Abstract
We investigated the role of the ubiquitin proteasome system (UPS), which allows proteins to be selectively degraded, during gametophyte development in Arabidopsis thaliana. Three mutant alleles altering the UPS were isolated in the Wassilewskija (Ws) accession: they affect the Regulatory Particle 5a (RPT5a) gene, which (along with RPT5b) encodes one of the six AAA-ATPases of the proteasome regulatory particle. In the heterozygous state, all three mutant alleles displayed 50% pollen lethality, suggesting that RPT5a is essential for male gametophyte development. However, a fourth mutant in the Columbia (Col) accession did not display such a phenotype because the RPT5b Col allele complements the rpt5a defect in the male gametophyte, whereas the RPT5b Ws allele does not. Double rpt5a rpt5b mutants showed a complete male and female gametophyte lethal phenotype in a Col background, indicating that RPT5 subunits are essential for both gametophytic phases. Mitotic divisions were affected in double mutant gametophytes correlating with an absence of the proteasome-dependent cyclinA3 degradation. Finally, we show that RPT5b expression is highly increased when proteasome functioning is defective, allowing complementation of the rpt5a mutation. In conclusion, RPT5 subunits are not only essential for both male and female gametophyte development but also display accession-dependent redundancy and are crucial in cell cycle progression.
INTRODUCTION
In higher plants, the postmeiotic haploid phase takes place in male and female gametophytes that develop in male and female reproductive organs (Boavida et al., 2005a). Both gametophytes are highly organized structures that fulfill specific requirements to allow sexual reproduction. Male gametophyte development requires two successive mitoses, the first one being asymmetric, leading to one large vegetative cell in which the two sperm cells are enclosed. In Arabidopsis thaliana, both mitoses take place in stamens before the pollen grains are released. Following pollination, the male gametophyte lands on the female organ (pistil) and extends a pollen tube that allows the delivery of the two sperm cells into the female gametophyte (McCormick, 2004). The mature female gametophyte (ovule) is an organ with seven cells and eight nuclei that results from three rounds of mitoses. Following maturation, the ovule stays embedded within sporophytic tissues. Eventually, the content of the two sperm cells is released into the female embryo sac, and double fertilization occurs with the two sperm nuclei migrating toward the egg and the central cells (Yadegari and Drews, 2004). After the respective karyogamies, the development of the 2n embryo and the 3n endosperm starts and a new sporophytic phase begins (Boavida et al., 2005b). Male and female gametophyte development differ considerably, but at the same time share the same fundamental hallmark of being haploid organs: it is therefore logical that they might require the same basal machinery and share a number of common regulators.
One important way of regulating cellular mechanisms is through the elimination of specific proteins: either abnormal (misfolded or damaged) proteins or regulatory proteins that are no longer needed or whose functions have to be turned off. In plants, as in yeast and animals, this regulation pathway acts via the ubiquitin/26S proteasome system (UPS) (Hershko and Ciechanover, 1998; Smalle and Vierstra, 2004). So far, the plant UPS system has been shown to be highly evolutionarily conserved with its yeast homolog. However, the number of genes encoding its components has increased through duplication, and the plant UPS includes a much higher number of genes (>1300 in Arabidopsis) than the yeast UPS (148) (Vierstra, 2003). It is therefore likely that this increase has been accompanied by redundancies and/or diversifications among genes. In Arabidopsis, a growing number of proteins that are targeted by the UPS have been characterized recently, and the UPS has been involved in many areas of plant development (Smalle and Vierstra, 2004), such as the cell cycle (Genschik et al., 1998), hormone regulation, including auxin, jasmonate, abscisic acid, and gibberellins (Lopez-Molina et al., 2001; Chini et al., 2007; Dreher and Callis, 2007; Thines et al., 2007), or response to light (Monte et al., 2007).
The UPS involves the specific attachment of a chain of ubiquitin, a highly conserved 76 amino acid protein, to the protein target by three sets of enzymes: E1, E2, and E3 (Hershko and Ciechanover, 1998). Once labeled, the target is recognized by the 26S proteasome, a 2-MD ATP-dependent enzymatic complex, whose composition in plants is highly similar to its yeast homolog. This complex can be subdivided into a 20S Core Protease (CP), which is a hollow cylinder where proteolysis takes place, and a 19S Regulatory Particle (RP) that can interact with either or both of the 20S CP extremities (Walz et al., 1998). The 19S RP is in charge of recognizing the target, unfolding it, opening the gate to the 20S chamber, and feeding in the target for its subsequent degradation. The 19S RP is itself composed of two subcomplexes, a lid and a base (Glickman et al., 1998). The base interacts with the 20S CP and is composed of six AAA-ATPases named RPT1 to RPT6 (RPT stands for Regulatory Particle Triple A ATPase) and three non-ATPase subunits (RPN1, 2, and 10, where RPN stands for RP Non-ATPase). The lid is composed of eight other RPN proteins. Consistent with the increase in the number of genes involved in the plant UPS, most of the RP subunits are encoded by two genes in Arabidopsis, including all the RPTs but RPT3 (Fu et al., 1999; Smalle and Vierstra, 2004). The proteasome ATPase function is provided by the six RPT subunits that assemble into a hexameric ring (Fu et al., 2001; Benaroudj et al., 2003). It has been shown in yeast that the six RPT AAA-ATPases perform different functions, suggesting that each one is involved in recognizing/interacting with specific subsets of proteins. All RPT subunits were shown to be essential for yeast development (Rubin et al., 1998). Interestingly, it was shown that with the exception of Sc rpt2, yeast RPT loss-of-function mutants could be restored by expressing their Arabidopsis RPT counterparts, suggesting that this specialization is evolutionarily conserved (Fu et al., 2001).
In Arabidopsis, several mutants affecting 19S RP subunits have been characterized. Several RPN subunit mutants were shown to display variable phenotypes with pleiotropic developmental defects. rpn10 and rpn12a display hormone response defects, mainly to abscisic acid and cytokinins, respectively (Smalle et al., 2002, 2003). As for the roles of AAA-ATPases, mutants have only been described in RPT2a (Ueda et al., 2004) and RPT5a and RPT5b genes (Cho et al., 2006; Huang et al., 2006). The rpt2a mutation was found to be the cause of the halted root phenotype, resulting from an inability of the plant to maintain its pool of stem cells. As a general feature, proteasome subunit mutant phenotypes were linked to an inability to degrade key developmental regulators. For example, the rpn8a mutation was shown to enhance adaxialization in an asymetric leaves2 (as2) background. A similar enhancement was observed when an as2 mutant was combined with several 19S RP subunits or 20S CP subunit mutants, suggesting that the whole 26S complex is involved in degrading a key, although uncharacterized, regulator of leaf polarity formation (Huang et al., 2006).
Several lines of evidence suggest that gametophyte development requires 26S proteasome function at least to control cell cycles: several male and female gametophytic mutants have been described that impair the anaphase promoting complex APC/C, an E3-ligase complex that targets mitotic cyclins for degradation during exit from mitosis (Capron et al., 2003b; Kwee and Sundaresan, 2003; Perez-Perez et al., 2008). Moreover, addition of chemical proteasome inhibitors severely impairs later stages of pollen development, including pollen tube germination (Speranza et al., 2001; Sheng et al., 2006). More recently, defects in ubiquitin-specific protease UPB3/UBP4 was shown to be essential for pollen development, illustrating another aspect of the involvement of the 26S UPS in male gametophyte development (Doelling et al., 2007). Putative involvement of the 26S UPS in gametophyte development is further supported by transcriptomic approaches that highlight the importance of UPS factors in male gametophyte transcriptomes (Becker et al., 2003; Honys and Twell, 2003, 2004; Pina et al., 2005). Also, a large-scale analysis of transposon-tagged mutants affected in female gametophyte development revealed that UPS factors are important candidates for proper embryo sac development (Pagnussat et al., 2005).
