Abstract
The 26S proteasome is an essential multicatalytic protease complex that degrades a wide range of intracellular proteins, especially those modified with ubiquitin. Arabidopsis thaliana and other plants use pairs of genes to encode most of the core subunits, with both of the isoforms often incorporated into the mature complex. Here, we show that the gene pair encoding the regulatory particle non-ATPase subunit (RPN5) has a unique role in proteasome function and Arabidopsis development. Homozygous rpn5a rpn5b mutants could not be generated due to a defect in male gametogenesis. While single rpn5b mutants appear wild-type, single rpn5a mutants display a host of morphogenic defects, including abnormal embryogenesis, partially deetiolated development in the dark, a severely dwarfed phenotype when grown in the light, and infertility. Proteasome complexes missing RPN5a are less stable in vitro, suggesting that some of the rpn5a defects are caused by altered complex integrity. The rpn5a phenotype could be rescued by expression of either RPN5a or RPN5b, indicating functional redundancy. However, abnormal phenotypes generated by overexpression implied that paralog-specific functions also exist. Collectively, the data point to a specific role for RPN5 in the plant 26S proteasome and suggest that its two paralogous genes in Arabidopsis have both redundant and unique roles in development.
INTRODUCTION
The 26S proteasome is an ATP-dependent protease complex responsible for most selective protein degradation in the nucleus and cytoplasm of eukaryotes. Its main role is to remove proteins first modified by ubiquitin (Ub), but nonubiquitylated proteins can be substrates as well (Voges et al., 1999; Hartmann-Petersen et al., 2003; Smalle and Vierstra, 2004; Vierstra, 2009). The holoprotease of ∼2.5 MD is composed of two functionally distinct and stable subcomplexes, a 14-subunit 20S core protease (CP) and an 18-or-more subunit 19S regulatory particle (RP). The CP is a broad-spectrum, ATP-, and Ub-independent peptidase. Its cylindrical shape is created by the assembly of four stacked heptameric rings of related α- and β-subunits in a symmetric α1-7β1-7β1-7α1-7 configuration (Groll et al., 1997). A central chamber houses the peptidase active sites provided by the β1, β2, and β5 subunits, which have peptidylglutamyl peptide-hydrolyzing, trypsin-like, and chymotrypsin-like activities, respectively. Together, these activities cleave proteins into short peptides that are then further disassembled to free amino acids by other proteases. Access to this chamber is restricted by a narrow gated channel, which allows only unfolded proteins to enter (Groll et al., 2000; Smith et al., 2007). This self-compartmentalized design spatially separates proteolysis from the cellular milieu and restricts degradation to only those proteins that are deliberately unfolded and imported.
The RP binds to each end of the CP and imparts both ATP dependence and substrate specificity to the holoenzyme, especially with respect to proteins bearing poly-Ub chains (Voges et al., 1999; Hartmann-Petersen et al., 2003). Following substrate identification, the RP disassembles the poly-Ub chains, opens the α-subunit ring gate, and directs entry of unfolded protein into the CP lumen for breakdown. The RP can be further dissected into two subparticles, the lid and base (Glickman et al., 1998; Fu et al., 2001). The base contains a ring of six RP triple-A-ATPase subunits (RPT1-6), which contacts the α-subunit ring and likely promotes target unfolding and transport, and three RP non-ATPase subunits (RPN1, RPN2, and RPN10). The lid binds to the ends of the base and contains eight additional RPN subunits (RPN3, 5-9, and 11-13) that assist in scaffolding, substrate recognition, and/or processing.
While it was originally thought that all subunits of the 26S proteasome work cooperatively to degrade substrates, a picture is emerging that the complex may employ individual subunits to generate a variety of nonoverlapping routes for substrate delivery. At least four RP subunits (RPT5, RPN1, RPN10, and RPN13) have been implicated in the recognition of ubiquitylated substrates (van Nocker et al., 1996a, 1996b; Fu et al., 1998b; Elsasser et al., 2002; Lam et al., 2002; Husnjak et al., 2008; Schreiner et al., 2008). Whereas RPN10 and RPN13 appear to be the main receptors for substrates bearing poly-Ub chains linked internally through Lys-48, RPN1 helps recognize ubiquitylated substrates noncovalently tethered to carrier proteins, such as RAD23 and DSK2. These carrier proteins contain both Ub-related and Ub-binding domains, thus allowing them to simultaneously dock with ubiquitylated proteins and Ub receptors.
We and others have also found that Arabidopsis thaliana mutants affecting individual RP subunits display a wide range of phenotypes consistent with each subunit participating in the removal of different substrates. For example, a mutation affecting RPN10 confers hypersensitivity to abscisic acid (ABA) by selectively stabilizing the ABA-INSENSITIVE5 (ABI5) protein (Smalle et al., 2003), a RPN12a mutation decreases cytokinin responsiveness (Smalle et al., 2002), inactivation of RPN1a arrests embryogenesis (Brukhin et al., 2005), and mutations affecting RPT2a inhibit root growth, presumably by stabilizing one or more proteins involved in root apical meristem (RAM) maintenance (Ueda et al., 2004). Similarly, yeast RPN3 has been implicated in the turnover of some substrates but not others (Bailly and Reed, 1999). Part of these functional distinctions may arise from the specific association of individual subunits with accessory proteins, some of which may be present at substoichiometric levels (Schmidt et al., 2005; Hanna and Finley, 2007).
In addition, plants and metazoans may also exploit isoform heterogeneity among individual CP and RP subunits to further expand the functional diversity of the holocomplex. In mammalian cells, for example, at least five CP types have been detected (Dahlmann et al., 2000). The most distinct is the immunoproteasome, which is created by substitution of the active-site β1, β2, and β5 subunits in the CP with catalytic variants that release peptides more favorable for antigen presentation (Kloetzel, 2001). Analyses of various plant genomes suggest that similar heterogeneity exists in the plant kingdom. In Arabidopsis, 10 of the 14 CP subunits and 15 of the 18 RP subunits are encoded by two loci (Fu et al., 1998a, 1999). In both rice (Oryza sativa) and Arabidopsis, most, if not all, of the CP and RP subunit pairs are expressed and assembled into the mature 26S particle, implying that plants generate a wide assortment of proteasome types (Shibahara et al., 2004; Yang et al., 2004). While sequence homology between isoforms would imply that many are functionally equivalent, some pairs are sufficiently divergent to suggest distinct functions (Fu et al., 1998a, 1999). Nonequivalence at least at the gene level was recently demonstrated by Brukhin et al. (2005), who showed that the Arabidopsis RPN1a and RPN1b gene pair, while expressing functionally equivalent proteins, act nonredundantly during embryogenesis.
To further understand the function(s) of each RP subunit and to explore the possibility that individual isoforms can imbue distinct specificities to the plant 26S proteasome, we analyzed genetically the two Arabidopsis isoforms of the RP lid subunit RPN5. Although the function(s) of RPN5 remain unclear, it is essential in yeast (Saccharomyces cerevisiae) (Yang et al., 2004) and is needed in a dosage-dependent manner for proper 26S proteasome assembly and localization in Schizosaccharomyces pombe (Yen et al., 2003). Here, we show that RPN5 is essential in Arabidopsis as plants missing both RPN5a and RPN5b are inviable. Whereas inactivation of RPN5b is phenotypically inconsequential, inactivation of RPN5a blocks pollen production and impairs embryo and seedling development in a manner partially distinct from those generated by mutants affecting other RP subunits (Smalle et al., 2002, 2003; Ueda et al., 2004; Brukhin et al., 2005). Biochemical studies suggest that part of this phenotype likely arises from a decreased stability of the holoproteasome complex, indicating that RPN5 is needed for complex integrity. The rpn5a phenotypes could be rescued by ectopic expression of either RPN5a or RPN5b, demonstrating that the two isoforms can act redundantly. However, plants overexpressing RPN5a but not RPN5b display an enhanced senescence phenotype, suggesting that nonredundant functions exist as well. Taken together, our results show that RPN5 has different functions from other 26S proteasome lid subunits and that the RPN5a and RPN5b isoforms may have both redundant and unique roles in Arabidopsis development.
RESULTS
The Arabidopsis RPN5a and RPN5b Genes
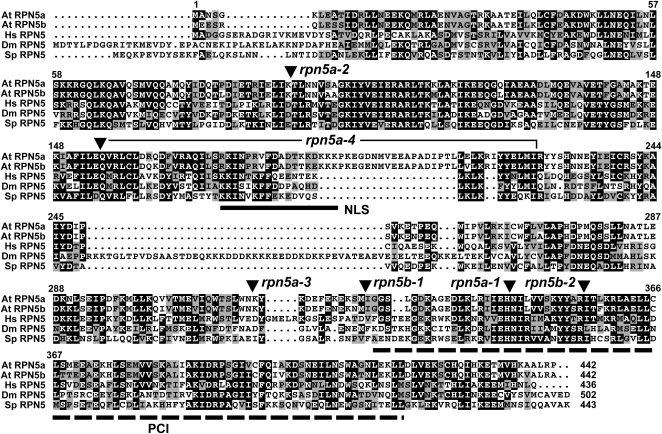
Arabidopsis contains two highly related genes on the distal arms of chromosome 5 that encode the RPN5 subunit, designated RPN5a and RPN5b (Yang et al., 2004). The two loci are within syntenic duplication blocks (http://bioinfo.genotoul.fr/PGCViewer/cgi-bin/PGCViewer.cgi.pl), suggesting that the pair arose from the whole-genome duplication proposed to occur ∼38 million years ago in Arabidopsis (Ermolaeva et al., 2003). The corresponding 51-kD proteins are colinear along their entire length of 432 residues with high amino acid sequence conservation to each other (95/93% amino acid similarity/identity) and to orthologs from Homo sapiens (60/42%), Drosophila melanogaster (47/32%), and S. pombe (52/35%) (Figure 1). Like orthologs from other species (Glickman et al., 1998), Arabidopsis RPN5a and RPN5b contain a PCI (for proteasome/COP9/initiation factor 3) domain near their C termini (residues 332 to 418 in RPN5a), previously shown to promote an interaction with RPN9 (Fu et al., 2001; Yang et al., 2004). Data from the yeast complex revealed that RPN5 also associates with RPN8 and possibly RPN6 and RPN11 to form a stable lid subcomplex important for RP assembly (Fu et al., 2001; Sharon et al., 2006; Isono et al., 2007). A possible bipartite nuclear localization signal is present in Arabidopsis RPN5a/b between residues 173 and 190. Notable differences within the RPN5 family include (1) varying length N-terminal extensions, (2) a 21–amino acid insertion (residues 188 to 209) in the two Arabidopsis paralogs, and (3) a downstream 45–amino acid insertion in D. melanogaster RPN5 (Figure 1). Despite these differences, the Arabidopsis RPN5a cDNA successfully complemented a yeast Δrpn5 strain, indicating that the plant protein is functionally orthologous to those from other kingdoms (Yang et al., 2004). RPN5 gene sequences are also available from a number of other plants species, including rice, sorghum (Sorghum bicolor), soybean (Glycine max), poplar (Populus trichocarpa), Physcomitrella patens, and Selaginella moellendorffii (Shibahara et al., 2002; see Supplemental Figure 1 online). All but S. moellendorffii contain a pair of RPN5 genes that associate phylogenetically with themselves and not with orthologs from other species, suggesting that the duplications that generated these pairs occurred at separate times during the evolution of higher plants.
Figure 1.
Amino Acid Sequence Comparison of RPN5 Proteins.
Identical and similar residues are shown in the black and gray boxes, respectively. Dots denote gaps. At, Arabidopsis thaliana; Hs, Homo sapiens; Dm, Drosophila melanogaster; Sp, Schizosaccharomyces pombe. The numbers refer to the amino acid position in At RPN5a. The residue length of each protein is shown at the end of the sequence. The locations of the PCI and NLS sequences are indicated by the dashed and solid lines, respectively. The positions of the various insertion mutants are indicated by the arrowheads. The bracket highlights the coding region that has been deleted in the rpn5a-4 allele.
