Abstract
Tendrils are contact-sensitive, filamentous organs that permit climbing plants to tether to their taller neighbors. Tendrilled legume species are grown as field crops, where the tendrils contribute to the physical support of the crop prior to harvest. The homeotic tendril-less (tl) mutation in garden pea (Pisum sativum), identified almost a century ago, transforms tendrils into leaflets. In this study, we used a systematic marker screen of fast neutron–generated tl deletion mutants to identify Tl as a Class I homeodomain leucine zipper (HDZIP) transcription factor. We confirmed the tendril-less phenotype as loss of function by targeting induced local lesions in genomes (TILLING) in garden pea and by analysis of the tendril-less phenotype of the t mutant in sweet pea (Lathyrus odoratus). The conversion of tendrils into leaflets in both mutants demonstrates that the pea tendril is a modified leaflet, inhibited from completing laminar development by Tl. We provide evidence to show that lamina inhibition requires Unifoliata/LEAFY-mediated Tl expression in organs emerging in the distal region of the leaf primordium. Phylogenetic analyses show that Tl is an unusual Class I HDZIP protein and that tendrils evolved either once or twice in Papilionoid legumes. We suggest that tendrils arose in the Fabeae clade of Papilionoid legumes through acquisition of the Tl gene.
INTRODUCTION
Many climbing plants use specialized organs called tendrils for support. Some tendrils explore the physical environment with characteristic circling movements (Darwin, 1875) followed by contact-induced coiling (Jaffe and Galston, 1968), permitting the plant to obtain support by grasping onto and entwining its neighbors. Plant tendrils may be derived from a variety of structures, such as leaf parts, whole leaves, or stems (Bell, 1991); for example, the grapevine tendril is a gibberellin-inhibited inflorescence (Boss and Thomas, 2002). Such diverse derivations, and the fact that tendrilled taxa are widespread in flowering plants (Darwin, 1875), suggest that tendrils are an example of convergent evolution. These novel organs enable plants to reach the canopy, where they can spread and maximize opportunities for pollination, photosynthesis, and seed dispersal with minimal energy investment in expensive supporting structures. Indeed, the climbing habit is associated with species richness compared with nonclimbing sister taxa (Gianoli, 2004) (see Supplemental Table 1 online), suggesting a selective advantage.
The three subfamilies of legumes, Caesalpinioideae, Mimosoideae, and Papilionoideae, together comprise >19,000 species, one of the largest flowering plant families (Lewis et al., 2005). Tendrils appear to have evolved independently at least once in each subfamily. In Bauhinia spp (Caesalpinioideae), tendrils arise at the base of the leaf, while tendrils form in the distal region of the leaf in Entada spp (Mimosoideae) and in peas, lentils, vetches, and chickpeas (Papilionoideae). All the economically important grain legume species are Papilionoids, which collectively provide approximately one-third of the total dietary protein needs of humans, as well as being used widely as animal feed. Under intensively planted field conditions, tendrils can form an interwoven network of support, conferring partial resistance to crop collapse or lodging. Therefore, a better understanding of tendril formation has the potential to aid agronomic performance and to provide insight on convergent morphological evolution.
Most legume leaves are compound (Lewis et al., 2005), with each leaf carrying one or more pairs of leaflets along the leaf axis. The leaf is further specialized in Papilionoid legume species belonging to the clades Cicereae (chickpeas) and Fabeae (peas, lentils, and vetches) where the organ formed at the terminal position of the leaf is a tendril, rather than a leaflet. Many species within the Fabeae are more extensively tendrilled, for example, garden pea (Pisum sativum) and sweet pea (Lathyrus odoratus) also produce pairs of tendrils in subterminal positions. A key regulator of the compound leaf trait in legumes is the meristem identity gene, Unifoliata (Uni), the ortholog of LEAFY (LFY) in Arabidopsis thaliana (Hofer et al., 1997). In garden pea, Uni maintains the meristematic potential of the compound leaf, enabling the sequential development of pairs of leaflets and tendrils in acropetal (first pair at the base and last pair at the tip) order. This role is shown by uni null mutants, which bear leaves composed of only a single leaflet (Hofer et al., 1997). A semidominant locus regulating tendril formation has long been known in garden pea (de Vilmorin, 1910; de Vilmorin and Bateson, 1911) and sweet pea (Punnett, 1923). Mutants in both species were originally called acacia (t) because of their tendril-less leaves; the locus was later renamed tendril-less (tl) in garden pea. It was not known if these were orthologous loci, and identification of the genes remained elusive. In this study, we employed an amplified fragment length polymorphism (AFLP) screening method to identify Tl as a Class I homeodomain leucine zipper (HDZIP) gene that confers the tendrilled trait on peas.
RESULTS
Generation of New tl Mutant Alleles and Identification of Tl
We used fast neutron (FN) mutagenesis to generate new tl deletion alleles in a conveniently dwarf, early flowering garden pea genotype. This allowed us to screen DNA samples for markers that distinguished wild-type plants from mutants. Compared with the wild-type leaf (Figure 1A), narrow, subterminal leaflets were found in place of tendrils in heterozygous FN mutants (Figure 1B), as expected for this semidominant mutation (de Vilmorin and Bateson, 1911; Marx, 1973), while the homozygous FN mutants displayed a classic homeotic transformation of tendrils into leaflets (Figure 1C). Tendril-less F1 progeny were obtained from tendril-less FN mutants crossed to lines carrying the tl-w type allele, confirming that the new FN mutants all carried allelic mutations. Notably, the wild-type, heterozygous (tl/Tl) and homozygous (tl/tl) garden pea phenotypes resembled phenotypes in sweet pea that were correspondingly wild-type (Figure 1D), heterozygous (Figure 1E), or homozygous (Figure 1F) at the t locus (Punnett, 1923), which suggested that tl and t might be orthologous loci.