As a consequence, it is very likely that mutations affecting essential components of the 26S proteasome would result in gametophytic mutant phenotypes. However, to our knowledge, none has been described so far. In most cases, only single proteasome subunit mutants have been assessed, and it is likely that, in these, the proteasome still functions through the redundant wild-type subunit. The most severe phenotype for a 19S subunit mutant has been described for rpn1a (Brukhin et al., 2005). Complete loss of function of the RPN1a subunit resulted in an embryo-lethality phenotype with development stopping at the globular stage. Interestingly, the rpn1b mutant did not display any phenotype nor did it enhance the rpn1a phenotype. Because male and female gametophytes carrying rpn1a or rpn1a rpn1b mutations could develop, it was concluded that the 26S proteasome might still be functional during gametogenesis even if a new pool of RPN1 subunits could no longer be provided. It could be that RPN1 is stable enough to maintain a pool sufficient to go through the development of both gametophytes. Furthermore, because the RPN1b gene did not complement the rpn1a mutant phenotype in embryos, these data also suggested that redundancy was far from being complete in duplicated RPT or RPN genes presumably because each copy may have evolved different expression patterns or idiosyncratic protein specificities. In this case, expression seems to be the important factor as the RPN1b gene complemented the rpn1a mutation when expressed under the control of the RPN1a promoter (Brukhin et al., 2005).
To clarify the UPS role during gametophyte development, we looked for insertional mutants affecting UPS components, searching for T-DNA segregation bias that is characteristic of gametophytic mutants. We isolated three alleles affecting RPT5a that all displayed a severe male gametophyte development defect. In pollen carrying the mutation, development stopped at the second pollen mitosis stage and pollen eventually degenerated. We showed that this mutation did not rely on RPT5a status alone but that the pollen lethality phenotype depended on the status of the RPT5b gene. These data suggested that although the Columbia (Col) RPT5b allele compensated for the rpt5a default, its Wassilewskija (Ws) counterpart was a weak allele and did not complement the rpt5a mutation in pollen. We also found that a rpt5a rpt5b mutant displayed the same male gametophyte defect and that a similar defect occurred during female gametophyte development. Finally, we showed that these defects affected proteasome functions in male and female gametophytes and that Col RPT5b redundancy with RPT5a is achieved following the upregulation of RPT5b in the absence of RPT5a through a proteasome-sensing feedback loop.
RESULTS
The Versailles T-DNA insertion collection, which has been developed in the Ws accession, was screened for putative gametophytic mutants, in which the T-DNA insertions are potentially affecting genes that are involved in the UPS. The T-DNA harbors a neomycin phosphotransferase gene NPTII, which confers kanamycin resistance to the plant. The basis for such screening is that if a single T-DNA is inserted in a gene that is essential for male (or female) gametophyte development, a 1:1 ratio of Kanr:Kans plants is expected instead of a 3:1 ratio among the selfed progeny of a plant heterozygous for the T-DNA insertion allele (Bonhomme et al., 1998).
rpt5a Mutations in the Ws Accession Cause Abortion of Pollen Development after Pollen Mitosis I
Three putative gametophytic mutant lines were isolated in the Ws background, all three carrying insertions in the RPT5a gene (At3g05530) that in Arabidopsis encodes, with RPT5b (At1g09100), the RPT5 proteasome subunit (Fu et al., 1999). Two alleles, named rpt5a-1 and rpt5a-2, contained a T-DNA insertion 62 and 115 bp upstream, respectively, of the predicted start codon, whereas in the third mutant allele, rpt5a-3, the T-DNA was inserted 52 bp downstream of the stop codon (Figure 1A). For all three alleles, fragments flanking the T-DNA left and right borders were cloned and sequenced to check the insertion location, and DNA gel blot experiments were performed and showed that each mutant had a single locus T-DNA insertion (see Supplemental Figure 1 online). The linkage between the kanamycin resistance gene and the T-DNA insertion was also checked for each allele (see Methods). Afterwards, plant kanamycin resistance was used as a marker for these rpt5a mutations.
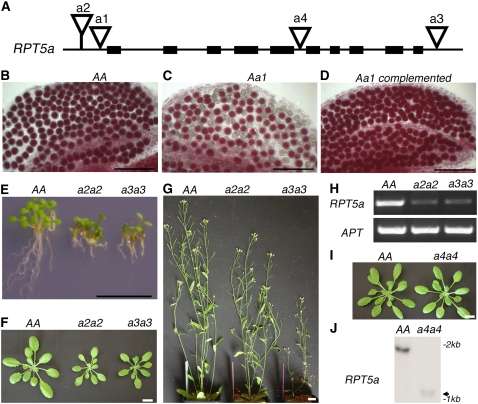
Figure 1.
rpt5a Mutant Phenotypes.
(A) Schematic representation of the RPT5a gene. Black boxes represent exons. T-DNA insertions in Ws accession (a1, a2, and a3 stand for rpt5a-1, rpt5a-2, and rpt5a-3, respectively) and in Col accession (a4 stands for rpt5a-4) are represented by open triangles.
(B) to (D) Mature pollen stained with Alexander's stain in anther locules from Ws wild-type AA plants (B), Aa1 heterozygous plants (C), or Aa1 heterozygous complemented with a RPT5a construct (D).
(E) Plantlet and root phenotype on 6-d-old wild-type Ws (AA) and homozygous a2a2 and a3a3 seedlings grown on vertical plates.
(F) and (G) Sporophytic phenotype on 20-d-old plants (F) and 40-d-old plants (G).
(H) RT-PCR amplification of RPT5a and APT mRNA extracted from 6-d-old seedlings. mRNAs were extracted from wild-type Ws plants (AA) and from homozygous a2a2 and a3a3 plants.
(I) Sporophytic phenotype on 20-d-old wild-type Col (AA) and a4a4 plants.
(J) RNA gel blot analysis of RNAs from wild-type Col (AA) and homozygous rpt5a-4/rpt5a-4 plantlets (a4a4) hybridized with a RPT5a full-length cDNA probe. The weak signal obtained for a4a4 mRNA is indicated by an arrow.
Bars = 100 μm in (B) to (D) and 1 cm in (E) to (G) and (I).
After selfing heterozygous rpt5a plants, the kanamycin segregation was scored for 10 independent sister lines, confirming the 1:1 ratio of Kanr:Kans for all three rpt5a alleles (Table 1). Male and female transmission rates were assessed by crossing rpt5a heterozygous plants to wild-type Ws plants and scoring percentages of kanamycin resistant plants in the progenies. Male and female transmission efficiencies (TEM and TEF, respectively) were calculated according to Howden et al. (1998) (Table 1). A 100% TE is expected for an allele that is normally transmitted. The female transmission efficiencies were found to be slightly or not affected (from 68 to 107%), but the male transmission efficiencies were strongly reduced for rpt5a-2 and rpt5a-3 (7 and 14%, respectively) and completely abolished for rpt5a-1. Therefore, these rpt5a mutations in the Ws accession trigger male gametophyte lethality.
Table 1.
Genetic Analysis of rpt5a Alleles
| Parent Genotype | Accession | FST Segregation in Selfing | % TEM | % TEF | rpt5a/ rpt5a Lines in Progeny |
|---|---|---|---|---|---|
| rpt5a-1/RPT5a | Ws | 1:1 | 0% (345) | 68% (647) | 0% (500) |
| rpt5a-2/RPT5a | Ws | 1:1 | 7% (326) | 107% (723) | 4.1% (1024) |
| rpt5a-3/RPT5a | Ws | 1:1 | 14% (324) | 96% (395) | 7.5% (1024) |
| rpt5a-4/RPT5a | Col | 3:1 | 79% (186) | 70% (183) | 20.3% (187) |
Flanking sequence tag (FST) segregation was determined either using the segregation ratio between Kanr and Kans seedlings (for rpt5a-1, rpt5a-2, and rpt5a-3) or by genotyping (rpt5a-4). Transmission efficiencies were calculated according to Howden et al. (1998). TE = number of progeny with T-DNA insertion/number of progeny without T-DNA insertion × 100. Total numbers of scored seedlings are indicated in parentheses.
Pollen viability was assessed by Alexander staining (Alexander, 1969) on rpt5a-1/RPT5a-1 plants, and 46% of the pollen grains were shown to be dead (n = 1517, Figure 1C). To determine at which stage the developmental defect occurred in rpt5a-1, the occurrence of pollen mitoses was checked following the staining of pollen nuclei with 4′,6-diamidino-2-phenylindole (DAPI) or propidium iodide. After meiosis, each microspore undergoes one asymetric mitosis (pollen mitosis I [PMI]) that generates a larger vegetative cell and a smaller generative cell (Figure 2B). After pollen mitosis II (PMII), the latter cell gives rise to the two sperm cells (Figure 2C). No differences could be found between Ws wild-type and heterozygous rpt5a-1 mutants up to PMI (Figures 2D and 2E). However, while all wild-type pollen grains proceeded to PMII, half of the rpt5a-1 pollen grains (44.7%, n = 170) remained blocked at a bicellular state (Figures 2F and 2G) before rapidly degenerating (Figure 2I). Alexander and propidium iodide staining performed on rpt5a-2/RPT5a and rpt5a-3/RPT5a plants confirmed the male gametophyte developmental defect as characterized in rpt5a-1/RPT5a (see Supplemental Figure 2 online).