RNA gel blot analyses demonstrated previously that both RPN5a and RPN5b are expressed in Arabidopsis (Yang et al., 2004). Measurement of transcript abundance by mining the GENEVESTIGATOR DNA microarray (Zimmermann et al., 2004), EST (www.Arabidopsis.org/), and Massively Parallel Signature Sequencing databases (www.mpss.udel.edu/at/) and the DNA microarray data set of Ma et al. (2005) showed that each gene is widely expressed in most Arabidopsis tissues (the exception being pollen). Normalized mRNA levels for RPN5a were consistently higher in almost all situations (Figure 2A). In support of this, 81 and 18 EST sequences were available for RPN5a and RPN5b, respectively, in the Arabidopsis EST collection (as of 10/10/2008).
Figure 2.
Expression Patterns of Arabidopsis RPN5a and RPN5b Genes and Proteins.
(A) Relative transcript abundance in various tissues determined from the GENEVESTIGATOR DNA microarray data set (Zimmermann et al., 2004). Sus, suspension culture; Yg, young; Mat, mature; Sen, senescent. Error bars represent the se from different arrays.
(B) Expression of RPN5apro:GUS (5a) and RPN5bpro:GUS (5b) in 12-d-old seedlings (bars = 2 mm), floral tissue (bars = 25 μm), root tips (bars = 100 μm), trichome socket cells (bars = 200 μm), and developing embryos (bars = 25 μm). Tissues were stained overnight in X-Gluc.
(C) Subcellular localization of RPN5a-GFP and RPN5b-GFP in the roots of rpn5a-1 seedlings as detected by confocal fluorescence microscopy (bars = 50 μm). The middle panels show a fourfold magnification of cells (bars = 10 μm). The panels below demonstrate the rescue of the rpn5a-1 mutant phenotype by the 35Spro:RPN5a-GFP and 35Spro:RPN5b-GFP transgenes (bars = 2 mm).
Widespread expression of RPN5a and RPN5b was confirmed by the analysis of promoter-β-glucuronidase (GUS) reporter fusions stably introduced into wild-type Columbia-0 (Col-0) plants. As shown in Figure 2B, ∼1.5-kb fragments upstream of the initiation ATG codon in the two genes drove indistinguishable GUS expression patterns in all tissues examined, including young seedlings, leaves, roots, flowers, and developing embryos. Strong staining was evident in the shoot and root apex, leaf and root vasculature, stamen, style, and budding lateral roots, with less staining detected in more mature tissues. Interestingly, the socket cells surrounding leaf trichomes stained strongly for GUS in each case, suggesting that both isoforms of RPN5, and likely the entire 26S proteasome, are important to this cell type (Figure 2B). In embryos, the RPN5a and RPN5b promoters drove GUS expression evenly throughout the globular and heart stages but became concentrated in the cotyledons at the torpedo and later stages of development (Figure 2B).
Arabidopsis Mutants Affecting the Expression of RPN5a and RPN5b
To examine the functions of RPN5a and RPN5b genetically, we assembled a collection of mutants disrupting the expression of each locus. Three independent T-DNA insertions (rpn5a-1, rpn5a-2, and rpn5a-3) and a single ethyl methanesulfonate–induced point mutation (rpn5a-4) were identified that altered the transcribed region of RPN5a, whereas two T-DNA insertions were found that altered the transcribed region of RPN5b (rpn5b-1 and rpn5b-2) (Figures 1 and 3A). Both the position/effect of the insertion/mutation and subsequent analysis of the corresponding transcript indicated that each mutation is a strong, and likely null, allele. The rpn5a-1 mutant was isolated by exon-trap mutagenesis, using the neomycin phosphotransferase (NPTII) gene that confers kanamycin resistance as the reporter (Babiychuk et al., 1997). Although the rpn5a-1 plants are kanamycin resistant, we could not confirm by RT-PCR or RNA gel blot analysis that a fusion mRNA containing the RPN5a and NPTII coding regions was produced. The insertion in rpn5a-3 caused the deletion of parts of exon 9 and intron 9. Despite containing only a single nucleotide substitution, the rpn5a-4 mutation is predicted to substantially affect the RPN5a coding region by altering the 3′-splice site within intron 5, which in turn could induce the deletion of the 6th exon during mRNA processing. The rpn5a-2, rpn5a-3, and rpn5b-2 alleles are in the Col-0 ecotype background, rpn5a-1 is in the C24 background, rpn5a-4 is in the Landsberg erecta (Ler) background, and rpn5b-1 is in the Wassilewskija (Ws) background.
Figure 3.
Molecular and Biochemical Descriptions of Arabidopsis rpn5a and rpn5b Mutants.
(A) Organization of the RPN5a and RPN5b genes and the positions of the T-DNA insertions. The bracket for rpn5a-4 defines the deleted region. White and black boxes indicate untranslated and coding regions, respectively. Lines denote introns.
(B) RNA gel blot analysis of 2-week-old wild-type seedlings, homozygous mutant seedlings, and wild-type seedlings expressing 35Spro:RPN5a or 35Spro:RPN5b transgenes. Equal loading of total RNA was confirmed by probing the blots with β-tubulin4 (TUB4).
(C) RNA gel blot analysis of RPN5a and RPN5b transcripts in 2-week-old rpn10-1, rpn12a-1, and rpt2a-2 mutant seedlings.
(D) Immunoblot detection of 26S proteasome subunits and Ub. Equivalent amounts of total protein (20 μg) were subjected to SDS-PAGE and immunoblot analysis with antibodies against RPN5a, RPN12a, PBA1, and Ub. Equal protein loads were confirmed by probing with anti-UBC1 antibodies. The positions of free Ub, poly-Ub chains, and Ub conjugates are indicated.
RNA gel blot analyses showed that the rpn5a-1 and rpn5b-2 mutations blocked detectable accumulation of the corresponding transcripts. Whereas the expected 1.6-kb RPN5a and 1.5-kb RPN5b transcripts could be easily detected in wild-type C24 and Col-0 plants, respectively, no transcripts from the affected loci were evident in the mutants (Figure 3B). Even prolonged exposure of the blots failed to reveal additional RPN5-hybridizing species, suggesting that the aberrant RPN5 mRNAs (if transcribed) are rapidly degraded. Although possible mRNAs generated from the rpn5a-2, rpn5a-3, rpn5a-4, and rpn5b-2 lines were not examined, it is likely that these mutants also fail to accumulate normal RPN5a and RPN5b transcripts, given that the mutations altered the respective coding regions and the phenotypic similarity of the alleles to rpn5a-1 and rpn5b-2 (see below). To help determine the effects of the mutations on protein accumulation, we measured immunologically the levels of the RPN5 proteins using anti-RPN5a antibodies (Smalle et al., 2002). These antibodies recognize recombinant RPN5a and RPN5b with equivalent sensitivities (see Supplemental Figure 2 online). In agreement with the stronger expression of RPN5a relative to RPN5b (see above), the amount of total RPN5 protein was unaffected in rpn5b-2 seedlings but was reduced in rpn5a-1 seedlings to approximately one-quarter the level of its corresponding wild type (Figure 3D).
rpn5a Mutants Have Pleiotropic Developmental Defects
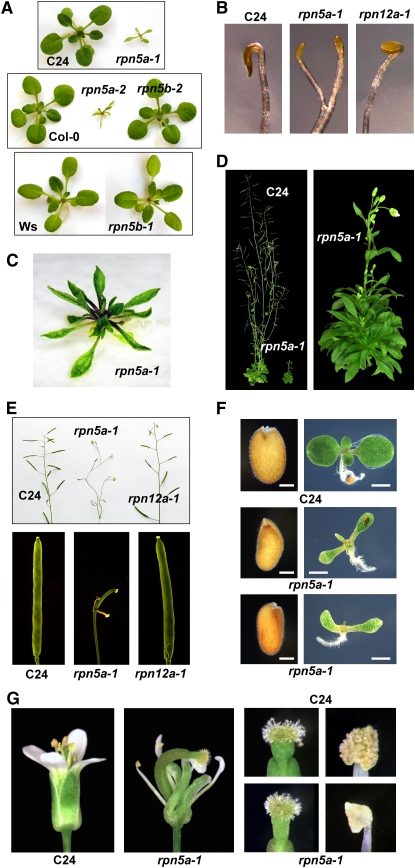
Consistent with the dominant expression of RPN5a, we found that seedlings homozygous for any of the four rpn5a mutations were developmentally challenged compared with their wild-type parents. As examples, young rpn5a-1 and rpn5a-2 seedlings grown in continuous light were severely stunted and generated smaller, highly lanceolate cotyledons and leaves (Figures 4A, 4C, and 4F). The reduction in cotyledon/leaf size could be due to reduced cell division (fewer cells) or reduced cell expansion (smaller cells), or a combination of both factors. By counting the number of cells in the first leaf after it reached maturity, we found that the rpn5a-3 and rpn5a-4 mutants had many fewer cells than the wild type (Table 1), indicating the mutants have reduced rates of cell proliferation. However, from analysis of adaxial pavement cells, we also found that the mutant cells were slightly larger than those in the wild type (Table 1). This is reminiscent of the compensation phenomenon observed in many mutants with reduced cell division, by which the plants attempt to reach a normal leaf size by increasing cell expansion (Horiguchi et al., 2006).
Figure 4.
Phenotype of rpn5a Mutants.
(A) Stunted growth of 2-week-old rpn5a seedlings compared with wild-type and rpn5b seedlings grown under continuous light. Each mutant is compared with its corresponding wild-type parent: C24 for rpn5a-1, Col-0 for rpn5a-2 and rpn5b-2, and Ws for rpn5b-1.
(B) Premature apical hook opening and increased petiole elongation of etiolated rpn5a seedlings compared with wild-type and rpn12a-1 seedlings. Seedlings were grown for 6 d in darkness.
(C) Stunted growth of 5-week-old rpn5a-1 seedling.
(D) Dwarfed influorescence of a 2-month-old rpn5a-1 plant compared with wild-type C24.
(E) Infertility of the rpn5a-1 mutant. Top panel, inflorescence of mature flowering plants. Bottom panel, close-up of developing siliques.
(F) Abnormal size and shape of hydrated rpn5a-1 seeds compared with a wild-type C24 sibling. Left panels, seeds before germination. Right panels, resulting 10-d-old seedlings. Bars = 0.1 mm (seeds) and 1 mm (seedlings).
(G) Abnormal floral development of rpn5a-1 plants. Left panels, whole flowers. Right panels, close-ups of the stigma and anthers from mature flowers.
Table 1.
Morphometric Analysis of rpn5a Mutants
| Genotype | Embryo Cortex Cell Numbera | Area of 1st Leaf (mm2) | Area of Adaxial Pavement Cells (μm2) | Number of Pavement Cells |
|---|---|---|---|---|
| Wild type | 50.0 ± 0.5 (28) | nd | nd | nd |
| rpn5a-1 | 16.2 ± 0.5 (41) | nd | nd | nd |
| Wild type | 52.5 ± 0.7 (30) | nd | nd | nd |
| rpn5a-2 | 11.0 ± 0.4 (29) | nd | nd | nd |
| Wild type | 48.0 ± 1.2 (22) | 9.0 ± 0.4 (15) | 2938 ± 19 (150) | 3067 ± 131 |
| rpn5a-3 | 14.3 ± 0.8 (33) | 2.4 ± 0.3 (10) | 4204 ± 94 (100)b | 582 ± 41 |
| Wild type | 50.6 ± 0.5 (32) | 9.7 ± 0.8 (16) | 3435 ± 108 (160) | 2924 ± 167 |
| rpn5a-4 | 11.2 ± 0.5 (36) | 3.5 ± 0.6 (12) | 3698 ± 120 (120)b | 915 ± 92 |
Numbers in parentheses indicate the number of individuals/leaves analyzed. The wild-type plants or embryos are siblings of the corresponding mutants. All data are expressed as the mean ± se. nd, not determined.
