB1, -2, and -3 were assessed by quantitative RT-PCR following treatment with heat and after wounding; HSP70 (At3g12580) and LOX2 (At3g45140), respectively, were used as positive controls for the effects of the treatments. Heat treatment showed decreased expression of PDCB2 after 2 h at 37°C and no change for PDCB1 or PDCB3, confirming the existing public data. There were no changes in the expression of PDCB genes in response to wounding (see Supplemental Figure 9 online).
DISCUSSION
Understanding the structure and function of Pds remains one of the major challenges in plant biology. By analogy with the nuclear pore complex, also mediating macromolecular trafficking, we might expect there to be many tens of proteins involved in Pd structure and function. Recent successes in identifying Pd proteins indicate that the identification of the constituent components of Pds need not remain the major hurdle in understanding macromolecular trafficking in plants (Lee et al., 2005; Sagi et al., 2005; Levy et al., 2007b; Thomas et al., 2008). However, the proteins identified so far show few examples of functional connectivity, indicating that many plasmodesmal proteins remain to be identified.
A key component in Pd flux control is callose deposited in the neighboring cell wall (Iglesias and Meins, 2000; Bucher et al., 2001; Rinne et al., 2005; Ueki and Citovsky, 2005). This is believed to exert a structural constriction around the neck of the Pd, reducing the Pd aperture and the physical space available for molecular trafficking. Our immunolocalization experiments based on transgenic expression of YFP-PDCB1 indicated that PDCB1 was associated closely with the neck region of Pds. PDCB1 and -2 (and by association, PDCB3) have been identified biochemically as GPI-anchor proteins (Elortza et al., 2003). These proteins, misassigned in Elortza et al. (2003) as 1,3-β-glucanases, were also assessed for their expression levels as EST abundance and as MPSS (massive parallel signature sequencing; http://mpss.udel.edu/grape/) outputs. Interestingly, the MPSS data record high expression levels for PDCB1 in Arabidopsis callus; Arabidopsis suspension culture was the source material for our original proteomic analysis (Bayer et al., 2006). The predicted and biochemically verified GPI nature of these proteins was supported by our indirect observations that their subcellular location was stabilized by an inhibitor of phospholipase C, an activity for which GPI-anchor proteins are very vulnerable (Svetek et al., 1999). The GPI anchoring of the PDCB proteins predicts a subcellular location with the C terminus anchored to the apoplastic face of the plasma membrane.
The mature PDCB1 and PDCB2 proteins, as fusions with thioredoxin, were assayed for biochemical interactions in vitro with callose using nondenaturing gel retardation assays. The smaller OLE-e10 protein had previously been analyzed (Barral et al., 2005) as a mature protein lacking both a secretory signal peptide and a GPI anchor. As the cloned sequence for this protein was based upon the isolated peptide sequences, it is not yet known whether the preprotein contains additional N- and C-terminal signals, although phylogenetically it clustered with the PDCB-related proteins lacking the GPI anchor (Figure 4). OLE-e10 appears to lack the unstructured extension seen in the PDCB proteins (i.e., it contains just the X8 domain). Like OLE-e10, thio-PDCB1 showed specific binding to 1,3-β-glucans (laminarin and hexalaminarin) and no binding to 1,4-β-glucans or a mixed glucan. This indicates that the structural association between X8 and polysaccharides is associated with higher-order structures, as hexalaminarin (six glucose residues) is able to form a single helical turn (Stone and Clarke, 1992). Our expression construct excluded the N- and C-terminal signals for secretion and GPI attachment but included the unstructured domain in the C-terminal half of the mature protein. We were unsuccessful in purifying the PDCB1 X8 peptide alone or as a thioredoxin fusion. By extrapolation to the OLE-e10 protein, we conclude that it is the X8 domain and not the C-terminal unstructured region that is responsible for callose binding activity. This is supported by the very similar predicted three-dimensional structure of the PDCB1 X8 domain to that of OLE-e9, a probable paralog of the olive OLE-e10 (Barral et al., 2004; Trevino et al., 2008). The subcellular location of PDCB and the callose binding activity points to a role where the N-proximal X8 domain is in a position to interact with callose and, through covalent linkage to GPI, to anchor the plasma membrane to the wall matrix at the neck of the Pds. PDCB1 contains 68 amino acids C-terminal to the X8 domain. Since this region lacks any predicted secondary structure (i.e., it is a random coil), it may function as a flexible spacer bringing the X8 domain closer to callose deposits in the wall. Appropriately, it contains a high proportion of Pro residues (Figure 1) to confer additional flexibility.
In this proposed structural role, it was somewhat surprising that single or double insertional mutant lines of PDCBs showed no growth phenotype. However, the absence of a mutant for PDCB1 and the fact that PDCB1, -2, and -3 show coregulation as overlapping tissue-specific expression in the vegetative/floral apex and mature vegetative tissues probably point to functional redundancy in this gene family.
