Abstract
Potassium deprivation leads to large reductions in plant growth and yields. How plants sense and transduce the stress signals initiated by potassium deprivation is poorly understood. Both ethylene production and the transcription of genes involved in ethylene biosynthesis increase when plants are deprived of potassium. To elucidate the role of ethylene in low potassium signaling pathways, we used both genetic and chemical approaches. Our results showed that ethylene is important in tolerance to low potassium and for changes in both root hair and primary root growth in Arabidopsis thaliana. We show that ethylene acts upstream of reactive oxygen species in response to potassium deprivation. The expression of High-Affinity K+ Transporter5 was used as a marker of potassium deprivation and was found to be dependent on ethylene signaling. In the ethylene insensitive2-1 (ein2-1) mutant, the ethylene-mediated low potassium responses were not completely eliminated, suggesting that some potassium deprivation–induced responses are either ethylene independent or EIN2 independent. Ethylene signaling is a component of the plant's response to low potassium that stimulates the production of reactive oxygen species and is important for changes in root morphology and whole plant tolerance to low potassium conditions.
INTRODUCTION
Soil nutrients are essential for plant growth and metabolism. Plant roots acquire nutrients from soils and have developed adaptive mechanisms to ensure nutrient acquisition under the varying nutritional conditions in soil. When plants are deprived of nutrients such as potassium, roots activate two important adaptive mechanisms for the uptake of nutrients that help support growth and survival. One adaptation involves deploying additional nutrient acquisition and remobilization systems, such as transporters (Ashley et al., 2006; Gierth and Maser, 2007) or channels (Lebaudy et al., 2007). The other adaptation involves changes in developmental processes of roots, including primary root growth, lateral root formation, and root hair elongation (Lopez-Bucio et al., 2003). Architectural changes in root systems, in response to nutrition deprivation, help plants to take up more nutrients by increasing the absorptive surface in specific regions of the soil.
Plants require potassium in large quantities for growth. Since potassium availability in plants can be affected by various environmental conditions, including drought (Liebersbach et al., 2004), soil density (Kuchenbuch et al., 1986), and the presence of competitors such as Na+ and NH4+ (Qi and Spalding, 2004; Rus et al., 2004), plants may frequently experience potassium deficiency during their life cycle. Potassium deprivation leads to large reductions of the growth and yield of plants, and the addition of potassium is required for the optimal yields. Therefore, studying plant responses to potassium deficiency together with nitrogen and phosphorus deficiency is of significant agricultural importance as these are the nutrients that are added to soils in the largest quantities to ensure high yields.
Although certain aspects of how plants respond to potassium deprivation at physiological and transcriptional levels are known, the details of the signaling pathway in response to low potassium are poorly characterized (Schachtman and Shin, 2007). It was reported that potassium starvation activates K+ uptake in plant roots by regulating the activity of potassium transport proteins (Ashley et al., 2006). Among the many potassium channels and transporters in Arabidopsis thaliana, it is well known that Arabidopsis K+ Transporter1 (AKT1), an inwardly rectifying potassium channel, and HAK5, a low K+-inducible potassium transporter, are major contributors to K+ uptake in response to low K+ conditions (Gierth et al., 2005; Gierth and Maser, 2007; Schachtman and Shin, 2007). Several independent studies showed that the transcription of HAK5 is strongly and rapidly activated in response to potassium deprivation in Arabidopsis and other plant species (Shin and Schachtman, 2004; Gierth et al., 2005). Recently, the two calcium binding proteins Calcineurin B-Like1 (CBL1) and CBL9 and their target CBL-Interacting Protein Kinase23 (CIPK23), a Ser/Thr protein kinase, were shown to form a protein complex in response to low concentrations of potassium in soil that activates the AKT1 potassium channel by phosphorylation (Li et al., 2006; Xu et al., 2006a; Cheong et al., 2007; Lee et al., 2007). This study implicated calcium in the low potassium signaling pathway.
Reactive oxygen species (ROS) are known to be involved in low potassium signaling pathways and have also been suggested to be an upstream regulator of calcium signaling (Li et al., 2006; Lebaudy et al., 2007). Previously, we demonstrated that H2O2 concentrations increase in plant roots in response to potassium deprivation and that low K+- inducible HAK5 expression is dependent on the ROS production (Shin and Schachtman, 2004). We also found that roots deprived of K+ induce the expression of genes involved in ethylene biosynthesis and signaling and produce approximately twofold higher levels of ethylene (Shin and Schachtman, 2004). These results suggested that ethylene may be involved in the low potassium signaling pathway.
Ethylene is a gaseous plant hormone that regulates a wide range of cellular and developmental processes, including cell expansion, senescence, leaf abscission, seed germination, and fruit ripening (Guo and Ecker, 2004; Chen et al., 2005; Etheridge et al., 2006). Ethylene biosynthesis and signaling have been well characterized at the molecular level, providing many genetic tools that can be used to dissect how ethylene signaling interacts with the plant's response to potassium deprivation. Briefly, we describe some of the different molecules involved in ethylene biosynthesis and signaling. In many cases, mutants in these processes are available for testing specific hypotheses. The 1-aminocyclopropane-1-carboxylic acid (ACC) synthases produce ACC, which is converted into ethylene by ACC oxidases. ETHYLENE OVERPRODUCER (ETO) proteins negatively regulate ethylene biosynthesis, and eto mutants produce 10 to 50 times more ethylene than the wild type (Woeste et al., 1999; Chae et al., 2003). In Arabidopsis, there are five ethylene receptors (ETHYLENE RESPONSE1 [ETR1], ETR2, ETHYLENE RESPONSE SENSOR1 [ERS1], ERS2, and ETHYLENE INSENSITIVE [EIN4]) that act negatively and redundantly in the perception of ethylene. Some gain-of-function mutants, including etr1-1, etr1-3, and ein4, show ethylene-insensitive phenotypes due to the constitutive activation of receptors by the mutation of ethylene binding sites. Ethylene receptors interact directly with CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), a Raf-like Ser/Thr protein kinase. CTR1 is also a negative regulator in ethylene signaling, and ctr1-1 shows constitutive ethylene responses (Clark et al., 1998; Gao et al., 2003). EIN2, which is similar to Nramp metal transporters, acts downstream of CTR1 and plays a major role in ethylene responses. Loss-of-function mutants such as ein2-1 and ein2-5 show complete ethylene insensitivity, indicating that EIN2 is an essential positive regulator of the pathway (Chen et al., 2005). Downstream of EIN2, EIN3/EIN3-Like transcription factors stimulate the expression of ethylene response target genes leading to ethylene responses of plants (Guo and Ecker, 2004).
Although ethylene is known to be an important component in low phosphorus or iron signaling (Lopez-Bucio et al., 2003; Zaid et al., 2003; Zhang et al., 2003), it is not clear whether ethylene plays a role in low K+ signaling. Using chemical and genetic approaches, we found that ethylene signaling plays an important role in low K+-induced plant responses, such as primary root growth inhibition, root hair elongation, ROS production, and HAK5 expression. Furthermore, we established that ethylene acts upstream of ROS in response to potassium deprivation. Ultimately, we have shown that ethylene signaling mediates the adaptive changes that increase the tolerance of Arabidopsis to low potassium conditions.
RESULTS
Role of Ethylene in Plant Growth under Low K+ Stress
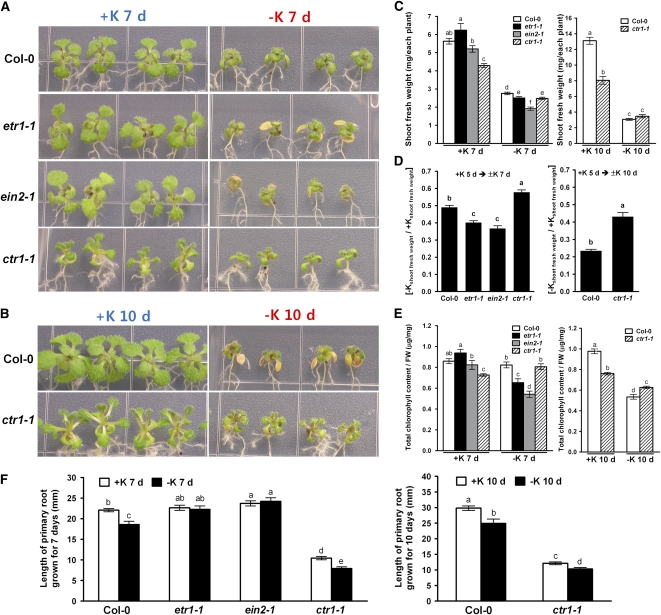
A previous study showed that potassium deprivation in Arabidopsis roots stimulates ethylene production and the upregulation of genes that are involved in ethylene biosynthesis and signaling (Shin and Schachtman, 2004). To investigate the possible role of ethylene in response to low potassium, we analyzed the low K+ stress sensitivity of ethylene-insensitive mutants (etr1-1 and ein2-1) and a constitutive ethylene response mutant (ctr1-1). After 7 d of K+ deprivation, the leaves of the ethylene-insensitive mutants were more chlorotic than those of the wild type (Figure 1A), and after 10 d of K+ deprivation, the constitutive response mutants were less chlorotic than those of the wild type (Figure 1B). Shoot fresh weights (Figure 1C) decreased in response to K+ deprivation in both the insensitive mutants and the wild type. However, the relative decrease in shoot fresh weight due to low K+ stress was significantly greater for the ethylene-insensitive mutants etr1-1 and ein2-1 and significantly less for ctr1-1 compared with the wild type (Figure 1D). Leaf chlorophyll content was also significantly lower under K+-deficient conditions in the insensitive mutants compared with the wild type or the ctr1-1 mutant (Figure 1E). There was no difference between wild-type and ctr1-1 plants grown under K+-deficient conditions for 7 d, except that ctr1-1 had a higher relative shoot fresh weight (Figure 1D). However, after 10 d of K+ deprivation, ctr1-1 appeared to be more tolerant than the wild type, based on the relative shoot fresh weight and total chlorophyll content (Figures 1B, 1D, and 1E). Under low K+ stress, primary root growth was inhibited in wild-type plants and ctr1-1 but not in etr1-1 or ein2-1 (Figure 1F). These results highlight the physiological role of ethylene signaling in plant responses to K+ deprivation.
