Abstract
Although the genetic regulation of recombination in allopolyploid species plays a pivotal role in evolution and plant breeding, it has received little recent attention, except in wheat (Triticum aestivum). PrBn is the main locus that determines the number of nonhomologous associations during meiosis of microspore cultured Brassica napus haploids (AC; 19 chromosomes). In this study, we examined the role played by PrBn in recombination. We generated two haploid × euploid populations using two B. napus haploids with differing PrBn (and interacting genes) activity. We analyzed molecular marker transmission in these two populations to compare genetic changes, which have arisen during meiosis. We found that cross-over number in these two genotypes was significantly different but that cross-overs between nonhomologous chromosomes showed roughly the same distribution pattern. We then examined genetic recombination along a pair of A chromosomes during meiosis of B. rapa × B. napus AAC and AACC hybrids that were produced with the same two B. napus genotypes. We observed significant genotypic variation in cross-over rates between the two AAC hybrids but no difference between the two AACC hybrids. Overall, our results show that PrBn changes the rate of recombination between nonhomologous chromosomes during meiosis of B. napus haploids and also affects homologous recombination with an effect that depends on plant karyotype.
INTRODUCTION
Polyploidy has played a pervasive and prominent role in the evolution of plants (Otto, 2007). It is estimated that 30 to 80% of extant flowering plants are polyploid (Masterson, 1994; Ramsey and Schemske, 1998; Rieseberg and Willis, 2007) and that almost all angiosperms have experienced at least one round of whole-genome duplication during their evolution (De Bodt et al., 2005; Cui et al., 2006). Some of the world's most important crop plants, such as wheat (Triticum aestivum), cotton (Gossypium hirsutum), and oilseed rape (Brassica napus), are allopolyploids (i.e., they originated as hybrids [followed by chromosome doubling] and contain different sets of related but not completely homologous chromosomes, called homoeologs). Given its importance, polyploidy has attracted a great deal of interest, and rapid progress has been made in many fields (Comai, 2005; Udall and Wendel, 2006; Chen, 2007).
The genetic regulation of recombination in allopolyploid species is a pivotal issue in evolution and agronomy but has been largely underexplored in recent years, except in wheat. Cross-over (CO) suppression between homoeologous chromosomes is required to ensure proper chromosome segregation and fertility; therefore; this mechanism is a determining factor in polyploid speciation. Otherwise, complex meiotic configurations would lead to unbalanced gametes, aneuploid progenies, and, hence, impaired fertility (Ramsey and Schemske, 2002). In most polyploid species, CO formation is restricted to homologs by the activity of genes that affect different processes throughout premeiotic interphase and meiotic prophase (Jenczewski and Alix, 2004). These same genetic systems hamper the incorporation of beneficial traits into crop plants from their wild relatives because they suppress introgressions that rely on the formation of cross-overs between nonhomologous chromosomes (Able and Langridge, 2006; Martinez-Perez and Moore, 2008).
Wheat is the only species in which a large number of continuing studies are devoted to characterizing loci that suppress COs between homoeologous chromosomes (Pairing homeologous loci). The main regulator, Ph1, was discovered 50 years ago (Riley and Chapman, 1958; Sears and Okamoto, 1958) and only recently characterized at the molecular level (Griffiths et al., 2006; Al-Kaff et al., 2007). However, the very peculiar nature of the Ph1 locus, which consists of a cluster of cyclin-dependant kinases (cdk-like genes) and a segment of subtelomeric heterochromatin, does not readily explain the multiple cytological effects attributed to Ph1 (Feldman, 1993; Mikhailova et al., 1998; Martinez-Perez et al., 2003; Prieto et al., 2005; Corredor et al., 2007). On the other hand, Ph2, another suppressor of homoeologous associations (Upadhya and Swaminathan, 1967; Mello-Sampayo, 1971), was shown to affect synaptic progression (Martinez et al., 2001) but the genes responsible for the phenotype are still to be identified (Sutton et al., 2003).
Recently, evidence was obtained that a system regulating homoeologous associations might also exist in B. napus (Jenczewski et al., 2003). B. napus (AACC; 2n = 38) is a young allopolyploid species formed by the hybridization of ancestors of Brassica oleracea (CC; 2n = 18) and Brassica rapa (AA; 2n = 20) (U, 1935; Palmer et al., 1983). B. napus haploid plants (AC; 19 chromosomes), which contain half the somatic chromosome number of euploid B. napus and thus no longer have homologous chromosomes, were first demonstrated to undergo complete meiosis to produce viable gametes containing one copy of each of the 19 B. napus chromosomes (Morinaga and Fukushima, 1933; Olsson and Hagberg, 1955; Tai and Ikonen, 1988). Haploids produced from different B. napus varieties were then shown to display very different meiotic behavior at Metaphase I (MI) (Olsson and Hagberg, 1955; Renard and Dosba, 1980; Attia and Röbbelen, 1986; Jenczewski et al., 2003); haploids produced from some varieties (such as Darmor-bzh) display a few univalents and a lot of nonhomologous associations at MI, whereas haploids produced from other varieties (such as Yudal) display mostly univalents and only a few nonhomologous associations. These differences are inherited in a Mendelian fashion consistent with the presence of a major locus, called PrBn (for Pairing Regulator in B. napus) (Jenczewski et al., 2003), that was subsequently localized to a C-genome chromosome (in addition to four to six other additive or epistatic quantitative trait loci; Liu et al., 2006).
In the absence of homologous chromosomes, the associations observed at MI during meiosis in B. napus haploids could involve (1) homoeologous A and C chromosomes that have diverged for 4 million years (Inaba and Nishio, 2002); (2) chromosomes containing regions of intragenomic or intergenomic homology, which arose by whole-genome duplications in the common ancestor of B. rapa and B. oleracea 13 to 17 million years ago (Parkin et al., 2003, 2005; Lysak et al., 2005); or (3) chromosomes carrying segmental duplications that occurred subsequent to these polyploidy events (Parkin et al., 2005; Yang et al., 2006). As in wheat haploids (Martinez et al., 2005), differences in meiotic behavior among B. napus haploids may therefore indicate genotypic variation in the effectiveness of CO suppression among nonhomologous chromosomes.
In this study, we aimed to further characterize the mode of action of PrBn (and the genes it interacts with), notably their effect on both the rate and distribution of meiotic COs between related genomes in B. napus haploids and hybrids. We first analyzed the transmission of molecular markers in two haploid × euploid F1 populations to compare the rate and distribution of chromosomal rearrangements generated by meiosis in two B. napus haploids showing different numbers of univalents at MI (and that therefore differ in PrBn and interacting genes activity). This survey was required to ascertain that the differences observed at MI between B. napus haploids were not simply due to genotypic variation for (1) achiasmatic associations that are commonplace in polyhaploids and interspecific hybrids (Orellana, 1985; Zhang et al., 1999) or (2) an idiosyncratic distribution of meiotic COs among and along chromosomes that may have resulted in fragile chiasmatic associations (Lamb et al., 1996). We then analyzed genetic recombination during meiosis between a pair of A chromosomes in B. rapa × B. napus triploid and tetraploid hybrids to understand if PrBn, and the genes it interacts with, lead to variations in the number of CO between almost homologous chromosomes. Altogether, our results demonstrate that PrBn suppresses both homoeologous and homologous recombination, with a magnitude that depends on the karyotype of the plants. Our study thus confirms that PrBn is another model to compare and contrast with wheat Pairing homeologous genes.
RESULTS
The Number of Chromosomal Rearrangements Varies between the Progenies of Darmor-bzh and Yudal Haploids
We assayed parental marker transmission within and between two haploid × euploid F1 populations. The first population was obtained by crossing five haploid Darmor-bzh plants (that display five univalents at MI on average) with one euploid Yudal plant. The second was generated from nine haploid Yudal plants (that display 12 univalents at MI on average) crossed with one Darmor-bzh euploid plant(Figure 1, experiment 1). A total of 150 (population one) and 141 (population two) markers, including 86 codominant markers that allowed direct comparisons (see Supplemental Figure 1 online), were used to genotype these two F1 populations and compare the number of chromosomal rearrangements that were generated by meiosis of Darmor-bzh and Yudal haploids that differ in PrBn (and interacting genes) activity. We anticipated that meiotic COs would produce chromosomal rearrangements during the first meiotic division, which would then be subsequently transmitted to the progenies of each B. napus haploid by unreduced gametes generated by an equational division of the sister chromatids (see Nicolas et al., 2007).