Cells were counted along a cortex cell file of the embryo from the root tip to the cotyledons.
Significant differences with the wild type (t test, P < 0.01 for cortex cell number of the embryo and analysis of variance; P = 0.021 for number of leaf pavement cells).
When grown under long days (16 h light/8 h dark), rosettes from all the homozygous rpn5a mutant lines produced a large cluster of aberrantly shaped leaves (Figure 4C) and slowly generated a disorganized short inflorescence (Figure 4D). Most rpn5a flowers displayed morphological defects in one or more floral whorls (Figure 4G). Whereas the stigma and carpel appeared wild-type in most flowers at least superficially, the anthers failed to produce and dehisce pollen at anthesis, implying a defect in male gametogenesis (Figure 4G). The resulting siliques from rpn5a flowers were substantially shorter and unexpanded and contained no seeds in most cases (Figure 4E). The few seeds that were produced were brown and shriveled and failed to germinate, suggesting that embryogenesis may also be blocked by inactivation of RPN5a. To test if some stigmas/carpels were functional, we attempted to cross-pollinate homozygous rpn5a-1 flowers with wild-type pollen. These flowers also failed to produce seeds, suggesting that female gametogenesis and/or fertilization may be blocked as well. Additionally, when germinated and grown in the dark, the rpn5a-1 mutant seedlings displayed a partial photomorphogenic phenotype that included premature opening of the apical hook and petiole elongation, a phenotype not seen with wild-type C24 or homozygous rpn10-1 and rpn12a-1 seedlings (Figure 4B). Side-by-side comparisons indicated that the rpn5a mutations confer a stronger repression on seedling growth than the previously described rpn10-1 or rpn12a-1 mutations (Figures 4B and 4E; Smalle et al., 2002, 2003).
In contrast with homozygous rpn5a plants, homozygous rpn5b plants were not compromised phenotypically. The development of etiolated and green seedlings and time to flowering in short and long days for homozygous rpn5b-1 or rpn5b-2 plants were indistinguishable from their wild-type Col-0 and Ws parents, respectively (Figure 4A). rpn5b-1 and rpn5b-2 plants were also completely fertile and showed normal transmission of the mutant alleles through the pollen and egg, demonstrating that the RPN5b locus is not essential for male or female gametogenesis and embryogenesis when the RPN5a locus is wild-type.
Previously, we showed that expression of 26S proteasome genes in Arabidopsis, like their counterparts in yeast, are coordinately upregulated when the abundance or activity of the complex drops (Yang et al., 2004; Kurepa et al., 2008). For example, when the activity of the 26S proteasome is depressed by mutations in RPN10 or RPN12a, a global increase in many other subunit transcripts and their corresponding proteins can be observed. A similar increase in subunit expression was found in rpn5a-1 but not rpn5b-2 plants. Whereas the levels of the PBA1 transcript and corresponding protein were not altered in the rpn5b-2 mutant, their levels were substantially elevated in the rpn5a-1 mutant compared with its wild-type C24 parent (Figures 3B and 3D). Similarly, the abundance of RPN5b mRNA increased in the rpn5a-1 background, but a commensurate increase was not seen for the RPN5a mRNA in the rpn5b-2 background (Figure 3B). Despite this differential effect, both RPN5 paralogs responded to 26S proteasome deficiencies. As shown in Figure 3C, mutations affecting RPN10 and RPN12a increased the transcript abundance for both RPN5a and RPN5b.
General inhibition of 26S proteasome activity in plants, either by proteasome inhibitors or loss of the RPN10 subunit, has been shown previously to increase the level of ubiquitylated proteins presumably awaiting breakdown (Woffenden et al., 1998; Girod et al., 1999; Smalle et al., 2003). The rpn5a mutations appear to induce the same effect. Slightly elevated levels of free Ub, free poly-Ub chains, and high molecular mass polyubiquitylated proteins were evident in homozygous rpn5a-1 seedlings, but not in homozygous rpn5b-2 seedlings (Figure 3D).
rpn5a Mutants Disrupt Embryogenesis
To inspect the reproductive defect further, we examined progeny from selfed heterozygous rpn5a-1 plants. In contrast with the complete infertility of homozygous rpn5a-1 plants, the heterozygotes developed normal shaped siliques packed with seeds. Significant seed abortion (empty spaces or abnormal seeds in the silique) was not evident, implying that the female gametogenesis is normal and that the resulting homozygous diploid rpn5a-1 zygote can proceed through embryogenesis.
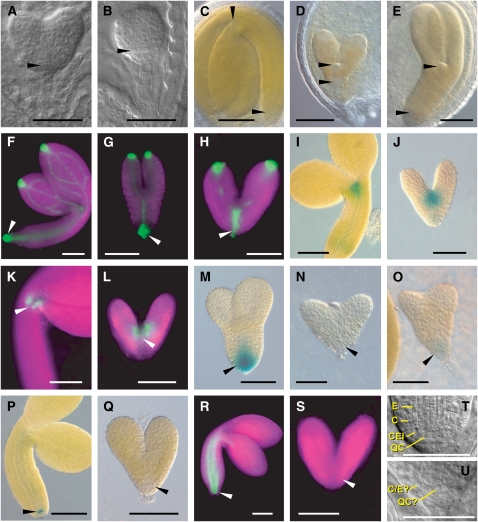
However, inspection of seeds generated from selfed rpn5a-1 heterozygous plants indicated that embryogenesis was abnormal. These populations contained a mixture of larger plump seeds, which likely have the wild-type or heterozygous rpn5a-1 genotype, together with smaller, misshaped seeds (Figure 4F). Invariably, the seedlings that germinated from these misshaped seeds displayed the lanceolate rpn5a-1 phenotype, which were later confirmed to be homozygous rpn5a-1 progeny (Figure 4F). Microscopic inspection of embryos dissected from self-pollinated heterozygous rpn5a-1, rpn5a-2, rpn5a-3, and rpn5a-4 plants revealed that all the embryos (up to 25% of which should be homozygous for each mutation) were indistinguishable very early in development. However, at the early- to mid-globular stages, a population of presumed rpn5a homozygous embryos emerged that developed slower than their wild-type or heterozygous siblings. For example, when wild-type embryos reached the early-heart stage, the likely homozygous rpn5a-4 embryos were still in the globular stage (Figures 5A and 5B). Cells derived from the hypophysis, which gives rise to the quiescent center (QC) and part of the RAM (Dolan et al., 1993), were particularly affected and had abnormal shapes and/or division patterns (Figures 5A, 5B, 5T, and 5U).
Figure 5.
Phenotype and Expression Patterns of Wild-Type and Homozygous rpn5a Embryos.
For each set of embryos, the wild type and mutant are shown at approximately the same developmental age. In all cases, the wild-type embryos are from heterozygous siliques of the same line as the corresponding mutant. In the fluorescent microscopy images, the tissue is highlighted by chlorophyll autofluorescence (false-colored magenta). In (A) and (B), arrowheads point to the abnormal shape of the cells in the position of the QC in rpn5a-1. In (C) to (U), arrowheads locate the RAM and SAM in the wild-type embryos or the equivalent position in the mutants. C, cortex; E, endodermis; QC?, cells in the QC position; C/E?, single cell file of unknown identity in the position of the cortex/endodermis. Bars = 50 μm.
(A) and (B) Young embryos of the wild type (A) and the rpn5a-4 mutant (B) at the heart stage.
(C) Mature embryos of the wild type and the rpn5a mutants.
(D) Example of the short embryo phenotype (rpn5a-2).
(E) Example of a long embryo phenotype (rpn5a-1).
(F) to (H) DR5rev:GFP expression in the wild type at the bent cotyledon stage (F) and rpn5a-4 mutant embryos with normal expression (G) and high expression that extended apically (H).
(I) and (J) STMpro:GUS expression in the wild type at the bent cotyledon stage (I) and rpn5a-4 (J).
(K) and (L) CUC1 (M0233 line) expression in the wild type at the bent cotyledon stage (K) and rpn5a-4 (L).
(M) to (O) PIN4pro:GUS expression in the wild type at the torpedo stage (M) and rpn5a-4 with no expression (N) or weak expression (O).
(P) and (Q) QC25pro:GUS expression in the wild type at the bent cotyledon stage (P) and rpn5a-4 (Q).
(R) and (S) SCRpro:GFP expression in the wild type at the bent cotyledon stage (R) and rpn5a-4 (S).
(T) and (U) Close-up of the root meristem in the wild type at the late heart stage (T) and rpn5a-3 (U) embryos.
After the late heart-shape stage, most of the rpn5a-2, rpn5a-3, and rpn5a-4 embryos had fewer hypocotyl cells and limited cotyledon development (designated as the short phenotype; Figures 5C and 5D). For example, the number of cortex cells between the root tip and cotyledons in the mature mutant embryos was substantially less (∼50 cells for wild-type embryos versus 11 to 14 cells for the short-phenotype embryos produced by the four rpn5a alleles; Table 1). This low number was similar to wild-type embryos at the early torpedo stage (11.1 ± 0.3 cells, n = 20). Within the mutant collection, the rpn5a-1 embryos were noticeably different in that cell division/expansion continued in the hypocotyl and cotyledons of some to produce a larger embryo, although still much smaller than wild-type embryos at maturity (designated as the long phenotype; Figure 5E). The shoot apical meristem (SAM) of both the short- and long-phenotype rpn5a-1 embryos was similar (or in some cases larger) in size to that of wild-type C24 (Figures 5C to 5E). When we crossed the short rpn5a-2 or rpn5a-4 alleles to several Col-0 backgrounds, the long phenotype appeared and segregated in subsequent selfed populations. Consequently, it is possible that the difference between the short and long phenotypes was caused by a modifier in the Col-0 background. The nature of this possible modifier is not known, but genetic analyses suggest that it is not the ERECTA (ER) locus as both the long and the short phenotype were observed in homozygous ER and er plants.
In total, ∼25% of the rpn5a embryos from individual siliques segregated for the short/long mutant phenotypes (rpn5a-1: 20.2% [n = 203], rpn5a-2: 23.1% [n = 350], rpn5a-3: 28.4% [n = 81], and rpn5a-4: 24.9% [n = 630], with a total of 23.9% [n = 1264]), indicating that the rpn5a mutations act recessively during embryogenesis. However, only 10 to 20% of the germinating seeds from these populations displayed the stunted seedling phenotype expected of homozygous rpn5a-1 plants, with an increased number of nongerminated seeds relative to the wild type. This difference implies that some homozygous rpn5a embryos, while completing much of embryogenesis, do not produce viable seedlings.
To further characterize rpn5a embryos, we analyzed the expression patterns of a number of genes important to embryogenesis in the short phenotype embryos from selfed heterozygous rpn5a-4 plants. Auxin responsiveness was examined by assaying the expression of DR5rev:GFP (for green fluorescent protein), which strongly responds to the endogenous pools of the hormone (Friml et al., 2003). As shown in Figures 5F to 5H, the patterns of DR5rev:GFP expression were similar between the wild type and rpn5a-4, with high levels evident in the cotyledon and root tips and the provasculature of the cotyledons (Figures 5F to 5H). While the wild-type and some of the short phenotype embryos (24%, n = 33) had moderate levels of DR5rev:GFP in the provasculature of the hypocotyl, with a maximum just below the QC (Figure 5H, arrowhead), most (76%) of the mutants showed much stronger provasculature expression that extended toward the apical meristem (Figure 5H). In either case, auxin responsiveness was clearly not reduced by the rpn5a-4 mutation.