Figure 1.
Tl Determines Tendril Organ Identity.
(A) Wild-type garden pea leaf.
(B) Heterozygous Tl/tl garden pea leaf.
(C) Homozygous tl/tl garden pea leaf.
(D) Wild-type sweet pea leaf.
(E) Heterozygous T/t sweet pea leaf.
(F) Homozygous t/t sweet pea leaf.
(G) Tendril-less phenotype of garden pea leaves on a homozygous mutant plant identified in a reverse genetics screen of an EMS-mutagenized TILLING population.
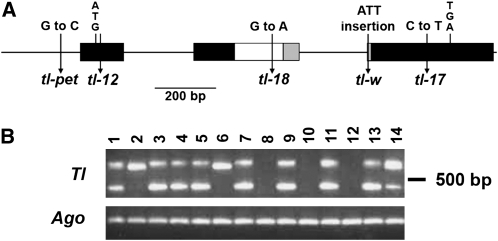
Genomic DNA from five new FN tl lines was pooled and screened for AFLP markers (Vos et al., 1995) that differentiated the mutant pool from the wild type. A 218-bp marker, absent from all five FN alleles and present in the wild type, was sequenced, and primers derived from this were used to screen a ninefold redundant P. sativum cv Cameor HindIII BAC library. BAC genomic DNA sequencing revealed that the marker spanned the second intron-exon junction of a gene encoding a 237–amino acid Class I HDZIP protein (Figure 2A). The entire coding sequence was deleted from all tl FN mutants used in the AFLP screen, indicating that deletions larger than 1 kb occur in this mutagenized population and that a loss of function causes the tendril-less phenotype. A reverse genetics screen for this gene in an ethane methanesulfonate (EMS)–mutagenized targeting-induced local lesions in genomes (TILLING) population (Dalmais et al., 2008) confirmed the identity of Tl. An M2 plant carrying a W117STOP TILLING lesion, designated tl-18 (Figure 2A; see Supplemental Table 2 online), subsequently yielded tendril-less M3 progeny homozygous for the mutation (Figure 1G).
Figure 2.
Analysis of Tl Allelic Variants.
(A) Gene structure represented as boxes for exons and lines for introns. The white box indicates the coding region for the homeodomain, and the gray box represents the leucine zipper region. The positions of the ATG start and TGA stop codons are shown. Vertical arrows indicate nucleotide changes present in various tl alleles.
(B) Tl gene expression in wild-type accessions and mutant alleles analyzed by PCR after reverse transcription of RNA extracted from 3-week-old shoot tips. Tl gene-specific primers flanking the second intron were predicted to amplify a 424-bp product from cDNA and a 645-bp product from genomic DNA present in the samples. Lane 1, wild-type Witham Wonder; lane 2, tl-pet JI 32; lane 3, wild-type JI 1194; lane 4, tl-w JI 1197; lane 5, wild-type JI 516; lane 6, tl-12 JI 1373; lane 7, wild-type JI 2224; lane 8, tl-13 JI 3128; lane 9, wild-type JI 3131; lane 10, tl-16 JI 3130; lane 11, wild-type JI 2282; lane 12, tl-10 FN 2086/3; lane 13, wild-type Torsdag 5839; lane 14, tl-17 MC1a/1. Primers specific for the garden pea Argonaute gene (Ago) were used as controls for PCR amplification.
Characterization of Previously Described tl Mutant Alleles
Five tl mutants described in the literature had been observed or generated in a variety of pea genotypes (see Supplemental Table 2 online), so we determined first whether each mutant and its reported progenitor were related using a fingerprinting technique (Ellis et al., 1998). Sequence-specific amplification polymorphism displays confirmed that alleles tl-7, tl-13, tl-16, and tl-17 were maintained in stocks near-isogenic to their specified progenitor wild-type lines. The progenitor of the original spontaneous tl mutant allele, tl-w, is unknown (de Vilmorin and Bateson, 1911); however, as a result of backcrossing (Marx, 1973), line JI 1197 carrying tl-w is near-isogenic to wild-type line JI 1194, and this was also confirmed by fingerprinting.
Sequencing revealed that tl-w contains a 3-bp insertion encoding an additional Ile residue at position 149, within the leucine zipper domain (Figure 2A). We predict that this transcribed allele (Figure 2B) produces a nonfunctional protein, altered in its capacity for homo- or heterodimerization via its leucine zipper. We investigated three independent tl deletion alleles that are not transcribed (Figure 2B) and phenotypically resemble the tl-w type line. Alleles tl-13 and tl-16 (Vassileva, 1979) are radiation-induced complete gene deletions, and tl-12 (Figure 2A), a spontaneous allele, carries a 1908-bp deletion, including the promoter and the first six amino acids of coding sequence (see Supplemental Table 2 online). Allele tl-17 (Figure 2A) carries an EMS-induced C/T transition, resulting in a Q226STOP mutation, which would truncate the C terminus by 12 amino acids. This allele is transcribed (Figure 2B), yet the phenotype of the mutant is similar to the deletion alleles, suggesting that the C-terminal domain may have an important function in stabilizing the protein.
The spontaneous allele tl-pet (Lamm, 1957) differs from other tl alleles in that distal leaflets are borne on elongated stalks (see Supplemental Figure 1 online). We were unable to identify the progenitor of tl-pet, but compared with all other alleles, it carries a unique G/C transversion in the promoter (Figure 2A). The altered nucleotide corresponds to the second position of a putative GGTCCAT auxin-responsive cis-regulatory element (Lescot et al., 2002), 115 bp from the ATG corresponding to the start codon and 33 bp upstream of a predicted TATA box (Bucher, 1990). Transcription of this allele is severely impaired (Figure 2B), suggesting that Tl may be an auxin-regulated gene. Auxin inhibitor studies in pea (DeMason and Chawla, 2004) and patterns of auxin transport during primordium formation suggest that auxin plays a role in regulating primordium type and polarity (Heisler et al., 2005). Studies in compound-leaved Cardamine hirsuta showed that pinformed1 mutants, which fail to accumulate auxin in their leaf rachis, fail to separate leaflet from rachis correctly (Barkoulas et al., 2008). An inability to respond appropriately to auxin may account for the stalked leaflet phenotype of tl-pet mutant leaves.