Arabidopsis proteins with X8 domains fall into two main groups: (1) 1,3-β-glucanase enzymes (24) that have an ancillary X8 domain, half of which also have a predicted GPI anchor; and (2) proteins (30) annotated to have the X8 domain as their only functional structure in the Pfam database ((Finn et al., 2006). This latter group is divided into those with additional sequence and a GPI anchor (i.e., the 11 PDCB1-like proteins) or PDCB-like proteins lacking the GPI anchor (Figure 4). Smaller proteins (e.g., OLE-e10; <20 kD) with just a X8 domain were not found in the Arabidopsis database. Of the 11 PDCB1-like proteins, we tested five for their targeting to Pds as YFP-tagged proteins. These ranged from 18 to 70% amino acid sequence similarity to PDCB1. The fact that all these proteins targeted to Pds suggest that all 11 proteins with an equivalent structure could be Pd proteins. Because of the association of the 1,3-β-glucanase enzymatic activity and X8 domains with callose, we would predict that at least some of the 1,3-β-glucanases with an X8 domain would also be targeted to and retained at Pd, although this remains to be tested. The Pd-associated 1,3-β-glucanase identified previously (Levy et al., 2007b) has a GPI anchor but lacks an X8 domain. It is probably not the case, however, that all X8 domain proteins are targeted to Pds. Callose is a structural component of Pds formed on the phragmoplast (Samuels et al., 1995; Hong et al., 2001), and it is associated with pollen development (Turner et al., 1998; Parre and Geitmann, 2005) and cotton hair formation (Ruan et al., 2004). In addition, it also acts as part of a complex defense response to attempted pathogen invasion (Hauck et al., 2003; Hamiduzzaman et al., 2005) where Pds are unlikely to play a direct role.
Considering that PDCB1 has an apoplastic location and lacks any known catalytic domain, it was surprising that overexpression of PDCB1 led to an increase in callose accumulation and that this correlated with a decrease in molecular diffusion in our GFP-movement assay. This was especially the case since we had no reason to believe that PDCB1 should exert a regulatory role on the expression of callose synthase or 1,3-β-glucanases. Alternatively, some posttranslational role affecting callose turnover through inhibition of 1,3-β-glucanases or activation of callose synthases is conceivable. Strictly, it is possible that both the increased callose accumulation and the associated decreased cell-to-cell movement could reflect indirect stress responses to overexpression of PDCB1. Certainly, there was a negative impact of overexpression seen in the homozygous transgenic lines. However, we cannot say whether this was due to the deleterious effects of reduced cell-to-cell trafficking or some indirect consequence of excess callose or PDCB1 accumulation. However, the failure to observe these effects following overexpression of another unrelated GPI-anchor protein makes the latter unlikely.
The location of YFP-PDCB1 at the neck region of Pds (as an indicator of the location of the native PDCB1 protein) is very different from that of many viral movement proteins that have a tendency to accumulate in the central symplastic cavity region of complex Pds or as physical tubules that mediate the passage of virus particles from cell to cell (Ding et al., 1992b; Itaya et al., 1998; Laporte et al., 2003). For the former group, accumulation often leads to an increase in trafficking. In our case, increased PDCB1 accumulation and callose deposition were negatively correlated with GFP diffusion in plants overexpressing PDCB1. This is in accord with the view that callose deposition at the neck region is important for modulating molecular movement through Pds.
Callose deposition and turnover is very dynamic and responsive to developmental and environmental cues. Hence, we tested whether PDCB genes, as potential regulators of callose accumulation, responded to external stresses. Of the large number of candidate treatments, we selected heat and wounding to validate the extensive public expression data set. In both cases, our analyses supported the public data, leading to the conclusion that PDCB genes are not the primary intermediaries in between the application of external stress and callose deposition.
From our limited understanding of the control of callose synthesis and turnover, we would expect to see a close physical association of Pds with callose synthase and 1,3-β-glucanase. Of these, however, only a 1,3-β-glucanase has been identified previously as a Pd-targeted protein (Levy et al., 2007b). Our identification of PDCBs as callose binding proteins with the potential to modify cell-to-cell trafficking introduces a potential novel player involved in the control of callose-mediated regulation of Pd function. In this respect, the identification of the PDCB family provides valuable tools for unpicking essential processes that mediate cell-to-cell communication through Pds.