Figure 1.
Growth and Cholorophyll Content of Arabidopsis Col-0 and Ethylene Mutants under Complete Nutrient Conditions and K+-Deficient Conditions.
(A) Plants were grown under complete nutrient conditions (+K, 5 mM K+) for 5 d and then transferred to medium with either complete nutrients (+K) or no potassium (−K) for 7 d.
(B) Plants were grown under complete nutrient conditions (+K, 5 mM K+) for 5 d and then transferred to either full nutrients (+K) or no potassium (−K) for 10 d.
(C) Shoot fresh weights of plants shown in (A) and (B). Values are mean ± se from 10 seedlings per replicate (n = 5 replicates).
(D) Ratio of shoot fresh weights of plants grown on complete medium for 5 d and then transferred to medium without K+ to those of plants transferred to K+ sufficient conditions, as shown in (C).
(E) Total chlorophyll content of plants shown in (A) and (B). The total chlorophyll content was expressed as micrograms per milligram shoot fresh weight. Values are mean ± se from 10 seedlings per replicate (n = 5 replicates).
(F) Primary root length of plants shown in (A) and (B). Root length was measured 7 or 10 d after K+ deprivation (n = 50 roots ± se).
Means that have different letters at the top of each bar are significantly different (P < 0.05; t test).
Ethylene Biosynthesis and Action Blockers Reduce the K+ Starvation-Induced ROS Production in Roots
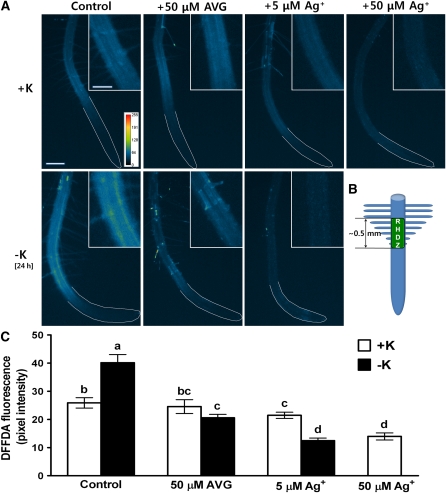
Potassium starvation induces an increase in ROS in Arabidopsis roots, which has been implicated in regulating gene transcription in response to low concentrations of K+ (Shin and Schachtman, 2004). To test whether there is a relationship between ethylene and low K+-induced ROS production, we used several methods to block ethylene signaling in roots. Two different ethylene inhibitors were used. Silver ion (Ag+) is an ethylene action blocker, and 2-aminoethoxyvinyl-glycine (AVG) inhibits ACC synthase (ACS) and ethylene biosynthesis. For the localization and measurement of ROS in roots, we employed a membrane-permeable fluorescent dye called 5-(and 6-) carboxy-2',7'-difluorodihydrofluorescein diacetate (DFFDA), an improved photostable version of 2′7′-dichlorodihydrofluorescein diacetate. Under complete nutrient conditions, only small amounts of ROS were detected, mainly in the root hair–differentiation zone (RHDZ; Figures 2A and 2C). Under K+-deficient conditions, ROS increased significantly in the RHDZ (see inset in Figure 2A and quantified data in Figure 2C), especially in what appears to be the epidermal cells that contain root hairs, but this localization needs to be confirmed using known epidermal cell markers. The low K+-induced ROS production in RHDZ was inhibited by the presence of AVG (50 μM; 2-h treatment) and almost completely blocked by Ag+ (5 μM; 2-h treatment), which inhibits ethylene perception (Figures 2A and 2C). The ROS concentrations detected under complete nutrient conditions were reduced by higher concentrations of Ag+ (50 μM; 2-h treatment), showing that ethylene stimulates ROS production under the complete nutrient conditions, but to a lesser degree than under nutrient-deprived conditions.
Figure 2.
Ethylene Inhibitors Reduce Low K+-Induced ROS Production.
(A) Pseudocolor images of ROS fluorescence are shown for K+-sufficient (+K, 1.75 mM K+) roots and for K+-starved (no K+) roots with or without Ag+ and AVG. Roots were deprived of K+ for 24 h and treated with ethylene inhibitors for 2 h. ROS images were collected following staining of 3-d-old roots with 20 μM DFFDA. Yellow and red colors indicate higher ROS production (see scale inset in pixel intensity). Insets show enlargements of the root hair differentiation zone. White lines were drawn by hand to outline the root. Bar = 200 μm; bar in inset = 100 μm.
(B) Diagram showing the RHDZ, which is the region where ROS was quantified in (C).
(C) Quantified data from representative images shown in (A) (n = 7 to 10 images of individual seedlings ± se).
Results from one of three independent experiments are shown here. Means that have different letters at the top of each bar are significantly different (P < 0.05; t test).
Ethylene Acts Upstream of ROS in Arabidopsis Roots
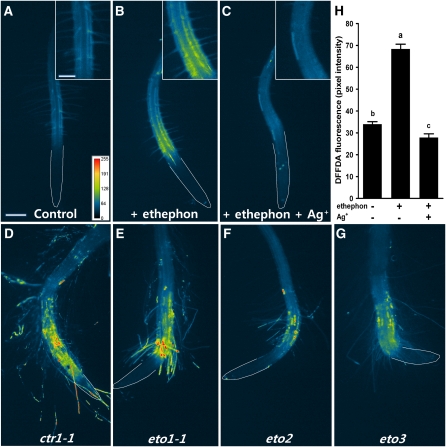
Our finding that low K+-induced ROS production is blocked by ethylene inhibitors suggested that ethylene may act upstream of ROS. To determine whether ethylene acts upstream of ROS, we investigated the effect of ethephon, a chemical that is metabolized into ethylene, on the ROS production in Arabidopsis roots. When roots were treated with ethephon, ROS production increased significantly in the RHDZ (Figures 3B and 3H). This ethephon-induced ROS production was greatly reduced by Ag+ (Figures 3C and 3H). Moreover, constitutive ethylene response mutants, such as ctr1-1, eto1-1, eto2, and eto3, had higher ROS levels than the wild type (Figures 3D to 3G).
Figure 3.
Ethephon Treatment of Col-0 and Constitutive Ethylene Response Mutants under Complete Nutrient Conditions Resulted in Higher Levels of ROS Production.
Pseudocolor images of ROS fluorescence are shown for K+-sufficient wild-type roots (1.75 mM K+) in the absence (A) or presence (B) of ethephon or ethephon plus Ag+ (C). Three-day-old roots were treated with 50 μM ethephon and/or 50 μM Ag+ for 3 h, followed by staining with 20 μM DFFDA. Fluorescence images of ctr1-1 (D), eto1-1 (E), eto2 (F), and eto3 (G) roots are shown. As indicated on the pesudocolor scale (in pixel intensity), yellow and red colors indicate a high ROS production. Inset shows enlargement of RHDZ. White lines were drawn by hand to show the boundary of roots. (H) shows the quantified data of (A) to (C) (n = 10 seedlings ± se). Means that have different letters at the top are significantly different (P < 0.05; t test). Bar = 200 μm; bar in inset = 100 μm.