Tl-Related Genes in Other Species
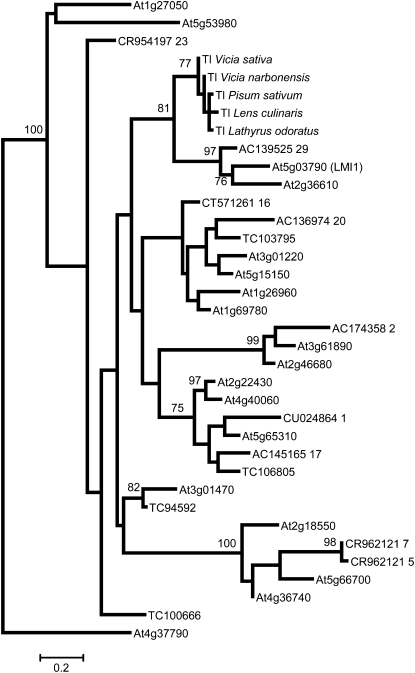
Class I HDZIP genes that play a role in diverse developmental processes have been described (Ariel et al., 2007; Komatsuda et al., 2007). Therefore, the relationship between Tl and other Class I HDZIP sequences in nontendrilled model plant species is of interest. A maximum likelihood tree based on aligned Class I HDZIP domains (Figure 3) shows that Tl and its immediate relatives (see below) are most closely related to At2g36610 and At5g03790 from Arabidopsis and AC139525_29 from Medicago truncatula. At2g36610 encodes an unusual plant Class I HDZIP protein that contains an eight–amino acid insertion between helix 1 and helix 2 of the homeodomain (see Supplemental Figure 2 online), followed by an exceptionally short C terminus. These features mean that it is less likely to resemble a Tl progenitor sequence than At5g03790 and AC139525_29. At5g03790, also known as LATE MERISTEM IDENTITY1 (LMI1), is a gene identified as a direct promoter binding target of LFY (William et al., 2004) that acts together with LFY to promote floral meristem identity in Arabidopsis (Saddic et al., 2006).
Figure 3.
Phylogenetic Tree of Legume and Arabidopsis Class I HDZIP Sequences.
Maximum likelihood tree with bootstrap support values >70% for tree branches shown. Arabidopsis sequences are labeled as locus identifiers beginning with At, Medicago sequences are labeled as database accession numbers beginning with CR, AC, CU, CT, or TC for transcript contig reports in the Medicago truncatula Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago).
A clear LMI1 ortholog with conserved functions in leaves and flowers has not been identified in pea or closely related legumes, but one candidate is AC139525_29, which we derived from Medicago BAC sequence data after manual editing to remove sequence corresponding to a 1249-nucleotide second intron. Spliced full-length transcripts were confirmed by sequencing products obtained from PCR on reverse transcribed cDNA. Although AC139525_29 is a potential LMI1 ortholog, it is not well supported as a Tl ortholog because it shares only 62% open reading frame nucleotide sequence identity and maps (http://www.medicago.org/genome/cvit_blast.php) to a nonsyntenic position on Medicago chromosome 1 (highest TBLASTN similarity score of 1e-40).
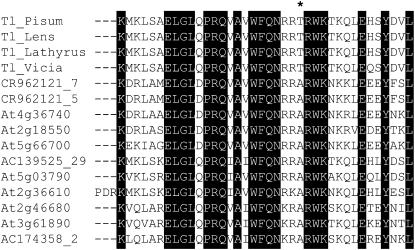
To identify a possible molecular basis for differences in Tl gene function compared with other Class I HDZIPs, we compared their HDZIP regions (Figure 4). The legume Tl sequences are distinguished by their shorter leucine zippers, which contain only four Leu residues, whereas most other Class I HDZIPs have zippers comprised of five or six hydrophobic residues. Residues in DNA binding helix 3 of the homeodomain are identical or conservatively substituted in all sequences compared, except for a Thr replacement for Ala at position 123 of Tl (Figure 4).
Figure 4.
Novelty of the Tl Gene.
Deduced amino acid sequences of selected Class I HDZIP domains showing a Thr residue (asterisk) characteristic of Tl genes from tendrilled legume species garden pea (P. sativum), lentil (L. culinaris), sweet pea (L. odoratus), and narbon bean (V. narbonensis).
Tl Is Expressed in Tendril Primordia
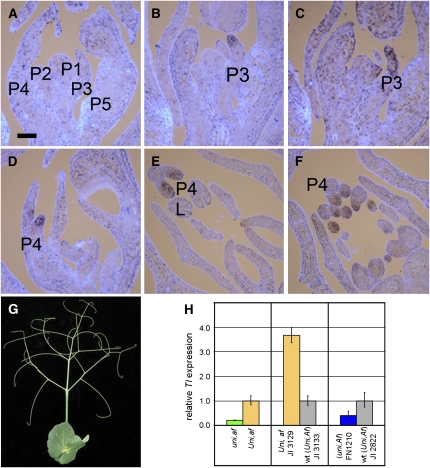
Surgical experiments have shown that terminal tendril fate in pea leaves is not determined at leaf initiation (plastochron 1, Figure 5A) but is acquired later, between plastochrons 3 and 5 and subsequent to the specification of all other lateral organs (Gould et al., 1994). In situ hybridization experiments revealed that Tl mRNA accumulates in terminal tendril primordial cells at plastochron 3 (Figure 5B), the earliest stage at which tendril fate is determined, and continues to be expressed there until at least plastochron 4 (Figure 5D). Tl mRNA was detected in plastochron 4 tendril primordia but not in adjacent leaflet primordia (Figure 5E). Tl transcripts did not accumulate in other vegetative organs, the shoot apex, or developing flowers.