METHODS
Construction of YFP-PDCB Fusion Proteins
The coding sequence (cds) for YFP was cloned as a translational fusion between the N-terminal signal sequence and the rest of the cds for PDCB, using overlap-PCR and Gateway cloning technology (Tian et al., 2004). Forward and reverse primers for YFP (also called pCitrine; Tian et al., 2004) and two sets of PDCB primers (P1/P2 and P3/P4) were used (see Supplemental Table 1 online for primer sequences). P1 and P2 amplified the N-terminal signal sequence. P1 contained sequences that partially overlapped the Gateway primers and a Kozak translational consensus start site, and P2 contained a region overlapping YFP. P3 and P4 amplified the rest of the cds. P3 contained a region overlapping YFP; P4 contained a stop codon and partially overlapping sequence with the Gateway primers. All fragments were amplified separately and then joined by overlap-PCR in two stages. Fragments were amplified from a cDNA clone of At5g61130 (Uniclone U20590; ABRC [Liu et al., 1998]) and YFP using EX taq (Takara Bio). The first PCR reaction using primers P1 and YFPRV, fused YFP to the N terminus of At5g61130 but downstream of the N-terminal signal sequence. A second PCR used gene-nonspecific Gateway primers to reconstitute the attB sites and fuse the N-YFP to the rest of the coding sequence. The PCR product was recombined into the Gateway vector pDONR207 (Invitrogen) using BP Clonase II mixture (Invitrogen) and transformed into Escherichia coli DH5α. This strategy was repeated for the homologs At5g08000, At1g18650, At1g69295, and At3g58100. The sequence of the resulting pDONR constructs was verified. Coding sequences for YFP-tagged PDCB and related proteins were transferred from pDONR into the Gateway binary destination vector pB7WG0.2 using LR Clonase II, resulting in 35Spro:YFP fusions. The constructs were transformed into Agrobacterium tumefaciens GV3101 to be used for transformation of Arabidopsis thaliana Col-0 or transient expression in Nicotiana benthamiana.
Construction of PDCB1pro:Reporter Lines
The PDCB1pro:YFP-PDCB1 construct was created by overlap-PCR of two fragments, YFP-PDCB1 and PDCB1pro. YFP-PDCB1 was amplified from the pDONR:YFP-PDCB1 using primers P4 and At5g61130promotorCDSFW. PDCB1pro (1.5 kb) was amplified from genomic DNA using primers at5g61130promotorFW and at5g61130promotorRV. The PDCB1pro:YFP-PDCB1 construct was inserted into the promotorless Gateway vector pEarleyGate301 (Earley et al., 2006).
The PDCBpro:GUS constructs were created by amplification of the PDCB promoter regions from genomic DNA using the following primers sets: PDCB1pro (1.5 kb), at5g61130promotorFW/At5g61130proAttB2RV; PDCB3pro (2 kb), At1g18650promotorFW/At1g18650proAttB2RV; and pPDCB2pro (1.7 kb), at5g08000promotorFW/At5g08000proAttB2RV. The three promoter regions were cloned into the vector pBGWFS7 (VIB-Gent University) using Gateway technology.
Analysis of PDCB Expression
Transgenic PDCBpro:GUS seeds were sown onto Murashige and Skoog (MS) plates. After 11 d, when the first true leaves had fully expanded, the seedlings were removed and washed in water. GUS assay solution (10 mM EDTA, 0.1% Triton X, 1 mg/mL X-Gluc, 100 mM sodium phosphate, pH 7, 0.5 mM potassium ferrocyanide, and 0.5 mM potassium ferricyanide) was vacuum infiltrated into the seedlings. After overnight incubation in the dark at 37°C, the seedlings were washed several times in 70% ethanol to clear the tissues. Images were taken using a Zeiss Stemi SV11 microscope adapted with a Cannon Powershot A640 camera.
To validate the public expression data set (https://www.genevestigator.ethz.ch), wild-type seedlings were heat treated by incubating whole seedlings grown on solid MS media for 2 h at 37°C or were wounded by injuring the leaf tissue using broad tweezers; wounded tissue was harvested after 1 h. Total RNA was extracted from treated leaf samples and subjected to quantitative RT-PCR following cDNA synthesis using Superscript II (Invitrogen). The analysis followed the SYBR Green Jumpstart (Sigma-Aldrich) protocol, and data were collected using an Opticon Monitor 2.02.24 (MJ Research). Biological (triplicate) and technical (triplicate) replicates were analyzed statistically using the software REST (relative expression software tool; Pfaffl, et al., 2002). Significance values were set at P value < 0.05 and expression values with more than twofold change. In all cases, elongation factor 1B α-subunit 1 (eEF1α) (At5g12110) was used as a reference gene. Positive controls for wounding and heat shock were LOX2 (At3g45140) and HSP70 (At3g12580).
PDCB Expression in Planta
Overnight cultures of A. tumefaciens containing 35Spro:YFP fusions were washed and resuspended in 10 mM MgCl2 + 100 mM acetosyringone at A600 = 0.9. The solution was infiltrated into expanded leaves of 5-week-old N. benthamiana plants. Arabidopsis Col-0 plants were transformed using the flower dip method as described in by Clough and Bent (1998).
PDCB Visualization
The localization of fluorescently tagged proteins was assessed using a Leica DM IRB inverted confocal microscope (Leica DM IRB); YFP fluorescence was viewed with excitation and emission wavelengths of 488 nm and 510 to 555 nm, respectively. Fully expanded leaves from 2-month-old transgenic Arabidopsis Col-0 plants were viewed. Before plasmolysis, leaves were pressure infiltrated with a 10 mM solution of O-Tricyclo[5.2.12,6]dec-9-yl dithiocarbonate potassium salt, which inhibits phospholipase C, so preventing stress-induced cleavage of GPI linkages (Muller-Decker, 1989). This was left for 4 h before the leaves were plasmolyzed by infiltration with a solution of 35% glycerol. Plasmolyzed leaves were viewed immediately.