To evaluate the patterning of the apical region, we examined the expression of SHOOTMERISTEMLESS (STM) and CUP-SHAPED COTYLEDONS1 (CUC1) that regulate transcription in the SAM and between the SAM and the cotyledons, respectively (Long et al., 1996; Takada et al., 2001). The expression patterns of the STMpro:GUS reporter (Figures 5I and 5J) and the GFP enhancer-trap line M0233 reflecting CUC1 expression (Cary et al., 2002) (Figures 5K and 5L) in the short phenotype embryos were indistinguishable from the wild type (n = 20), suggesting that the patterning of the apical region is normal in rpn5a-4 embryos. For the root pole, we analyzed the PIN4 auxin efflux carrier (PIN4pro:GUS; Friml et al., 2002), which is expressed in the QC and surrounding cells (Figures 5M to 5O), the QC marker QC25pro:GUS (Jenik et al., 2005) (Figures 5P and 5Q), and the transcription factor SCARECROW (SCRpro:GFP; Wysocka-Diller et al., 2000), which is expressed in the QC, cortex/endodermis initial (CEI), and endodermis along the hypocotyl (Figures 5R and 5S). Neither SCRpro:GFP nor QC25pro:GUS was expressed at detectable levels in the short phenotype embryos (n = 31 and 33, respectively). In the case of PIN4pro:GUS, most of the short embryos also failed to express detectable levels. However, in a few (6 of the 90 embryos inspected), weak GUS staining was seen near the root tip (Figure 5O). Because SCR is responsible for the periclinal divisions of the CEIs that lead to the formation of separate cortex and endodermal cell files (Wysocka-Diller et al., 2000), we reexamined the cells in this region microscopically (Figures 5T and 5U). Of the 30 rpn5a-4 embryos examined, 19 appeared to have only one cell file in the position of the cortex/endodermis instead of two files normally seen in the wild type. The other 11 mutant embryos had two cell files with irregular shapes and unclear identity and no obvious CEI. These phenotypes are reminiscent to those of scr (Wysocka-Diller et al., 2000) and suggest that an active RPN5a locus is needed to properly specify the QC and CEIs.
Both RPN5a and RPN5b Are Essential for Gametogenesis
To determine the functional importance of RPN5a and RPN5b together, we attempted to generate double homozygous rpn5a-2 rpn5b-2 plants. Here, an rpn5a-2 heterozygous plant was crossed to an rpn5b-2 heterozygous plant (both in the Col-0 background), and double heterozygous progenies were identified in the F1 progeny by genomic PCR. These double heterozygous plants appeared wild-type phenotypically, indicating that plants missing one copy of RPN5a and one copy of RPN5b are not even subtly compromised compared with plants missing both copies of RPN5a.
A RPN5a/rpn5a-2 RPN5b/rpn5b-2 heterozygote was then self-crossed. Microscopy examination of 1106 resulting embryos found 950 (85.9%) that appeared wild-type and 156 (14.1%) that had the aberrant short phenotype (6:1 ratio), with no more severe phenotypes, embryo arrest, or seed abortion evident (Figure 5D). The ovules from the double heterozygous carpels also appeared wild-type. Given the absence of aborted ovules and the 6:1 segregation ratio (wild type:rpn5a-2), we conclude that rpn5a-2 rpn5b-2 pollen grains are inviable, but rpn5a-2 rpn5b-2 ovules are normal. Following germination, the seedlings in this selfed population were genotyped for segregation of the various mutant allele combinations. Surprisingly, analysis of 140 individuals failed to identify plants that were homozygous for both rpn5a-2 and rpn5b-2 or plants homozygous for one mutation and heterozygous for the other (rpn5a-2/+ rpn5b-2/rpn5b-2 or rpn5a-2/rpn5a-2 rpn5b-2/+) (Table 2). The absence of these allele combinations is consistent with the inviability of ab pollen combined with a scenario where the Aabb and aaBb embryos arrest development prematurely or where the resulting seeds do not germinate. We also found that the rpn5a-2 single homozygous mutant (aaBB) segregated at one-fifth the frequency of the rpn5b-2 single homozygous mutant (AAbb), which likely reflects the reduced viability seen for homozygous rpn5a-2 seeds (see above). To further support the need for both RPN5a and RPN5b, we analyzed a large population of progeny from a double heterozygous parent (n = 1270) to find rpn5a-2/rpn5a-2 RPN5b/rpn5b-2 plants. Sixty-eight of the progeny displayed the rpn5a-2 phenotype, indicating that they were homozygous for the rpn5a-2 mutation. Of these 68, all were genotyped as homozygous wild type for the RPN5b loci. Taken together, the segregation data indicate that Arabidopsis requires (1) both RPN5a and RPN5b for pollen production, (2) at least one wild-type RPN5a locus for normal embryogenesis, and (3) at least two of the four RPN5 loci to produce viable seeds with the inactivation of both RPN5a loci alone attenuating seed viability.
Table 2.
Aberrant Segregation of rpn5a rpn5b Double Mutants
| Genotype | Number | % | Expected %a | |
|---|---|---|---|---|
| Genotypes of progeny from selfed rpn5a-2/+ rpn5b-2/+ (AaBb) parentb | ||||
| AABB | 15 | 11 | 6 | 10 |
| AABb | 32 | 24 | 13 | 20 |
| AAbb | 15 | 11 | 6 | 10 |
| AaBB | 32 | 24 | 13 | 20 |
| AaBb | 38 | 28 | 25 | 30 |
| aaBB | 3 | 2 | 6 | 10 |
| Aabb | 0 | 0 | 13 | 0 |
| aaBb | 0 | 0 | 13 | 0 |
| aabb | 0 | 0 | 6 | 0 |
Left column: percentage expected if all genotypes are viable. Right column: percentage expected if rpn5a rpn5b pollen and rpn5a-2/+ rpn5b-2/rpn5b-2 and rpn5a-2/rpn5a-2 rpn5b-2/+ embryos are inviable.
Total individuals genotyped = 140.
Inactivation of RPN5a Destabilizes the 26S Proteasome
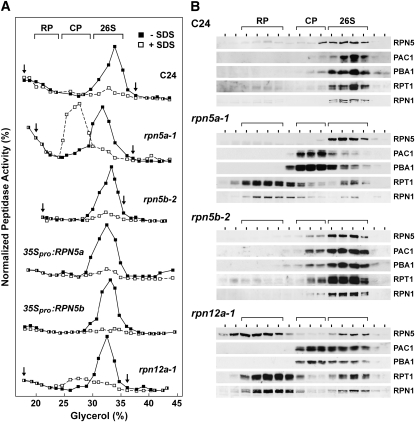
A temperature-sensitive allele of the single yeast gene encoding RPN5 impairs 26S proteasome assembly at restrictive temperatures, thus elevating the accumulation of free CP and the RP lid and base subcomplexes (Isono et al., 2007). To test if the rpn5a alleles have a similar destabilizing effect in Arabidopsis, we subjected proteasome-enriched fractions from wild-type C24 and Col-0, rpn5a-1, and rpn5b-1 seedlings to glycerol gradient centrifugation, which can resolve free CP and RP from the 26S proteasome (Glickman et al., 1998; Yang et al., 2004). The sedimentation profiles of the active CP and 26S complexes were then determined by peptidase activity toward the established proteasome substrate Suc-LLVY-AMC in the absence (26S proteasome) or presence of 0.02% SDS (CP) (Yang et al., 2004). The profiles of the CP, RP, and 26S complexes were also revealed by immunoblot analysis with the antibodies against PAC1 and PBA1 (α3 and β1 subunits of the CP), RPT1 and RPN1 (subunits of the RP base), and RPN5 (subunit of the RP lid). A single proteasome complex was detected by both immunoblotting and peptidase assay in C24 wild-type seedlings (Figures 6A and 6B), which based on sedimentation position represents the 26S holoprotease (Yang et al., 2004). A similar profile was seen for rpn5b-2 seedlings, indicating that the composition of the complex was unaffected by inactivation of RPN5b, presumably because ample RPN5a was present. By contrast, a dramatic change in the profile was evident for proteasomes enriched from rpn5a-1 seedlings, which reflect a destabilization of the complex. Not only was the total amount of peptidase activity (−SDS) lower in the mutant, but a new slower sedimenting particle in addition to the 26S particle appeared that represented the CP based on its coelution with the CP subunits PAC1 and PBA1 and its strong peptidase activity in the presence of SDS. Another complex also appeared that contained the RP subunits RPN1 and RPT1, which likely represented the free RP. None of the residual RPN5b protein was in this free RP fraction, consistent with a role for RPN5 in stabilizing RP-CP binding (Isono et al., 2007).
Figure 6.
Instability of the 26S Proteasome from the rpn5a-1 Mutant.
26S proteasomes were enriched from 10-d-old liquid-grown seedlings of the indicated genotypes and subjected to centrifugation in a 10 to 40% glycerol gradient. The general location of the 26S proteasome and the 20S CP and 19S RP subcomplexes are indicated by the brackets.
(A) Assay of gradient fractions for CP peptidase activity in the absence or presence of 0.02% SDS using the substrate Suc-LLVY-AMC. The activity scale for each profile was adjusted to generate a near equal height for the peak activity. The arrows delineate the range of fractions that were used for the immunoblot analysis (B).
(B) Immunoblot analysis of gradient fractions for the presence of various 26S proteasome subunits.
To test whether this 26S proteasome instability was specific for rpn5a mutants or could be observed for mutants affecting other RP subunits, we examined the sedimentation profile of proteasomes enriched from rpn12a-1 plants. This exon-trap mutation expresses a fusion of full-length RPN12a with NPTII that can integrate into the 26S complex (Smalle et al., 2002). While homozygous rpn12a-1 plants display a pleiotropic phenotype, including a dwarfed growth habit and cytokinin insensitivity, they are not as phenotypically compromised as rpn5a plants and are fertile (Smalle et al., 2002). Glycerol gradient centrifugation of proteasomes from rpn12a-1 plants revealed that they were also partially dissociated (Figures 6A and 6B). Although only a small peak of CP was measured by peptidase activity, free RP (RPN1 and RPT1) and CP (PAC1 and PBA1) complexes were evident in addition to the 26S particle. While part of the RPN5 pool was in the 26S fraction, most appeared to be associated with the free RP.
Complementation of rpn5a-1
To confirm that the rpn5a phenotypes were caused by inactivation of RPN5a and to help address whether the corresponding RPN5a and RPN5b proteins have equivalent activities, we attempted to rescue homozygous rpn5a-1 seedlings with RPN5a or RPN5b transgenes. These transgenes either expressed the proteins under the control of the near constitutive cauliflower mosaic virus (CaMV) 35S promoter (35Spro:RPN5a and 35Spro:RPN5b), their own promoters (RPN5apro:RPN5a and RPN5bpro:RPN5b), or the promoter from their paralogs (RPN5apro:RPN5b and RPN5bpro:RPN5a). Because homozygous rpn5a-1 plants are sterile, the transgenes were first introduced into the heterozygous background, and then homozygous rpn5a-1 plants containing the transgenes were identified in selfed F2 populations (Figure 7A).
Figure 7.
Rescue of the rpn5a Mutant Phenotype with RPN5a and RPN5b.
rpn5a-1 mutants in the C24 background were transformed with the RPN5a or RPN5b genomic coding regions expressed under the control of the CaMV 35S promoter (35Spro), the RPN5a promoter (5apro), or the RPN5b promoter (5bpro).
(A) Genotyping of complementation lines. DNA was isolated from 10-d-old seedlings and PCR amplified with primers specific for wild-type RPN5a gene, the T-DNA in rpn5a-1 (rpn5a-1 T-DNA), or the RPN5a and RPN5b transgenes (trans). Plants homozygous (homo) and heterozygous (het) for the rpn5a-1 T-DNA insertion were included for comparison.