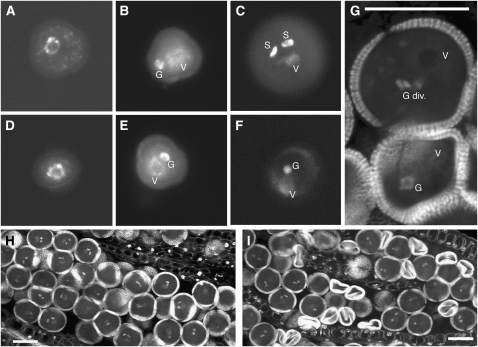
Figure 2.
Mitotic Progression of Wild-Type and rpt5a-1 Mutant Pollen.
Pollen grains from wild-type plants ([A] to [C] and [H]) and rpt5a-1/RPT5a plants ([D] to [G] and [I]) were stained with DAPI ([A] to [F]) or propidium iodide ([G] to [I]). Bars = 20 μm in (G) to (I).
(A) and (D) Uninucleate microspores before Pollen Mitosis I (PMI)
(B) and (E) Bicellular pollen grains (BCP) following PMI presenting one large vegetative nucleus (V) and one dense generative nucleus (G).
(C) and (F) Pollen grains at the tricellular pollen stage. PMII has occurred normally in wild-type grains (C) and in half of the grains from rpt5a-1/RPT5a plants, showing one large vegetative nucleus (V) and two dense spermatic nuclei (S), whereas the other half of the pollen grains from the mutant look binuclear (F).
(G) Optical section in a rpt5a-1/RPT5a anther showing a transition from BCP to tricellular pollen stage (top) with a dividing generative nucleus (G div.) next to a degenerating BCP grain (bottom).
(H) and (I) Optical section through a wild-type (H) and a rpt5a-1/RPT5a anther (I) at mature pollen stage, showing a uniform population of mature pollen grains (H) and a mix of mature and collapsed pollen grains (I).
For final confirmation that the pollen defect was caused by the RPT5a mutation, a 4500-bp genomic fragment encompassing RPT5a 1945 bp upstream and 99 bp downstream of the coding sequence was amplified and used to complement the rpt5a-1 mutant. Transgenic lines harboring the complementation construct showed a wild-type pollen phenotype in rpt5a-1 heterozygous plants (Figure 1D) and allowed recovery of homozygous rpt5a-1. Male transmission to the progeny was restored (rpt5a-1 male transmission efficiency to the progeny in a line homozygous for the complementation construct was 67%, n = 329). These data show that RPT5a is essential for male gametophyte development and that the rpt5a-1 to -3 mutations affect male gametophyte development before PMII, leading to pollen death.
rpt5a Mutants in the Ws Accession Are Also Affected in Sporophyte Development
As expected for a fully penetrant gametophytic mutation, no homozygous rpt5a-1 plants could be recovered among progeny resulting from the selfing of rpt5a-1/RPT5a plants (n = 500). Conversely, as a result of the residual mutant male transmission, a few plants that were homozygous for rpt5a-2 or rpt5a-3 were recovered (Table 1). Both rpt5a-2/rpt5a-2 and rpt5a-3/rpt5a-3 plants showed reduced development, including small roots compared with wild-type plants (Figure 1E), smaller rosettes (Figure 1F), as well as dwarfism and low fertility (Figure 1G). Expression studies confirmed that rpt5a-2 and rpt5a-3 are only partial mutations as some residual RPT5a mRNA could be detected by RT-PCR in both homozygous lines (Figure 1H). Finally, an allelism test was performed by crossing rpt5a-3/RPT5a plants as male parent to rpt5a-2/RPT5a plants. As expected given the rpt5a-3 reduced male transmission, 7.6% of dwarf plants (n = 339) were found in the progeny. The genotypes of the dwarf plants were all rpt5a-2/rpt5a-3, proving that the phenotype was caused by the mutations in RPT5a.
The rpt5a Male Gametophyte Mutant Phenotype Is Accession Dependent
A fourth line for which RPT5a is interrupted by a T-DNA insertion at the beginning of intron 5 was isolated from the SALK collection in a Col background (Alonso et al., 2003) (Figure 1A). We have renamed this mutation as rpt5a-4 that has so far been referred to as rpt5 (Huang et al., 2006). In contrast with other rpt5a alleles, heterozygous rpt5a-4/RPT5a lines segregated about one-quarter of plants homozygous for rpt5a-4, and these rpt5a-4/rpt5a-4 plants displayed no obvious sporophytic phenotype (Figure 1I). None of the rpt5a-4 plants displayed any defects in pollen development (Table 1; see Supplemental Figure 2 online). To check if the T-DNA might have been removed during RPT5a mRNA maturation, the RPT5a mRNA expression was checked in homozygous rpt5a-4 plants, and it was found that only its 5′ part is expressed (Figure 1J) ruling out this hypothesis. Therefore, either the N-terminal part of RPT5a is sufficient for its function, or the rpt5a mutation is complemented in Col but not in Ws. To test the latter hypothesis, Ws rpt5a-1/RPT5a plants were crossed with Col plants using the wild-type plants as a pollen donor. The pollen viability was then checked in the rpt5a-1/RPT5a F1 plants that were found to display consistently 25% dead pollen (Figure 3A). The reverse experiment was performed by introgressing the Col rpt5a-4 allele (that showed no pollen defect in Col) into Ws and showed that this cross also generated F1 plants with 25% dead pollen (Figure 3A).
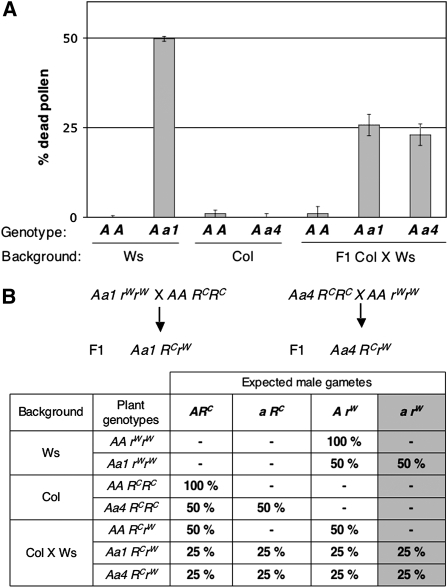
Figure 3.
rpt5a Male Gametophyte Phenotype Is Accession Dependent.
(A) Percentage of dead pollen in mature anthers as determined by Alexander staining and subsequent counting. For each genotype, 300 grains were counted per plant. Values are shown as means on six independent plants (±sd). Genotypes are indicated as in Figure 1, and genetic backgrounds are indicated below.
(B) Working model explaining how a restorer gene complements the rpt5a pollen phenotype and generation of the two F1 introducing the a1 and a4 mutations into a Col × Ws background. This hypothesis states that a gene, unlinked to RPT5a, behaves as a restorer allele in Col (RC), whereas the Ws allele (rW) does not restore the wild-type phenotype. The table gives the expected male gametes in the parents and in the F1 generations. For each genotype, the percentages of all expected gametes are indicated; hence, if the hypothesis is correct, the percentage of dead pollen in Figure 3A should be indicated by the last gray column.
This suggested either that a Ws locus unlinked to RPT5a contributes to pollen lethality together with rpt5a or that its Col allele can restore the rpt5a pollen viability (Figure 3B). If so, there should be plants that are homozygous for the rpt5a mutation among F2 plants resulting from a rpt5a-1/RPT5a × Col cross. Indeed, 25 rpt5a-1/ rpt5a-1 plants were isolated from 79 kanamycin resistant F2 plants, whereas no homozygous rpt5a-1plants had been found in the initial Ws background (n = 500, Table 1). Finally, as a direct test for the presence of one locus in Col complementing male transmission in rpt5a-1 male gametophytes, reciprocal crosses were performed between wild-type Ws plants and F1 rpt5a-1/RPT5a × Col plants. Kanamycin resistance was scored in progeny. The slight female transmission defect was eliminated after introgression (rpt5a-1 TEF = 104%, n = 675), while the male gametophyte transmission was restored to TEM = 35% (n = 1086, 50% expected). The same experiment was performed using Col instead of Ws as the wild type with similar results (TEF = 86%, n = 537 and TEM = 42%, n = 210).