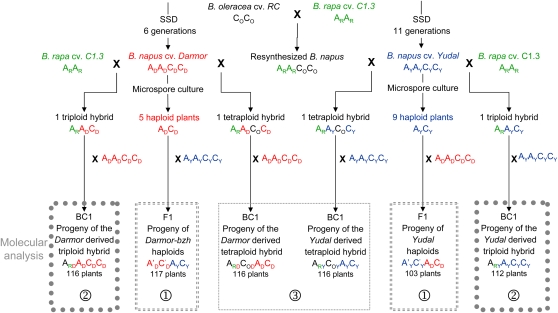
Figure 1.
Genealogy of Plant Material.
AD, AY, and AR designate the A genome from B. napus cv Darmor-bzh, B. napus cv Yudal, and B. rapa cv Chicon (C1.3), respectively. Likewise, CD, CY, and CO designate the C genome from B. napus cv Darmor-bzh, B. napus cv Yudal, and B. oleracea cv RC, respectively. ARD and ARY indicate that chromosomes from B. rapa recombined with those of Darmor and Yudal, respectively. COD and COY indicate that chromosomes from B. oleracea recombined with those of Darmor and Yudal, respectively. A′DC′D and A′YC′Y indicate that chromosome rearrangements partially reshuffled the A and C genomes of Darmor-bzh and Yudal, respectively. (1) The two haploid progenies were used to compare the frequencies at which chromosome rearrangements were generated during meiosis of haploid Darmor-bzh (ADCD) versus haploid Yudal (AYCY). (2) and (3) Progenies of triploid and tetraploid hybrids were used to compare the effects of Darmor versus Yudal genotypes on the frequency of homologous recombination on A7/R7. [See online article for color version of this figure.]
Losses and duplications of parental markers were observed in the two F1 haploid × euploid populations, confirming that chromosomal rearrangements were generated during meiosis in Darmor-bzh and Yudal haploids. Very significant differences were observed between the two F1 populations regardless of the method used to score rearrangements (see Methods). For example, the average frequency of haploid parent allele loss was higher in the progeny of Darmor-bzh haploids than in the progeny of Yudal haploids (method 1 using codominant markers only: 5% versus 1.5%; P < 0.01%). Likewise, a total of 342 chromosomal rearrangements were detected in the 117 progeny of Darmor-bzh haploids compared with only 111 rearrangements in the 103 progeny of Yudal haploids (method 2 using codominant and dominant markers: P < 0.01%). We obtained the same results when comparisons were performed using only the codominant markers; this confirmed that variations in the number of rearrangements did not result from slight variations in genome coverage between the two genetic backgrounds (see Supplemental Figure 1 online). Analysis with codominant markers also showed that euploid parent alleles were missing at almost equal frequencies in both populations (0.17% in euploid Yudal versus 0.24% in euploid Darmor-bzh), but their numbers were too low for sound statistical comparisons.
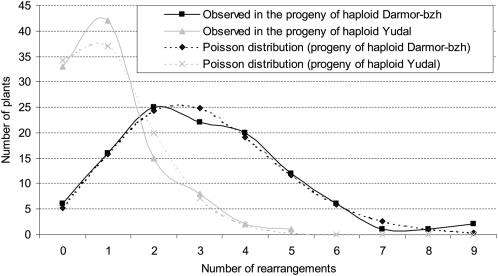
Significant differences were also observed in the number of chromosomal rearrangements per individual offspring (Figure 2). For example, rearrangements were absent in only six plants (out of 117) in the progeny of Darmor-bzh haploids compared with 34 plants (out of 103) in the progeny of Yudal haploids (χ2 = 26.1; P < 0.01%). Furthermore, among the offspring of Yudal haploids, a maximum of five missing regions was observed, whereas up to nine were missing in two offspring of Darmor-bzh haploids. In both cases, the distribution of the number of rearrangements per individual matched a Poisson distribution (Figure 2); this indicated that there was no, or only weak, selection against the gametes and offspring that carried these rearranged chromosomes. Therefore, the observed differences between the two populations likely reflect significant differences in the frequency at which these rearrangements were generated during meiosis.
Figure 2.
Distribution of the Number of Rearrangements per Individual Offspring in the Progeny of Darmor-bzh versus Yudal Haploids.
Two Poisson distributions (diamond for Darmor-bzh and cross for Yudal) are used to model the number of rearrangements per plant. The average number of missing regions per plant (per progeny) is used as the average rate for each of the two distributions.
To confirm this, we compared the numbers of rearrangements observed in the two F1 populations with those expected from the cytological survey described by Jenczewski et al. (2003). We first estimated that a total of 405 and 181 chromosomal rearrangements should be observed among the 117 and 103 plants surveyed in the progenies of Darmor-bzh and Yudal haploids, respectively, if (1) every chromosome association observed at MI was chiasmatic (i.e., triggered rearrangements that segregated to half the gametes; see Nicolas et al., 2007) and (2) there was no gametic/zygotic selection against rearranged chromosomes. In the two populations, these estimates were significantly higher than the total number of rearrangements detected with molecular markers spanning 75% of the genome (Darmor-bzh: χ2 = 11; P < 0.001; and Yudal: χ2 = 24; P < 0.001). Resampling procedures were used to extrapolate the total number of rearrangements that would have been detected if 100% of the genome was covered (see Supplemental Figure 2 online). These extrapolated numbers roughly matched expectations from the cytological survey; in fact, we estimated that ∼425 and ∼137 rearrangements should have been observed in the progenies of Darmor-bzh and Yudal haploids, respectively, if 100% of genome was covered. Given the substantial variability obtained across simulated data sets (see Supplemental Figure 2 online), these estimates are reasonably close to expectation.
Thus, the higher number of bivalents/multivalents observed at MI in Darmor-bzh compared with Yudal haploids is closely paralleled by an increase in the number of chromosomal rearrangements, which could have arisen either from genome-wide or chromosome specific differences in recombination frequency. To determine which of the two possibilities occurred, we compared the distribution of rearrangements among linkage groups between the two F1 populations.
The Difference between Darmor-bzh and Yudal Haploids Is Not Due to Chromosome-Specific Differences in Recombination
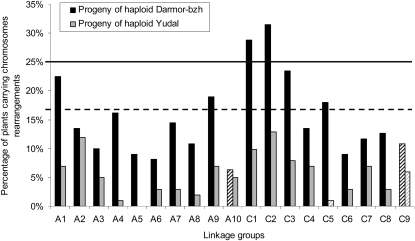
We observed that all linkage groups were rearranged at least once in the two populations (except A5 in the progeny of Yudal haploids; Figure 3), demonstrating that all chromosomes were able to recombine in both genotypes. Significant differences in the number of chromosomal rearrangements were found for 11 out of 17 linkage groups (two groups were not included in this comparison because they showed extensive coverage differences between the two progenies). On average, chromosomes were rearranged three times more often in the progeny of Darmor-bzh versus Yudal haploids; the difference was higher for A4 and A8, but lower for C7 and A2. A2 was the only linkage group for which the same frequency of rearrangements was seen in the two populations (12 to 13% of rearranged plants; Figure 3).
Figure 3.
Percentage of Plants with Rearrangements for Each Chromosome in the Progenies Darmor-bzh and Yudal Haploids.
The solid line indicates the expected proportion of plants with rearrangements when chromosomes systematically undergo a cross-over during haploid meiosis. The dashed line indicates the lowest limit of the confidence interval around this expected proportion when α = 5%. Striped black and gray bars (such as for C9) indicate the linkage groups that were poorly covered in Darmor-bzh and Yudal, respectively.