(B) Levels of RPN5 protein in the complementation lines. Crude protein extracts were prepared from 10-d-old seedlings and subjected to SDS-PAGE and immunoblot analysis with antibodies against RPN5a, RPN1a, RPN10, RPN12a, and PBA1. Analysis with anti-HSP70 antibodies was used to confirm equal protein loads.
(C) Phenotypic rescue of rpn5a-1 mutants. Pictures of 14-d-old plants homozygous for the rpn5a-1 mutation and homozygous for the RPN5a or RPN5b transgenes.
Introduction of the 35Spro:RPN5 transgenes strongly rescued the aberrant morphological phenotypes associated with rpn5a plants in at least five independently transformed rpn5a-1 lines (Figure 7C). Importantly, the fertility of these plants was restored and the enhanced accumulation of other 26S proteasome subunits (PBA1, RPN1, and RPN12a) seen in the rpn5a-1 background was abolished (Figure 7B). A similar strong morphological and biochemical rescue was also observed when RPN5a and RPN5b were expressed by their own promoters or by the promoter from their paralog (Figures 7B and 7C). That the RPN5b promoter was effective is consistent with the similar tissue expression profiles of the two promoters (Figures 2A and 2B). Transgenic expression of the RPN5a protein in the rpn5a-1 background returned the immunodetectable levels of RPN5 back to normal in all cases (Figure 7B). However, for the RPN5b transgenes, the immunodetectable levels were not restored, which may reflect an inability of the plants to accumulate RPN5b to the same level as RPN5a. Together, the data imply that the RPN5b protein can functionally replace RPN5a and suggest that at least some of the rpn5a phenotypes are caused by insufficient RPN5 protein, which cannot be generated from the native RPN5b locus by itself.
Overexpression of RPN5a and RPN5b Affects Arabidopsis
To examine the effects of RPN5 overexpression, we introduced the 35Spro:RPN5a and 35Spro:RPN5b transgenes into wild-type C24 plants. RNA gel blot analyses confirmed that each transgene substantially elevated the level of their respective mRNAs (Figure 3B) but surprisingly did not detectably increase the abundance of the corresponding proteins (Figure 8D). Regardless, these transgenes phenotypically altered the plants in a way that implied that RPN5a and RPN5b also have unique functions in Arabidopsis. Both sets of overexpression lines generated an early flowering phenotype, which could be seen under continuous light or short days (8 h light/16 h dark) (Figures 8A and 8B). (This phenotype provided confirmation that the 35Spro:RPN5a and 35Spro:RPN5b transgenes and resulting proteins were active.) Early flowering subsequently reduced the fecundity of the plants given their small size at bolting. 35S-driven overexpression of two other RP subunits, RPN12a and RPN10 (as confirmed by immunoblotting; Figure 8D), did not induce early flowering, demonstrating that this phenotype is not common to the overexpression of any RP subunit (Figure 8A).
Figure 8.
Effects of Overexpression of RPN5a and RPN5b on Arabidopsis Development.
The RPN5a and RPN5b cDNAs were expressed under the control of the CaMV 35S promoter in wild-type Col-0 seedlings. mRNA levels are shown in Figure 2B.
(A) Phenotype of 3-week-old 35Spro:RPN5a and 35Spro:RPN5b plants grown in continuous light.
(B) Early flowering of 35Spro:RPN5a and 35Spro:RPN5b plants grown in continuous light compared with Col-0 seedlings overexpressing two other RP subunits (35Spro:RPN10 and 35Spro:RPN12a).
(C) Premature leaf senescence of 35Spro:RPN5a plants grown in continuous light. Pictured are two independent lines with differing degrees of severity next to wild-type Col-0.
(D) Levels of the corresponding RP proteins in the RPN5a-, RPN5b-, RPN10-, and RPN12a-overexpressing plants shown in (A). Crude protein extracts were prepared from 7-d-old green plants and subjected to immunoblot analysis with antibodies against RPN5a, RPN10, RPN12a, and PBA1. Analysis with anti-UCH1 antibodies was used to confirm equal protein loads.
In addition, overexpression of RPN5a specifically generated a slow growth, chlorotic, and early senescence phenotype (Figures 8B and 8C). However, the plants remained fertile. The severity of the phenotype varied among independent transformants (Figure 8C) and was dosage dependent, with homozygous 35Spro:RPN5a plants displaying a more severe phenotype compared with their hemizygous siblings (data not shown). Some of the T2 plants and potential T3 progeny were even more compromised than the initial transformants.
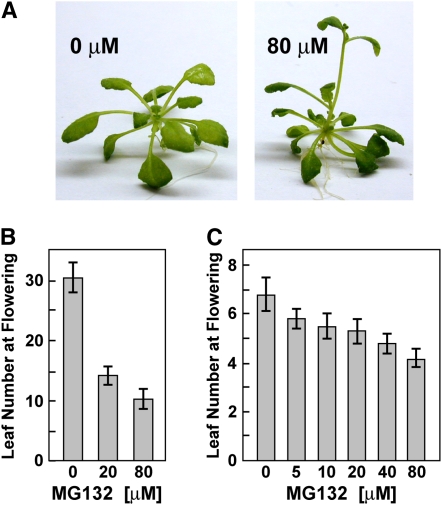
The early flowering phenotype of 35Spro:RPN5a and 35Spro:RPN5b plants suggested that excessive amounts of either RPN5 isoform adversely affect 26S proteasome function with the stress prematurely inducing reproduction. In support of this, we found that extended exposure of Arabidopsis seedlings to the proteasome inhibitor MG132 also significantly accelerated flowering (Figure 9). MG132 competitively blocks the chymotryptic activity of the CP and is effective at nanomolar concentrations against the purified plant 26S complex (Yang et al., 2004). At high concentrations (200 μM), MG132 impaired Arabidopsis seedling growth, presumably by globally inhibiting protein breakdown. However, at less toxic concentrations (≤80 μM), MG132 induced early flowering (as determined by number of rosette leaves before bolting) without a strong effect on seedling growth. For example, wild-type C24 plants treated with 80 μM MG132 flowered 20 d earlier (30.5 ± 2.8 d versus 10.5 ± 1.7 d) in short days, while in continuous light, they flowered ∼2.6 d earlier (6.8 ± 0.7 d versus 4.2 ± 0.4 d) (Figures 9B and 9C).
Figure 9.
Acceleration of Flowering Time by Exposure of Seedlings to MG132.
(A) Wild-type Col-0 plants grown without or with 80 μM MG132 for 4 weeks under a short-day photoperiod.
(B) and (C) Leaf number at the onset of flowering for seedlings grown under a short-day photoperiod (B) or under continuous light (C) with or without exposure to various concentrations of MG132. Each bar represents the average (±sd) of 10 seedlings.
More specifically, the early senescence phenotype of RPN5a-overexpressing plants suggested that elevated levels of the RPN5a protein relative to RPN5b could adversely affect the activity or integrity of the 26S proteasome. Whereas the levels of the RP subunits RPN10 and RPN12a were unaltered, the level of CP subunit PBA1 was slightly elevated in the 35Spro:RPN5a lines (Figure 8D). In addition, only a slight increase in Ub conjugates was observed in the 35Spro:RPN5a lines, suggesting that 26S proteasome activity was not dramatically compromised (Figure 3D). The integrity of the 26S particle also appeared normal. The profile of the 26S proteasome from 35Spro:RPN5a plants after glycerol gradient centrifugation was indistinguishable from wild-type and 35Spro:RPN5b plants with respect to both peptidase activity and the association of CP and RP subunits (Figure 6).
Intracellular Localization of RPN5a and RPN5b
One possible reason for the nonoverlapping functions of RPN5a and RPN5b is that each preferentially accumulates in different compartments within the cell (e.g., nucleus versus cytoplasm). To test this possibility, we introduced transgenes under the control of the 35S promoter, which express in-frame fusions of RPN5a and RPN5b to the N terminus of GFP, and visualized their subcellular localization by confocal fluorescence microscopy. To demonstrate that the GFP fusions are functional in planta, we transformed them into heterozygous rpn5a-1 seedlings, selected for plants homozygous for the rpn5a-1 mutation and at least hemizygous for the GFP transgene, and examined the plants for complementation. As shown in Figure 2C, both the RPN5a-GFP and RPN5b-GFP reporters successfully rescued most, if not all, aspects the rpn5a-1 phenotype.
Microscopy analysis of root tips revealed that the RPN5a-GFP and RPN5b-GFP fusions have indistinguishable intracellular distributions. Fluorescence was highest in the nucleus with substantial but lower fluorescence in the cytoplasm (Figure 2C), a distribution similar to that reported by Kwok et al. (1999) for the RPN6 subunit detected in Arabidopsis protoplasts immunologically. This nuclear enrichment became more pronounced in more mature cells further away from the root tip. Interestingly, close examination of the nuclear patterns revealed a strong peripheral signal with a dark central area that most likely represents the nucleolus (Figure 2C). Often near the middle of this nucleolar area was a dot of fluorescence, which could represent a subnucleolar compartment. Identical patterns were seen for the RPN5b-GFP fusions introduced into wild-type Col-0 plants and rpn5a-1 plants, demonstrating that the absence of RPN5a did not trigger a non-native distribution of the RPN5b-GFP protein.
DISCUSSION
The 26S proteasome plays a central role in the selective turnover of most cytoplasmic and nuclear proteins in eukaryotes, especially those proteins first marked by ubiquitylation. The RP subparticle is central to the substrate specificity of this complex, but at present, the functions of many of its subunits remains unclear, particularly those in the lid. In plants, analysis of these functions may be further complicated by the presence of isoform pairs for most RP subunits, some of which may have distinct activities (Yang et al., 2004). To provide more insight into the role(s) of each RP subunit and to examine the potential that RP paralogs have nonredundant functions in plants, we investigated the RPN5 subunit encoded by two genes in Arabidopsis. RPN5, like almost all other 26S proteasome subunits, is essential in S. cerevisiae (the exceptions are RPN9 and RPN10) (Yang et al., 2004; Isono et al., 2007). In S. pombe, rpn5Δ mutants are viable, but the loss of RPN5 stabilizes several 26S proteasome substrates and impairs assembly of the complex and its localization to the nucleus (Yen et al., 2003).
Here, we showed that RPN5 is essential in Arabidopsis from the analysis of a set of strong, and likely null, alleles affecting RPN5a and RPN5b. Double mutants disrupting both loci could not be generated. Segregation analysis of progeny from double heterozygous selfed plants failed to even find plants homozygous for one paralog and heterozygous for the other. These transmission defects appear to arise at several developmental steps. First, homozygous rpn5a plants fail to produce pollen, while rpn5a rpn5b pollen is inviable. Homozygous rpn5a embryos can be produced from heterozygous parents, but their development is stunted leading to the production of smaller embryos with fewer cells. While the resulting homozygous rpn5a plants retain a modest level of RPN5 due to the transcriptional activity of the RPN5b gene, they have reduced 26S proteasome activity/integrity, as seen by fractionation of the complex, by the increased accumulation of Ub conjugates, and by the upregulation of other 26S proteasome subunits, a phenomenon widespread among eukaryotes and indirectly caused by attenuated proteasome level/activity (Xie and Varshavsky, 2001; Meiners et al., 2003; Yang et al., 2004; Kurepa et al., 2008).