Figure 5.
Expression Domains of Tl.
(A) Longitudinal section of a 2-week-old wild-type garden pea shoot showing the first five emerging compound leaf primordia, hybridized to a control sense strand Tl probe. Bar = 100 μm.
(B) Adjacent section hybridized to an antisense Tl probe, showing Tl expression at the tip of a plastochron 3 leaf primordium.
(C) Adjacent section showing Uni expression in a plastochron 3 leaf primordium.
(D) Longitudinal section showing Tl expression in emerging tendrils on a plastochron 4 leaf primordium.
(E) Transverse section showing Tl expression in emerging tendrils on a plastochron 4 leaf primordium.
(F) Transverse section of an af mutant genotype, JI 1195, in which tendrils replace leaflets, showing Tl expression in all emerging tendril positions. L, leaflet primordium; P1 to P5, plastochron 1 to plastochron 5 of leaf primordium development.
(G) Excessively tendrilled leaf phenotype of af mutant garden pea cultivar Kahuna, in which tendrils replace leaflets.
(H) Quantitative PCR analysis of Tl gene expression. Segregating af individuals in an F2 population derived from the cross JI 2171 × JI 1195 were confirmed as Uni/Uni (orange bar, left panel) or uni/uni (green bar) homozygotes by DdeI digestion of amplified Uni PCR products. Relative Tl expression was measured by performing PCR reactions in triplicate with standard deviations shown. Separate experiments were performed to compare Tl expression in af line JI 3129 and progenitor wild-type line JI 3133 (orange versus gray bar, center panel). Reactions were performed in quadruplicate with standard deviations shown. To facilitate comparison between panels, equivalent genotypes have the same color. The effect of Uni alone in an Af background was similarly measured separately by comparing FN 1210 with its progenitor JI 2822 (blue versus gray bar, right panel).
The transcription start site of Tl was mapped by 5′ rapid amplification of cDNA ends (RACE) PCR to a CA dinucleotide within a CCANTG LFY binding motif (William et al., 2004), 49 nucleotides upstream of the ATG corresponding to the start codon. Uni is the garden pea ortholog of the Arabidopsis gene LFY (Weigel et al., 1992; Hofer et al., 1997), and it plays a role in maintaining the meristematic potential of the compound leaf, enabling pairs of leaflets, followed by pairs of tendrils, to develop. Uni can fully complement Arabidopsis lfy mutants; therefore, it must share LFY activities (Maizel et al., 2005; Wang et al., 2008), including promoter binding. We postulated that if the predicted LFY binding motif represented an actual Uni binding motif, then Tl transcription would be dependent on Uni. We tested this hypothesis genetically by generating uni tl double mutants and found that unifoliolate uni single mutant and uni tl double mutant phenotypes were indistinguishable in a segregating population (t test, P = 0.63; see Supplemental Table 3 online). This shows that uni is epistatic to tl and that these two genes most likely act in the same developmental pathway. Next, we examined the expression patterns of the two genes in situ, predicting that if Uni regulates transcription initiation by binding to the CCANTG motif, then the two genes would have overlapping domains of expression. Adjacent tissue sections hybridized with a Uni probe (Figure 5C), showed accumulation of Uni mRNA at leaf initiation and also later, in the distal region of plastochron 3 primordia, where Tl expression was observed (Figure 5B). This confirmed results from earlier work showing that Uni expression can be detected as late as plastochron 4 (Gourlay et al., 2000). The expression of Uni and Tl therefore overlaps both temporally, during plastochrons 3 and 4, and spatially, in the distal region of leaf primordia, where tendril initiation occurs.
Finally, we tested the dependence of Tl expression on Uni by quantitative PCR. Uni expression is known to be upregulated in the afila (af) genotype (Gourlay et al., 2000), a prolifically tendrilled mutant of pea used widely in agriculture (Figure 5G). Our expectation that Tl mRNA would also accumulate to higher levels in this genotype due to the increased number of tendril primordia was confirmed by in situ hybridization (Figure 5F) and quantitative PCR (Figure 5H). The af genotype was used as a sensitive reporter of Tl expression in further quantitative PCR analyses. Tl mRNA levels were shown to be reduced fivefold in a uni,af genotype compared with a Uni,af genotype (Figure 5H), indicating that Tl transcription is positively regulated by Uni. Similar results were obtained in a uni mutant genotype compared with its corresponding wild type (JI 2822), where Tl expression was reduced 2.5-fold in the mutant. Notably, Tl expression was not abolished completely in uni af mutants or uni mutants, suggesting that a basal level of Tl transcription can occur, even in the absence of Uni.
Tl Is Present in Other Tendrilled Legumes
The phenotypic similarity of the tendril-less garden pea and sweet pea mutants (Figure 1) suggested that tl and t might be orthologous loci. To test for cosegregation of t and sweet pea Tl, an F2 population of 185 individuals was generated from a cross between sweet pea cultivar America and a tendril-less, homozygous t/t mutant (Punnett, 1923). Garden pea Tl gene primers that flanked the HDZIP region were used to amplify a PCR marker suitable for scoring the F2 population. A presence-absence polymorphism distinguished the wild-type and t/t parents, which suggested that the mutant carried a gene deletion. Absence of the marker cosegregated with the tendril-less phenotype of all 45 homozygous t/t F2 segregants, indicating that it is likely that tl and t are orthologous loci and that these closely related legumes share the same genetic mechanism for the regulation of tendril formation.