We then observed that for six linkage groups in the progeny of Darmor-bzh haploids, the proportion of plants with rearrangements was not significantly different from that expected if the corresponding chromosome systematically formed a chiasmatic bivalent at meiosis (i.e., 25%; Figure 3). By contrast, this proportion was observed for none of the linkage groups tested in the progeny of Yudal haploids (Figure 3). It thus appears that at least six chromosomes could systematically form cross-overs during meiosis in Darmor-bzh haploids, while this was never the case for any chromosome during meiosis of Yudal haploids. Randomization tests then demonstrated that the highest frequencies of linkage group rearrangements observed in the progenies of Darmor-bzh and Yudal haploids would not be obtained if rearrangements occurred at random, with equal probabilities for all linkage groups. This means that some linkage groups were rearranged more often than expected by chance in these two populations.
Chromosomal Rearrangements Occur in Similar Patterns in the Progenies of Darmor-bzh and Yudal Haploids
We showed that the number of rearrangement breakpoints per linkage group in the progeny of Darmor-bzh haploids was positively and significantly correlated to the number of rearrangement breakpoints per linkage group in the progeny of Yudal haploids (R2 = 0.7; P < 1%). This showed that the same linkage groups were rearranged at a proportionally high, intermediate, or low frequency in the two populations.
We compared the proportion of rearrangements that entailed concurrent losses and duplications of homoeologous haploid parent alleles (i.e., for markers located in homoeologous regions) between the two populations to assess the relative frequency of homoeologous recombination (for methods, see Nicolas et al., 2007). Table 1 summarizes the results obtained for homoeologous regions located on five different pairs of homoeologous chromosomes. The low number of rearrangements detected in the progeny of Yudal haploids resulted in huge locus-to-locus variations that are certainly due to sampling variability and hampered statistical comparisons. Averaging over loci, which seldom has biological significance, suggested that ∼55% of rearrangements were derived from COs between homoeologous chromosomes in the two F1 populations (χ2 = 0.06, P = 0.81; Table 1).
Table 1.
Comparison of the Proportion of Chromosomal Rearrangements Derived from Homoeologous Recombination between the Progenies of Darmor-bzh versus Yudal Haploids
| Linkage Groups | Locus Showing Loss of HP Allelea | Homoeologous Linkage Groups | Corresponding Homoeologous Locus Showing Duplicated HP Allelesb | No. of Chromosomal Rearrangements That Entailed Concurrent Loss and Duplication of HP Allelesb
|
No. of Chromosomal Rearrangements That Did Not Entail Concurrent Loss and Duplication of HP Alleles
|
Chromosomal Rearrangements That Entailed Concurrent Loss and Duplication of HP Alleles (%)
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Darmor-bzh | Yudal | Darmor-bzh | Yudal | Darmor-bzh | Yudal | ||||
| N1 | CB10081b | N11 | CB10081a | 11 | 2 | 5 | 2 | 69 | 50 |
| Ol12F11b | Ol12F11a | 3 | 0 | 2 | 0 | 60 | nd | ||
| N11 | CB10081a | N1 | CB10081b | 5 | 2 | 6 | 1 | 45 | 67 |
| Ol12F11a | Ol12F11b | 4 | 0 | 2 | 1 | 67 | 0 | ||
| N14 | Ol11D12 | N4 | CB10335 | 6 | 0 | 2 | 0 | 75 | nd |
| E35M67ab | CB10347 | 0 | 0 | 0 | 2 | nd | 0 | ||
| N2 | CZ5b703024b (Y)/Brass037 (D) | N12 | CZ5b703024a | 2 | 6 | 6 | 2 | 25 | 75 |
| Na12H09a (Y)/Brass037 (D) | Na12H09b | 1 | 2 | 0 | 0 | 100 | 100 | ||
| N12 | CZ5b703024a | N2 | CZ5b703024b | nd | 5 | nd | 2 | nd | 71 |
| Na12H09b | Na12H09c | nd | 2 | nd | 1 | nd | 67 | ||
| N3 | CB10021a | N13 | Ol10B08 | 0 | 0 | 4 | 4 | 0 | 0 |
| CB10415-IH08a | JLP011 | 0 | 0 | 1 | 1 | 0 | 0 | ||
| N18 | JLP042 | N9 | CZ0b687858 | 6 | 1 | 2 | 2 | 75% | 33% |
| N9 | Ol12F02b | N18 | Brass031 | 0 | 0 | 1 | 0 | 0% | nd |
| All loci | 38 | 20 | 31 | 18 | 55% | 53% | |||
Alleles from the haploid parent.
Exact correspondence between homoeologous loci was ascertained when marker assays detected multiple loci mapping to colinear blocks (Ol12F11a/b and CB10081a/b). When these markers were not available, we used markers anchored in Arabidopsis thaliana to find the most likely homoeologs within colinear blocks (see Supplemental Data Set 1 online).
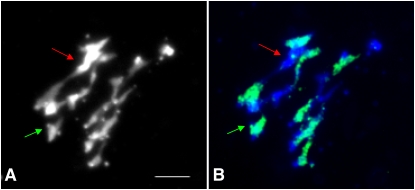
MI chromosomes were labeled with the BoB014O06 probe, which specifically hybridizes to all C-genome chromosomes, to determine the relative proportion of autosyndetic (A-A or C-C) versus allosyndetic (A-C) bivalents. The average meiotic behavior of Yudal haploids at MI is shown in Table 2. We observed that autosyndesis was as proportionally commonplace at MI in Yudal (Figure 4) as in Darmor-bzh haploids (Table 2; χ2 = 0.99, P = 0.32) and represented ∼30% of the bivalents observed. We then compared the distribution of COs along chromosomes in the two F1 populations.
Table 2.
Comparison of Allosyndesis and Autosyndesis at MI between Haploid Darmor-bzh and Yudal
| Average Meiotic Behaviora
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Autosyndesis
|
Allosyndesis
|
||||||||
| No. of PMCs | Ia | IC | IIAA | IICC | IIAC | III+IV | (IIAA+ IICC)/IItot | IIAC/IItot | |
| Darmor-bzhb | 37 | 3 (96) | 2.2 (66) | 0.8 (31) | 0.7 (25) | 5.2 (203) | 0.14 (50) | 0.22 | 0.78 |
| Yudal | 20 | 5.3 (106) | 5.1 (103) | 0.8 (16) | 0.4 (8) | 3 (60) | 0 | 0.29 | 0.71 |
IA and IC indicate univalents belonging to the A and C genomes, respectively; IIAA and IICC indicate autosyndetic bivalents formed between a pair of A or a pair C chromosomes, respectively; IIAC indicates allosyndetic bivalents formed between A and C chromosomes; IIAA + IICC + IIAC = IItot; III and IV indicate trivalents and quadrivalents, respectively.
Total number of chromosomes present as IA, IC, IIAA, IICC, IIAC, III, and IV, respectively, are indicated in parentheses.
Data published by Nicolas et al. (2007).
Figure 4.
Detection of Autosyndesis at Metaphase I Using BAC–Fluorescence in Situ Hybridization on Pollen Mother Cells from Haploid Yudal.
Staining with 4′,6-diamidino-2-phenylindole was used on anther meiocytes to establish the meiotic behavior of every Yudal pollen mother cell (PMC) at MI (A) and then combined with BoB014O06 fluorescence in situ hybridization (FISH) signals (green), which identifies C chromosomes (B) to distinguish autosyndetic and allosyndetic associations. Three bivalents (two autosyndetic, which are indicated by red and green arrows between A and C chromosomes, respectively, and one allosyndetic) and 13 univalents were observed in the cell presented. Bar = 5 μm.