In both S. cerevisae and S. pombe, rpn5 mutants display cell division defects, consistent with the importance of ubiquitylation and the 26S proteasome to numerous checkpoints of the cell cycle (Yen et al., 2003; Isono et al., 2007). Both the defects in male gametogenesis and altered cell division patterns in the Arabidopsis embryo collectively point to a critical role of RPN5 during meiosis and mitosis in plants as well. Mutations in other RP subunits (RPN1, RPN10, RPN12a, and RPT2a) also generate similar slow growth phenotypes that have been connected (directly or indirectly) to altered expression of cell cycle–related factors (Smalle et al., 2002, 2003; Ueda et al., 2004; Brukhin et al., 2005). Consequently, this defect likely reflects a general impairment of 26S proteasome activity.
In addition to a developmental delay, homozygous rpn5a embryos display altered expression of key embryonic pattern genes. Expression of STM and CUC1 appears normal, suggesting that SAM identity and maintenance is unaffected. Similarly, the expression patterns of the auxin-responsive marker DR5rev:GUS was only subtly altered, indicating that perception of this hormone, which is central to embryonic development (Friml et al., 2003), is only marginally affected by diminished RPN5 levels. By contrast, the expression patterns of PIN4, QC25, and SCR were notably altered in the rpn5a backgrounds. SCR is required to specify the QC (acting in parallel with auxin) (Aida et al., 2004) and to direct the periclinal division of the CEI daughter cell (Wysocka-Diller et al., 2000). The aberrant expression pattern of SCR in particular could explain the root phenotype of rpn5a embryos that includes no obvious QC or periclinal divisions of the CEI daughter cells and an abnormal root pole. However, scr embryos are otherwise normal phenotypically (Sabatini et al., 2003), indicating that the loss of SCR expression in the rpn5a backgrounds cannot not fully explain the reduced cell divisions in the hypocotyl and embryonic root. Whatever the mechanism, our results link the 26S proteasome to SCR transcriptional regulation. Cytokinin has been recently reported to be required for proper SCR expression and RAM formation in the embryo (Muller and Sheen, 2008). It is interesting to note that several RP mutants (rpn12a, rpn10, and hlr/rpt2a) have altered cytokinin responses (Smalle et al., 2002, 2003; Kurepa et al., 2008), suggesting a more general connection among the 26S proteasome, cytokinins, and embryonic root patterning.
After germination, homozygous rpn5a seedlings display a host of other developmental defects, including partial deetiolation of seedlings grown in the dark, aberrant cotyledon/leaf development, severely dwarfed rosettes and infloresences, delayed flowering, abnormal flowers, and a block in pollen production resulting in complete infertility. The low expression of RPN5a and RPN5b in pollen relative to other reproductive tissues may explain the particular sensitivity of these cells to loss of the RPN5 subunit. Ovules from homozygous rpn5a plants also failed to be fertilized by wild-type pollen, indicating a defect in female gametogenesis and/or fertilization in the absence of RPN5a. These defects were found in four different rpn5a alleles and could be rescued by introducing RPN5a-expressing transgenes into the rpn5a-1 background, confirming that the problems were caused by inactivation of RPN5a specifically. We also noticed that homozygous rpn5a seeds likely have reduced germination rates and that rpn5a/+ rpn5b rpn5b and rpn5a rpn5a rpn5b/+ plants could not be generated. These defects imply that RPN5 gene dosage is important and that insufficient levels of RPN5 in embryos can also reduce or block embryo maturation, seed viability, and/or germination.
Some of the phenotypic defects associated with rpn5a mutants are notably different from those reported for mutants affecting other Arabidopsis RP subunits. For example, we previously showed that mutants affecting the single genes encoding RPN10 and RPN12 also display a dwarfed growth habit, but these plants remain at least partially fertile and do not display inappropriate deetiolated development (Smalle et al., 2002, 2003). In fact, rpn12a-1 plants show a delay in deetiolation after extended darkness instead of the acceleration seen in rpn5a plants. The lanceolate leaf phenotype can also be seen in rpn10-1 and rpn12a-1 plants but is not as severe as that in rpn5a plants (this report; Smalle et al., 2002, 2003). Like the rpn5a mutants, the rpn12a-1 mutant still accumulates the wild-type-sized protein, suggesting that the phenotypes reflect inadequate levels of RPN12a and not the presence of an altered form. For rpn10-1 seedlings, an altered form accumulates that is missing the Ub- binding site responsible for its recognition of ubiquitylated proteins (Smalle et al., 2003). The mutant is also hypersensitive to ABA due to a strong stabilization of the ABI5 transcription factor. Arabidopsis rpt2a mutants (also known as halted root) are also developmentally defective (Ueda et al., 2004). Homozygous rpt2a plants are fully fertile, but like rpn5a plants, they have altered RAM and SAM function. Finally, RPN1a is also essential for embryogenesis (Brukhin et al., 2005). However, unlike rpn5a mutants, null homozygous rpn1a mutants cannot complete embryogenesis and arrest at the globular stage. The embryo arrest could be rescued with either RPN1a or RPN1b expressed under the control of the RPN1a promoter, suggesting that the corresponding proteins are functionally equivalent.
Why rpn5a mutants generate a set of phenotypes distinct from other rpn mutants in Arabidopsis is not yet clear. One strong possibility is that these severe phenotypes reflect the unique position of RPN5 within the RP, which in turn affects the turnover rates of a subset of 26S proteasome targets. Interaction maps generated by yeast two-hybrid analysis and mass spectrometry currently place RPN5 at the center of a subcomplex within the RP lid that also includes RPN8 and RPN9, and possibly RPN6 and RPN12 (Fu et al., 2001; Sharon et al., 2006; Isono et al., 2007). Binding to RPN9 in particular is directed by the PCI domain in RPN5 (Fu et al., 2001). That both rpn5a and rpn12a mutants destabilize 26S proteasome integrity supports the presence of this RP subcomplex. Additionally, RPN5 may also contact RPN3 within a second lid subcomplex containing RPN7 and SEM1 (Sharon et al., 2006). As a consequence, RPN5 may play a scaffolding role central to stabilizing the entire lid. Inadequate amounts of this subunit in turn could generate lid complexes not only missing RPN5 but also a host of other RPN subunits and the collection of accessory proteins that they may recruit to the RP.
The high amino acid sequence identity of RPN5a and RPN5b (93%) strongly suggested that these paralogs are functionally equivalent. This was confirmed by our complementation studies, which showed that the expression of both proteins could nearly fully rescue homozygous rpn5a-1 plants. That either promoter could rescue the phenotype likely reflected the strongly overlapping expression of the promoters as shown using promoter-GUS fusions. However, estimates of expression strengths by DNA microarrays and EST representation imply that the RPN5a promoter is substantially stronger than that for RPN5b in planta. This relative strength of the RPN5a locus is also supported by the failure of the native RPN5b locus to rescue the rpn5a-1 phenotype even though its expression is upregulated in the rpn5a backgrounds but could when a second RPN5b locus was provided by the RPN5bpro:RPN5b transgene. The expression difference, in turn, could easily generate a greater deficiency of the RPN5 protein in the rpn5a backgrounds and thus explain why rpn5a but not rpn5b mutants are phenotypically compromised.
As with a number of other 26S proteasome subunits (Xie and Varshavsky, 2001; Yang et al., 2004), the levels of RPN5a/b are tightly controlled by the abundance/activity of the complex in both plants and yeasts (Isono et al., 2007; this report). When RPN5a was inactivated, the transcript level of its paralog, RPN5b, increased. Furthermore, transcript levels for both RPN5a and RPN5b increased when other subunits (RPN10 and RPN12a) were compromised. However, unlike the compensation situation in S. pombe where overexpression of one RPN5 paralog dampens expression of the other (Yen et al., 2003), overexpression of RPN5a or RPN5b in Arabidopsis failed to diminish transcript abundance of either isoform. Thus, while Arabidopsis can respond to insufficient RPN5, it does not appear to respond to excess RPN5.
In addition to their functional equivalence, RPN5a and RPN5b have indistinguishable intracellular locations in Arabidopsis as detected using functional GFP fusions. In young cells of the root, both isoforms are present in the cytoplasm and nucleus consistent with the importance of ubiquitylation and the 26S proteasome for protein turnover in these compartments. As the cells away from the root tip elongated and matured, most of the RPN5 pool became concentrated in the nucleus. Similar to distributions reported in yeasts (Yen et al., 2003; Isono et al., 2007), Arabidopsis RPN5 was most abundant at the nuclear periphery with much lower levels evident in the presumed nucleolus. Interestingly, we often observed a punctate area inside the nucleolus that was enriched for the RPN5-GFP protein. The significance of this dot is currently unknown. We have detected similar nucleolar foci using GFP fusions to other 26S proteasome subunits (A.J. Book, unpublished data). Consequently, these foci could represent an aggregation site for unincorporated subunits. Or more intriguingly, they could represent a subnucleolar region involved in 26S proteasome assembly (Yen et al., 2003), a nucleolar site actively engaged in protein breakdown, or a storage site for excess proteasomes as has been detected recently in stationary yeast cultures (Laporte et al., 2008).
In addition to inactivating RPN5a, we also found that increased expression of either RPN5a or RPN5b has negative consequences in Arabidopsis seedlings, implying that too much RPN5 is cytotoxic. In a similar fashion, Isono et al. (2007) found that overexpression of RPN5 in S. pombe is also problematic and intensifies the phenotypes of other RP mutants. Why overexpression of either RPN5a or RPN5b induces early flowering in Arabidopsis is not yet clear. It could reflect a general stress response of the plants or the critical role for ubiquitylation and the 26S proteasome in flowering time, given their roles in the breakdown of the flowering time regulator CONSTANS and several circadian clock components (Mas et al., 2003; Imaizumi et al., 2005; Jang et al., 2008). That the phenotype was not replicated by overexpression of two other RPN subunits but was phenocopied by exposing the plants to the proteasome inhibitor MG132 suggests that overexpression of RPN5 specifically alters 26S proteasome capacity. One possibility is that excess free RPN5 inhibits lid assembly by binding its contacts within the lid (e.g., RPN3, RPN6, RPN9, and RPN12; Fu et al., 2001; Sharon et al., 2006; Isono et al., 2007) or yet to be described RPN5-associating proteins in a way that interferes with their subsequent incorporation into the RP or transport into the nucleus.
While all data point to RPN5a and RPN5b being functionally equivalent, our observations that overexpression of RPN5a also generates an early senescence phenotype suggest that nonoverlapping functions for these paralogous genes exist as well. It remains unclear why RPN5a overexpression triggers early senescence. The notion that the different phenotypes are caused by RPN5a and RPN5b accumulating in different compartments is discounted by our data showing that these paralogs have indistinguishable intracellular distributions. Another possibility is that the ratio of RPN5a and RPN5b is important to Arabidopsis. By expressing large amounts of RPN5a, the occupancy of RPN5b in the 26S proteasome could be commensurately reduced. However, as plants missing rpn5b are phenotypically normal, the effect cannot be simply due to the lack of RPN5b in the holo-26S proteasome. That the same 35S promoter was used to express both RPN5a and RPN5b eliminates differential expression patterns as the cause. However, we did notice that RPN5b overexpression was unable to restore RPN5 protein to wild-type levels as detected immunologically. Consequently, it is possible that the different phenotypes are caused by the different degrees of RPN5 overexpression. Moderate overexpression of RPN5 could induce early flowering as seen in both the 35Spro:RPN5a and 35Spro:RPN5b lines, but at excessive levels, a highly stressed, early senescence phenotype appears. A final possibility is that the RPN5a protein in particular becomes associated with a specific group of regulatory proteins and/or substrates in Arabidopsis. By binding to the excess amount of free RPN5a in the 35Spro:RPN5a lines, these proteins could be prevented from associating with the 26S proteasome, thereby either blocking their function or preventing their breakdown. Clearly, compositional analysis of 26S proteasomes bearing RPN5a or RPN5b specifically is now needed to test this scenario.