To correlate PDCB1 location with callose, leaves as above were pressure infiltrated with a 2:3 v/v mixture of 0.1% aniline blue (Fluka) and 1 M glycerol at pH 9.5 and viewed after 10 min using excitation and emission wavelengths of 405 nm and 460 to 500 nm, respectively. Leaf areas within the infiltrated area were selected at random. To quantify callose accumulation in the PDCB1, YFP-PDCB1, and YFP-PDCB3 overexpressing lines, confocal images were subjected to intensity analysis performed under nonsaturating excitation levels using the image processing software IMAGEJ version 1.31 (http://rsb.info.nih.gov/ij). This was performed in two complementary ways. Either the gray values of individual aniline blue stained foci were compared between the oe1 lines and wild-type Col-0, or the size of the foci (in pixels) was compared. For each treatment, a total of 100 data sets were collected from 25 separate images. For each image, the four brightest fluorescent foci were used. As the data did not show a normal distribution, statistical analysis was done using a Mann-Whitney Hugh test using MINITAB version 13 software.
Bioinformatics
The identification of the sequences for N-terminal secretion and GPI signals was predicted using Signal P (Emanuelsson et al., 2007) and DGPI (http://129.194.185.165/dgpi/DGPI_demo_en.html), respectively. The domains of PDCB were identified using Pfam domains (http://pfam.sanger.ac.uk/) and Prosite (http://www.expasy.ch/prosite/). Tertiary structure of the PDCB1 X8 domain was predicted using Fugue (Shi et al., 2001) and ESyPred3D (Lambert et al., 2002). Potential PDCB promoter regions were identified with the Promoter 2.0 Prediction Server (Knudsen, 1999), TF Search (Heinemeyer et al., 1998), and Softberry (Solovyev and Solovyev, 2004).
Phylogenetic Analysis
Homologs of the PDCB protein were identified using BLAST (Altschul et al., 1990). The homologs were aligned using ClustalW (Notredame et al., 2000; www.ebi.ac.uk/tools/clustalw/) and were manually annotated using Genedoc (www.cris.com/ketchup/genedoc.shtml; http://www.nrbsc.org/gfx/genedoc/index.html). The phylogenetic tree and bootstrap values were obtaining using Phylip version 3.67 (Felsenstein, 1989; http://evolution.genetics.washington.edu/phylip.html). Bootstrap values were the product of 1000 trials.
PDCB1 Expression in E. coli
The cds of the mature PDCB1, -2, or -3, or thioredoxin (lacking the sequences for the N- and C-terminal signals), were cloned between the NcoI and EcoRI sites of pET32a (Invitrogen) after PCR amplification using pET32NcoIFW and pET32EcoRIRV primers. These primers included the NcoI and EcoRI restriction sites for cloning and a translational stop codon in the reverse primer. The resulting clone was transformed into the E. coli DH5α, and the insert was verified by DNA sequencing. This clone was transformed into E. coli BL21 DES pLysE strain for protein expression. Expression from the pET32a vector results in a protein fused to an N-terminal thioredoxin protein with increased PDCB solubility. Between the thioredoxin and the PDCB sequence, there was a 6xHis tag to aid purification. As a control, empty pET32a expressing thioredoxin-6xHis alone was processed in parallel.
For protein expression and purification, bacterial cultures were grown with shaking at 37°C to OD 0.7 to 1 and then induced using 0.5 mM Isopropyl-beta-thio galactopyranoside (IPTG). The temperature was reduced to 21°C and the bacteria left to grow overnight. The resulting bacteria were collected by centrifugation, resuspended in buffer (50 mM sodium phosphate, pH 8, 300 mM sodium chloride, 20 mM imidazole), and disrupted using a French press. After centrifugation at 30,000g, the soluble fraction was retained. His-tagged protein was purified from the soluble fraction by fast protein liquid chromatography (Amersham) using a 1-mL HiTrap Chelating HP column (Amersham). The PDCB-containing fractions were further purified using an Amersham Hiload 16/60 Superdex 200 gel filtration column, and the resulting PDCB protein was concentrated using a Centricon YM-30 (Millipore) concentrator.