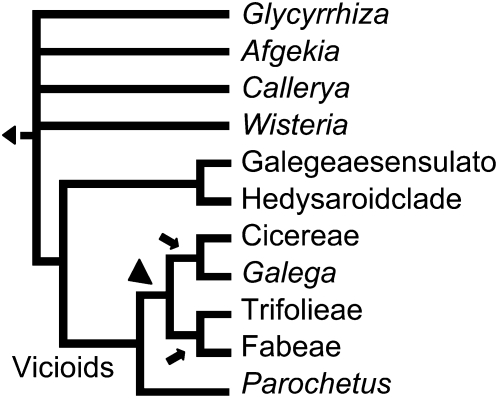
Tendrils are characteristic of the Papilionoid clade Fabeae, to which peas (Pisum spp and Lathyrus spp), vetches (Vicia spp), and lentils (Lens spp) belong (Figure 6). Various wild species of chickpea (Cicer spp) are also tendrilled but are contained within a separate clade, the Cicereae (Figure 6). Since all other Papilionoid legumes are untendrilled, this phylogeny suggests that tendrils arose either once (marked with an arrowhead), with at least two independent losses, or, twice (marked with arrows) independently. Garden pea Tl gene primers were able to amplify HDZIP PCR products from tendrilled sister taxa representatives, common vetch (Vicia sativa), narbon bean (Vicia narbonensis), and lentil (Lens culinaris). The sequences of these products comprise a single distinct Tl-like clade (Figure 3). Alignment of these sequences (see Supplemental Figure 2 online) identified the striking common feature of a Thr substitution for Ala in the presumed DNA binding domain (Figure 4), suggesting that the mechanism for tendril formation may be shared more widely within the Fabeae clade. Efforts to amplify a Tl ortholog from wild tendrilled chickpea species, Cicer anatolicum and Cicer canariensis, were unsuccessful. We were also unsuccessful in attempts to amplify Tl orthologs from nontendrilled taxa that are closely related, but outside the Fabeae clade, such as Medicago and clover (Trifolieae clade), and no clear ortholog is present in the Medicago genome sequence. We concluded that either these species do not have Tl orthologs, or the sequence is too diverged to detect. Our evidence supports the possibility that the evolution of tendrils in the Fabeae was dependent on acquisition of Tl, but it remains an open question whether the same acquisition event, an independent acquisition of Tl, or an independent event altogether, led to tendrils in the Cicereae (Figure 6).
Figure 6.
Phylogenetic Tree of the Inverted Repeat–Lacking Clade of Papilionoid Legumes.
Tree adapted from http://www.tolweb.org/IRLC_%28Inverted_Repeat-lacking_clade%29/60358. Arrows point to possible independent origins of tendrils in Fabeae and Cicereae clades. Arrowhead points to possible single origin of tendrils in Papilionoid legumes.
DISCUSSION
All Papilionoid legumes, apart from taxa in the Fabeae and Cicereae clades, lack tendrils. We propose that the wild-type Tl allele (such as that carried by present-day tendrilled peas) arose as a semidominant mutation that suppressed leaflet blade development in a tendril-less progenitor legume, either in the Fabeae clade ∼18 million years ago, or in the Fabeae-Cicereae clade ∼33 million years ago (Lewis et al., 2005). We propose that this distinct allele survived the constraints of selection by providing a novel phenotype, permitting adaptation to a new and advantageous climbing growth habit.
Tendrils have arisen many times in flowering plants (Darwin, 1875; Bell, 1991). In grapevine, for example, the tendril is a gibberellin-inhibited inflorescence, as shown by a dwarf, tendril-less GIBBERELLIN-INSENSITIVE1 mutant (Boss and Thomas, 2002). In legumes, they are recently acquired specializations of the leaf, and here we have shown that pea tendril formation likely involves interaction between the Class I HDZIP gene Tl and the meristem identity gene Uni/ LFY.
The closest related gene to Tl in Arabidopsis, LMI1, was reported to have additional LFY-independent roles in leaf development, including promotion of leaf margin serrations and suppression of blade outgrowth from the petiole (Saddic et al., 2006). Thus, certain aspects of Tl and LMI1 function appear to be similar, such as blade suppression and transcriptional regulation by LFY. It is not clear, however, that regulation involves transcription initiation in both cases, and the suppression of leaf blade outgrowth probably involves different mechanisms in pea and Arabidopsis, since this process is largely Uni dependent in the former, but LFY independent in the latter. Other aspects of Tl, such as its semidominant inheritance and its tightly delimited expression domain in the leaf, are not characteristic of LMI1.
Tl encodes an unusual Class I HDZIP protein, and in all the tendrilled species, we have examined this protein has a Thr replacement for Ala at position 123 of the DNA binding helix 3 of the homeodomain. The closest relative identified in Medicago does not seem to be a good candidate for a Tl ortholog. This suggests one of three possibilities: (1) within the lineage leading to the Fabeae, after its divergence from the Trifoleae, a gene duplication occurred generating LMI1 paralogs, one of which we see as Tl but the other has not yet been found or is lost. (2) The Tl gene and the Medicago gene AC139525_29 are both LMI1 orthologs, but the Tl-like genes in tendrilled taxa relatives have evolved a new, presumably additional, function and structure. (3) The common ancestor of the tendrilled legumes and Medicago had duplicate LMI1 homologs. One paralog retained LMI1 structure and function while the other diverged as Tl. In the Medicago lineage, the Tl-like paralog was lost. This ancestral duplication may be preserved in some Cicer species. In all three of these possible histories, Tl has a novel sequence and function.