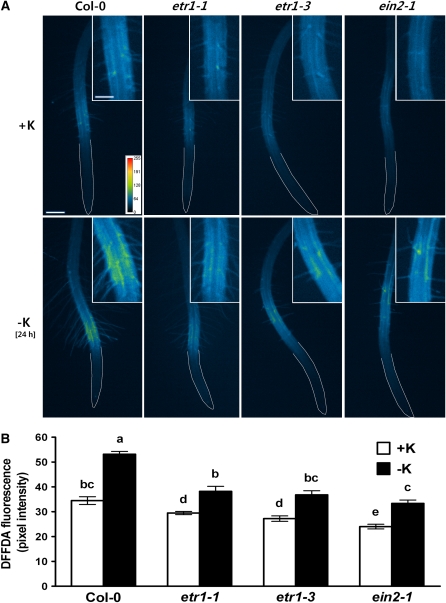
To determine whether the ETR1 and EIN2 proteins are important components in low K+-induced ROS production, we also measured ROS levels in etr1-1, etr1-3, and ein2-1 under K+-deficient conditions. Our data show that K+ deprivation stimulated ROS production in these mutants, but to a lesser extent than it did in the wild type Columbia (Col-0) (Figures 4A and 4B).
Figure 4.
K+ Deficiency Induces Increased ROS in Ethylene-Insensitive Mutants.
(A) Pseudocolor images of ROS fluorescence are shown for etr1-1, etr1-3, and ein2-1 roots grown on K+-sufficient (+K, 1.75 mM K+) and K+-deficient (no K+) media. Roots were deprived of K+ for 24 h. ROS was visualized by staining 3-d-old roots with 20 μM DFFDA. Yellow and red colors indicate higher ROS production (see pseudocolor scale). Inset shows enlargement of RHDZ. White lines show the boundary of roots. Bar = 200 μm; bar in inset = 100 μm.
(B) Quantified data from images as shown in (A) (n = 10 seedlings ± se).
Means that have different letters at the top are significantly different (P < 0.05; t test).
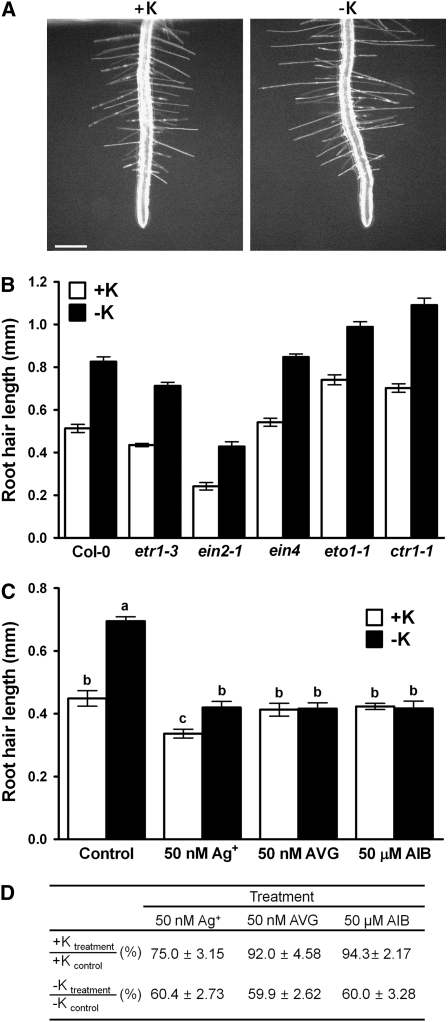
K+ Deprivation Stimulates Root Hair Elongation in Col-0 and All Ethylene Mutants Tested, but Not in the Presence of Ethylene Inhibitors
Previously, it was reported that K+ deprivation enhances ROS and ethylene production in roots (Shin and Schachtman, 2004). Since ROS and ethylene are known to be essential for root hair elongation (Tanimoto et al., 1995; Pitts et al., 1998; Foreman et al., 2003), we tested whether K+ deprivation could affect root hair elongation. We measured root hair length of plants that were grown on K+-sufficient and K+-deficient medium (Figure 5). K+ deprivation stimulated root hair elongation in the Col-0 wild type (Figures 5A and 5B; P < 0.05). We also tested several ethylene mutants to determine which components of the ethylene signaling pathway might be involved in low K+-induced root hair elongation. K+ deprivation induced root hair elongation in all ethylene mutants tested (Figures 5A and 5B). To determine whether ethylene inhibitors reduce the low K+-induced root hair elongation, we measured root hair length of plants grown on plates containing K+-sufficient and K+-deficient medium with or without Ag+, AVG, and aminoisobutyric acid (AIB; an ethylene biosynthesis inhibitor that inhibits ACC oxidase). The low K+-induced root hair elongation was inhibited (Figures 5C and 5D; P < 0.05) by very low concentrations of Ag+ (50 nM; 22-h treatment), AVG (50 nM; 22-h treatment), and AIB (50 μM; 22-h treatment). Since ectopic root hair formation is known to be a specific indicator of ethylene activity (Tanimoto et al., 1995), we tested whether K+ deprivation induces ectopic root hair formation. K+ deprivation did not cause ectopic root hair formation in Arabidopsis (see Supplemental Table 1 online).
Figure 5.
K+ Deprivation Induces Root Hair Elongation in the Wild Type and All Ethylene Mutants Tested but Not in the Presence of Ethylene Inhibitors.
(A) Light microscopy images showing root hair length of wild-type Col seedlings grown under K+-sufficient (+K, 1.75 mM K+) and K+-deficient (−K, no K+) conditions. Three-day-old roots deprived of K+ for 28 h were used for the measurement of root hair length in (A) and (B). Bar = 0.5 mm.
(B) Root hair length of seedlings grown under K+-sufficient (+K) and K+-deficient (−K) conditions (n = 8 to 10 seedlings, means ± se). As in the wild type, K+ deprivation stimulated root hair elongation in all of the ethylene mutants tested (P < 0.05; t test).
(C) Root hair length of Col-0 in the absence or presence of 50 nM Ag+, 50 nM AVG, and 50 μM AIB under K+-sufficient (+K, 5 mM K+) or K+-deficient (−K, no K+) conditions (n = 10 seedlings, means ± se). Root hair length of 3-d-old Col-0 roots was measured 22 h after K+ deprivation and ethylene inhibitor treatment. Different letters indicate that means are significantly different (P < 0.05; t test).
(D) Root hair length when treated with Ag+, AVG, or AIB as a percentage of untreated control root hair length under either complete nutrient or K+-deprived conditions. The ratio (expressed as a percentage) of root hair lengths of treated plants grown on complete medium to those of untreated control plants, for example, [+Ktreatment]/[+Kcontrol] (%), and the ratio of root hair lengths of treated plants to those of untreated control plants grown under low potassium conditions, for example, [−Ktreatment]/[−Kcontrol] (%), were calculated from data shown in (C). Data are means ± se.
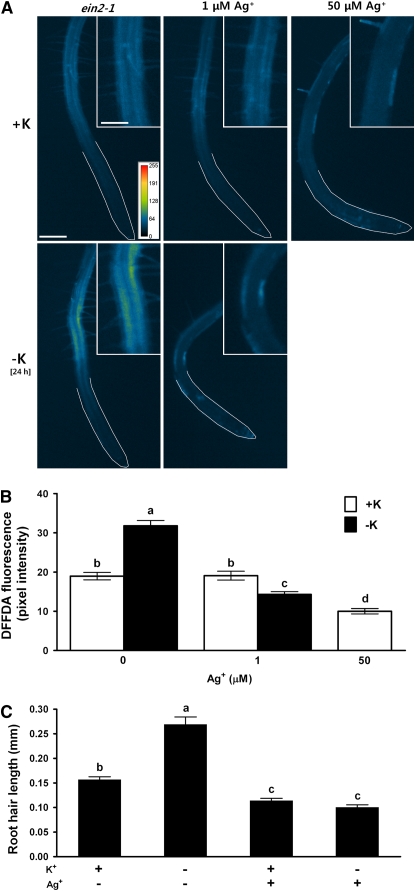
Low K+-Induced ROS Production and Root Hair Elongation in ein2-1 Are Blocked by Ag+
EIN2 is considered to be a central component in ethylene signaling because ein2-1 and ein2-5 mutants are completely ethylene insensitive (Chen et al., 2005). That low K+ still stimulates ROS production and root hair elongation in ein2-1 suggested the existence of other signaling pathways or an EIN2-independent ethylene pathway that may play an important role in low K+ signaling. To determine whether low K+-induced ROS production and root hair elongation in ein2-1 can be further reduced by ethylene inhibitors, we treated ein2-1 roots with Ag+ under K+-sufficient and K+-deficient conditions. ROS production and root hair elongation induced by K+ deprivation were blocked by Ag+ in ein2-1 roots (Figures 6A to 6C). Moreover, even under complete nutrient conditions, ein2-1 roots treated with Ag+ had significantly lower concentrations of ROS and shorter root hairs than roots not treated with Ag+, indicating the presence of an ethylene- or EIN2-independent pathway.
Figure 6.
Ag+ Reduces Low K+-Induced ROS Production and Low K+-Induced Root Hair Elongation in ein2-1.
(A) ROS production increases in ein2-1 under low potassium conditions, and this increase is blocked by Ag+. Pseudocolor images (pixel intensity) of ROS fluorescence are shown for K+-sufficient (+K, 1.75 mM K+) roots and for K+-starved (−K, no K+) roots with or without Ag+ (1, 50 μM; 2-h treatment). Three-day-old ein2-1 roots were used 24 h after K+ deprivation. Bar = 200 μm; bar in inset = 100 μm.