Gel Retardation Experiments
Thioredoxin-PDCB binding to complex carbohydrates was assessed using gel retardation assays as described by Barral et al. (2005). Briefly, nondenaturing PAGE gels (12% [w/v] polyacrylamide gels in 0.3 M Tris buffer, pH 8.0) were prepared containing 0.5 to 4 mg/mL of different polysaccharides (i.e., laminarin, hexalaminarin, lichenan, and carboxymethyl (CM) cellulose) in the separating gel. Lichenan (Sigma-Aldrich) was mixed with water at room temperature to dissolve low molecular weight sugars, which were then discarded. The remaining polysaccharide was solubilized in water by heating at 80°C for 30 min. CM cellulose (Sigma-Aldrich) also required heating at 80°C for 30 min for solubilization. Then, 1.5 μg of thioredoxin-PDCB and thioredoxin alone were loaded onto the gel and electrophoresed at 120 V for 3 h at room temperature. BSA (2 μg; Sigma-Aldrich) was run as an additional negative control. The proteins were visualized using Coomassie Brilliant Blue staining (Novexin; instant blue)
Immunogold Labeling
Polyclonal antibodies (York Bioscience) for PDCB1 were raised in rabbits using protein acquired by expression of 6xHis-tagged PDCB1 from the pDEST17 vector (Invitrogen). The cDNA was PCR amplified to eliminate the N-terminal signal peptide and GPI-anchor signal region, using the primers pDEST17FW and pDEST17RV. The clone was transformed into E. coli BL21 DES pLysE strain for protein expression. The insoluble protein was isolated from washed inclusion bodies by solubilization in 8 M urea and purified using affinity chromatography as described above for thioredoxin-PDCB1. Immunogold labeling for PDCB1 was achieved using anti-PDCB1 antibody diluted 1/300 and a rat anti-rabbit antibody conjugated to 25-nm gold particles (Aurion). Sections were processed following a short IGL protocol based on that described by Aurion. In brief, all solutions were filtered except the Aurion blocking buffer. All of the steps were performed on clean Parafilm, unless otherwise stated. Grids were floated section side down on drops of 50 mM glycine/PBS for 15 min and then immersed in Aurion blocking buffer for a further 30 min. Sections were then equilibrated with two drops of 0.1% BSA-C/PBS/0.1% Tween 20 (Solution A) over 5 min. The primary antibody was diluted in solution A, and grids were floated on 18- to 20-μL drops for 1 h at 37°C. The grids were then washed in solution A five times over 10 min and then floated on secondary antibody diluted in solution A for a further 1 h at 37°C. Grids were washed with PBS and water for 5 min each and then stained for 1 h by floating on drops of 2% uranyl acetate on clean dental wax. After washing in water five times and air drying, labeled sections were examined using a Jeol 1200Ex transmission electron microscope.
Immunoblotting
Crude protein extracts from transgenic and wild-type plants were separated on 10% SDS-PAGE gels, and the separated proteins were electroblotted onto Immobilon-P (Millipore). PDCB1-specific proteins were revealed using the anti-PDCB1 antibody, diluted 1:4000, and alkaline phosphatase-conjugated anti-rabbit secondary antibodies (1:8000; Sigma-Aldrich) and incubation with the chromogenic substrates (5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium).
PDCB Mutant Lines and GFP Cell-to-Cell Diffusion Assay
Arabidopsis Col-0 insertional mutant lines were obtained for At5g08000 (PDCB2) and At1g18650 (PDCB3) (Salk_034932 and Salk_043611.56.00.x, respectively). Confirmation that these were knockout lines was obtained by sequencing both ends of the insertion sites and by RT-PCR to analyze the specific transcribed RNAs; no RNA was detected from three replicate plants for each line (see Supplemental Figure 5 online). A combined knockout was also generated by crossing. We also attempted to generate PDCB1 RNAi lines in the pdcb2 pdcb3 double mutant background using the artificial miRNA strategy laid out by Schwab et al. (2006). Sequences for predicted target sites at coding nucleotides 241 to 261 (5′-CCTGGCTCTTGTAATTTCGAT-3′) and 286 to 306 (5′-GATCCCAGTTATACAGGTTGT-3′) were included in hairpin constructs inserted into the transformation vector pGreenII containing CaMV 35Spro.
Particle bombardment followed published methods (Levy et al., 2007b), using 5 μg of each DNA plasmid that were mixed and precipitated onto 1-μm gold particles (Bio-Rad). Fully expanded Arabidopsis leaves on plates (MS medium + 0.8% agar) were bombarded twice. GFP diffusion, counted as the numbers of secondary cells surrounding the primary target site (shown by RFPer), was analyzed 24 and 48 h after bombardment by confocal microscopy. Statistical nonparametric Mann-Whitney analysis was performed using Graph Prism software (GraphPad Software).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At5g61130 (PDCB1), At5g08000 (PDCCB2), At1g18650 (PDCB3), At3g58100, At1g69295, and At5g44130 (FLA13).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confocal Series through Arabidopsis Leaf Epidermal Pavement Cells Expressing 35S:YFP.PDCB1.
Supplemental Figure 2. Confocal Micrograph of N. benthamiana Tissues Transiently Expressing 35S:YFP.PDCB1.
Supplemental Figure 3. Confocal Microscopy of Unplasmolyzed Arabidopsis Leaf Spongy Mesophyll Cells Expressing 35S:YFP.PDCB1 Viewed at Increasing Power Settings.
Supplemental Figure 4. Arabidopsis Leaf Epidermal Pavement Cells Expressing 35S:YFP.PDCB1 Viewed after Plasmolysis without Pretreatment with the Phospholipase C Inhibitor.