Whatever the mechanism, the unique phenotype of the RPN5a-overexpressing plants points to a specific role for the RPN5a gene/protein in Arabidopsis development. An intriguing possibility is that it reflects an additional layer of substrate specificity to the 26S proteasome, generated either through unique activities conferred by the various isoforms or through their ability to specifically interact with accessory proteins. As examples of the latter, the UBP6 deubiquitylating enzyme and the arsenite-inducible RNA-associated protein were recently shown to preferentially associate with the 26S proteasome as ways to ameliorate Ub limitation stress by enhancing Ub recycling or stabilize the complex during arsenite exposure, respectively (Stanhill et al., 2006; Hanna et al., 2007). By tapping into gene/isoform diversity, plants may deliberately tailor the isoform composition of the 26S proteasome to better handle distinct sets of targets or allow for sophisticated modes of regulation.
METHODS
Plant Materials and Growth Conditions
The rpn5a-1 allele present in the Arabidopsis thaliana C24 background was generated by exon-trap mutagenesis (Babiychuk et al., 1997). The rpn5a-2, rpn5b-2, and rpt2a-2 alleles in Col-0 background were created by T-DNA insertional mutagenesis and were obtained from the ABRC (SALK lines 010840, 127791, and 005596, respectively). The rpn5b-1 mutant in the Ws background was isolated by a PCR-based screen of the T-DNA insertional lines generated at the University of Wisconsin at Madison. The rpn5a-3 mutant (emb2107) in the Col-0 background was identified in a screen for embryo-defective mutants (ABRC stock CS16219) (McElver et al., 2001). The rpn5a-2 and rpn5b-2 lines contained a single NPTII gene associated with the T-DNA that conferred kanamycin resistance, while the rpn5a-3 and rpn5b-1 lines contained a single PHOSPHINOTRICIN ACETYLTRANSFERASE gene within the T-DNA that conferred BASTA herbicide resistance. The rpn5a-4 allele was isolated in a mixed Ws/Ler background during a screen for embryo-defective mutants following ethane methanesulfonate mutagenesis (Joy, 2001) and was named previously mariposa. This point mutation removed a FalI restriction site at base pair 1398 in the genomic coding region.
Gene-specific primer pairs used for PCR genotyping of the T-DNA insertions were rpn5a-1 (P1 and P2), rpn5a-2 (P3 and P4), rpn5a-3 (P5 and P6), rpn5b-1 (P7 and P8), and rpn5b-2 (P9 and P10). Left-border primers used to amplify the T-DNA insertions were P11 for rpn5a-1, P12 for rpn5a-2 and rpn5b-2, P13 for rpn5b-1, and P14 for rpn5a-3 (see Supplemental Table 1 online for all primer sequences used in this study). The mutants were backcrossed three times to the respective wild-type parents and then made homozygous by selfing. For the rpn5a-4 mutant, it was backcrossed to wild-type Ler, and the mutant allele was then tracked in the progeny by phenotype.
The rpn5a and rpn5b mutations were combined by fertilizing heterozygous rpn5b-2 flowers with pollen from an rpn5a-2 heterozygous plant. Kanamycin-resistant F1 progeny were genotyped for both T-DNA insertions. Double heterozygous plants were then self-crossed and the F2 progeny were genotyped for both insertions by genomic PCR. The rpn12a-1 and rpn10-1 mutant lines in C24 background were described previously (Smalle et al., 2002, 2003).
Unless otherwise noted, plants were grown under sterile conditions on solid Gamborg's B-5 growth medium (GM) with 2% sucrose and 0.7% agar (Gibco BRL). To examine flowering time in response to MG132 (benzyloxycarbonyl-Leu-Leu-Leu-al) (Sigma-Aldrich), seeds were directly sown on solid GM containing the inhibitor. After stratification, the plates were placed under continuous irradiation at 22°C for 3 weeks or under a short-day photoperiod of 8 h light and 16 h darkness for 4 weeks. To examine etiolated development, wild-type and mutant seedlings were grown in the dark for 6 d. For the analysis of embryos, plants were grown on soil.
Sequence Alignment and Phylogenetic Analysis
Amino acid sequences were aligned using CLUSTALX MAC V.1.6b (Thompson et al., 1997) and displayed by MACBOXSHADE V.2.11 (Institute of Animal Health). A phylogenetic tree of full-length protein sequences (aligned with ClustalW (see Supplemental Data Set 1 online) was generated in MEGA3.1 (Kumar et al., 2004) by the neighbor-joining method using the Poisson distance method, pairwise deletion of gaps, and the default assumptions that the substitution patterns among lineages and substitution rates among sites were homogeneous.
Analysis of RPN5 Expression Patterns
Promoter-GUS coding region fusions were generated with PCR-amplified fragments from genomic Col-0 DNA that ended immediately upstream of the translation start sites for RPN5a and RPN5b (primers P15 and P16 for RPN5apro and P17 and P18 for RPN5bpro). The fragments (1521 bp for RPN5a and 1401 bp for RPN5b) were subcloned into the Gateway pENTR/ D-TOPO entry vector (Invitrogen Life Technologies) and then recombined into pMDC163 destination vector, which included the GUS coding region (Curtis and Grossniklaus, 2003). The fusions were introduced into wild-type Col-0 plants by the floral dip method using the Agrobacterium tumefaciens strain GV3101 (Smalle et al., 2002). Hygromycin-resistant seedlings were screened for GUS expression by histochemical assay with the substrate 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid (X-Gluc) (Emborg et al., 2006). Three independent insertion lines were examined for each construction to avoid artifacts generated by the site of transgene insertion.
Microscopy and Histochemistry
Embryos and leaves were examined on a Leica DMRB microscope using either fluorescence or differential interference contrast optics and imaged with ProgRes MF cool or C5 cameras (Jenoptik). For embryo morphology, developing seeds were dissected out of the siliques and mounted in Hoyer's solution (70% chloral hydrate, 4% glycerol, and 5% gum arabic). For the analysis of leaves, 21-d-old plants were fixed overnight in 3:1 ethanol:acetic acid, cleared with 70% ethanol, and mounted in Hoyer's solution. The slides were scanned on a flatbed scanner, and leaf area was measured using ImageJ V1.33 (National Institutes of Health). Cell areas were determined under the microscope with the ProgRes Capture Pro software (Jenoptik). For the analysis of embryo gene expression, embryos were squeezed from the seeds into 10% glycerol and analyzed by fluorescence microscopy (for GFP) or stained with 100 mM potassium phosphate, pH 7.0, 1 mM EDTA, 1% Tween 20, 2.5 mM potassium ferri/ferrocyanide, and 1 mg/mL X-Gluc at 37°C for several hours (for GUS). The reporter genes tested for embryonic expression were described previously (Cary et al., 2002; Jenik et al., 2005).
RPN5a-GFP and RPN5b-GFP fusions expressed under the control of the 35S promoter were created by a two-step PCR amplification. First, the coding regions for RPN5a/RPN5b and GFP were amplified by two reactions that generate overlapping products (primers P19 and P20 for RPN5a, P21 and P22 for RPN5b, and P23 and P24 for GFP). These products were denatured and annealed together and then used as a template in a second PCR reaction to generate the full-length RPN5a/RPN5b-GFP fusion constructions. These constructions were sequence verified, cloned into the pCAMBIA 3301 vector (Curtis et al., 2002) harboring the 35S promoter, and transformed into Arabidopsis (both C24 and rpn5a-1 heterozygous mutants) as described above. T2 progeny containing the transgenes were identified by Basta resistance and for the presence of the rpn5a-1 mutation by PCR genotyping. Fluorescence confocal microscopy of 10-d-old root tips expressing either RPN5a-GFP or RPN5b-GFP was conducted with a Zeiss 510-Meta scanning laser confocal microscope using 488-nm light excitation (Thompson et al., 2005).
RNA Gel Blot Analyses
Total RNA was extracted from 3-week-old seedlings using the Trizol reagent (Gibco BRL) and 10 μg of RNA was subjected to gel blot analyses according to Smalle et al. (2002). 32P-labeled riboprobes were synthesized with T7 or SP6 RNA polymerase using the appropriate linearized plasmids and the Riboprobe Gemini II core system (Promega). The RPN5a template used the EST clone 245J16 linearized with PstI. The RPN5b template used the pJSRPN5b plasmid linearized with SphI. This plasmid was created by PCR amplification of the RPN5b coding frame from the cDNA using the primers P25 and P26 and insertion of the product into pGEM-T (Promega). The β-TUBULIN4 and PBA1 templates were used as controls. Signals were detected using Classic Autoradiography Film (MidSci) as described (Smalle et al., 2002).
rpn5a-1 Complementation
The RPN5a and RPN5b genomic coding regions plus their 3′ untranslated regions were PCR amplified using the primers P27 and P28 for RPN5a, and P29 and P30 for RPN5b. The PCR products were recombined into the pDONR221 plasmid using Gateway Technology (Invitrogen Life Sciences). RPN5a and RPN5b promoter sequences were subcloned into pGEM-T using the primers P31 and P32 for RPN5apro, and P33 and P34 for RPN5bpro. Following digestion with NotI, the promoter fragments were gel purified and ligated upstream of the coding regions. These entry clones were then recombined into complementation vector pMDC99 (Curtis and Grossniklaus, 2003). To drive expression by the 2xCaMV 35S promoter, the RPN5a and RPN5b genomic coding sequences were recombined into pMDC32 (Curtis and Grossniklaus, 2003). All constructions were introduced into heterozygous rpn5a-1 and wild-type Col-0 and C24 seedlings as described above and selected by hygromycin resistance. Plants homozygous for the rpn5a-1 allele and the various RPN5a/b transgenes were identified by PCR genotyping and hygromycin resistance in selfed F2 populations and then tested for phenotypic rescue.
Immunoblot Analysis
Immunoblot analysis was performed according to Smalle et al. (2002). Antibodies against RPN5a, RPT1a, RPT2a, PBA1, PAC1, RPN10, RPN12a Ub, and UCH2 were as described (van Nocker et al., 1996a, 1996b; Smalle et al., 2002; Yang et al., 2004). Signal was detected using classic autoradiography film. Exposure times were adjusted to remain in the linear range of the film. Antigenicity of the anti-RPN5a antibodies against RPN5a and RPN5b was tested on recombinant proteins expressed in pDEST17 (Invitrogen) with an N-terminal 6His tag and a C-terminal Flag tag. Immunoblot analysis with anti-Flag antibodies (Sigma-Aldrich) was used to confirm equal loading.
RPN5a and RPN5b Overexpression
The RPN5a cDNA was PCR amplified from the EST clone 245J16 (using the primers P35 and P36 that were designed to contain an SpeI site at both ends). The RPN5b cDNA was PCR amplified directly from a seedling cDNA library using the primers P37 and P38 that were designed to contain an XbaI site at both ends. The resulting products were digested with either SpeI or XbaI and inserted downstream of the CaMV 35S promoter into the similarly digested T-DNA vector pGSVE9 conferring hygromycin resistance (Smalle et al., 2002). The plasmids introduced into the A. tumefaciens strain AtC58RifR(pMP90) were inserted into heterozygous rpn5a-1 and wild-type C24 seedlings as above. Presence of the 35Spro:RPN5a and 35Spro:RPN5b transgenes and the rpn5a-1 mutation were followed by hygromycin and kanamycin resistance, respectively. RPN12a and RPN10 overexpression constructs were as described (Smalle et al., 2002, 2003). Self-crossed T3 progenies homozygous for both kanamycin and hygromycin resistance were assessed for phenotypic rescue of rpn5a-1.