(B) Quantified data of images shown in (A) (n = 10 to 15 images of roots ± se).
(C) Root hair length of ein2-1 plants in the absence (−) or presence (+) of 150 nM Ag+ under K+-sufficient (+K, 5 mM K+) and K+-deficient (−K, no K+) conditions. Root hair length of 3-d-old ein2-1 roots was measured 22 h after K+ deprivation and Ag+ treatment (n = 10 seedlings, means ± se). Means that have different letters at the top are significantly different (P < 0.05; t test).
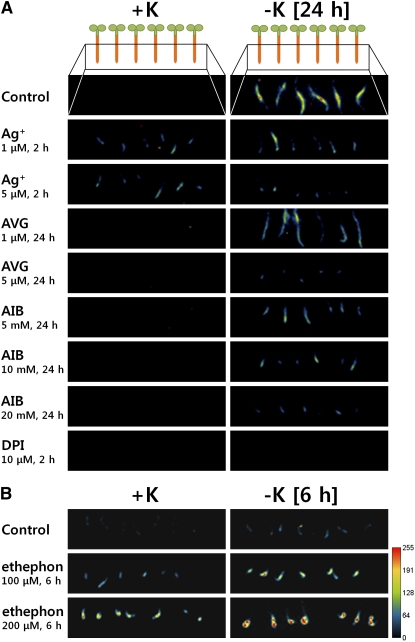
Ethylene Inhibitors and Producers Alter K+ Deprivation-Induced HAK5pro:LUCIFERASE Expression
HAK5 is a high-affinity K+ transporter that is a major contributor to K+ uptake under very low K+ conditions (Gierth and Maser, 2007; Qi et al., 2008). It has been established that K+ deprivation strongly induces HAK5 expression (Ahn et al., 2004; Armengaud et al., 2004; Shin and Schachtman, 2004; Gierth et al., 2005). To determine whether ethylene signaling is important for HAK5 expression, we tested the effect of ethylene inhibitors and producers on the low K+-induced HAK5 expression in roots. To monitor HAK5 expression by the activity of a HAK5 promoter in vivo, a homozygous transgenic line carrying HAK5pro:LUCIFERASE (HAK5pro:LUC) constructs was used. HAK5pro:LUC was strongly induced 24 h after K+ deprivation (Figure 7A). The low K+-induced HAK5pro:LUC expression was almost completely inhibited by Ag+, AVG, and AIB. Diphenylene iodonium (DPI), an inhibitor of NADPH oxidases, also completely inhibited low K+-induced HAK5pro:LUC expression (Figure 7A), confirming that HAK5 expression is ROS dependent (Shin and Schachtman, 2004). HAK5pro:LUC expression was greatly enhanced in roots treated with ethephon under both complete nutrient conditions and K+-deficient conditions (Figure 7B). Moreover, ACC, an ethylene precursor, also induced HAK5pro:LUC expression under complete nutrient conditions (see Supplemental Figure 1 online).
Figure 7.
Ethylene Inhibitors Reduce Low Potassium-Induced Expression of HAK5, whereas Chemicals That Stimulate Ethylene Production Increase HAK5 Expression in Arabidopsis Roots, even under Nutrient-Sufficient Conditions.
Pseudocolored luminescence images of homozygous transgenic Arabidopsis seedlings transformed with HAK5pro:LUC. Orange part of the illustration at the top shows the root region that was imaged for (A) and (B). Three-day-old seedlings were incubated with ethylene inhibitors (Ag+, AVG, and AIB) or an NADPH oxidase inhibitor (DPI) for the times indicated in the figure, or with an ethylene producer (ethephon) for 6 h, under K+-sufficient (+K, 5 mM K+) and K+-deficient (−K, no K+) conditions. Yellow and red colors indicate a high luciferase expression. Experiments were performed three times, and data from one experiment are shown here.
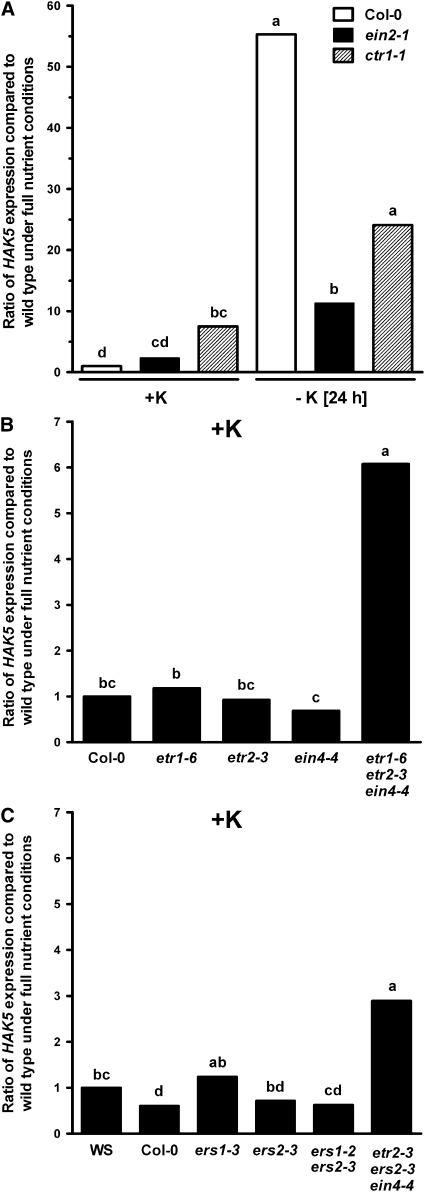
HAK5 Expression Is Altered in Ethylene Mutants under Complete Nutrient and K+-Deficient Conditions
To further determine which components of ethylene signaling are important for low K+-induced HAK5 expression, quantitative real-time RT-PCR was used to measure the HAK5 mRNA level in the wild type and ethylene mutants under complete nutrient and K+-deficient conditions. HAK5 expression was induced by K+ deprivation in the wild type, as has been previously shown (Ahn et al., 2004; Figure 8A). However, the induction of HAK5 expression was significantly reduced in ein2-1, an ethylene-insensitive mutant, compared with the wild type. In the constitutive ethylene response mutant ctr1-1, HAK5 expression was higher under complete nutrient conditions compared with the wild type. However, K+ deprivation could still enhance HAK5 expression in both mutants. These results indicate that CTR1 and EIN2 are partially involved in low K+-induced HAK5 expression.
Figure 8.
HAK5 Expression Is Altered in Ethylene Mutants Grown under Complete Nutrient Conditions and K+-Deficient Conditions.
(A) HAK5 expression levels in ein2-1 and ctr1-1 mutants under K+-sufficient (+K, 5 mM K+) and K+-deficient (no K+) conditions relative to those in the wild type grown on complete medium (set to 1).
(B) and (C) Relative HAK5 expression levels in the combinatorial loss-of-function mutants of five ethylene receptors under K+-sufficient (+K, 5 mM K+) conditions.
Three-day-old seedlings were deprived of K+ for 24 h. The expression data were obtained by quantitative RT-PCR. A β-tubulin gene was used as a reference gene. The fold change of HAK5 level in each sample was compared with the HAK5 level of the wild-type control under K+-sufficient conditions. Different letters above the bars indicate values that are significantly different (n = 3, P < 0.05; t test). Col-0 was used as a control for the ein2-1, ctr1-1, etr1-6, etr2-3, ein4-4, and etr1-6 etr2-3 ein4-4 plants, whereas Wassilewskija (WS) was used as a control for the ers1-3, ers2-3, and ers1-2 ers2-3 plants. The triple mutant etr2-3 ers2-3 ein4-4 is in the background of Col-0 and Wassilewskija. All loss-of-function mutants are predicted to be null mutants, except for ers1-2, which is known to have a decreased level of full-length transcripts (Binder, 2008).
In Arabidopsis, CTR1 and EIN2 act downstream of five ethylene receptors (ETR1, ETR2, ERS1, ERS2, and EIN4) in the ethylene signaling pathway (Guo and Ecker, 2004; Chen et al., 2005). Since ethylene receptors, together with CTR1, are known to be negative regulators in ethylene signaling, we hypothesized that loss-of-function mutants of ethylene receptors may have higher HAK5 expression due to constitutive activation of ethylene signaling. To test this hypothesis, several loss-of-function mutants of five ethylene receptors, including single, double, or triple mutants, were used to study HAK5 expression under full nutrient conditions. There was no difference in HAK5 expression between the wild type and single loss-of-function mutants of five ethylene receptors under full nutrient conditions (Figures 8B and 8C). However, a significant increase in HAK5 expression was detected in triple loss-of-function mutants, such as etr1-6 etr2-3 ein4-4 and etr2-3 ers2-3 ein4-4, when grown under complete nutrient conditions (Figures 8B and 8C). Under low K+ conditions, HAK5 expression also increased in the triple loss-of-function mutants compared with Col-0 (see Supplemental Figure 2 online).