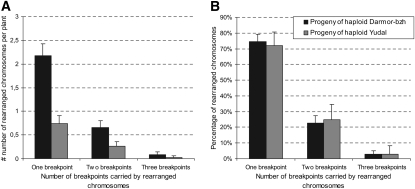
A wide variety of distal and interstitial rearrangements were observed in both progenies, which were classified according to the number of breakpoints they caused on linkage groups. The mean number of chromosomes per plant carrying single, double, and triple breakpoints was higher in the progeny of Darmor-bzh haploids than in the progeny of Yudal haploids (Figure 5A). By contrast, the relative proportion of rearranged chromosomes in each class was strikingly similar in the two F1 populations (χ2 = 0.28, P = 0.85; Figure 5B). In other words, once a linkage group was rearranged in any of the two populations, there was the same chance, irrespective of the haploid genotype, that this rearrangement entailed one (∼70 to 75%), two (∼25%), or three (∼3%) breakpoints.
Figure 5.
Typology of Chromosome Rearrangements in the Darmor-bzh and Yudal Haploid Progenies.
The mean number (A) and relative proportion (B) of rearranged linkage groups showing one, two, or three breakpoints are compared between the Darmor-bzh and Yudal haploid progenies. Error bars indicate confidence intervals at α = 5%.
We compared the positions of COs along chromosomes between the two progenies, approximated by the positions of single breakpoints. We compared the genetic size of the missing distal regions between the two progenies. Linkage groups or regions with extensive coverage differences between the two progenies or that displayed only a few rearrangements were excluded from this comparison. The relative genetic size of the missing distal regions appeared to be slightly but significantly higher in the Darmor-bzh progeny than in Yudal, regardless of the way the breakpoints were located within the rearranged intervals (Table 3). The margin was small, however, suggesting that CO distribution along chromosomes was very similar during meiosis of Darmor-bzh and Yudal haploids.
Table 3.
Relative Genetic Size of Distal Rearrangements Compared between the Progenies of Darmor-bzh versus Yudal Haploids
| Position of Breakpoints within the Rearranged Intervalsa | Mean Genetic Size of the Rearrangements, as an Estimate Based on the Inferred Position of the Rearrangement Endpoint, in the Progeny of:
|
||
|---|---|---|---|
| Darmor-bzh Haploids | Yudal Haploids | P Valueb | |
| (1) At the position of the first removed marker | 26 cM | 20 cM | 0.045 |
| (2) In the middle of the interval | 32.5 cM | 26.5 cM | 0.038 |
| (3) At the position of the last present marker | 39 cM | 33 cM | 0.048 |
The missing distal regions that are considered to stretch away from the breakpoint as far as the end of the linkage group.
P value from a t test.
Comparison of Homologous Recombination in Triploid and Tetraploid Hybrids
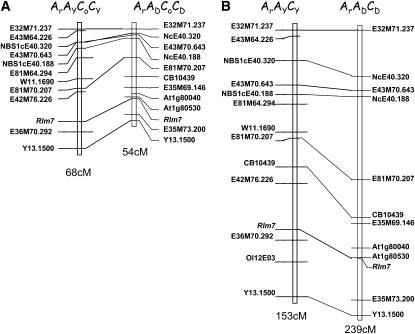
Finally, genetic experiments were performed to compare the frequency of meiotic COs on a pair of homologous chromosomes (linkage group A7) between two tetraploid hybrids produced by crossing the same resynthesized B. napus to Darmor and Yudal, respectively (Figure 1, experiment 3). Using seven markers common to the two maps (Figure 6A),we found that the total genetic size of the linkage group was slightly (1.25-fold) but not significantly higher (P = 0.002 > α′ α=0.01 = 0.001) in the progeny of the tetraploid produced with Yudal compared with Darmor. Likewise, no significant difference was observed when we compared the number of cross-overs per interval (P = 0.2).
Figure 6.
Map Comparison of Linkage Group A7 in Progenies of the Two Tetraploid and the Two Triploid Hybrids.
ARAYCYCO and ARADCDCO are two tetraploid hybrids (A) produced by crossing the same resynthesized B. napus to Yudal and Darmor, respectively. ARAYCY and ARADCD are two triploid hybrids (B) produced by crossing the same plant of B.rapa to Yudal and Darmor, respectively. Genetic distances are given in centimorgans (cM).
The frequency of homologous recombination between two triploid hybrids, produced by crossing the same B. rapa plant with Darmor and Yudal, respectively (Figure 1, experiment 2), was then compared on the same A7 linkage group. We observed a highly significant heterogeneity in genetic map distances between the progeny of the triploid hybrid produced with Darmor (ARADCD) versus Yudal (ARAYCY) (Morton test extended to multiple loci; P < 10−5; Figure 6B). The proportion of COs calculated for every interval between adjacent markers on A7 was higher in the ARADCD hybrid than the ARAYCY hybrid, although no significant differences were observed for COs in the vicinity of the centromere (close to marker E43M70.643; Pouilly et al., 2008) or at one end of the linkage group. Using 12 and 14 markers in the ARADCD and ARAYCY progenies, respectively, we observed that the mean number of exchange points per chromatid was 1.7 (ranging from 0 to 4) in the ARADCD hybrid and 1.0 (ranging from 0 to 3) in the ARAYCY hybrid.
These results demonstrate that changing the genotype of the B. napus variety used to produce hybrids had a clear effect on the frequency of homologous recombination across A7 for the triploid hybrids but no effect for the tetraploid hybrids.
DISCUSSION
Natural euploid B. napus displays predominantly 19 bivalents at MI and an almost strict disomic inheritance; this shows that the vast majority of COs is formed between homologous chromosomes. Evidence for rare homoeologous exchanges have nonetheless been obtained in several B. napus cultivars (Parkin et al., 1995; Sharpe et al., 1995; Lombard and Delourme, 2001; Osborn et al., 2003; Piquemal et al., 2005; Udall et al., 2005), but their frequency remains very low. By contrast, the slightly irregular meiosis of resynthesized B. napus generates a higher proportion of homoeologous exchanges when compared with natural B. napus (Parkin et al., 1995; Sharpe et al., 1995; Udall et al., 2005; Lukens et al., 2006; Gaeta et al., 2007; A.M. Chèvre, unpublished results). These results collectively indicate that an element of meiotic correction has been selected during the evolution of B. napus. Evidence was recently obtained that a major locus, called PrBn, largely determines the number of bivalents and multivalents that form at meiosis in B. napus haploids (AC; n = 19) (Jenczewski et al., 2003; Liu et al., 2006). In this study, we showed that PrBn controls the frequency, but not the distribution, of COs between homoeologous chromosomes in haploid plants and between homologous chromosomes during meiosis of triploid ArAnC plants. We also showed that PrBn has no effect on the level of homologous recombination in tetraploid ArAnCC hybrids, suggesting that its effect on recombination depends on plant karyotype.
Our study shows that the different meiotic behaviors observed between Darmor-bzh and Yudal haploids are caused by a threefold difference in the number of COs that are formed between nonhomologous chromosomes. Given that these two meiotic phenotypes are genetically determined by PrBn and the genes it interacts with, we conclude that these loci have an effect on recombination between nonhomologous chromosomes.
We first showed that the number of rearranged chromosomes in the progenies of Darmor-bzh and Yudal haploids matched the number of bound chromosomes at MI in each of the two haploid genotypes. These findings indicate that achiasmatic associations between nonhomologous chromosomes (Orellana, 1985; Zhang et al., 1999) do not (widely) occur during meiosis of Darmor-bzh and Yudal haploids and incidentally extends the conclusion from our previous study (Nicolas et al., 2007) to the whole-genome level (for Darmor-bzh) and the Yudal genotype. It appears that most rearrangements in the progenies of B. napus haploids stem from COs between nonhomologous chromosomes and notably between homoeologous chromosomes (Table 1).
We then proved that the contrasted meiotic behaviors between Darmor-bzh and Yudal haploids were not caused by a difference in the number of chromosomes that are susceptible to recombine (because of preexisting chromosomal rearrangements, for example). Indeed, all chromosomes were able to recombine during meiosis in Yudal haploids (Figure 3) even if probably none of them systematically underwent a CO. We found that the chance, on average, of most chromosomes in Yudal haploids recombining was three times less than in Darmor-bzh haploids, indicating an overall reduction in recombination between nonhomologous chromosomes.