Supplemental Figure 5. Alignment of the X8 Domain Amino Acid Sequences for Proteins with Structural Similarity to PDCB1.
Supplemental Figure 6. RNA Analysis for pdcb2 and pdcb3 Mutant Lines.
Supplemental Figure 7. GFP Diffusion in Arabidopsis Knockout Lines Is Unchanged in pdcb2 and -3 Knockout Lines and in a Transgenic Line Expressing the Unrelated GPI-Anchor Protein FLA13 (At5g44130).
Supplemental Figure 8. Heat Map Representation of the Public Expression Data for the PDCB Genes after Treatment of Arabidopsis with a Range of Stress Conditions.
Supplemental Figure 9. Quantitative RT-PCR Data for Changes in Expression for PDCB1, -2, and -3 after Plant Treatment with Heat or Wounding.
Supplemental Data Set 1. Sequences Used to Generate the Phylogeny in Figure 4.
Supplementary Material
Acknowledgments
We thank Jamie Ashby and Christophe Ritzenthaler for critical review of the manuscript prior to submission. We also thank Grant Calder for providing technical support on the confocal microscope, Vitaly Citovsky for providing us with pCitrine plasmid, and Rosalia Rodriguez for a sample of the OLE-e10 protein. Gateway plasmids were provided by the Flanders Interuniversity Institute for Biotechnology, Belgium, and Invitrogen. C.S. was in receipt of a UK Biotechnology and Biological Science Research Council Studentship. The John Innes Centre is grant-aided by the Biotechnology and Biological Science Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andrew J. Maule (andy.maule@bbsrc.ac.uk).
The online version of this article contains Web-only data.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Samaj, J., Napier, R., and Volkmann, D. (1999). Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J. 19 481–488. [DOI] [PubMed] [Google Scholar]
- Barral, P., Batanero, E., Palomares, O., Quiralte, J., Villalba, M., and Rodriguez, R. (2004). A major allergen from pollen defines a novel family of plant proteins and shows intra- and interspecies cross-reactivity. J. Immunol. 172 3644–3651. [DOI] [PubMed] [Google Scholar]
- Barral, P., Suarez, C., Batanero, E., Alfonso, C., Alche, J.D., Rodriguez-Garcia, M.I., Villalba, M., Rivas, G., and Rodriguez, R. (2005). An olive pollen protein with allergenic activity, Ole-e10, defines a novel family of carbohydrate-binding modules and is potentially implicated in pollen germination. Biochem. J. 390 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, E., Thomas, C.L., and Maule, A.J. (2004). Plasmodesmata in Arabidopsis thaliana suspension cells. Protoplasma 223 93–102. [DOI] [PubMed] [Google Scholar]
- Bayer, E., Bottrill, A., Walshaw, J., Vigouroux, M., Naldrett, M., Thomas, C., and Maule, A. (2006). Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6 301–311. [DOI] [PubMed] [Google Scholar]
- Beffa, R., and Meins, F. (1996). Pathogenesis-related functions of plant beta-1,3-glucanases investigated by antisense transformation - A review. Gene 179 97–103. [DOI] [PubMed] [Google Scholar]
- Blackman, L.M., and Overall, R.L. (1998). Immunolocalisation of the cytoskeleton to plasmodesmata of Chara corallina. Plant J. 14 733–741. [Google Scholar]
- Botha, C.E.J., and Cross, R.H.M. (2000). Towards reconciliation of structure with function in plasmodesmata - Who is the gatekeeper? Micron 31 713–721. [DOI] [PubMed] [Google Scholar]
- Bucher, G.L., Tarina, C., Heinlein, M., Di Serio, F., Meins, F., and Iglesias, V.A. (2001). Local expression of enzymatically active class I beta-1,3- glucanase enhances symptoms of TMV infection in tobacco. Plant J. 28 361–369. [DOI] [PubMed] [Google Scholar]
- Burgess, J. (1971). Observations on structure and differentiation in plasmodesmata. Protoplasma 73 83–95. [Google Scholar]
- Canto, T., and Palukaitis, P. (2005). Subcellular distribution of mutant movement proteins of Cucumber mosaic virus fused to green fluorescent proteins. J. Gen. Virol. 86 1223–1228. [DOI] [PubMed] [Google Scholar]
- Cantrill, L.C., Overall, R.L., and Goodwin, P.B. (1999). Cell-to-cell communication via plant endomembranes. Cell Biol. Int. 23 653–661. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium tumefaciens mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Ding, B., Haudenshield, J.S., Hull, R.J., Wolf, S., Beachy, R.N., and Lucas, W.J. (1992. b). Secondary plasmodesmata are specific sites of localization of the Tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4 915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., Kwon, M.O., and Warnberg, L. (1996). Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 10 157–164. [Google Scholar]
- Ding, B., Turgeon, R., and Parthasarathy, M.V. (1992. a). Substructure of freeze-substituted plasmodesmata. Protoplasma 169 28–41. [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Elortza, F., Nuhse, T.S., Foster, L.J., Stensballe, A., Peck, S.C., and Jensen, O.N. (2003). Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics 2 1261–1270. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protocols 2 953–971. [DOI] [PubMed] [Google Scholar]