DISCUSSION
Despite the importance of potassium for plant growth and agricultural productivity, root sensing and the subsequent signal transduction pathways in response to K+ deprivation have not been completely elucidated. It has been established that K+ deprivation induces the expression of the potassium transporter HAK5 and activates the potassium channel AKT1 via CIPK23 (Armengaud et al., 2004; Shin and Schachtman, 2004; Gierth et al., 2005; Li et al., 2006; Xu et al., 2006a; Cheong et al., 2007; Lee et al., 2007). Low K+ also stimulates ROS production in roots, and the induced expression of HAK5 is dependent on ROS production (Shin and Schachtman, 2004). K+ deficiency was shown to increase ethylene production, suggesting the involvement of ethylene in low K+ signaling pathways (Shin and Schachtman, 2004). In this study, using both genetic and chemical approaches, we determined that ethylene signaling acts upstream of ROS when plants are deprived of potassium. This work adds a component to potassium signaling that is near the top of the transduction cascade and provides strong evidence linking it to downstream components, including ROS and HAK5 expression. Our data also revealed that both the primary root response and the root hair response to low potassium involve ethylene-mediated pathways. Finally, we also show that ethylene is important in plant tolerance to low potassium conditions.
Ethylene as a Key Player for Biotic and Abiotic Stress Signaling
Plants under biotic or abiotic stress, such as pathogen attack, salt, wounding, drought, heat, flooding, low phosphorus (Borch et al., 1999), and low iron (Romera et al., 1999), produce higher levels of ethylene than nonstressed plants. It is now becoming clear that various ethylene-regulated stress responses are essential for stress tolerance and the survival of plants. For example, the submergence tolerance in rice (Oryza sativa) is determined by the levels of ethylene and the activity of SUBMERGENCE1A (SUB1A), which is induced by ethylene (Xu et al., 2006b; Perata and Voesenek, 2007; Fukao and Bailey-Serres, 2008). In addition, ethylene-insensitive mutants (etr1-1, ein2-1, and ein4-1) are more sensitive to high salt concentrations than the wild type, whereas a constitutive ethylene response mutant (ctr1-1) is more tolerant, supporting the important role of ethylene in salt stress signaling (Achard et al., 2006; Cao et al., 2007). Ethylene production increases when plants are deprived of K+ (Shin and Schachtman, 2004), and in this work, we showed that ethylene-insensitive mutants are more sensitive to low K+ in terms of leaf chlorosis and shoot growth than is the wild type. These findings implicate ethylene as an important component in the plant response to low K+ stress.
Although ethylene appears to be important in a wide range of biotic and abiotic stresses, the role of ethylene in each specific stress may be different. Specificity in response may be determined by enzymes such as ACS, which is a key player in ethylene synthesis. The Arabidopsis genome encodes nine ACS isoforms that can form functional heterodimers as well as functional homodimers (Tsuchisaka and Theologis, 2004b). Each isoform of ACS is temporally and spatially differentially expressed (Tsuchisaka and Theologis, 2004a) and is also regulated by different stresses and hormones (Wang et al., 2002). For example, ACS2 and ACS7 are transcriptionally upregulated by salt stress (Achard et al., 2006), while ACS6 is induced by ozone, touch, or ethylene treatment. In rice, it was suggested that ethylene synthesis via ACS1 and ACS5 regulates cell division and elongation during submergence (Zarembinski and Theologis, 1997; Zhou et al., 2002). In the case of potassium deprivation, the ACC synthases ACS2 and ACS6 are induced in response to K+ deprivation (see data in Genevestigator database, https://www.genevestigator.ethz.ch/; Zimmermann et al., 2004). The complexity of ACS regulation by various stress signals suggests that ethylene may play distinct functional roles in response to different types of stress.
Ethylene Acts Upstream of ROS in Roots
We showed that ethephon induces ROS production in the RHDZ and that ethylene mutants have altered ROS levels in the RHDZ (Figures 3 and 4). These findings indicate that ethylene can act upstream of ROS in Arabidopsis roots. However, we cannot exclude the possibility that there might be positive feedback regulation so that ethylene-induced ROS may further stimulate ethylene production (Wang et al., 2002). The ethylene-induced ROS production in roots was partially NADPH oxidase dependent since DPI almost completely blocked the ethephon-induced ROS production (see Supplemental Figure 3 online). Other reports have shown a relationship between ethylene and ROS in several signaling and developmental processes. ROS acts upstream of ethylene in wounding and UV-B stress responses and in the root nodulation process of a semiaquatic legume, Sesbania rostrata (Wang et al., 2002; D'Haeze et al., 2003). In guard cells of Arabidopsis, ethylene stimulates ROS production via Respiratory Burst Oxidase HomologF (RBOHF) NADPH oxidase, leading to stomatal closure, indicating that ethylene acts upstream of ROS in this specialized cell type (Desikan et al., 2006). Although most of the results of this previous study support our current findings, in that study, ethephon-induced ROS production was completely blocked in guard cells of etr1-1, etr1-3, and rbohF, which is in contrast with our findings in roots where ethephon still induces ROS production in etr1-1, etr1-3, and rbohF plants (see Supplemental Figure 4 online).
Ethylene Signaling Plays an Important Role in Low K+ Signaling Pathways
Since HAK5 is known to be induced by low potassium (Armengaud et al., 2004; Shin and Schachtman, 2004; Gierth et al., 2005), we used the expression of this gene as a marker to further understand the upstream components of the signal transduction pathway involved in K+ starvation-induced signaling. Our data showed that HAK5pro:LUC expression in response to low concentrations of K+ was almost completely blocked by three ethylene inhibitors (i.e., Ag+, AVG, and AIB), which act on different targets to inhibit ethylene signaling, while HAK5pro:LUC expression was greatly induced by two ethylene producers (namely, ethephon and ACC; Figure 7). These results suggest that HAK5 expression under low concentrations of K+ is ethylene dependent. We also showed that low K+-induced HAK5 expression was significantly reduced in ein2-1 (Figure 8A). The ctr1-1 and the triple loss-of-function mutants of ethylene receptors had higher HAK5 expression than the wild type under full nutrient conditions due to the constitutive activation of ethylene signaling (Figures 8B and 8C). However, there was no difference in HAK5 expression between the wild type and any of the five single loss-of-function mutants of ethylene receptors and a double mutant (ers1-2 ers2-3), which has residual levels of ERS1 expression under complete nutrient conditions (Binder, 2008). These findings are consistent with the observation that constitutive ethylene responses are observed in triple or quadruple loss-of-function mutants of ethylene receptors but not in single loss-of-function mutants (Hua and Meyerowitz, 1998; Cao et al., 2007). Our data connect HAK5 expression with the low K+ signaling pathway via components of the ethylene signaling pathway.
We showed in this work that K+ deprivation inhibits primary root growth while it promotes root hair elongation (Figures 1F and 5B). Similar morphological changes in root systems have also been noted in ethylene-treated plants or constitutive ethylene response mutants. Root growth inhibition is one of the classical ethylene triple responses (Wang et al., 2002), and ethylene stimulates root hair elongation (Pitts et al., 1998). Our findings demonstrate a role for ethylene in low K+-induced inhibition of primary root growth and promotion of root hair elongation. Moreover, K+ deprivation inhibits primary root growth in the wild type, but not in the etr1-1 and ein2-1 mutants, suggesting that the primary roots of the mutants etr1-1 and ein2-1 may lack the ability to sense or transduce the low K+ stress conditions and that low K+-induced root growth inhibition is ETR1 and EIN2 dependent. The ethylene-mediated morphological change of primary roots might be caused by the ethylene mediated regulation of stem cell division and activity in roots (Ortega-Martinez et al., 2007).
Exogenous ethylene treatment and mutations of CTR1 and ETO1 induce ectopic root hair formation, indicating that ethylene may also be involved in root hair initiation (Tanimoto et al., 1995; Masucci et al., 1996; Cho and Cosgrove, 2002). Therefore, we tested whether K+ deprivation causes ectopic root hair formation and found that it does not. Neither ethylene-insensitive mutants, such as etr1-1, ein4, and ein2-1, nor plants treated with 1-Methylcyclopropene (1-MCP) have reduced root hair numbers, indicating that endogenous ethylene is not required for root hair initiation under normal conditions (Cho and Cosgrove, 2002). Likewise, endogenous ethylene produced by K+ deprivation may act on some specific tissues (e.g., it may regulate root hair elongation). Alternatively, root hair formation may require a higher ethylene level than does the root hair elongation process (Cho and Cosgrove, 2002). K+ deprivation induces about a twofold increase of ethylene in Arabidopsis plants (Shin and Schachtman, 2004), and it is possible that the ethylene produced by K+ deprivation may not be sufficient to induce ectopic root hair formation.