Comparison of the HDZIP regions of Tl-like proteins (see Supplemental Figure 2 online) showed that the legume Tl sequences have shorter leucine zippers that may optimize Tl for homodimerization or specific heterodimerization partners. The Thr replacement for Ala at position 123 of Tl in the DNA binding helix 3 of the homeodomain (Figure 4) is in the equivalent position to Met-54 of Antennapedia, a well-studied example of a DNA-bound eukaryotic homeodomain protein (Fraenkel and Pabo, 1998). The hydrophobic side chain of Met-54 points into the major groove and makes contact with the DNA recognition motif. This suggests that Tl is either impaired significantly in DNA binding or is optimized for a different target from other Class I HDZIPs. Plant Class IV HDZIP proteins have a Thr-containing, rather than an Ala-containing, Helix 3 DNA binding motif, and Outer Cell Layer1 (Ingram et al., 1999) from maize (Zea mays) in particular has conserved adjacent Arg residues like Tl (see GenBank accession Y17898). This suggests that binding targets may exist for the Tl homeodomain in peas. If binding targets existed in a nontendrilled progenitor, then the origin of Tl may have established a new network of interactions that promoted tendril formation.
A Tl-instigated developmental pathway transforms lateral organ primordia from their default leaflet fate to a tendril fate where vascular bundles surround a central pith and adaxial polarity is suppressed (Tattersall et al., 2005). This transformation could occur if Tl interfered, either positively, to provide alternative binding sites to a heterodimeric partner protein as suggested above or in a dominant-negative manner to prevent DNA binding of a partner protein. Both models can explain the semidominant effect of Tl because partner proteins would either acquire abaxial polarizing activity or be prevented from exerting their adaxial polarizing activity due to competitive dimerization with Tl in lateral primordia. The resulting tendril would be interpreted as an abaxialized leaflet. Putative partners might include Class IV HDZIP proteins or pea orthologs of the Class III HDZIP proteins PHABULOSA, PHAVOLUTA, and REVOLUTA, which are known to play a role in the establishment of lateral organ adaxial identity in Arabidopsis (McConnell et al., 2001; Emery et al., 2003). Identification of Tl partners is essential to test these models and to gain further understanding of the tendrilled trait in crop legume species.
METHODS
Plant Material
All garden pea (Pisum sativum), lentil (Lens culinaris), vetch (Vicia sativa), and chickpea (Cicer arietinum) lines were obtained from the John Innes Pisum Germplasm collection, apart from garden pea lines MC1a/1 from J.W. and M3 4092-1 from C.L. Sweet pea (Lathyrus odoratus) cv America was obtained from Chiltern Seeds (http://www.chilternseeds.co.uk/), and the t/t line was commercially available as Snoopea. Plants were grown in 16 h daylength in John Innes No. 1 compost with 30% extra grit. DNA was prepared from leaves according to Ellis et al. (1998).
Mutagenesis
A total of 1400 seeds of line JI 2822 were subjected to 20 Grays FN irradiation at Oak Ridge National Laboratory. Irradiated M1 plants were self-fertilized, and M2 families of up to four plants were screened for tendril-less phenotypes. Mutants were backcrossed to JI 2822 to generate lines FN 1081/6, FN 1132/1, FN 1167/3, FN 1347/6, FN 1484/1, FN1770/4, and FN 2086/3.
Fingerprinting
Wild-type progenitor and tl mutant pairs were analyzed as described (Ellis et al., 1998). TaqI-digested genomic DNA ligated to a TaqI adaptor was used as a template for PCR with a 33P-labeled primer matching the polypurine tract of the Pea Dispersed Repeat1 retrotransposon and a TaqI primer with two selective bases (AA). Reactions were loaded side by side and separated by gel electrophoresis on a 4.5% denaturing polyacrylamide gel. Dried gels were displayed using a Typhoon 8600 PhosphorImager (Molecular Dynamics, GE Healthcare UK).
Marker Screening
AFLP marker screening was performed as described (Vos et al., 1995), except that enzyme PstI was substituted for EcoRI. Genomic DNA (0.5 μg) was digested with PstI and MseI, and adapters were ligated. PstI adapter 1 and 2 together with MseI adapter 1 and 2 sequences are given in Supplemental Table 4 online. The ligation reaction was diluted 10-fold, and 2 μL was used in a 20-μL preamplification reaction using PstI and MseI primers (see Supplemental Table 4 online) with one selective base. Twenty cycles of preamplification PCR in 20-μL reactions, containing 15 ng of each primer, were performed according to Vos et al. (1995). Reactions were diluted 10-fold, and 1 μL was used as template in 10-μL AFLP reactions containing 15 ng 6-carboxyfluorescein–labeled PstI primer with two bases of selection and 15 ng MseI primer with three bases of selection. After 35 cycles of PCR, fragments were separated by capillary electrophoresis and displayed using GeneMapper v3.7 software (Applied Biosystems).
Sequencing
Sequencing was performed using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) at the John Innes Centre Genome Laboratory. Genomic DNA sequence was obtained from P. sativum cv Cameor BAC clone 129B19 and wild-type and tl mutant lines using the following primers: 34R9, 34R8, 34F8REV, 34F7REV, TLHD5′, TLHD5′REV, TLHDF1, TLHDF2, 34cDNA5′, TLHD3′nest, and TLHD3′ (see Supplemental Table 4 online).
Cloning
An 892-bp cDNA clone, c21, was amplified from JI 2822 cDNA using primers TLHDcDNA5′ and 34cDNA3′ (see Supplemental Table 4 online) and cloned into a TopoTA vector (Invitrogen). This sequence has been deposited under accession number 1119567 in GenBank. A 2371-bp clone, 34g40, was obtained by PCR amplification from JI 2822 genomic DNA using primers TLHD5′nest and TLHD3′nest2 (see Supplemental Table 4 online) using Phusion Hot Start High-Fidelity DNA Polymerase (Finnzymes). The product was A-tailed with Taq polymerase before insertion into a TopoTA vector (Invitrogen). This sequence has been deposited under accession number 1119577 in GenBank.