- Escobar, N.M., Haupt, S., Thow, G., Boevink, P., Chapman, S., and Oparka, K. (2003). High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 15 1507–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP- Phylogeny interference package (version 3.2). Cladistics 5 164–166. [Google Scholar]
- Finn, R.D., et al. (2006). Pfam: Clans, web tools and services. Nucleic Acids Res. 34 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, K.L., Paquette, A.J., Nakajima, K., and Benfey, P.N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14 1847–1851. [DOI] [PubMed] [Google Scholar]
- Gomez, G., Torres, H., and Pallas, V. (2005). Identification of translocatable RNA-binding phloem proteins from melon, potential components of the long-distance RNA transport system. Plant J. 41 107–116. [DOI] [PubMed] [Google Scholar]
- Griesbeck, O., Baird, G.S., Campbell, R.E., Zacharias, D.A., and Tsien, R.Y. (2001). Reducing the environmental sensitivity of yellow fluorescent protein. Mechanisms and applications. J. Biol. Chem. 276 29188–29194. [DOI] [PubMed] [Google Scholar]
- Hamiduzzaman, M.M., Jakab, G., Barnavon, L., Neuhaus, J.M., and Mauch-Mani, B. (2005). Beta-aminobutyric acid-induced resistance against downy mildew in grapevine acts through the potentiation of callose formation and jasmonic acid signaling. Mol. Plant Microbe Interact. 18 819–829. [DOI] [PubMed] [Google Scholar]
- Hauck, P., Thilmony, R., and He, S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A.E., Kel, O.V., Ignatieva, E.V., Ananko, E.A., Podkolodnaya, O.A., Kolpakov, F.A., Podkolodny, N.L., and Kolchanov, N.A. (1998). Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 26 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., Zhang, Z., Olson, J.M., and Verma, D.P.S. (2001). A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 13 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, A.R., Heppler, M.L., Ju, H.J., Krishnamurthy, K., Payton, M.E., and Verchot-Lubicz, J. (2004). Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328 185–197. [DOI] [PubMed] [Google Scholar]
- Hughes, J.E., and Gunning, B.E.S. (1980). Glutaraldehyde-induced deposition of callose. Can. J. Bot. 58 250–258. [Google Scholar]
- Iglesias, V.A., and Meins, F. (2000). Movement of plant viruses is delayed in a beta-1,3-glucanase- deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 21 157–166. [DOI] [PubMed] [Google Scholar]
- Itaya, A., Woo, Y.-M., Masuta, C., Bao, Y., Nelson, R.S., and Ding, B. (1998). Developmental regulation of intercellular protein trafficking through plasmodesmata in tobacco leaf epidermis. Plant Physiol. 118 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.-Y., Rim, Y., Wang, J., and Jackson, D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19 788–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, S. (1999). Promotor 2.0: For the recognition of PolII promotor sequences. Bioinformatics 15 356–361. [DOI] [PubMed] [Google Scholar]
- Kurata, T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132 5387–5398. [DOI] [PubMed] [Google Scholar]
- Lambert, C., Leonard, N., De Bolle, X., and Depiereux, E. (2002). ESyPred3D: Prediction of proteins 3D structures. Bioinformatics 18 1250–1256. [DOI] [PubMed] [Google Scholar]
- Laporte, C., Vetter, G., Loudes, A.-M., Robinson, D.G., Hillmer, S., Stussi-Garaud, C., and Ritzenthaler, C. (2003). Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15 2058–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.-Y., Taoka, K.-i., Yoo, B.-C., Ben-Nissan, G., Kim, D.-J., and Lucas, W.J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 10 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, A., Erlanger, M., Rosenthal, M., and Epel, B.L. (2007. b). A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J. 49 669–682. [DOI] [PubMed] [Google Scholar]
- Levy, A., Guenoune-Gelbart, D., and Epel, B. (2007. a). β-1,3-Glucanases: Plasmodesmal gate keepers for intercellular communication. Plant Signal. Behav. 2 288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liarzi, O., and Epel, B.L. (2005). Development of a quantitative tool for measuring changes in the coefficient of conductivity of plasmodesmata induced by developmental, biotic, and abiotic signals. Protoplasma 225 67–76. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. (1998). The univector plasmid-fusion system, a method for rapid construction of recombinant DNA with restriction enzymes. Curr. Biol. 8 1300–1309. [DOI] [PubMed] [Google Scholar]
- Lodish, H., Berk, A., Zipursky, L.S., Matsudaira, P., Baltimore, D., and Darnell, J. (2000). Molecular Cell Biology. (New York: W.H. Freeman and Company).