These data show that although the RPT5a gene is essential for male gametophyte development in Ws accession, the rpt5a mutant phenotype is complemented by a single locus, which is unlinked to RPT5a in Col accession.
RPT5bCol, but Not RPT5bWs, Complements the rpt5a-1 Defect in Male Gametophyte Development
In Arabidopsis, the RPT5 subunit is encoded by two genes, RPT5a and RPT5b. RPT5a and RPT5b are highly homologous, displaying 94% amino acid identity. RPT5b is located on chromosome 1 and RPT5a on chromosome 3, hence the loci are unlinked. Therefore, RPT5b was an obvious candidate as the rpt5a-complementing locus if the hypothesis stating that RPT5bCol complements the rpt5a mutation whereas RPT5bWs does not is correct. A derived cleaved amplified polymorphic sequence (dCAPS) marker in RPT5b intron 7 was designed to discriminate RPT5bCol from RPT5bWs (Figure 4C, SNP#2). There was no segregation bias between RPT5bCol and RPT5bWs wild-type alleles in the progeny of Col × Ws F1 (RPT5bCol TEM and TEF were assessed as being 122 and 127%, n = 83 and 75, respectively). Because rpt5a-2 and rpt5a-3 mutants occasionally yield plants that are homozygous for the rpt5a mutation, the study focused on the strong allele rpt5a-1 for which, in a Ws background, no homozygous plants were ever isolated.
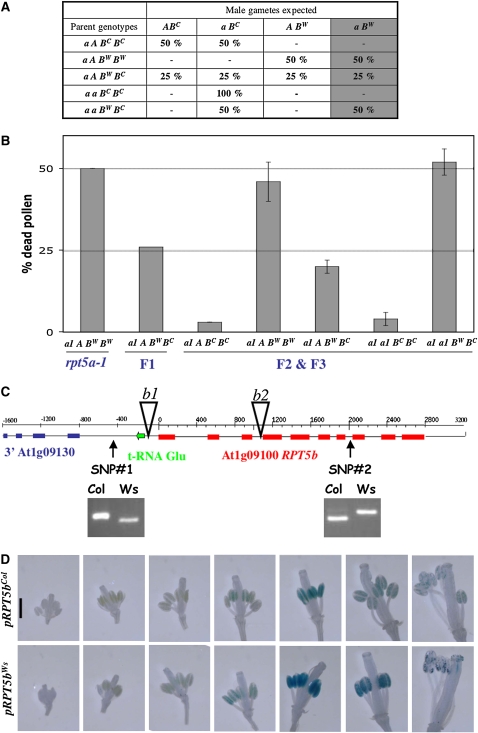
Figure 4.
RPT5bCol Complements the rpt5a-1 Male Gametophyte Default.
(A) Hypothesis for redundancy between RPT5a and RPT5b. A and a represent wild-type and mutant alleles of RPT5a, respectively, and BC and BW represent the Col and Ws RPT5b alleles, respectively. A gray background indicates putative male gametophyte lethality of the a BW allele combination. As in Figure 3, for each genotype, the percentages of all expected gametes are indicated; hence, if the hypothesis is correct, the percentage of dead pollen in Figure 4B should be indicated by the last gray column.
(B) Dead pollen was scored as in Figure 3. For reference, results for Aa1 heterozygous mutant and its F1 to Col are copied from Figure 3. Other values were scored on genotypes originating from the F2 or F3 progenies from the Aa1 × Col. Values are shown as means on six independent plants (±sd).
(C) Structure of the RPT5b locus. The two SNPs identified between Col and Ws accessions are presented as genotyped with dCAPS markers (see Methods). T-DNA insertion rpt5b-1 and rpt5b-2 (b1 and b2) are represented by open triangles.
(D) GUS expression as driven by Col and Ws RPT5b 800-bp promoters, respectively. All flowers from an inflorescence are presented from the youngest to the most mature. Sepals and petals were removed.
The RPT5b status was indirectly assessed in gametophytes carrying a rpt5a-1 mutation. To do so, progeny resulting from backcrosses of rpt5a-1/RPT5a RPT5bCol/RPT5bWs to wild-type Ws plants were analyzed. The rpt5a-1/RPT5a plants in these progeny were genotyped for their RPT5b status. If the restoring locus is unlinked to RPT5b, the progeny should segregate 50% RPT5bWs/RPT5bCol plants and 50% RPT5bWs/RPT5bWs. Alternatively, if this ratio was unbalanced in the rpt5a-1/RPT5a progenies, this would mean that one RPT5b allele would have been preferentially selected in rpt5a-1 viable gametes. Among the rpt5a-1/RPT5a progenies, 56% RPT5bWs/RPT5bCol plants and 44% RPT5bWs/RPT5bWs were obtained when using the mutant parent (rpt5a-1/RPT5a RPT5bCol/RPT5bWs) as female (n = 99), while 98% RPT5bWs/RPT5bCol plants and 2% RPT5bWs/RPT5bWs plants were observed when using the mutant as male (n = 230). These results show that while the rpt5a-1 mutation is transmitted through the female gametophyte regardless of the RPT5b status, restoration of male gametophyte viability in rpt5a-1 is strongly linked to RPT5bCol.
Next, the rpt5a-1 male defect was complemented with a RPT5bCol construct (pBIB:RPT5bCol). A 4-kb Col genomic fragment encompassing the RPT5b gene, including 800 bp upstream of the initiation codon and 500 bp of 3′ untranslated region, was subcloned into a binary vector and transformed into the original mutant. rpt5a-1 male transmission was directly tested on T1 plants. The pBIB:RPT5bCol construct successfully restored rpt5a-1 male transmission efficiencies to between 30 and 100% when the transformants were used as male donor.
From these results, it was hypothesized that rpt5a-1/RPT5bCol male gametes were viable but that the rpt5a-1/RPT5bWs combination was lethal. To confirm this hypothesis, pollen viability was characterized by Alexander staining in segregating F2 and F3 from the rpt5a-1/RPT5a × Col introgression. The results fitted this hypothesis (Figures 4A and 4B). All plants with a RPT5bCol/RPT5bCol genotype had almost no dead pollen (as in wild-type plants) whether the RPT5a allele was mutated or not. Consistent with this observation, the rpt5a-1 male transmission was fully restored in a RPT5bCol/RPT5bCol background (TEM = 89%, n = 154).
To understand the basis of the difference between the RPT5bCol allele and the RPT5bWs allele in complementing rpt5a-1, a 4800-bp genomic fragment was sequenced in both Col and Ws to search for polymorphisms between the two loci. Only two single nucleotide polymorphisms (SNPs) were found between both sequences: one in the promoter, the other being located at the end of intron 7 (Figure 4C). The RPT5b coding sequence of both accessions was found to be identical.
In a wild-type background, we have no evidence of any effect of the two SNPs on differential RPT5b expression or splicing between the Col and Ws alleles (see Supplemental Figure 3 online). Using transgenic lines expressing β-glucuronidase (GUS) fusions, it was shown that both the Col and the Ws RPT5b promoters drive a similar pattern of expression: detectable expression was only found at a late stage of pollen development (Figure 4D), suggesting the promoter SNP did not significantly alter the promoter expression pattern.
rpt5a rpt5b Double Mutants Are Affected in Both Male and Female Gametophyte Development
To examine in detail the redundancy between RPT5a and RPT5b genes, the double mutant in a Col background was analyzed. The aim was to assess whether the rpt5a rpt5b double mutant would reproduce at least the rpt5a RPT5bWs male gametophyte defect.