Finally, we found that the higher number of univalents observed in Yudal haploids cannot result from premature bivalent separation, a phenomenon that was previously observed for COs that were too close to telomeres (Lamb et al., 1996). Indeed, the placement of single exchange events was so similar in the two progenies (Table 3), and the difference between the genetic and cytological data so small, that susceptible chiasma configurations are unlikely to play a significant role in generating the observed univalents at MI.
This study thus highlights differences in the number of COs between Darmor-bzh and Yudal haploids but not their distribution among chromosomes. For example, the number of rearrangement breakpoints per linkage group detected in the progeny of Yudal haploids was highly and positively correlated with the number in the progeny of Darmor-bzh haploids. Likewise, the relative proportions of autosyndesis (i.e., bivalents between pairs of A or pairs of C chromosomes) versus allosyndesis (i.e., bivalents between A and C chromosomes) were found to be very similar in Darmor-bzh and Yudal haploids (Table 2). We also showed that COs were preferentially formed between homoeologous chromosomes in the two progenies (Table 1). These results demonstrate that CO distribution was not random (see randomization test) and followed roughly the same rules during meiosis in the two genotypes. It notably appears that in none of the genotypes were chromosomes randomly scattered in the meiotic nucleus and recombined by chance only when pairs of chromosomes happened to lie close to each other.
It is tempting to further interpret our data and infer that the stringency at which divergence is scrutinized by the meiotic machinery is the same in Darmor-bzh and Yudal haploids. However, for this assumption to be made, a clear and precise understanding of the level of divergence between all the regions that recombined during meiosis of Darmor-bzh and Yudal haploids is required. At present, this information is not available, notably because only two complete sets of paralogous regions were compared at the sequence level in either B. oleracea or B. rapa (Town et al., 2006; Yang et al., 2006). Similarly, it is difficult to interpret our results concerning autosyndesis in Darmor-bzh and Yudal haploids because it is not possible to determine if autosyndesis results from the occurrence of COs between regions that were duplicated 13 to 17 million years ago in each of the parental genomes and are now quite divergent, or conversely if it is caused by CO formation between recent and still very similar segmental duplications that occurred after the divergence between the rapa/oleracea lineages (Yang et al., 2006). Finally, comparisons among Ph1-active and Ph1-defective (ph1b) wheat haploids demonstrated that the same trend for CO distribution among nonhomologous chromosomes can be obtained (Jauhar et al., 1991 and references therein) regardless of the fact that Ph1 increases the stringency at which recombination occurs (Dubcovsky et al., 1995; Luo et al., 1996). As a consequence, conclusions cannot be made from this data concerning the way divergence is scrutinized by Darmor-bzh or Yudal haploids.
In this study, we showed that variations in COs between nonhomologous chromosomes among B. napus haploids are closely paralleled by a significant difference in recombination between homologous chromosomes in two triploid ArAnC hybrids produced using Darmor-bzh or Yudal genotypes (Figure 6B). Using the same plants, Leflon et al. (2006) observed that the frequency of homoeologous recombination was slightly but proportionally higher in triploid hybrids produced with Darmor-bzh (5.5%) compared with Yudal (3.3%).
In a set of very similar experiments, Dubcovsky et al. (1995) and Luo et al. (1996) observed that recombination between wheat chromosomes 1A and 5A and Triticum monococcum chromosomes 1Am and 5 Am was severely repressed when Ph1 is active and concluded that this reduction was due to the highly discriminatory activity of Ph1 that could recognize minor differences between the A and Am related chromosomes. The same interpretation does not appear to apply in our case because the Ar and An genomes of B. rapa and B. napus, respectively, have not been diverging for very long (presumably <10,000 years) and are therefore very closely related (Rana et al., 2004; Parkin et al., 1995; Parkin and Lydiate, 1997). Likewise, it is very unlikely that changes in the frequency of homoeologous recombination are responsible for the difference in the number of recombinant A chromosomes between the two triploid ArAnC hybrids (Zwierzykowski et al., 1999). In both hybrids, only 8 to 9% of PMCs had trivalents or tetravalents, and no less than 70% of PMCs showed nine univalents and 10 bivalents that mainly consisted of pairs of A chromosomes (Leflon et al., 2006). Instead, it is likely that the significant difference in recombination detected between the two ArAnC hybrids is due to variations in CO formation between homologous chromosomes. Given that PrBn, and its interacting partners, determine the number of COs that are formed between nonhomologous chromosomes in B. napus haploids, we hypothesize that these loci also affect the number of COs between homologous chromosomes in the two triploid ArAnC hybrids.
Does this mean that the overall capacity for forming CO is different between Darmor-bzh and Yudal? At least two observations do not support this hypothesis. One is that we did not observe a clear genotypic effect on the level of homologous recombination between the two tetraploid hybrids (Figure 6A). The second issue is that the relative proportion of rearrangements that entailed one, two, or three breakpoints was strikingly similar in the progenies of Darmor-bzh and Yudal haploids (Figure 5B), suggesting that only first cross-over formation between nonhomologous chromosomes differed between the two haploid genotypes. These two results are unexpected if the two genotypes only differ in the number of COs they are able to form and clearly demonstrate that CO variations depend on plant karyotype (for other relevant examples, see Nicolas et al., 2008).
The underlying cause of this karyotypic effect on CO variation is unknown, but at least two hypotheses deserve further investigation. First, a dosage effect by genes on the B. napus C genome, notably PrBn, can be postulated; one copy of the gene(s) carried by the Yudal C genome would lead to fewer COs than one copy of the gene(s) carried by the Darmor-bzh C genome, but two copies of gene(s) carried by the C genomes would confer the same numbers of COs irrespective of the genotype (this may explain why there is no difference in the loss of markers from the euploid parent of each genotype). Dosage effects were shown to be commonplace among the genes regulating chromosome pairing and recombination in polyploid species (Naranjo and Palla, 1982; Ceoloni et al., 1986; Jauhar, 1975; Moore, 2002). Second, the presence of chromosomes that remain unsynapsed during meiosis (in haploids and triploids, but less frequently in tetraploids) may trigger changes in the progression/completion of important steps of meiosis that are genotype dependant (see Discussion in Carlton et al., 2006; Martinez-Perez and Moore, 2008). Clearly, these mechanisms are speculative and should be tested directly by further appropriately designed experiments.
Meiosis impacts the evolution of polyploid species by (1) contributing to fertility, (2) enabling sexual propagation, and (3) generating, through meiotic errors, large-scale chromosomal variation upon which genetic drift and/or selection can act (Leitch and Leitch, 2008). Overall, our results clearly show that PrBn, and the genes it interacts with, regulate the number of COs between nonhomologous (usually homoeologous) chromosomes without changing their distribution throughout the genome during meiosis in B. napus haploids. Our findings also suggest that these loci may confer similar mechanisms, between homologous chromosomes, in triploid hybrids but not in tetraploid hybrids. Although new genetic and cytological experiments are required to decipher the causes of map length heterogeneities between triploid and tetraploid hybrids, it appears that the role played by PrBn in suppressing recombination depends on a plant's chromosomal composition. Given that cloning PrBn will certainly require a large amount of tedious and time-consuming work, the most effective way forward to gain further insights into the mode of action of this locus could be via an accurate and comparative cytological description of the different meiotic stages that occur during meiosis in haploids with different numbers of univalents at MI.