RT-PCR
Total RNA was extracted from shoot apices dissected from 3-week-old seedlings using a Qiagen RNeasy Plant Mini kit (Qiagen Sciences). DNA was removed from 80 μg of RNA samples by digestion with 30 Kunitz units DNaseI (GE Healthcare) in 100 μL 1× One-Phor-All buffer. Two micrograms of RNA was reverse transcribed with Moloney Murine Leukemia Virus reverse transcriptase (Invitrogen) from an oligo(T) primer. One microliter of first-strand cDNA was used in 20-μL PCR reactions containing 0.25 μM primers TLHDF1 and 34F1 for 40 cycles with an annealing temperature of 56°C. Primers flanking introns 19, 20, and 21 of a pea ARGONAUTE1 cDNA clone were used in control reactions. Primers PsAGO1 and PsAGO2, flanking introns 19, 20, and 21 of a pea ARGONAUTE1 cDNA clone were used in control reactions, see Supplemental Table 4 online for primer sequences.
Quantitative PCR
cDNA was synthesized from total RNA prepared as above, except that DNaseI (Ambion) treatment was performed according to the manufacturer's instructions. RNA was quantified by spectrophotometry (NanoDrop Technologies), and 5 μg of RNA was reverse transcribed using SuperScript RNaseH reverse transcriptase (Invitrogen). Twenty nanograms of cDNA was used as template in 10-μL PCR reactions containing 1× SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) and 0.2 μM forward and reverse primers for 40 cycles with an annealing temperature of 60°C. Samples were amplified on a Chromo4 Real Time PCR machine (Bio-Rad) and analyzed with MJ Opticon monitor software V3.1. A 94-bp Tl amplicon spanning intron 2 was amplified with primers PsTlF and PsTlspanR. A 104-bp control amplicon corresponding to pea actin was amplified with primers PsActF and PsActR5′ with primer PsActF sequence modified from GenBank accession number U81047 (see Supplemental Table 4 online for primer sequences).
RACE-PCR
RNA ligase-mediated 5′ RACE was performed on 10 μg of shoot apex total RNA. A FirstChoice RLM-RACE kit (Ambion) was used according to the manufacturer's protocol. Nested PCR was performed using 5′ RACE primers supplied and Tl gene-specific primers 34F6 and 34F6adj. A single amplified product was sequenced directly. RNA was reverse transcribed, and two rounds of 3′ RACE PCR were performed according to the manufacturer's protocol using primers supplied and Tl gene-specific primers 34cDNA5′ and 34PstextR1 (see Supplemental Table 4 online for primer sequences). Heterogeneous products were cloned into a TopoTA vector and sequenced.
RNA in Situ Hybridization
A 400-bp Tl cDNA 3′ fragment was amplified using primers 34cDNA5′ and 34cDNA3′ (see Supplemental Table 4 online) and cloned into a TopoTA vector (Invitrogen) to generate clone 34/19 lacking the HDZIP region for use as an in situ hybridization probe. Digoxygenin-labeled antisense probes were generated from NotI-digested clone 34/19 transcribed with T3 RNA polymerase and an EcoRI-digested Uni cDNA clone transcribed with T7 RNA polymerase. Control sense probes were generated from PmeI-digested clone 34/19 transcribed with T7 RNA polymerase and a XhoI-digested Uni cDNA clone transcribed with T3 RNA polymerase. Sectioning, hybridization, and microscopy were performed as described previously (Hofer et al., 1997).
Phylogenetic Analysis
The deduced amino acid sequences of 35 Arabidopsis thaliana, Medicago truncatula, and other legume Class I HDZIP genes were aligned using ClustalW version 2.0.5 (see Supplemental Figure 2 online for alignment). Residues 132 to 237 of the HDZIP region were selected to estimate maximum likelihood trees using PROML in PHYLIP version 3.67 (http://evolution.genetics.washington.edu/phylip.html) with the Jones-Taylor-Thornton probability model of change between amino acids and the Class II HDZIP At4g37790 defined as an outgroup. Bootstrap support was obtained from 100 replicates for majority-rule consensus tree branches.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: EU938524 (P. sativum Tl mRNA, complete coding sequence), EU938525 (P. sativum Tl gene, complete coding sequence), EU938526 (L. odoratus Tl gene), EU938527 (L. culinaris Tl gene), EU938528 (Vicia narbonensis Tl gene), and EU938529 (V. sativa Tl gene). The pea ARGONAUTE1 sequence is available as accession number EF108450. The pea actin sequence corresponds to accession number U81047.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Tendril-Less Leaf Phenotype of Allele tl-pet.
Supplemental Figure 2. ClustalW2 Sequence Alignment Used to Estimate Tree in Supplemental Figure 2.
Supplemental Table 1. Comparison of Species Richness in Legume Lineages.
Supplemental Table 2. Confirmed tendril-less Alleles.
Supplemental Table 3. Epistasis of uni over tl.
Supplemental Table 4. Primer and Adapter Sequences.
Supplemental Data Set 1. Text File Corresponding to the Alignment in Supplemental Figure 2.