Both the Col allele rpt5a-4 and the rpt5a-1 mutation were used. The latter had been introgressed into Col (rpt5a-1/rpt5a-1 RPT5bCol/RPT5bCol) and no longer displayed any pollen mutant phenotype.
rpt5b-1 was isolated as a mutant affecting RPT5b (Alonso et al., 2003). The line carrying this mutation has been described as having a complete loss of function due to a T-DNA insertion in RPT5b first intron (Cho et al., 2006). However, characterizing this line, we found that the T-DNA was in fact located 90 bp upstream of the coding sequence and that the rpt5b-1 allele is not a null allele (Figure 4C). RT-PCR experiments show that residual gene expression is driven from the T-DNA left border, presumably by the cauliflower mosaic virus 35S promoter that is part of this construct (see Supplemental Figure 4 online). Due to promoter swapping, the mutant allele should misexpress RPT5b mRNA, especially considering that the cauliflower mosaic virus 35S promoter is poorly expressed in male gametophytes (Wilkinson et al., 1997). Another allele, rpt5b-2, in which two T-DNAs are inserted into the third intron of RPT5b, was isolated from the SAIL collection (Sessions et al., 2002). It is a null allele in which only the first three exons are transcribed (Figure 4C; see Supplemental Figure 4 online). Similarly to the rpt5a-4 mutant in Col, rpt5b-1 and rpt5b-2 mutants did not display any pollen defects, and heterozygous rpt5b/RPT5b plants segregated to give one-quarter of homozygous plants with no developmental defect (see Supplemental Figure 5 online). Neither rpt5b-1 nor rpt5b-2 displayed any male or female gametophyte transmission defect (all transmission efficiencies rated between 88 and 113%; see Supplemental Figure 4 online).
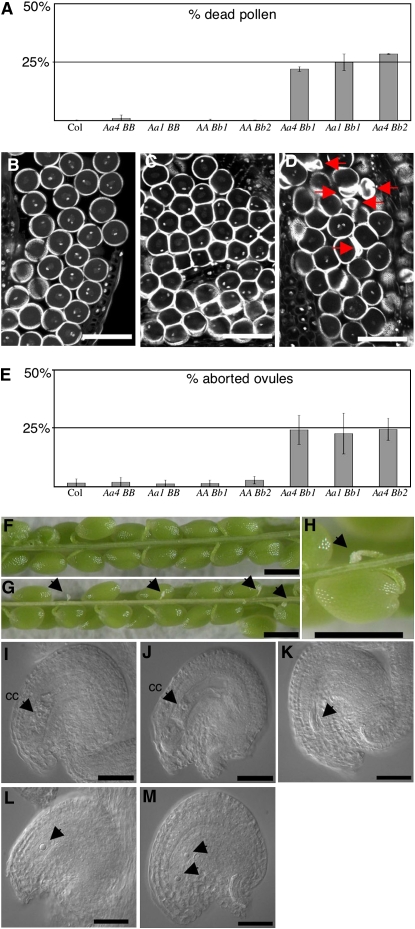
To assess the effect of a simultaneous loss of function in both RPT5a and RPT5b in gametophytes, a rpt5a-4/RPT5a rpt5b-1/RPT5b hybrid was generated and backcrossed to wild-type Col to check for male or female transmission of both mutant alleles together. PCR genotyping of the progenies allowed the determination of the gamete genotypes (Table 2). Those data fitted with the hypothesis of a double mutant lethal phenotype, whereas independent male and female transmission of each independent mutation was not affected. To confirm this lethality, male and female gametophyte developments of these plants were characterized. As controls, plants that carried one of each mutation at a heterozygous stage were analyzed (rpt5a-4/RPT5a RPT5b/RPT5b and RPT5a/RPT5a rpt5b-1/RPT5b plants, respectively). Very little dead pollen was detected following Alexander staining in control plants, while rpt5a-4/RPT5a rpt5b-1/RPT5b plants showed 25% dead pollen (Figure 5A). Similar results were obtained when combining rpt5a-1 and rpt5b-1 mutations and rpt5a-4 and rpt5b-2 mutations (Figure 5A). By staining stamens with propidium iodide, it was shown that the developmental defect occurred after PMI (Figures 5B to 5D; see Supplemental Figure 6 online). In conclusion, the rpt5a rpt5b male developmental defect was found to be highly similar to the rpt5a RPT5bWs combination.
Table 2.
Gametophytic Transmission of rpt5a-4 and rpt5b-1 Alleles in a Backcross from RPT5a/rpt5a-4 RPT5b/rpt5b-1 Plants to Wild-Type Col Plants
| Gamete Genotypes | rpt5a-4 RPT5b | rpt5a-4 rpt5b-1 | RPT5a RPT5b | RPT5a rpt5b-1 | Total Numbers |
|---|---|---|---|---|---|
| Expected if normal | 25% | 25% | 25% | 25% | |
| Expected if gametophytic | 33% | 0% | 33% | 33% | |
| Tmale | 30% | 0% | 31% | 39% | 124 |
| Tfemale | 36% | 0% | 36% | 28% | 120 |
Segregation values between different allele combinations are depicted if mutant alleles are transmitted in a Mendelian way (normal) or not (gametophytic). Tmale and Tfemale are percentage of male and female, respectively, T-DNA transmission in progenies of backcrosses between double heterozygous mutant plants and the wild type.
Figure 5.
rpt5a rpt5b Mutants Are Male and Female Gametophytic Lethal.
(A) Scoring of pollen lethality by Alexander staining of mature anthers followed by counting of dead pollen. B, b1, and b2 stand for wild-type RPT5b and mutant rpt5b-1 and rpt5b-2 alleles, respectively. All crosses are set in Col background. Values are shown as means on six independent plants (±sd).
(B) to (D) Optical sections through propidium iodide–stained anthers show mature pollen nuclei of Aa4BB (B), AABb1 (C), and Aa4Bb1 (D) plants. In single heterozygous mutants, pollen develop until PMII (B, C), while in double heterozygous mutants, some pollen grains degenerate, as indicated by arrows (D). Because of section planes, only one of the two sperm cell nuclei can be seen in some pollen grains.
(E) Scoring of ovule lethality in mature siliques using the same plants as in (A). Percentage of aborted ovules per siliques is represented. Values are shown as means on 10 independent siliques (±sd).
(F) to (H) All seeds develop in Col siliques (F), while Aa4Bb1 display 25% white structures that are aborted ovules ([G] and [H], designated by arrows).
(I) DIC image of a mature wild-type ovule at the time of fertilization. It features an enlarged central cell nucleus (cc).
(J) to (M) DIC image of mature ovule in Aa4Bb1 plants. At the time of fertilization, in siliques, 75% of the ovules present the feature of well-developed embryo sacs, such as an enlarged central cell nucleus (cc) and a large vacuole (J). Other embryo sacs display an altered structure and are blocked at one- (L) or two-nuclei stages (M); arrows indicate the nuclei. In a few cases, the whole embryo sac has degenerated ([K], arrow).
Bars = 40 μm in (B) to (D), 100 μm in (F) to (H), and 25 μm in (I) to (M).
The abolition of the female transmission of rpt5a-4 rpt5b-1 gametes was quite unexpected and suggested the involvement of RPT5 subunits in embryo sac development. The female gametophyte development was analyzed in rpt5a-4/RPT5a rpt5b-1/RPT5b and in the respective single heterozygous. When mature siliques were opened in Col wild type and in single mutants, they revealed very few aborted seeds (Figures 5E and 5F). By contrast, rpt5a-4/RPT5a rpt5b-1/RPT5b siliques consistently displayed 25% aborted ovules (Figures 5E, 5G, and 5H). Similar results were obtained in rpt5a-1/RPT5a rpt5b-1/RPT5b and rpt5a-4/RPT5a rpt5b-2/RPT5b siliques. The nucleus of the female macrospore undergoes three rounds of mitosis resulting in an eight-nuclei cell. The two polar nuclei fuse in later development resulting in a characteristic seven-nuclei structure. To check at which stage mutant embryo sacs arrested, 2-mm-long pistils that should only contain mature ovules ready to be fertilized were analyzed, and ovules were cleared and observed using differential interference contrast (DIC) microscopy. The rpt5a-4/RPT5a RPT5b/RPT5b and RPT5a/RPT5a rpt5b-1/RPT5b plants developed mainly wild-type ovules (<1.5% aborted ovules, n = 500). By comparison, rpt5a-4/RPT5a rpt5b-1/RPT5b siliques contained 26.3% aborted ovules (n = 609). Most of those ovules showed arrested embryo sac development as they contained only one (Figure 5L) or two (Figure 5M) nuclei, while the surrounding tissues (integuments and nucellus) have adopted a curved structure that is characteristic of mature development. In some cases, the entire embryo sac had degenerated (Figure 5K).