We also determined the effect of ethylene inhibitors on the low K+-induced ROS production (Shin and Schachtman, 2004) that is likely, in part, driving the root hair elongation response. Our data showed that ethylene inhibitors blocked the low K+-induced ROS production and root hair elongation in RHDZ (Figures 2 and 5C), suggesting that ethylene signaling acts upstream of these two processes. However, the root hair low K+-induced responses were not blocked in any of the ethylene-insensitive mutants tested (Figure 5B), which conflicts with the inhibitor data (Figures 5C and 5D). When ctr1-1 was treated with a high concentration of AVG for a long time period, root hair formation could be abolished even though the ctr1-1 constitutive ethylene responses should occur in the absence of the effect that AVG has on the inhibition of ethylene synthesis, suggesting that AVG has some side effects on root hair development (Cho and Cosgrove, 2002). To minimize the side effects of inhibitors, we used nanomolar concentrations of AVG and Ag+ and micromolar concentrations of AIB, which are a 100 to 1000 lower than concentrations used in other published work (Cho and Cosgrove, 2002; Desikan et al., 2006). These low concentrations had little if any effect on root hair elongation under control conditions, but we found that the low concentrations of ethylene inhibitors effectively inhibited low K+-induced root hair elongation (Figure 5C). Furthermore, the ethylene inhibitors used all have different targets and all inhibited the low K+-induced root hair response, which may further support the conclusions that a block in ethylene signaling reduced the low K+-induced root hair elongation. Ethylene is a positive regulator in root hair elongation (Pitts et al., 1998; Cho and Cosgrove, 2002), and ROS production is essential for root hair elongation (Foreman et al., 2003). Taken together, these results suggest that low K+-induced ethylene production increases ROS levels in RHDZ, which promotes root hair elongation that in turn is important for nutrient uptake (Bates and Lynch, 2000; Ahn et al., 2004; Shin et al., 2005).
Other Pathways May Be Involved in Low K+ Signaling Pathways
Our data also suggest the possibility that ethylene- and/or EIN2-independent pathways are involved in low K+-induced plant responses. This conclusion is based on our data showing that K+ deprivation induced ROS production, root hair elongation, and HAK5 expression in ein2-1 (Figures 4, 6, and 8A), which is known to be an ethylene-insensitive mutant (Chen et al., 2005). Jasmonic acid (JA) is another hormone that may be involved in low K+ signaling. A microarray study showed that the transcripts of many JA-related genes, including JA biosynthesis genes, were induced in response to low K+ stress (Armengaud et al., 2004). Auxin signaling may also play roles in low K+ signaling. In response to low concentrations of iron, both ethylene and auxin are known to be involved in root responses (Lopez-Bucio et al., 2003). Auxin has also been shown to be involved in lateral root development under K+-deficient conditions (Shin et al., 2007). As in the case of sugar sensing (Rolland et al., 2006), it is likely that mineral nutrient sensing will involve multiple hormone pathways that alter physiological responses and developmental pathways to provide plants with a program for adaptation to low nutrient environments.
We showed that low K+-induced ROS production and root hair elongation in ein2-1 were almost completely blocked by Ag+ (Figure 6). Moreover, low K+-induced HAK5 expression was also blocked by Ag+, but it was still enhanced in ein2-1 (Figure 8A). These findings may suggest that an ethylene-dependent EIN2-independent pathway may exist or that the responses we have studied are only partially dependent on ethylene. An EIN2-independent pathway may function through Arabidopsis Response Regulator2 (ARR2), which is a transcriptional regulator that mediates cytokinin signaling (Hwang et al., 2002). However, ARR2 also plays a role in ethylene signaling (Hass et al., 2004; Desikan et al., 2006). It was proposed that ARR2 may function via a novel branch of the ethylene signaling pathway that does not include CTR1 or EIN2 (Hass et al., 2004; Chen et al., 2005).
We have provided evidence that ethylene signaling plays an important role in the morphological and functional changes of Arabidopsis roots that are initiated by K+ deprivation. In the future, it will be important to identify genes induced by ethylene signaling in response to low K+. Ethylene-responsive factor (ERF) genes are possible candidates that are implicated in biotic and abiotic stress responses and in many hormonal responses (Nakano et al., 2006). ERF1 and ERF4 are transcription factors that act downstream of ethylene signaling, followed by the regulation of stress and defense response genes (Etheridge et al., 2006). Ethylene-inducible SUB1A, which is essential for submergence tolerance of rice, also belongs to the ERF family. The expression of two expansin genes that are important for root hair development are regulated by ethylene (Cho and Cosgrove, 2002), and these may be regulated by ERFs in Arabidopsis, as is the case for Sub1A (Fukao et al., 2006).
DELLA genes are key players for plant growth in response to environmentally activated hormonal signals. Under nutrient deprivation, plant growth decreases, and, as we have shown, primary root growth is inhibited. The accumulation of DELLA proteins in response to external stress signals inhibits plant shoot and root growth. Ethylene regulates plant growth via DELLA function (Achard et al., 2003, 2006). In addition, ethylene-mediated salt tolerance is DELLA dependent (Achard et al., 2006), and the DELLA signaling pathway is also involved in phosphate starvation responses, such as root architectural changes and anthocyanin accumulation (Jiang et al., 2007). Given the involvement of DELLA in both ethylene and nutrient responses, DELLA genes might play a role during K+ deprivation via ethylene signaling to inhibit growth as a general mechanism for plant survival, until more mineral nutrients become available. Further investigation will be required to determine whether ERF or DELLA genes are involved in the low K+ signaling pathway.
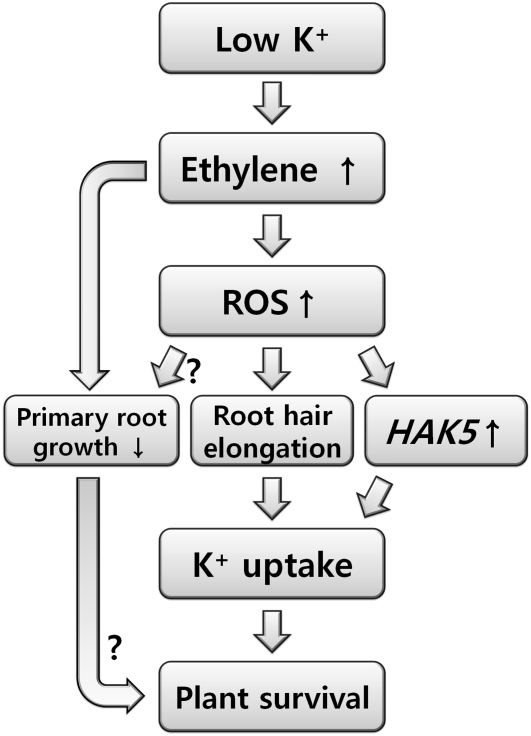
In conclusion, the data presented here provide evidence that ethylene signaling plays an important role in low K+-induced plant responses. These responses and the hierarchy in the low K+ signal transduction pathway are summarized in Figure 9. In Arabidopsis, low K+ induces ethylene production (Shin and Schachtman, 2004), which stimulates ROS production in the RHDZ. This results in root hair elongation and the induction of the high-affinity K+ uptake transporter HAK5. The low K+-induced ethylene production also inhibits primary root growth. Inhibitor studies suggest that low K+-induced root hair elongation is likely to be mediated by ethylene signaling; however, this is not supported by the analysis of the available ethylene signaling mutants. Our findings show that ethylene-mediated plant responses increase tolerance to potassium-deprived conditions through changes in root morphology and physiological changes. A major question still to be resolved is how the nutrient stress is perceived and what factors act upstream of ethylene in this nutrient signal transduction pathway.
Figure 9.
A Schematic Model for Low K+ Signaling.
The activation of ethylene signaling induced by K+ deprivation positively regulates ROS production in roots. ROS stimulates root hair elongation and HAK5 expression, which enhances K+ uptake and stress tolerance. Up arrow, increase; down arrow, decrease; question mark, unclear.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana mutants used in this study were in the Col background, except for ers1-3, ers2-3, and ers1-2 ers2-3, which were in the Wassilewskija background. Seeds of etr1-1 (CS237), etr1-3 (CS3070), ein2-1 (CS3071), ctr1-1 (CS8057), ein4 (CS8053), and eto1-1 (CS3072) were obtained from the ABRC. eto2 and eto3 seeds were kindly provided by Elizabeth Vierling (University of Arizona). etr1-6, etr2-3, ein4-4, ers1-3, ers2-3, ers1-2 ers2-3, etr1-6 etr2-3 ein4-4, and etr2-3 ein4-4 ers2-3 were kindly provided by Sara E. Patterson and Brad M. Binder (University of Wisconsin). Seeds were sterilized in 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 and then planted on 10-cm-diameter sterile plates containing nutrient medium [2 mM Ca(NO3)2, 0.5 mM phosphoric acid, 0.75 mM MgSO4, 50 μM H3BO3, 10 μM MnCl, 2 μM ZnSO4, 1.5 μM CuSO4, 0.075 μM NH4Mo7O24, and 74 μM Fe-EDTA, pH 5.6, with Ca(OH)2], 2% sucrose, and 0.8% SeaKem agarose (Cambrex). After stratification of the seeds at 4°C for 2 to 3 d, the plates were transferred to the growth chamber at 22°C with a 16-h daylength at 200 μmol·m−2s−1. Seedlings were grown vertically. Three- to fifteen-day-old seedlings were used throughout the study. For potassium-sufficient (+K) medium, 1.75 mM KCl, unless indicated otherwise, was added to the medium described above. For potassium-deficient (−K) medium, KCl was removed from the nutrient solution.