METHODS
Plant Material
The production of haploid plants (AC; 19 chromosomes) from Brassica napus cv Darmor-bzh, a dwarf winter B. napus cultivar (ADCD), and B. napus cv Yudal, a spring korean line (AYCY), was described by Jenczewski et al. (2003). The production and selection of haploid × euploid progenies were described by Nicolas et al. (2007). Briefly, haploid plants were used as female parents with the male euploids providing haploid pollen (Figure 1). A first F1 population was produced by crossing five Darmor-bzh haploid plants with one Yudal euploid plant; in this study, we refer to it as “the progeny of Darmor-bzh haploids.” A second F1 population was obtained by crossing nine Yudal haploid plants with one Darmor-bzh euploid plant; in this study, we refer to it as “the progeny of Yudal haploids.” Genetic variation was not expected among the different haploid plants that were used to produced each of the two progenies because these haploids were isolated from almost homozygous euploid lines that were recovered after more than six (Darmor-bzh) and 15 generations (Yudal) of single seed descent, respectively. A total of 117 and 103 plants carrying 38 chromosomes, which were derived from unreduced gametes produced by the haploid plants (see Nicolas et al., 2007), were sorted in the progenies of Darmor-bzh and Yudal haploids, respectively. These two progeny populations were used to compare the frequency and distributions of chromosomal rearrangements generated during meiosis of haploid B. napus (Figure 1).
Two digenomic triploid hybrids (ARADCD and ARAYCY) were produced by crossing one single Brassica rapa cv Chicon, C1.3 (ARAR; 2n = 20) plant with Darmor (ADADCDCD; 2n = 38) and Yudal (AYAYCYCY; 2n = 38), respectively (Figure 1). One single ARADCD and one single ARAYCY hybrid were then backcrossed as female to Darmor and Yudal, respectively, and two progeny populations of 116 and 112 plants were generated. The same B. rapa plant, C1.3, was crossed, as male, with a Brassica oleracea doubled haploid line, RC (COCO; 2n = 18) (Figure 1). The resulting interspecific hybrid was colchicine doubled to produce one resynthesized B. napus (ARARCOCO). The resynthesized B. napus was then crossed as female to Darmor and Yudal; two digenomic tetraploid hybrids (ARADCDCO and ARAYCYCO) were obtained. One single ARADCDCO and one single ARAYCYCO hybrid were backcrossed as female to Darmor and Yudal, respectively, and two progenies of 116 plants were generated. The two progenies of digenomic triploid hybrids and the two progenies of digenomic tetraploid hybrids were used to compare the rate of homologous recombination on one pair of A chromosomes (linkage group A7).
FISH with a Genome-Specific BAC Clone
FISH analyses were performed using the BAC clone BoB014O06 as described by Leflon et al. (2006) and Nicolas et al. (2007). Fluorescence images were captured using a CoolSnap HQ camera (Photometrics) on an Axioplan 2 microscope (Zeiss) and analyzed using MetaVue (Universal Imaging).
Molecular Analysis
Genomic DNA was extracted from young leaves according to Lombard and Delourme (2001). PCR and electrophoresis was performed using the same protocols as described by Nicolas et al. (2007) for Random Amplified Polymorphic DNA, Single Sequence Repeat, and specific markers and Leflon et al. (2007) for Amplified Fragment Length Polymorphism markers.
Detection of Chromosomal Rearrangements in the Haploid Progenies with Molecular Markers
Chromosomal rearrangements generated during meiosis of haploid B. napus were detected and quantified as described by Nicolas et al. (2007). Most rearrangements were detected by identifying F1 plants in the progenies of Darmor-bzh and Yudal haploids that lacked the haploid parent alleles at a set of loci. Chromosomal rearrangements were then scored in three different ways: (1) by scoring the presence/absence of haploid parent alleles at every locus independently from the others, (2) by considering the concurrent loss of linked loci as a consequence of a single rearrangement, thereby quantifying the number of chromosomal rearrangements, and (3) by considering the number of breakpoints on linkage groups, which occurred when one marker was present while the marker located on the other side of the same interval was absent. Duplications were scrutinized for a subset of loci by combining a quantitative estimation of HP allele peak area (electrophoresis with sequencer ABI Prism 3130xl analyzed with GeneMapper v3.7; Applied Biosystems) with maximum likelihood analyses (Nicolas et al., 2007).
We used the chromosome/linkage group nomenclature that was recently proposed as a reference by the Multinational Brassica Genome Project Steering Committee where B. napus N1-N19 nomenclature is replaced by A1-A10 and C1-C9 designations (http://www.brassica.info/information/lg_assigments.htm; see Delourme et al. [2006] for correspondence to former DY maps).
All the markers used in this study were from published maps (Foisset et al., 1996; Lombard and Delourme, 2001; Delourme et al., 2006; Liu et al., 2006) (see Supplemental Figure 1 online). We used 86 codominant markers that cover 62% of the genetic reference map with an average density of one marker per 17 cM; using these markers, the rate of chromosomal rearrangements in the two progenies could be compared at exactly the same physical position and therefore any bias for chromosomal rearrangements along chromosomes due to local variations, in particular for COs, was avoided. These data set were completed using dominant markers so that a total of 150 and 141 loci were examined in progenies of Darmor-bzh and Yudal haploids, respectively. By combining dominant and codominant markers, 75% of the genetic reference map in the two progenies was covered with an average density of one marker per 12 cM. With the exception of three linkage groups (A10, C5, and C9) that were not considered for comparative analyses, up to 50% of all linkage groups was covered in the two progenies and most had similar genome coverage between the haploid progenies (see Supplemental Figure 1 online).
Analysis of Genetic Recombination in the Two Triploid and Two Tetraploid Hybrids
The different segregating populations were genotyped with 11 polymorphic markers positioned along linkage group R7/A7 (A genome). This linkage group was chosen because it has been the focus of considerable interest due to the presence of two resistance genes to the fungus Leptosphaeria maculans (Leflon et al., 2007) and therefore includes the highest number of polymorphic molecular markers that can be used to compare recombination between the different genetic backgrounds.
Statistical Analyses
All statistical analyses were performed using R version 2.2.1 software (R Development Core Team, 2006). χ2 or exact fisher tests were computed using the function chisq.test or fisher.test, respectively, while Student's tests were performed using the t.test function (package stats).
Estimation of the Number of Rearrangements Expected Given the Number of Chromosomes Associated at MI
The data described by Jenczewski et al. (2003) were used to estimate the total number of rearrangements that would be observed in the Darmor-bzh and Yudal haploids progenies if (1) every chromosome association observed at MI triggered rearrangements that segregated to half the gametes (Nicolas et al., 2007) and (2) there was no gametic/zygotic selection against rearranged chromosomes. We first estimated the probabilities of detecting X rearrangements in the progeny of a haploid given that there were Y bivalents at MI as the binomial distribution probabilities: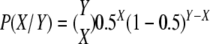 (Nicolas et al., 2007). Then, all P(X/Y) probabilities were multiplied by the probability that a PMC showed Y bivalents at MI, inferred from the data summarized by Jenczewski et al. (2003): P(X) = P(X/Y) × P(Y). Average numbers of rearrangements per plant were finally estimated by summing P(X)*X for all X, which directly gave the expected total number of rearrangements in the two progenies.
(Nicolas et al., 2007). Then, all P(X/Y) probabilities were multiplied by the probability that a PMC showed Y bivalents at MI, inferred from the data summarized by Jenczewski et al. (2003): P(X) = P(X/Y) × P(Y). Average numbers of rearrangements per plant were finally estimated by summing P(X)*X for all X, which directly gave the expected total number of rearrangements in the two progenies.
Comparison of the Observed Rearrangement Distributions among Linkage Groups with Those Expected if Rearrangements Occurred at Random
For each genotype, randomization tests were performed on the rearrangement repartition. The null hypothesis was that the rearrangements occurred at random in each haploid, with equal probabilities for all linkage groups (LGs). Random sample sets (n = 1000) were simulated by setting the same number of rearrangements for each haploid but drawing their repartition among the LGs at random. The LG rearrangement frequencies were calculated for each simulated sample set. Then, the observed quantiles of these frequencies were compared with their distribution over the simulated sample sets to determine whether the observed LG rearrangement frequencies were compatible with a random distribution.