In conclusion, the rpt5a-4 rpt5b-1 female gametophyte development is arrested at the one or two nuclei stage so that the developmental defect coincides with the first or second mitosis of the megagametogenesis. Similar results were obtained by combining the rpt5a-1 with rpt5b-1 mutations (see Supplemental Figure 7 online).
Proteasome Functions Are Impaired in rpt5a rpt5b Gametophytes
While all RPT subunits have been shown to be essential for yeast RP function in proteasome-mediated protein degradation (Rubin et al., 1997), similar results are yet to be put forward for other organisms. We wanted to look at whether the lack of RPT5 subunits in the mutant gametophytes affected protein degradation. Given how mitosis was similarly affected in male and female rpt5 mutants, we decided to look at whether the degradation of common proteasome targets might be affected in these two developmental pathways.
Mitotic cyclin A and B are well-known substrates of the proteasome. These cyclins are expressed during G2 phase and degraded at the end of mitosis through the E3 ligase complex APC/C (Genschik et al., 1998; Capron et al., 2003a). As a gene reporter system, the pAL101 transgene that expresses a fusion of the N-terminal part of cyclin A3 to the GUS gene under the control of a gametophyte-specific promoter has been previously used (Capron et al., 2003b). With such a system, no GUS staining can be detected when the cycA3:GUS chimera is degraded, whereas GUS staining occurs if the proteasome degradation machinery is affected. For instance, GUS expression was detected in ovules upon mutation of the APC/C component APC2 (Capron et al., 2003b).
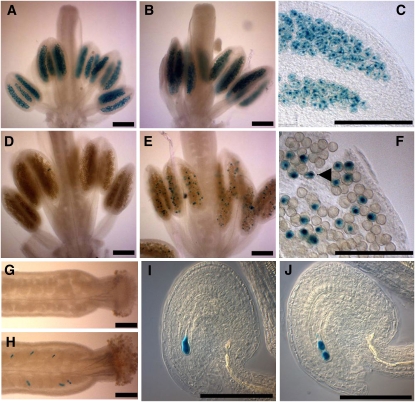
GUS expression pattern was first assessed in the pollen of pAL101/pAL101 plants as this pattern has not been previously investigated. These plants expressed GUS activity only transiently in all pollen grains at an early stage (pistil size 0.8 mm) that corresponds to the tetrad stage (Figures 6A and 6C). This signal appeared to be nuclear located. However, although the GUS protein is known to be highly stable, the GUS staining disappeared suddenly in all older stamens, suggesting that the cycA3:GUS chimera is indeed actively degraded (Figure 6D). The pAL101 plants were then crossed to double heterozygous mutants, and rpt5a-4/RPT5a rpt5b-1/RPT5b pAL101/pAL101 plants were selected in the F3. The same GUS staining pattern as in a wild-type background was found in young pollen grains (Figure 6B). But, in contrast with the wild-type background, a fraction of pollen still strongly expressed GUS at later stages (Figures 6E and 6F). In comparison with earlier stages, some GUS staining was found in the cytoplasm of collapsed pollen grains, suggesting they are the affected grains with a rpt5a-4 rpt5b-1 pAL101 genotype.
Figure 6.
rpt5a rpt5b Gametophytic Mutants Are Defective in cycA3 Degradation.
(A) to (F) cycA3:GUS staining in pollen from plants homozygous for the pAL101 construct (cyclinA3-GUS reporter construct) ([A] to [F]). At late meiosis/tetrad stage ([A] to [C] for enlargement: pistil length = 0.8 mm), all pollen grains are stained. GUS staining disappears at later stages of male gametophyte development (pistil length = 1.2 mm) during pollen mitosis in wild-type AABB background (D), whereas some staining remains in some Aa4Bb1grains ([E] and [F]). A GUS-stained degenerating pollen grain is indicated by an arrow in (F).
(G) to (J) cycA3:GUS staining in ovule. No GUS staining accumulated in the AABB wild-type background (G), whereas some strong staining could be seen in Aa4Bb1 pistils (H). Staining appeared as staining one (I) or two nuclei (J) of the arrested embryo sac.
Bars = 200 μm in (A) to (F) and 50 μm in (G) to (J).
As previously stated (Capron et al., 2003b), no GUS staining was detected in pAL101/pAL101 female gametophytes from wild-type plants (Figure 6G), but when GUS expression was monitored in rpt5a-4/RPT5a rpt5b-1/RPT5b pAL101/pAL101 pistils, a strong staining was found in one- and two-cell arrested ovules in those plants (Figures 6H to 6J).
This suggests that the proteasome substrate CycA3 is not degraded in both male and female rpt5a-4 rpt5b-1 gametophytes and therefore that specific proteasome functions are altered in those mutants. This shows that not only that similar developmental pathways are affected in rpt5 gametophyte development but also that similar targets are affected.
RPT5b Complements Loss of RPT5a Expression in Gametophytes through the Proteasome Feedback Loop
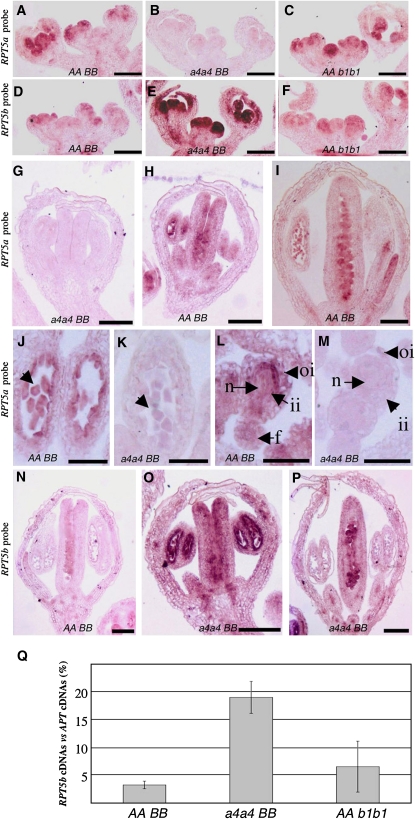
Eventually, the RPT5a and RPT5b expression patterns in Arabidopsis were investigated to see whether these genes could replace each other. RT-PCR experiments confirmed data from the Genevestigator DNA microarray database (http://www.genevestigator.ethz.ch) showing that both genes are expressed in all tested tissues, with RPT5a being more strongly expressed than RPT5b (see Supplemental Figure 8 online). To look more precisely at the expression patterns of those genes, mRNA in situ hybridizations were performed in Col inflorescences. First, the RPT5a expression pattern was analyzed using a specific probe. To check that the RPT5a probe could not cross-hybridize with RPT5b mRNA, it was tested that it gave no signal at all on rpt5a-4/rpt5a-4 inflorescences that present a RPT5a knockout and that do not display any sporophytic or gametophytic phenotype (Figures 7B, 7G, 7K, and 7M). In wild-type Col, RPT5a was expressed in meristems and young flower primordia. Expression was particularly high in the third and fourth whorls of primordia that will give rise to male and female reproductive organs, respectively (Figure 7A). At later stages of flower development, RPT5a signal was found to be particularly strong in anthers, both in tapetum and young pollen and in ovule primordia and ovules (Figures 7H, 7I, 7J, and 7L). Finally, the RPT5a expression pattern was not affected in the rpt5b-1/rpt5b-1 mutant (Figure 7C).
Figure 7.
RPT5 Genes Are Expressed in Flower Primordia, and RPT5b Is Upregulated in rpt5a.
mRNA in situ hybridization with an RPT5a probe ([A] to [C] and [G] to [M]) or an RPT5b probe ([D] to [F] and [N] to [P]). All sections are longitudinal cross sections. Bars = 100 μm in (A) to (I) and (N) to (P) and 50 μm in (J) to (M).
(A) to (F) Hybridization on young Col inflorescences ([A] and [D]), on rpt5a-4/rpt5a-4 ([B] and [E]), or on rpt5b-1/rpt5b-1 ([C] and [F]) inflorescences.
(G) to (I) Hybridization with a RPT5a probe on rpt5a4/rpt5a4 stage 6 flower (G) and stage 6 and 10 Col wild-type flower ([H] and [I], respectively).
(J) and (K) Hybridization with a RPT5a probe on Col wild-type (J) and on rpt5a4/rpt5a4 (K) anther locules from stage 6 flowers. Young pollen grains are indicated with an arrow.
(L) and (M) Hybridization with a RPT5a probe on Col wild-type (L) and rpt5a4/rpt5a4 (M) ovules on stage 10 flowers. oi, outer integuments; ii, inner integuments; n, nucellus; f, funiculus.