Chemicals
Ethephon, AVG, AIB, ACC, and DPI were from Sigma-Aldrich. Silver nitrate was from the PhytoTechnology Laboratory. The DPI stock solution was prepared in 100% DMSO, while other chemicals were dissolved in water.
Phenotype Assay
Five-day-old seedlings grown on the complete nutrient (+K) plates were transferred to complete nutrient or K+-deprived plates. For each mutant, 10 seedlings per plate were grown, and five replicate plates were used. Seven or ten days after transfer, photographs and measurements of Col-0, etr1-1, ein2-1, and ctr1-1 were taken. Shoot weight and total chlorophyll content were measured by bulking 10 plants from each of five plates. Chlorophyll was extracted and assayed according to the procedure of Hiscox and Israelstam (1979). Statistical significance was evaluated with a Student's t test.
ROS Detection and Measurement
For the ROS study using ethylene-insensitive mutants, 2-d-old seedlings were floated on K+-sufficient and K+-deficient liquid nutrient solutions as described above. After 23 h and 40 min, roots were further incubated with 20 μM DFFDA (Invitrogen) for 20 min. After a brief wash with the medium that did not contain DFFDA, the roots were observed using fluorescence microscopy. All fluorescence images were captured and stored in grayscale using a Nikon SMZ1500 microscope and a Q-Imaging Retiga cooled 12-bit camera with 460- to 500-nm band-pass excitation and 510- to 560-nm band-pass emission. ROS fluorescence in the root hair elongation zone (∼0.5 mm) was quantified and converted into pseudocolor images by the NIH ImageJ software program (available at rsb.info.nih.gov/ij/). Background noise was subtracted from the fluorescence intensity value for quantification. The same microscopy parameters (i.e., UV exposure time, gain, contrast, etc.) were used to compare ROS signal intensity within specific experiments.
For the ROS study using ethylene inhibitors, 2-d-old seedlings of Col-0 and ein2-1 were floated on K+-sufficient and K+-deficient nutrient solutions for 24 h. The ethylene inhibitors were added to K+-sufficient and K+-deficient nutrient solutions 2 h before fluorescence images were obtained.
For the ROS study, 3-d-old seedlings of constitutive ethylene response mutants were floated on complete nutrient solution with or without ethephon and silver nitrate. Three hours later, fluorescence images of roots were obtained and analyzed for quantification.
Root Hair Length Measurement and Ectopic Root Hair Analysis
For the potassium deprivation experiments using ethylene-insensitive mutants, 2-d-old seedlings were transferred to plates containing K+-sufficient (1.75 mM K+) or K+-deficient (no K+) medium. After 28 h, photographs of roots were taken using a Nikon SMZ1500 microscope and a Q-Imaging Retiga cooled 12-bit camera. For each root, the length of the root hairs, in a 2-mm region starting 0.5 mm above the root hair differentiation zone (Figure 2B), was measured (n = 15 to 20 root hairs), avoiding root hairs that were growing into the medium, using NIH ImageJ software program. Eight to ten plants per genotype or treatment were used for root hair length measurements (n = 8 to 10).
For the potassium deprivation experiments, 2-d-old seedlings of wild-type (Col-0) and ein2-1 plants were transferred to plates containing K+-sufficient (5 mM K+) or K+-deficient (no K+) medium, with or without the indicated concentrations of ethylene inhibitors. After 22 h, photographs of roots were taken, and root hair lengths were measured as described above.
The number of root hairs was determined according to a standard method devised by Masucci et al. (1996) with some modifications. Briefly, plants were grown under complete nutrient conditions for 2 d and then transferred to either complete nutrient medium or to medium lacking potassium for 24 and 48 h. Roots were stained with 0.01% Toluidine Blue solution for 5 min at room temperature and placed on a glass microscope slide under a cover slip. For each root, five consecutive epidermal cells from the same cell file were observed, and a total of 20 cells from two hair cell files and the adjacent two nonhair cell files were counted. Ten roots (for a total of 200 cells) were scored. Any protrusion was scored as the presence of a root hair, regardless of the length.
In Vivo Study Using HAK5pro:LUC Lines
The HAK5 native promoter fused to a LUC reporter gene in the plant binary vector (Collier et al., 2005) was generated through modifying the construct of the HAK5 promoter fused with a β-glucuronidase reporter gene that was described by Qi et al. (2008). The β-glucuronidase reporter gene was exchanged with the LUC reporter gene that was amplified from the pSP:LUC+ vector (Promega). Arabidopsis plants were transformed with the HAK5pro:LUC transgene by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). A transgenic line homozygous for the HAK5pro:LUC transgene was used throughout the experiments.
For in vivo imaging of HAK5 expression, 3-d-old HAK5pro:LUC transgenic seedlings were floated on K+-sufficient (5 mM K+) and K+-deficient nutrient solution for 6 h (for ethephon treatment experiment) or 24 h for all other experiments. During the incubation, seedlings were treated with silver nitrate (1 or 5 μM) or DPI (10 μM) for 2 h, with ethephon (100 or 200 μM) for 6 h, and with AVG (1 or 5 μM) or AIB (5 or 10 mM) for 24 h before luminescence images were obtained. Then, seedlings were transferred onto plates containing K+-sufficient (5 mM K+) and K+-deficient medium and sprayed with 1 mM luciferin sodium salt (Gold Bio Technology) in 0.01% Triton X-100 solution. After placing the plates in the dark for 5 to 10 min, the plates were imaged in grayscale using a cooled CCD camera system (Roper Scientific). Finally, images were converted into pseudocolor images using the NIH ImageJ software. At least two independent experiments were performed, and similar results were obtained.
Quantitative Real-Time RT-PCR Analysis
Three-day-old seedlings of ethylene mutants were floated on K+-deficient or K+-sufficient nutrient solution. After 24 h, plants were harvested and total RNA was isolated by grinding whole seedlings in liquid nitrogen in the presence of Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA was quantified and treated with RQ RNase-free DNase I (Promega). DNase-treated RNA was tested for genomic DNA contamination, and the quality of total RNA was determined by agarose gel electrophoresis. Two and a half micrograms of DNA-free RNA was then reverse transcribed using the First-Stand Synthesis System (Invitrogen) for quantitative real-time RT-PCR analysis according to the manufacturer's instructions.
Quantitative real-time RT-PCR analysis was performed using the Bio-Rad iCycler iQ system (Bio-Rad Laboratories) and Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). HAK5 primers were used to amplify the endogenous HAK5 transcripts (Ahn et al., 2004). For the normalization of HAK5 transcripts, β-tubulin (TUB2) sense (5′-GCCAATCCGGTGCTGGTAACA-3′) and antisense (5′-CATACCAGATCCAGTTCCTCCTCCC-3′) was used as an internal control. The fold change of HAK5 transcript in each sample was calculated based on the efficiency calibrated model (Yuan et al., 2006) and compared with the HAK5 transcript level of the wild-type control under the K+-sufficient condition. Statistical differences in HAK5 transcript levels between samples were evaluated by a Student's t test using ΔΔCt values (Yuan et al., 2006). At least two experiments on independently grown plant material were performed to confirm the results. In each experiment, three biological replicates were used to generate means and statistical significance.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HAK5 (AT4G13420), ETR1 (AT1G66340), ETR2 (AT3G23150), ERS1 (AT2G40940), ERS2 (AT1G04310), EIN4 (AT3G04580), CTR1 (AT5G03730), EIN2 (AT5G03280), ETO1 (AT3G51770), ETO2 (AT5G65800), ETO3 (AT3G49700), and TUB2 (AT5G62690).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effects of ACC on HAK5 Expression in Arabidopsis Roots.
Supplemental Figure 2. Low K+ Induces HAK5 Expression in Triple Mutants of Ethylene Receptors.
Supplemental Figure 3. DPI inhibits Ethephon-Induced ROS Production in Roots.
Supplemental Figure 4. Ethephon Induces ROS Production in etr1-1, etr1-3, and rbohF Plants.
Supplemental Table 1. Root Hair Number under K+-Deficient and -Sufficient Conditions.
Supplementary Material
Acknowledgments
We thank Douglas Chalker, Robert Kranz, Barbara Kunkel, Craig Pikaard, Mark Running, and Dan Ruzicka for critical comments; Min Jung Kim for quantitative RT-PCR assistance; Howard Berg for microscopy assistance; and Sara E. Patterson and Brad M. Binder for providing loss-of function mutants of ethylene receptors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Daniel P. Schachtman (dschachtman@danforthcenter.org).
Online version contains Web-only data.