Supplementary Material
Acknowledgments
This research was funded by European Union FP6 Grain Legumes Integrated Project FOOD-CT-2004-506223 (A.D., C.L.S., J.H., and M.D.), by a Department for Environment, Food, and Rural Affairs funded Pulse Crop Genetic Improvement Network (AR0711) grant (C.M., L.T., and M.A.), by a grant-in-aid from the Biotechnology and Biological Sciences Research Council to the John Innes Centre (N.E.), by support from Genoplante, the French consortium for plant genomics and Genopole (A.B.), and by an Australian Research Council Discovery Project, DP0556508 (J.W.). We thank David Laurie and Liam Dolan for critically reading the manuscript. We thank Hilary Ford and Ruth Pothecary for plant care, Andrew Smooker for preparation of sweet pea DNA samples, Andrew Davis for photography, Grant Calder for advice on microscopy, and Liesl New for computational facilities.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Noel Ellis (noel.ellis@bbsrc.ac.uk).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ariel, F.D., Manavella, P.A., Dezar, C.A., and Chan, R.L. (2007). The true story of the HD-Zip family. Trends Plant Sci. 12 419–426. [DOI] [PubMed] [Google Scholar]
- Barkoulas, M., Hay, A., Kougioumoutzi, E., and Tsiantis, M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40 1136–1141. [DOI] [PubMed] [Google Scholar]
- Bell, A.D. (1991). Plant Form: An Illustrated Guide to Flowering Plant Morphology. (Oxford, UK: Oxford University Press).
- Boss, P.K., and Thomas, M.R. (2002). Association of dwarfism and floral induction with a grape 'green revolution' mutation. Nature 416 847–850. [DOI] [PubMed] [Google Scholar]
- Bucher, P. (1990). Weight matrix descriptions of four eukaryotic RNA polymerase-II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212 563–578. [DOI] [PubMed] [Google Scholar]
- Dalmais, M., Schmidt, J., Le Signor, C., Moussy, F., Burstin, J., Savois, V., Aubert, G., Brunaud, V., de Oliveira, Y., Guichard, C., Thompson, R., and Bendahmane, A. (2008). UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 9 R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C. (1875). The Movements and Habits of Climbing Plants. (London: John Murray).
- DeMason, D.A., and Chawla, R. (2004). Roles for auxin during morphogenesis of the compound leaves of pea (Pisum sativum). Planta 218 435–448. [DOI] [PubMed] [Google Scholar]
- de Vilmorin, P. (1910). Experiments on Mendelian inheritance (transl. from French). C. R. Acad. Sci. Paris 2 548–551. [Google Scholar]
- de Vilmorin, P., and Bateson, W. (1911). A case of gametic coupling in Pisum. Proc. R. Soc. Lond. B. Biol. Sci. 84 9–11. [Google Scholar]
- Ellis, T.H.N., Poyser, S.J., Knox, M.R., Vershinin, A.V., and Ambrose, M.J. (1998). Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol. Gen. Genet. 260 9–19. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Fraenkel, E., and Pabo, C.O. (1998). Comparison of X-ray and NMR structures for the Antennapedia homeodomain-DNA complex. Nat. Struct. Biol. 5 692–697. [DOI] [PubMed] [Google Scholar]
- Gianoli, E. (2004). Evolution of a climbing habit promotes diversification in flowering plants. Proc. R. Soc. Lond. B. Biol. Sci. 271 2011–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, K.S., Cutter, E.G., and Young, J.P.W. (1994). The determination of pea leaves, leaflets, and tendrils. Am. J. Bot. 81 352–360. [Google Scholar]
- Gourlay, C.W., Hofer, J.M.I., and Ellis, T.H.N. (2000). Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, COCHLEATA, AFILA, and TENDRIL-LESS. Plant Cell 12 1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hofer, J., Turner, L., Hellens, R., Ambrose, M., Matthews, P., Michael, A., and Ellis, N. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7 581–587. [DOI] [PubMed] [Google Scholar]
- Ingram, G.C., Magnard, J.L., Vergne, P., Dumas, C., and Rogowsky, P.M. (1999). ZmOCL1, an HDGL2 family homeobox gene, is expressed in the outer cell layer throughout maize development. Plant Mol. Biol. 40 343–354. [DOI] [PubMed] [Google Scholar]
- Jaffe, M.J., and Galston, A.W. (1968). Physiology of tendrils. Annu. Rev. Plant Physiol. 19 417–434. [Google Scholar]
- Komatsuda, T., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm, R. (1957). Three new genes in Pisum. Hereditas 43 541–548. [Google Scholar]
- Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., Rouze, P., and Rombauts, S. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, G.P., Schrire, B., Mackinder, B., and Lock, M. (2005). Legumes of the World. (Richmond, UK: Kew Publishing).
- Maizel, A., Busch, M.A., Tanahashi, T., Perkovic, J., Kato, M., Hasebe, M., and Weigel, D. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308 260–263. [DOI] [PubMed] [Google Scholar]
- Marx, G.A. (1973). Instability in flowering behaviour associated with inbreeding. Pisum Newsletter 5 29–30. [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Punnett, R.C. (1923). Linkage in the sweet pea. J. Genet. XIII 101–123. [Google Scholar]
- Saddic, L.A., Huvermann, B.R., Bezhani, S., Su, Y.H., Winter, C.M., Kwon, C.S., Collum, R.P., and Wagner, D. (2006). The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development 133 1673–1682. [DOI] [PubMed] [Google Scholar]
- Tattersall, A.D., Turner, L., Knox, M.R., Ambrose, M.J., Ellis, T.H.N., and Hofer, J.M.I. (2005). The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. Plant Cell 17 1046–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva, M. (1979). Induced mutagenesis in Pisum. I. Genetic studies on the acacia mutant. Genet. Sel. 12 396–408. [Google Scholar]
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., Vandelee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., and Zabeau, M. (1995). AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 23 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Chen, J., Wen, J., Tagede, M., Li, G., Liu, Y., Mysore, K.S., Ratet, P., and Chen, R. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146 1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- William, D.A., Su, Y.H., Smith, M.R., Lu, M., Baldwin, D.A., and Wagner, D. (2004). Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. USA 101 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.