(N) Hybridization with an RPT5b probe on a stage 10 Col wild-type flower.
(O) and (P) Hybridization with an RPT5b probe on a stage 6 and a stage 10 rpt5a4/rpt5a4 flower.
(Q) Quantitative RT-PCR of RPT5b cDNA compared with APT cDNA accumulation from wild-type Col (AABB), rpt5a-4/rpt5a-4 (a4a4BB), and rpt5b-1/rpt5b-1 (AAb1b1) inflorescences. Results represent data collected from two replicates on two independent biological samples (±sd).
Next, the RPT5b expression pattern was analyzed. The promRPT5bCol:GUS transgene expression pattern had revealed that the promoter was not expressed except in mature pollen (Figure 4D). Using mRNA in situ hybridization, a weak expression pattern, mimicking the RPT5a expression pattern, was detected in Col inflorescences as well as in the rpt5b-1/rpt5b-1 mutant (Figures 7D and 7F). This is consistent with weak expression of RPT5b and with the rpt5b-1 mutant not being a null allele. At later stages of flower development, a weak RPT5b signal was found in ovule primordia but overall was very difficult to distinguish (Figure 7N). More strikingly, the same RPT5b pattern of expression was found in the rpt5a-4/rpt5a-4 plants, but very strongly enhanced. As for RPT5a, from meristem onwards through all stages of development, RPT5b was highly expressed especially in whorls 3 and 4 and in male and female reproductive organs (Figure 7E). In a rpt5a-4/rpt5a-4 background, the RPT5b expression pattern was highly overexpressed in both reproductive organs, mimicking the RPT5a mRNA expression pattern (Figures 7O and 7P). This expression is ubiquitous both in sporophytic and gametophytic tissues, which is consistent with RPT5 being a general machinery factor. This RPT5b mRNA overaccumulation in rpt5a-4/rpt5a-4 plants was confirmed by quantitative RT-PCR (Figure 7Q).
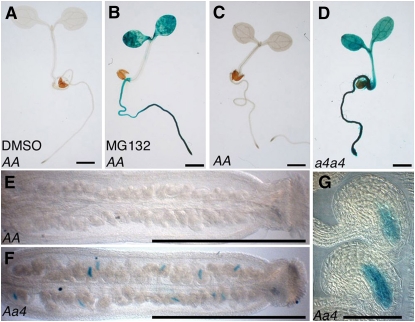
This upregulation is not completely unexpected and can be explained by the negative feedback mechanism that regulates the 26S proteasome composition. This feedback mechanism allows coordinated transcriptional overexpression of the genes encoding proteasome subunits upon proteasome function inhibition (Dohmen et al., 2007). To confirm that RPT5b is subjected to this feedback regulation, we first looked at whether the RPT5b promoter might be induced by a proteasome inhibitor, MG132. This was tested on young seedlings for which the addition of inhibitor is known to be efficient (Kurepa et al., 2008; Perez-Perez et al., 2008). The RPT5bCol promoter (promRPT5bCol), which drives no detectable GUS staining in seedlings, was induced by MG132 (Figures 8A and 8B).
Figure 8.
RPT5b Is Upregulated through the Proteasome Feedback Loop in the Sporophyte and Gametophytes.
GUS staining of Col plantlets that are transgenic for GUS constructs driven by the RPT5bCol promoter. Bars = 1 mm in (A) to (F) and 50 μm in (G).
(A) and (B) For the proteasome inhibitor experiment, 4-d-old Col plantlets were treated for 36 h without MG132 ([A]; DMSO) or with 100 μM MG132 (B).
(C) and (D) Six-day-old transgenics in Col wild-type (C) or a4a4 (D) background.
(E) and (F) GUS staining on 2-mm pistils from a Col wild-type (E) or a Aa4 ([F] and [G]) plant heterozygous for the promRPT5bCol:GUS transgene.
When the promRPT5bCol:GUS construct was introduced in a rpt5a-4/rpt5a-4 background, the RPT5b promoter was also found to be strongly induced (Figures 8C and 8D).
To gain insight into RPT5b behavior in gametophytic tissues only, the GUS staining of promRPT5bCol:GUS constructs was analyzed in a rpt5a-4/RPT5a background. The lack of RPT5a is then expected to occur in gametophytic tissues only. Consistent with this hypothesis, GUS staining was only found in some ovules in the rpt5a-4/RPT5a background (Figures 8E to 8G) and localized to the embryo sac, hence, the gametophytic tissues (Figure 8G). The promRPT5bWs:GUS was found to be similarly enhanced by a proteasome feedback loop in both sporophyte and gametophytes (see Supplemental Figure 9 and Supplemental Table 2 online). Overall, both Col and Ws RPT5b promoter are similarly enhanced by the proteasome feedback loop in both Col and Ws accessions (see Supplemental Figure 10 online).
These data shed new light on the redundancy between RPT5a and RPT5b and especially how RPT5b (i.e., RPT5bCol) complements for the lack of RPT5a. Rather than just RPT5b taking over RPT5a's place in a rpt5a mutant, it is more likely through the proteasome feedback loop that RPT5b expression is induced in rpt5a reproductive organs so that RPT5b is able to replace the missing RPT5a. This also explains why in a wild-type background, the RPT5b promoter is not expressed at stages where RPT5b is supposed to complement rpt5a, namely, during early pollen development and during female gametophyte development.
DISCUSSION
26S proteasome complexes are well conserved in eukaryotes, where they carry out a central role in protein degradation (Kurepa and Smalle, 2007). Therefore, loss of function of the 26S proteasome should have a tremendous impact on development. This was highlighted in yeast with special emphasis on the role of triple A ATPase-encoding genes (RPT subunits) as rpt active site mutants were either lethal or severely impaired growth (Rubin et al., 1997).
In Arabidopsis, a growing number of mutants affecting the 19S RP subunits have been described. Mutants were shown to display both pleiotropic and a highly diverse set of developmental defects (Kurepa and Smalle, 2007). So far the most severe phenotype described in Arabidopsis for a 26S proteasome subunit defect has been a complete embryonic lethality as described for the rpn1a mutant (Brukhin et al., 2005).
RPT5 Subunits Are Essential for Gametophyte Development and Affect Sporophyte Development
In Arabidopsis, the RPT5 subunit is encoded by two genes, RPT5a and RPT5b, that encode two highly similar proteins. We showed by RT-PCR experiments, reporter fusion studies and mRNA in situ hybridization that expression profiles of these two genes differ, with RPT5a being consistently more highly expressed than RPT5b. Notably, we found RPT5a to be highly expressed in male and female reproductive organs, including gametophytes. In partial agreement with these data, all three rpt5a mutant alleles in the Ws accession were impaired in male gametophyte development, whereas those mutations only had a weak effect on female gametophyte development. This could be related to each gametophytic phase having specific requirements at the level of proteasome function and/or, not exclusively, that RPT5b complements a defective RPT5a in the female phase but not in the male.
Further genetic analysis revealed that the male defect observed in rpt5a mutants was due to the fact that the wild-type Ws RPT5b gene does not complement the mutant phenotype. By contrast, in a Col background, rpt5a-4 null male gametophytes are totally viable due to the RPT5b Col allele. This study therefore is a case where mutant analysis is combined with natural variation, and it stresses how mutations in a gene can produce very different phenotypes depending upon the background (Koornneef et al., 2004). Interestingly, similar accession-dependent phenotypes for proteasome subunit mutants have been already reported as rpn8a mutations are responsible for the production of long and narrow leaves in the Landsberg erecta accession but not in Col (Huang et al., 2006). RPT5bWs might present a partial loss of function in gametophytes. This hypothesis is further strengthened by the study of combinations of rpt5a and rpt5b loss-of-function mutants in Col. Male and female gametophytes mutant for both rpt5a and rpt5b showed arrested development. These results show that RPT5 proteasome subunits are essential for both gametophyte developmental pathways and as each single mutation is viable, that RPT5a and RPT5b act in a redundant way.