Comparison of Homologous Recombination Rates
The genetic maps obtained in BC progenies were compared using Morton's likelihood ratio test for heterogeneity of the fraction of cross-overs among different families (Morton, 1956), extended to multiple linked loci (Lander et al., 1987):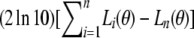 , where Li(θ) and Lp(θ) are the log-likelihood values for linkage groups with the same set of adjacent markers in population i and for data pooled from all n populations. This statistic is asymptotically distributed as a χ2 with n − 1 degrees of freedom, where n is the number of compared populations. As several markers were studied simultaneously, significance levels had to be reevaluated, and we used the approximate relationship 1 − (1 − α′)m≅α proposed by Lander and Botstein (1989), where m is the number of markers.
, where Li(θ) and Lp(θ) are the log-likelihood values for linkage groups with the same set of adjacent markers in population i and for data pooled from all n populations. This statistic is asymptotically distributed as a χ2 with n − 1 degrees of freedom, where n is the number of compared populations. As several markers were studied simultaneously, significance levels had to be reevaluated, and we used the approximate relationship 1 − (1 − α′)m≅α proposed by Lander and Botstein (1989), where m is the number of markers.
The heterogeneity of cross-over rates among populations was also assessed separately for every interval. χ2 tests were performed for every pair of loci, and a false discovery rate threshold (Benjamini and Hochberg, 1995) was applied to account for multiple comparisons.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Localization of the Markers Used in This Study and Coverage of the Frame Map Established for Each Linkage Group by Delourme et al. (2006).
Supplemental Figure 2. Resampling-Based Estimation of the Relationship between Genome Coverage and the Number of Rearranged Chromosomes Detected in the Progenies of Darmor-bzh or Yudal Haploids.
Supplemental Data Set 1. Markers Used in This Study.
Supplementary Material
Acknowledgments
We thank Jean-Claude Letanneur (Institut National de la Recherche Agronomique, Unité Mixte de Recherche 118, Amélioration des Plantes et de Biotechnologies Végétales, France) for his significant contribution to the production of plant material as well as Tomasz Ksiazczyk and Jolanta Maluszynska (University of Silesia, Poland) for providing a stab of BoB014O06. We also thank Harry Belcram, Karine Budin, and Boulos Chalhoub (Unité de Recherche en Génomique Végétale, Institut National de la Recherche Agronomique, Evry, France) for developing and screening Positional Functional Markers markers and Genoplante for funding their development. Mathilde Grelon, Christine Mézard, and Marta Cifuentes (Institut National de la Recherche Agronomique, Versailles, France) and Karine Alix (Unité Mixte de Recherche de Génétique Végétale du Moulon, France) are gratefully acknowledged for their critical reading and valuable comments on the manuscript, and Leigh Gebbie is acknowledged for English corrections. We also thank three anonymous reviewers for helpful comments on previous versions of the manuscript. Stéphane Nicolas was supported by a Centre Technique Interprofessionnel des Oléagineux Métropolitains and Institut National de Recherche Agronomique - Génétique et Amélioration des Plantes fellowship. This work was carried out with the financial support of the ANR- Agence Nationale de la Recherche–The French National Research Agency under the Programme Biodiversité project ANR-05-BDIV-015, Effet de la polyploïdie sur la biodiversité et l'évolution du génome des plantes.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eric Jenczewski (ejenczewski@versailles.inra.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Able, J.A., and Langridge, P. (2006). Wild sex in the grasses. Trends Plant Sci. 11 261–263. [DOI] [PubMed] [Google Scholar]
- Al-Kaff, N., Knight, E., Bertin, I., Foote, T., Hart, N., Griffiths, S., and Moore, G. (2007). Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: With deletion mutants and expression profiling. Ann. Bot. (Lond.) 101 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia, T., and Röbbelen, G. (1986). Meiotic pairing in haploids and amphihaploids of spontaneous versus synthetic origin in rape, Brassica napus L. Can. J. Genet. Cytol. 28 330–334. [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. [Ser A] 57 289–300. [Google Scholar]
- Carlton, P.M., Farruggio, A.P., and Dernburg, A.F. (2006). A link between meiotic prophase progression and crossover control. PLoS Genet. 2 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceoloni, C., Strauss, I., and Feldman, M. (1986). Effect of different doses of group-2 chromosomes on homoeologous pairing in intergeneric wheat hybrids. Can. J. Genet. Cytol. 28 240–246. [Google Scholar]
- Chen, Z.J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L. (2005). The advantages and disavantages of being polyploid. Nat. Rev. Genet. 6 836–846. [DOI] [PubMed] [Google Scholar]
- Corredor, E., Lukaszewski, A.J., Pachon, P., Allen, D.C., and Naranjo, T. (2007). Terminal regions of wheat chromosomes select their pairing partners in meiosis. Genetics 177 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, L., et al. (2006). Widespread genome duplications throughout the history of flowering plants. Genome Res. 16 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt, S., Maere, S., and Van de Peer, Y. (2005). Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20 591–597. [DOI] [PubMed] [Google Scholar]
- Delourme, R., et al. (2006). Genetic control of oil content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 113 1331–1345. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J., Luo, M., and Dvorak, J. (1995). Differentiation between homoeologous chromosomes 1A of wheat and 1Am of Triticum monococcum and its recognition by the wheat Ph1 locus. Proc. Natl. Acad. Sci. USA 92 6645–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. (1993). Cytogenetic activity and mode of action of the pairing homoeologous (Ph1) gene of wheat. Plant Breed. 33 894–897. [Google Scholar]
- Foisset, N., Delourme, R., Barret, P., Hubert, N., Landry, B.S., and Renard, M. (1996). Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled haploid progeny. Theor. Appl. Genet. 93 1017–1025. [DOI] [PubMed] [Google Scholar]
- Gaeta, R.T., Pires, J.C., Iniguez-Luy, F., Leon, E., and Osborn, T.C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, S., Sharp, R., Foote, T.N., Bertin, I., Wanous, M., Reader, S., Colas, I., and Moore, G. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439 749–752. [DOI] [PubMed] [Google Scholar]
- Inaba, R., and Nishio, T. (2002). Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor. Appl. Genet. 105 1159–1165. [DOI] [PubMed] [Google Scholar]
- Jauhar, P.P. (1975). Genetic control of diploid-like meiosis in hexaploid tall fescue. Nature 254 595–597. [DOI] [PubMed] [Google Scholar]
- Jauhar, P.P., Riera-Lizarazu, O., Dewey, W.G., Gill, B.S., Crane, C.F., and Bennett, J.H. (1991). Chromosome pairing relationships among the A, B, and D genomes of bread wheat. Theor. Appl. Genet. 82 441–449. [DOI] [PubMed] [Google Scholar]
- Jenczewski, E., and Alix, K. (2004). From diploids to allopolyploids: The emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23 21–45. [Google Scholar]
- Jenczewski, E., Eber, F., Grimaud, A., Huet, S., Lucas, M.O., Monod, H., and Chevre, A.M. (2003). PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, N.E., et al. (1996). Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat. Genet. 14 400–405. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., and Botstein, D. (1989). Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E., and Newburg, L. (1987). MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1 174–181. [DOI] [PubMed] [Google Scholar]
- Leflon, M., Brun, H., Eber, F., Delourme, R., Lucas, M., Vallée, P., Ermel, M., Balesdent, M., and Chèvre, A. (2007). Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus. Theor. Appl. Genet. 115 897–906. [DOI] [PubMed] [Google Scholar]
- Leflon, M., Eber, F., Letanneur, J.C., Chelysheva, L., Coriton, O., Huteau, V., Ryder, C.D., Barker, G., Jenczewski, E., and Chèvre, A.M. (2006). Pairing and recombination at meiosis of Brassica rapa (AA) × Brassica napus (AACC) hybrids. Theor. Appl. Genet. 113 1467–1480. [DOI] [PubMed] [Google Scholar]