- Lucas, W.J. (2006). Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344 169–184. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Bouchepillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its messenger-RNA through plasmodesmata. Science 270 1980–1983. [DOI] [PubMed] [Google Scholar]
- Maeda, H., Song, W., Sage, T.L., and DellaPenna, D. (2006). Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 18 2710–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule, A.J. (2008). Plasmodesmata: Structure, function and biogenesis. Curr. Opin. Plant Biol. 11 1–7. [DOI] [PubMed] [Google Scholar]
- Meikle, P.J., Bonig, I., Hoogenraad, N.J., Clarke, A.E., and Stone, B.A. (1991). The location of (1,3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1,3)-β-glucan-specific monoclonal antibody. Planta 185 1–8. [DOI] [PubMed] [Google Scholar]
- Muller-Decker, K. (1989). Interruption of TPA-induced signals by an antiviral and antitumoral xanthate compound: Inhibition of a phospholipase C-type reaction. Biochem. Biophys. Res. Commun. 162 198–205. [DOI] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D., and Heringa, J. (2000). T-Coffee: A novel method for multiple sequence alignments. J. Mol. Biol. 302 205–217. [DOI] [PubMed] [Google Scholar]
- Olesen, P. (1979). Neck constriction in plasmodesmata - Evidence for a peripheral sphincter-like structure revealed by fixation with tannic-Acid. Planta 144 349–358. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J. (2004). Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci. 9 33–41. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, L., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97 743–754. [DOI] [PubMed] [Google Scholar]
- Overall, R.L., and Blackman, L.M. (1996). A model of the macromolecular structure of plasmodesmata. Trends Plant Sci. 1 307–311. [Google Scholar]
- Parre, E., and Geitmann, A. (2005). More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol. 137 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W., Horgan, G.W., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt, S., Knight, A.E., Hodge, T.P., Baluska, F., Samaj, J., Volkmann, D., and Kendrick-Jones, J. (1999). Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J. 19 555–567. [DOI] [PubMed] [Google Scholar]
- Rinne, P.L., van den Boogaard, R., Mensink, M.G., Kopperud, C., Kormelink, R., Goldbach, R., and van der Schoot, C. (2005). Tobacco plants respond to the constitutive expression of the tospovirus movement protein NS(M) with a heat-reversible sealing of plasmodesmata that impairs development. Plant J. 43 688–707. [DOI] [PubMed] [Google Scholar]
- Ruan, Y.-L., Xu, S.-M., White, R., and Furbank, R.T. (2004). Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fibre elongation mediated by callose turnover. Plant Physiol. 136 4104–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi, G., Katz, A., Guenoune-Gelbart, D., and Epel, B.L. (2005). Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 17 1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, A., Giddings, T., Jr., and Staehelin, L. (1995). Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht, G., Brown, J.E., and Verchot-Lubicz, J. (2008). Plasmodesmata transport of GFP alone or fused to Potato virus X TGBp1 is diffusion driven. Protoplasma 232 143–152. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Blundell, T.L., and Mizuguchi, K. (2001). Fugue: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310 243–257. [DOI] [PubMed] [Google Scholar]
- Sivaguru, M., Fujiwara, T., Samaj, J., Baluska, F., Yang, Z., Osawa, H., Maeda, T., Mori, T., Volkmann, D., and Matsumoto, H. (2000). Aluminum-induced 1→3-beta-D-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol. 124 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, W., and Solovyev, I.A. (2004). PromH: Promotors identification using orthologous genomic sequences. Nucleic Acids Res. 31 3540–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, B.A., and Clarke, A.E. (1992). Chemistry and Biology of (1-3) Beta Glucans. (Victoria, Australia: La Trobe University Press).
- Svetek, J., Yadav, M.P., and Nothnagel, E.A. (1999). Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J. Biol. Chem. 274 14724–14733. [DOI] [PubMed] [Google Scholar]
- Thomas, C., Bayer, E., Ritzenthaler, C., Fernandez-Calvino, L., and Maule, A. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino, M.A., Palomares, O., Castrillo, I., Villalba, M., Rodriguez, R., Rico, M., Santoro, J., and Bruix, M. (2008). Solution structure of the C-terminal domain of Ole e9 a major allergen of olive pollen. Protein Sci. 17 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, A., Bacic, A., Harris, P.J., and Read, S.M. (1998). Membrane fractionation and enrichment of callose synthase from pollen tubes of Nicotiana alata link et Otto. Planta 205 380–388. [DOI] [PubMed] [Google Scholar]
- Turner, A., Wells, B., and Roberts, K. (1994). Plasmodesmata of maize root-tips - Structure and composition. J. Cell Sci. 107 3351–3361. [DOI] [PubMed] [Google Scholar]
- Ueki, S., and Citovsky, V. (2005). Identification of an interactor of cadmium ion-induced glycine-rich protein involved in regulation of callose levels in plant vasculature. Proc. Natl. Acad. Sci. USA 102 12089–12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, S., Deom, C.M., Beachy, R.N., and Lucas, W.J. (1989). Movement protein of Tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246 377–379. [DOI] [PubMed] [Google Scholar]
- Yoo, B.-C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.