Glycerol Gradient Fractionation
The 26S proteasome was partially purified from 10-d-old green Arabidopsis seedlings grown in liquid GM. Plants were frozen to liquid nitrogen temperatures, pulverized, and ground in 1.25 volumes of buffer A (20 mM Tris-HCl, pH 7.5, 10% glycerol, 2 mM ATP, 5 mM MgCl2, 1 mM DTT, 10 mM phosphocreatine, and 1 mg/mL creatine phosphokinase). Protein extracts were filtered through two layers of cheesecloth, clarified for 20 min at 30,000g, made 10% in polyethylene glycol 8000, and incubated for 30 min at 4°C. The precipitate was collected by centrifugation at 12,000g for 15 min and resuspended in 300 μL of buffer A. Following clarification, the resuspended pellet was loaded on top of a 11-mL 10 to 40% glycerol density gradient in Buffer A and centrifuged at 100,000g for 18 h at 4°C. Fractions (0.5 mL) were collected and assayed for proteasome activity with the substrate succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC) (Sigma-Aldrich) or subjected to SDS-PAGE and immunoblot analysis with various 26S proteasome subunit antibodies (Yang et al., 2004).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At5g09900 (RPN5a) and At5g64760 (RPN5b).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Comparison of RPN5 Protein Homologs.
Supplemental Figure 2. RPN5a and RPN5b Are Equally Recognized by Anti-RPN5a Antibodies.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Sequence Alignment in Figure 1.
Supplemental Data Set 2. Text File of the Sequence Alignment Used to Generate the Phylogenetic Tree in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Elena Babiychuk and Sergei Kushnir for providing the rpn5a-1 mutant, David Meinke for sharing unpublished data on emb2107, and the ABRC for providing the EST clones and SALK T-DNA lines. This work was supported by grants from the U.S. Department of Energy Basic Energy Sciences program (DE-FG02-88ER13968) and the National Science Foundation Arabidopsis 2010 Program (MCB-0115870) to R.D.V., a National Institutes of Health predoctoral fellowship to A.J.B. (5F31NS054563), a Louis and Elisa Thomsen Wisconsin Distinguished Predoctoral Fellowship to P.Y., a Wain International Travel Fellowship to J.H.H., and funds from Oberlin and Franklin and Marshall Colleges to P.D.J.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard D. Vierstra (vierstra@wisc.edu).
Online version contains Web-only data.
References
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.S., Amasino, R., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E., Fuangthong, M., Van Montagu, M., Inze, D., and Kushnir, S. (1997). Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl. Acad. Sci. USA 94 12722–12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, E., and Reed, S.I. (1999). Functional characterization of RPN3 uncovers a distinct 19S proteasomal subunit requirement for ubiquitin-dependent proteolysis of cell cycle regulatory proteins in budding yeast. Mol. Cell. Biol. 19 6872–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukhin, V., Gheyselinck, J., Gagliardini, V., Genschik, P., and Grossniklaus, U. (2005). The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17 2723–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary, A.J., Che, P., and Howell, S.H. (2002). Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J. 32 867–877. [DOI] [PubMed] [Google Scholar]
- Curtis, I.S., Nam, H.G., Yun, J.Y., and Seo, K.H. (2002). Expression of an antisense GIGANTEA (GI) gene fragment in transgenic radish causes delayed bolting and flowering. Transgenic Res. 3 249–256. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmann, B., Ruppert, T., Kuehn, L., Merforth, S., and Kloetzel, P.M. (2000). Different proteasome subtypes in a single tissue exhibiting different enzymatic properties. J. Mol. Biol. 303 643–653. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119 71–84. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Gali, R.R., Schwickart, M., Larsen, C.N., Leggett, D.S., Muller, B., Feng, M.T., Tubing, F., Dittmar, G.A., and Finley, D. (2002). Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4 725–730. [DOI] [PubMed] [Google Scholar]
- Emborg, T.J., Walker, J.M., Noh, B., and Vierstra, R.D. (2006). Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol. 140 856–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva, M.D., Wu, M., Eisen, J.A., and Salzberg, S.L. (2003). The age of the Arabidopsis thaliana genome duplication. Plant Mol. Biol. 51 859–866. [DOI] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jurgens, G., and Palme, K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Fu, H., Doelling, J.H., Arendt, C.S., Hochstrasser, M., and Vierstra, R.D. (1998. a). Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics 149 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Doelling, J.H., Rubin, D.M., and Vierstra, R.D. (1999). Structural and functional analysis of the six regulatory particle AAA-ATPase subunits from the Arabidopsis 26S proteasome. Plant J. 18 529–539. [DOI] [PubMed] [Google Scholar]
- Fu, H., Reis, N., Lee, Y., Glickman, M.H., and Vierstra, R.D. (2001). Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Sadis, S., Rubin, D.M., Glickman, M., van Nocker, S., Finley, D., and Vierstra, R.D. (1998. b). Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26S proteasome subunit Mcb1. J. Biol. Chem. 273 1970–1981. [DOI] [PubMed] [Google Scholar]
- Girod, P.A., Fu, H., Zryd, J.P., and Vierstra, R.D. (1999). Multiubiquitin chain binding subunit MCB1 (RPN10) of the 26S proteasome is essential for developmental progression in Physcomitrella patens. Plant Cell 11 1457–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H., Rubin, D.M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A., and Finley, D. (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94 615–623. [DOI] [PubMed] [Google Scholar]
- Groll, M., Bajorek, M., Kohler, A., Moroder, L., Rubin, D.M., Huber, R., Glickman, M.H., and Finley, D. (2000). A gated channel into the proteasome core particle. Nat. Struct. Biol. 7 1062–1067. [DOI] [PubMed] [Google Scholar]
- Groll, M., Ditzel, L., Lowe, J., Stock, D., Bochtler, M., Bartunik, H.D., and Huber, R. (1997). Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386 463–471. [DOI] [PubMed] [Google Scholar]
- Hanna, J., and Finley, D. (2007). A proteasome for all occasions. FEBS Lett. 581 2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, J., Meides, A., Zhang, D.P., and Finley, D. (2007). A ubiquitin stress response induces altered proteasome composition. Cell 129 747–759. [DOI] [PubMed] [Google Scholar]
- Hartmann-Petersen, R., Seeger, M., and Gordon, C. (2003). Transferring substrates to the 26S proteasome. Trends Biochem. Sci. 28 26–31. [DOI] [PubMed] [Google Scholar]
- Horiguchi, G., Ferjani, A., Fujikura, U., and Tsukaya, H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119 37–42. [DOI] [PubMed] [Google Scholar]
- Husnjak, K., Elsasser, S., Zhang, N., Chen, X., Randles, L., Shi, Y., Hofmann, K., Walters, K.J., Finley, D., and Dikic, I. (2008). Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. [DOI] [PubMed] [Google Scholar]
- Isono, E., Nishihara, K., Saeki, Y., Yashiroda, H., Kamata, N., Ge, L., Ueda, T., Kikuchi, Y., Tanaka, K., Nakano, A., and Toh-e, A. (2007). The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell 18 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S., Marchal, V., Panigrahi, K.C., Wenkel, S., Soppe, W., Deng, X.W., Valverde, F., and Coupland, G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P.D., Jurkuta, R.E., and Barton, M.K. (2005). Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell 17 3362–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy, R.E. (2001). Arabidopsis Embryonic Development: A Screen for Mutants Disrupted in Pattern Formation and Analysis of BOBBER, a Gene Required for Proper Specification of the Apical Region of the Arabidopsis Embryo. PhD dissertation (Madison, WI: University of Wisconsin).
- Kloetzel, P.M. (2001). Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2 179–187. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Kurepa, J., Toh, E.A., and Smalle, J.A. (2008). 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53 102–114. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Staub, J.M., and Deng, X.W. (1999). Characterization of two subunits of the Arabidopsis 26S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285 85–95. [DOI] [PubMed] [Google Scholar]
- Lam, Y.A., Lawson, T.G., Velayutham, M., Zweier, J.L., and Pickart, C.M. (2002). A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416 763–767. [DOI] [PubMed] [Google Scholar]
- Laporte, D., Salin, B., Daignan-Fornier, B., and Sagot, I. (2008). Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Ma, L., Sun, N., Liu, X., Jiao, Y., Zhao, H., and Deng, X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, P., Kim, W.Y., Somers, D.E., and Kay, S.A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570. [DOI] [PubMed] [Google Scholar]
- McElver, J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiners, S., Heyken, D., Weller, A., Ludwig, A., Stangl, K., Kloetzel, P.M., and Kruger, E. (2003). Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J. Biol. Chem. 278 21517–21525. [DOI] [PubMed] [Google Scholar]
- Muller, B., and Sheen, J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, S., Heidstra, R., Wildwater, M., and Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M., Hanna, J., Elsasser, S., and Finley, D. (2005). Proteasome-associated proteins: Regulation of a proteolytic machine. Biol. Chem. 386 725–737. [DOI] [PubMed] [Google Scholar]
- Schreiner, P., Chen, X., Husnjak, K., Randles, L., Zhang, N., Elsasser, S., Finley, D., Dikic, I., Walters, K.J., and Groll, M. (2008). Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, M., Taverner, T., Ambroggio, X.I., Deshaies, R.J., and Robinson, C.V. (2006). Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 4 e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara, T., Kawasaki, H., and Hirano, H. (2002). Identification of the 19S regulatory particle subunits from the rice 26S proteasome. Eur. J. Biochem. 269 1474–1483. [DOI] [PubMed] [Google Scholar]
- Shibahara, T., Kawasaki, H., and Hirano, H. (2004). Mass spectrometric analysis of expression of ATPase subunits encoded by duplicated genes in the 19S regulatory particle of rice 26S proteasome. Arch. Biochem. Biophys. 421 34–41. [DOI] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Babiychuk, E., Kushnir, S., Durski, A., and Vierstra, R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12a. Plant Cell 14 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Emborg, T.J., Babyichuk, E., Kushnir, S., and Vierstra, R.D. (2003). The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis thaliana growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Smith, D.M., Chang, S.C., Park, S., Finley, D., Cheng, Y., and Goldberg, A.L. (2007). Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhill, A., Haynes, C.M., Zhang, Y., Min, G., Steele, M.C., Kalinina, J., Martinez, E., Pickart, C.M., Kong, X.P., and Ron, D. (2006). An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell 23 875–885. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Thompson, A.R., Doelling, J.H., Suttangkakul, A., and Vierstra, R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, M., Matsui, K., Ishiguro, S., Sano, R., Wada, T., Paponov, I., Palme, K., and Okada, K. (2004). The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131 2101–2111. [DOI] [PubMed] [Google Scholar]
- van Nocker, S., Deveraux, Q., Rechsteiner, M., and Vierstra, R.D. (1996. a). Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc. Natl. Acad. Sci. USA 93 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker, S., Sadis, S., Rubin, D.M., Glickman, M., Fu, H., Coux, O., Wefes, I., Finley, D., and Vierstra, R.D. (1996. b). The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2009). The ubiquitin/26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. (in press). [DOI] [PubMed]
- Voges, D., Zwickl, P., and Baumeister, W. (1999). The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68 1015–1068. [DOI] [PubMed] [Google Scholar]
- Woffenden, B.J., Freeman, T.B., and Beers, E.P. (1998). Proteasome inhibitors prevent tracheary element differentiation in zinnia mesophyll cell cultures. Plant Physiol. 118 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller, J.W., Helariutta, Y., Fukaki, H., Malamy, J.E., and Benfey, P.N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127 595–603. [DOI] [PubMed] [Google Scholar]
- Xie, Y., and Varshavsky, A. (2001). RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proc. Natl. Acad. Sci. USA 98 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P.Z., Fu, H., Walker, J., Papa, C.M., Smalle, J., Ju, Y.-M., and Vierstra, R.D. (2004). Purification of the Arabidopsis 26 S proteasome - Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279 6401–6413. [DOI] [PubMed] [Google Scholar]
- Yen, H.C.S., Espiritu, C., and Chang, E.C. (2003). Rpn5 is a conserved proteasome subunit and required for proper proteasome localization and assembly. J. Biol. Chem. 278 30669–30676. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.