- Leitch, A.R., and Leitch, I.J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320 481–483. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Adamczyk, K., Manzanares-Dauleux, M., Eber, F., Lucas, M.-O., Delourme, R., Chevre, A.M., and Jenczewski, E. (2006). Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard, V., and Delourme, R. (2001). A consensus linkage map for rapeseed (Brassica napus L.): Construction and integration of three individual maps from DH populations. Theor. Appl. Genet. 103 491–507. [Google Scholar]
- Lukens, L.N., Pires, J.C., Leon, E., Vogelzang, R., Oslach, L., and Osborn, T. (2006). Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M.C., Dubcovsky, J., and Dvorak, J. (1996). Recognition of homeology by the wheat Ph1 locus. Genetics 144 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M.A., Koch, M.A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, M., Cuadrado, C., Laurie, D.A., and Romero, C. (2005). Synaptic behaviour of hexaploid wheat haploids with different effectiveness of the diploidizing mechanism. Cytogenet. Genome Res. 109 210–214. [DOI] [PubMed] [Google Scholar]
- Martinez, M., Cuñado, N., Carcelén, N., and Romero, C. (2001). The Ph1 and Ph2 loci play different roles in the synaptic behaviour of hexaploid wheat Triticum aestivum. Theor. Appl. Genet. 103 398–405. [Google Scholar]
- Martinez-Perez, E., and Moore, G. (2008). To check or not to check? The application of meiotic studies to plant breeding. Curr. Opin. Plant Biol. 11 222–227. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez, E., Shaw, P., Aragon-Alcaide, L., and Moore, G. (2003). Chromosomes form into seven groups in hexaploid and tetraploid wheat as a prelude to meiosis. Plant J. 36 21–29. [DOI] [PubMed] [Google Scholar]
- Masterson, J. (1994). Stomatal size in fossil plants - Evidence for polyploidy in majority of angiosperms. Science 264 421–424. [DOI] [PubMed] [Google Scholar]
- Mello-Sampayo, T. (1971). Genetic regulation of meiotic chromosome pairing by chromosome 3D of Triticum aestivum. Nature New Biol. 230 22–23. [DOI] [PubMed] [Google Scholar]
- Mikhailova, E.I., Naranjo, T., Shepherd, K., Wennekes-van Eden, J., Heyting, C., and de Jong, J.H. (1998). The effect of the wheat Ph1 locus on chromatin organisation and meiotic chromosome pairing analysed by genome painting. Chromosoma 107 339–350. [DOI] [PubMed] [Google Scholar]
- Moore, G. (2002). Meiosis in allopolyploids–The importance of ‘Teflon’chromosomes. Trends Genet. 18 456–463. [DOI] [PubMed] [Google Scholar]
- Morinaga, T., and Fukushima, E. (1933). Karyological studies on a spontaneous haploid mutant of Brassica napella. Cytologia (Tokyo) 4 457–460. [Google Scholar]
- Morton, N.E. (1956). The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. Am. J. Hum. Genet. 8 80–96. [PMC free article] [PubMed] [Google Scholar]
- Naranjo, T., and Palla, O. (1982). Genetic control of meiotic pairing in rye. Heredity 48 57–62. [Google Scholar]
- Nicolas, S.D., Leflon, M., Liu, Z., Eber, F., Chelysheva, L., Coriton, O., Chèvre, A.M., and Jenczewski, E. (2008). Chromosome 'speed dating' during meiosis of polyploid Brassica hybrids and haploids. Cytogenet. Genome Res. 120 331–338. [DOI] [PubMed] [Google Scholar]
- Nicolas, S.D., et al. (2007). Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175 487–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, G., and Hagberg, A. (1955). Investigations on haploid rape. Hereditas 41 227–237. [Google Scholar]
- Osborn, T.C., Butrulle, D.V., Sharpe, A.G., Pickering, K.J., Parkin, I.A.P., Parker, J.S., and Lydiate, D.J. (2003). Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana, J. (1985). Most of the homeologous pairing at metaphase in wheat-rye hybrids is not chiasmatic. Genetics 111 917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S.P. (2007). The evolutionary consequences of polyploidy. Cell 131 452–462. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D., Shields, C.R., Cohen, D.B., and Orton, T.J. (1983). Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor. Appl. Genet. 65 181–189. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A., and Lydiate, D.J. (1997). Conserved patterns of chromosome pairing and recombination in Brassica napus crosses. Genome 40 496–504. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A.P., Gulden, S.M., Sharpe, A.G., Lukens, L., Trick, M., Osborn, T.C., and Lydiate, D.J. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin, I.A.P., Sharpe, A.G., Keith, D.J., and Lydiate, D.J. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38 1122–1131. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A.P., Sharpe, A.G., and Lydiate, D.J. (2003). Patterns of genome duplication within the Brassica napus genome. Genome 46 291–303. [DOI] [PubMed] [Google Scholar]
- Piquemal, J., Cinquin, E., Couton, F., Rondeau, C., Seignoret, E., Doucet, I., Perret, D., Villeger, M.J., Vincourt, P., and Blanchard, P. (2005). Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor. Appl. Genet. 111 1514–1523. [DOI] [PubMed] [Google Scholar]
- Pouilly, N., Delourme, R., Alix, K., and Jenczewski, E. (2008). Repetitive sequence-derived markers tag centromeres and telomeres and provide insights into chromosome evolution in Brassica napus. Chromosome Res. 16 683–700. [DOI] [PubMed] [Google Scholar]
- Prieto, P., Moore, G., and Reader, S. (2005). Control of conformation changes associated with homologue recognition during meiosis. Theor. Appl. Genet. 111 505–510. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2006). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Ramsey, J., and Schemske, D.W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29 467–501. [Google Scholar]
- Ramsey, J., and Schemske, D.W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33 589–639. [Google Scholar]
- Rana, D., Boogaart, T., O'Neill, C.M., Hynes, L., Bent, E., Macpherson, L., Park, J.Y., Lim, Y.P., and Bancroft, I. (2004). Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40 725–733. [DOI] [PubMed] [Google Scholar]
- Renard, M., and Dosba, F. (1980). Etude de l'haploidie chez le colza (Brassica napus L. var oleifera Metzger). Ann. Amel. Pl. 30 191–209. [Google Scholar]
- Rieseberg, L.H., and Willis, J.H. (2007). Plant speciation. Science 317 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, R., and Chapman, V. (1958). Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182 713–715. [Google Scholar]
- Sears, E.R., and Okamoto, M. (1958). Intergenomic chromosome relationships in hexaploid wheat. In Proceedings of the Tenth International Congress of Genetics, Montreal, Canada. pp. 258–259.
- Sharpe, A.G., Parkin, I.A.P., Keith, D.J., and Lydiate, D.J. (1995). Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38 1112–1121. [DOI] [PubMed] [Google Scholar]
- Sutton, T., Whitford, R., Baumann, U., Dong, C., Able, J.A., and Langridge, P. (2003). The Ph2 pairing homoeologous locus of wheat (Triticum aestivum): Identification of candidate meiotic genes using a comparative genetics approach. Plant J. 36 443–456. [DOI] [PubMed] [Google Scholar]
- Tai, W., and Ikonen, H. (1988). Incomplete bivalent pairing in dihaploids of Brassica napus L. Genome 30 450–457. [Google Scholar]
- Town, C.D., et al. (2006). Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U, N. (1935). Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap. J. Bot. 7 389–452. [Google Scholar]
- Udall, J.A., Quijada, P.A., and Osborn, T.C. (2005). Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics 169 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall, J.A., and Wendel, J.F. (2006). Polyploidy and crop improvement. Crop. Sci. 46 3–14. [Google Scholar]
- Upadhya, M.D., and Swaminathan, M.S. (1967). Mechanism regulating chromosome pairing in Triticum. Biol. Zentralbl. Suppl. 86 239–255.
- Yang, T.-J., et al. (2006). Sequence-Level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Pickering, R., and Murray, B. (1999). Direct measurement of recombination frequency in interspecific hybrids between Hordeum vulgare and H. bulbosum using genomic in situ hybridization. Heridity 83 304–309. [DOI] [PubMed] [Google Scholar]
- Zwierzykowski, Z., Lukaszewski, A.J., Naganowska, B., and Lesniewska, A. (1999). The pattern of homoeologous recombination in triploid hybrids of Lolium multiflorum with Festuca pratensis. Genome 42 720–726. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.