References
- Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., and Harberd, N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94. [DOI] [PubMed] [Google Scholar]
- Achard, P., Vriezen, W.H., Van Der Straeten, D., and Harberd, N.P. (2003). Ethylene regulates arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S.J., Shin, R., and Schachtman, D.P. (2004). Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud, P., Breitling, R., and Amtmann, A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136 2556–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley, M.K., Grant, M., and Grabov, A. (2006). Plant responses to potassium deficiencies: A role for potassium transport proteins. J. Exp. Bot. 57 425–436. [DOI] [PubMed] [Google Scholar]
- Bates, T.R., and Lynch, J.P. (2000). Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 87 958–963. [PubMed] [Google Scholar]
- Binder, B.M. (2008). The ethylene receptors: Complex perception for a simple gas. Plant Sci. 175 8–17. [Google Scholar]
- Borch, K., Bouma, T.J., Lynch, J.P., and Brown, K.M. (1999). Ethylene: A regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ. 22 425–431. [Google Scholar]
- Cao, W.H., Liu, J., He, X.J., Mu, R.L., Zhou, H.L., Chen, S.Y., and Zhang, J.S. (2007). Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, H.S., Faure, F., and Kieber, J.J. (2003). The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.F., Etheridge, N., and Schaller, G.E. (2005). Ethylene signal transduction. Ann. Bot. (Lond.) 95 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, Y.H., Pandey, G.K., Grant, J.J., Batistic, O., Li, L., Kim, B.G., Lee, S.C., Kudla, J., and Luan, S. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52 223–239. [DOI] [PubMed] [Google Scholar]
- Cho, H.T., and Cosgrove, D.J. (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K.L., Larsen, P.B., Wang, X., and Chang, C. (1998). Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 95 5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Collier, R., Fuchs, B., Walter, N., Kevin Lutke, W., and Taylor, C.G. (2005). Ex vitro composite plants: An inexpensive, rapid method for root biology. Plant J. 43 449–457. [DOI] [PubMed] [Google Scholar]
- Desikan, R., Last, K., Harrett-Williams, R., Tagliavia, C., Harter, K., Hooley, R., Hancock, J.T., and Neill, S.J. (2006). Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 47 907–916. [DOI] [PubMed] [Google Scholar]
- D'Haeze, W., De Rycke, R., Mathis, R., Goormachtig, S., Pagnotta, S., Verplancke, C., Capoen, W., and Holsters, M. (2003). Reactive oxygen species and ethylene play a positive role in lateral root base nodulation of a semiaquatic legume. Proc. Natl. Acad. Sci. USA 100 11789–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge, N., Hall, B.P., and Schaller, G.E. (2006). Progress report: Ethylene signaling and responses. Planta 223 387–391. [DOI] [PubMed] [Google Scholar]
- Foreman, J., Demidchik, V., Bothwell, J.H., Mylona, P., Miedema, H., Torres, M.A., Linstead, P., Costa, S., Brownlee, C., Jones, J.D., Davies, J.M., and Dolan, L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446. [DOI] [PubMed] [Google Scholar]
- Fukao, T., and Bailey-Serres, J. (2008). Ethylene-A key regulator of submergence responses in rice. Plant Sci. 175 43–51. [Google Scholar]
- Fukao, T., Xu, K., Ronald, P.C., and Bailey-Serres, J. (2006). A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z., Chen, Y.F., Randlett, M.D., Zhao, X.C., Findell, J.L., Kieber, J.J., and Schaller, G.E. (2003). Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 278 34725–34732. [DOI] [PubMed] [Google Scholar]
- Gierth, M., and Maser, P. (2007). Potassium transporters in plants–Involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 581 2348–2356. [DOI] [PubMed] [Google Scholar]
- Gierth, M., Maser, P., and Schroeder, J.I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2004). The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 7 40–49. [DOI] [PubMed] [Google Scholar]
- Hass, C., Lohrmann, J., Albrecht, V., Sweere, U., Hummel, F., Yoo, S.D., Hwang, I., Zhu, T., Schafer, E., Kudla, J., and Harter, K. (2004). The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 23 3290–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox, J.D., and Israelstam, G.F. (1979). A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 57 1332–1334. [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 261–271. [DOI] [PubMed] [Google Scholar]
- Hwang, I., Chen, H.C., and Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., Gao, X., Liao, L., Harberd, N.P., and Fu, X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbuch, R., Claassen, N., and Jungk, A. (1986). Potassium availability in relation to soil-moisture. 1. Effect of soil-moisture on potassium diffusion, root-growth and potassium uptake of onion plants. Plant Soil 95 221–231. [Google Scholar]
- Lebaudy, A., Very, A.A., and Sentenac, H. (2007). K+ channel activity in plants: Genes, regulations and functions. FEBS Lett. 581 2357–2366. [DOI] [PubMed] [Google Scholar]
- Lee, S.C., Lan, W.Z., Kim, B.G., Li, L., Cheong, Y.H., Pandey, G.K., Lu, G., Buchanan, B.B., and Luan, S. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. USA 104 15959–15964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Kim, B.G., Cheong, Y.H., Pandey, G.K., and Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K+ response in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 12625–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebersbach, H., Steingrobe, B., and Claassen, N. (2004). Roots regulate ion transport in the rhizosphere to counteract reduced mobility in dry soil. Plant Soil 260 79–88. [Google Scholar]
- Lopez-Bucio, J., Cruz-Ramirez, A., and Herrera-Estrella, L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6 280–287. [DOI] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA 2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122 1253–1260. [DOI] [PubMed] [Google Scholar]
- Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Martinez, O., Pernas, M., Carol, R.J., and Dolan, L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317 507–510. [DOI] [PubMed] [Google Scholar]
- Perata, P., and Voesenek, L.A.C.J. (2007). Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends Plant Sci. 12 43–46. [DOI] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16 553–560. [DOI] [PubMed] [Google Scholar]
- Qi, Z., Hampton, C.R., Shin, R., Barkla, B.J., White, P.J., and Schachtman, D.P. (2008). The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 59 595–607. [DOI] [PubMed] [Google Scholar]
- Qi, Z., and Spalding, E.P. (2004). Protection of plasma membrane K+ transport by the salt overly sensitive1 Na+-H+ antiporter during salinity stress. Plant Physiol. 136 2548–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57 675–709. [DOI] [PubMed] [Google Scholar]
- Romera, F.J., Alcantara, E., and De la Guardia, M.D. (1999). Ethylene production by Fe-deficient roots and its involvement in the regulation of Fe-deficiency stress responses by strategy I plants. Ann. Bot. (Lond.) 83 51–55. [Google Scholar]
- Rus, A., Lee, B.H., Munoz-Mayor, A., Sharkhuu, A., Miura, K., Zhu, J.K., Bressan, R.A., and Hasegawa, P.M. (2004). AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136 2500–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman, D.P., and Shin, R. (2007). Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 58 47–69. [DOI] [PubMed] [Google Scholar]
- Shin, R., Berg, R.H., and Schachtman, D.P. (2005). Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 46 1350–1357. [DOI] [PubMed] [Google Scholar]
- Shin, R., Burch, A.Y., Huppert, K.A., Tiwari, S.B., Murphy, A.S., Guilfoyle, T.J., and Schachtman, D.P. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, R., and Schachtman, D.P. (2004). Hydrogen peroxide mediates plant root response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 101 8827–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto, M., Roberts, K., and Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8 943–948. [DOI] [PubMed] [Google Scholar]
- Tsuchisaka, A., and Theologis, A. (2004. a). Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136 2982–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka, A., and Theologis, A. (2004. b). Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl. Acad. Sci. USA 101 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K.L., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.): S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste, K., Ye, C., and Kieber, J.J. (1999). Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of ACC synthase. Plant Physiol. 119 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Li, H.D., Chen, L.Q., Wang, Y., Liu, L.L., He, L., and Wu, W.H. (2006. a). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360. [DOI] [PubMed] [Google Scholar]
- Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., Ismail, A.M., Bailey-Serres, J., Ronald, P.C., and Mackill, D.J. (2006. b). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442 705–708. [DOI] [PubMed] [Google Scholar]
- Yuan, J.S., Reed, A., Chen, F., and Stewart, C.N., Jr. (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics 7 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaid, H., El Morabet, R., Diem, H.G., and Arahou, M. (2003). Does ethylene mediate cluster root formation under iron deficiency? Ann. Bot. (Lond.) 92 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarembinski, T.I., and Theologis, A. (1997). Expression characteristics of OS-ACS1 and OS-ACS2, two members of the 1-aminocyclopropane-1-carboxylate synthase gene family in rice (Oryza sativa L. cv. Habiganj Aman II) during partial submergence. Plant Mol. Biol. 33 71–77. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.J., Lynch, J.P., and Brown, K.M. (2003). Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. J. Exp. Bot. 54 2351–2361. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., de Almeida Engler, J., Rouan, D., Michiels, F., Van Montagu, M., and Van Der Straeten, D. (2002). Tissue localization of a submergence-induced 1-aminocyclopropane-1-carboxylic acid synthase in rice. Plant Physiol. 129 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.