This is, to our knowledge, the first report of mutants affecting a 19S RP subunit showing such a dramatic and early phenotype. But it might also be that such gametophyte development defects have been overlooked in the past. RPN10/rpn10-1 plants show a partial rpn10-1 male gametophyte lethality (Smalle et al., 2003). Similarly, Huang et al. (2006) reported that a double mutant rpn8a rnp8b could not be recovered. Determination of the exact stages where such mutants are affected, if they are, will prove useful in comparing the requirements for the diverse subunits in 19S RP. It remains that some 19S RP subunits, such as RPN1, must be stable enough to allow the proteasome to function through the haploid phase or that those subunits might be dispensable during gametophyte development.
Because severe rpt5a rpt5b mutations affected the development of both gametophytes, the study of sporophyte development was not possible for those double mutants. However, partial loss of RPT5 function was observed with plants homozygous for weak rpt5a mutations. Interestingly, these showed a general defect in all stages of sporophytic development. The first and most striking phenotype was a much reduced root development. Such a phenotype has previously been described for rpt2a mutant (halted root) as well as rpn10 and rpn12 homozygous mutants (Ueda et al., 2004; Kurepa et al., 2008). The fact that different subunit mutations result in a similar phenotype strongly suggests that the whole RP proteasome complex is required for normal sporophytic growth, presumably through the degradation of several protein targets that may be involved in stem cell maintenance or hormone signaling pathways (Ueda et al., 2004).
rpt5 Mutant Gametophytes Are Deficient in Mitosis
In this study, we show that the lack of both RPT5a and RPT5b subunits leads to severe developmental defects in male and female gametophyte development. rpt5a rpt5b gametophytes were affected during the postmeiotic mitoses that take place during the development of both gametophytes (PMII and female megagametophyte, respectively). Using a reporter construct, we showed that proteasome protein degradation functions were affected in the rpt5a rpt5b gametophytes. Therefore, a tempting hypothesis is that similar proteasome targets of both male and female gametophytes could no longer be degraded in the double mutant. Indeed, we found that a construct harboring a cycA3 construct accumulated in the mutant gametophytes. The timing of cyclin degradation is essential for correct progression through the cell cycle (Pines, 2006).
Our results are consistent with the phenotypes of the plants that present a defect in the anaphase-promoting complex (APC/C) mutants. APC/C is an E3 ubiquitin-ligase complex that specifically targets cyclins and cohesins to 26S proteasome degradation and therefore regulates the exit from mitosis. Mutations affecting either genes encoding APC/C subunits APC2 and APC6 have been shown to trigger a female gametophyte lethality phenotype that correlates with an inability to degrade mitotic cyclins (Capron et al., 2003b; Kwee and Sundaresan, 2003). More recently, it has been shown that the CDC27 subunit of the APC/C, which is encoded by two genes CDC27A and HOBBIT, is also necessary for female gametophyte development and to a lesser extent male gametophyte development (Perez-Perez et al., 2008). So it seems reasonable to assume that defects in the 26S proteasome recapitulate the phenotypes triggered by defects in APC/C and block cells during mitosis by stabilizing cyclins. Furthermore, recent data show that the mitotic cell cycle progression that is essential for gametophyte development is regulated through proteasome function: several E3-ligases have been isolated that act to target cyclin-dependent kinase inhibitors whose degradation are essential for full gametophyte development (Kim et al., 2008; Liu et al., 2008). In rpt5a rpt5b gametophytes, it is primarily the later mitoses that were affected during both developmental phases while the first mitoses took place normally. This might be either because of some residual RPT5 activity in double mutants or because the defect appeared only once the first mitosis had taken place.
Finally, although gametophyte development is blocked at the mitotic stage, we have no evidence that this developmental defect is subordinated to a mitosis defect. As pointed out by Doelling et al. (2007), mutants for a large number of candidate genes are affected at this stage and any of them are likely targets whose lack of degradation could account for a defect in male/female gametophyte development (Doelling et al., 2007).
Basis of RPT5a and RPT5b Redundancy
A large number of the 19S RP subunits are encoded by two genes (Kurepa and Smalle, 2007). The question arises whether these paralogs are functionally redundant and/or differentially expressed. So far it has been shown that RPN1a and RPN1b proteins are functionally redundant but that RPN1a and RPN1b genes are not because they have different expression patterns. It was shown that the RPN1b gene did complement rpn1a when expressed under the RPN1a promoter but not under its own promoter (Brukhin et al., 2005).
In this study, we were able to show that duplicated genes RPT5a and RPT5b are partially redundant, especially in female gametophytes. However, in male gametophytes, this redundancy is accession dependent. The RPT5b Col allele complemented for a rpt5a mutation during male gametophyte development, whereas the Ws RPT5b allele did not. The same was true during sporophyte development. It is striking that rpt5a-4/rpt5a-4 rosettes developed as wild type in a Col background, whereas weaker rpt5a-2/rpt5a-2 or rpt5a-3/rpt5a-3 mutations led to short roots and smaller rosettes in a Ws background. These sporophytic phenotypes were suppressed by introgressing the rpt5a-2 and rpt5a-3 mutations into a RPT5bCol background (see Supplemental Figure 11 online). This shows that the differences between RPT5b Col and Ws alleles, however subtle, are not only male gametophyte specific but are also found at all stages of sporophytic development.
Expression studies revealed that, in a wild-type background, although RPT5a and RPT5b expression patterns overlapped, RPT5b was very weakly expressed compared with RPT5a. We showed that RPT5b was highly expressed when proteasome function was defective, whether upon addition of proteasome inhibitor or in an RPT5a-deficient mutant background. This upregulation is not completely unexpected and can be explained by the negative feedback mechanism that regulates the 26S proteasome composition. This mechanism was first demonstrated in yeast (Xie and Varshavsky, 2001). It implies that when the proteasome function is altered (through lack of subunits, proteasome inhibition, or stress), a regulator that is usually proteasome targeted is no longer degraded and boosts the transcription of genes encoding proteasome subunits. The existence of a similar mechanism has been suggested in Arabidopsis, but a homolog of the yeast regulator, Rpn4p, could not be isolated (Yang et al., 2004; Kurepa and Smalle, 2007).
Moreover, it has been shown in yeast that Rpn4p acts as a transcription factor by binding to a highly conserved nonamer located in 26S proteasome subunit promoters (Mannhaupt et al., 1999). Using transgenic plants in which an 800-bp RPT5b promoter was fused to the reporter GUS gene, we showed that the determinant for the feedback upregulation of RPT5b was present in these 800 bp: a strong GUS signal was observed upon proteasome function inhibition, either using a proteasome inhibitor (MG132) or rpt5a mutation. However, we could not find any conserved sequences in promoters from genes encoding Arabidopsis 26S subunits (data not shown), suggesting that the proteasome feedback loop, although conserved in plants, has evolved differently as it has in mammalian cells (Meiners et al., 2003). It is worth noting that not all Arabidopsis subunits are subjected to such a strong upregulation, and, for example, RPN1b upregulation is not sufficient to complement a rpn1a defect (Brukhin et al., 2005). Similarly, we did not detect any RPT5a mRNA accumulation in rpt5b mutants, presumably because of the low expression level of RPT5b, its disappearance does not affect RPT5 mRNA levels to trigger the proteasome feedback loop. Interestingly, microarray data presented by Kurepa and Smalle (2007) showed that in the presence of the proteasome inhibitor MG132, RPT5b was the gene most subjected to this proteasome feedback upregulation. It could be that this RPT5 redundancy through the proteasome feedback loop might instead be an exception.
Besides acquiring different expression patterns, duplicated genes may have developed distinct functions (Taylor and Raes, 2004). So one crucial question remains: whether duplicated RPT and RPN subunits might, while retaining mainly overlapping functions, develop a specificity that would contribute toward proteasome function plasticity. In yeast, RPT5 has been shown to be involved in specific functions, such as binding ubiquitinated substrates (Lam et al., 2002) and, together with RPT2, in the 20S CP gate opening (Smith et al., 2007). In Arabidopsis, it was shown that the glucose sensor hexokinase HXK1 could interact with RPT5b in the nucleus but not with its close homolog RPT5a (Cho et al., 2006). Given that RPT5b is poorly expressed in all tissues, but is induced upon proteasome defects, it is very tempting to speculate that RPT5b could be expressed under stress conditions and carry new specificities even participating in a stress proteasome status.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Col and Ws accessions were used as control wild-type plants. rpt5a-1 rpt5a-2 and rpt5a-3 mutant alleles were isolated in the Versailles collection of T-DNA insertion mutants (Ws accession) as EAT54 (FLAG_218D02), DTM5 (FLAG_130B03), and DYI110 (FLAG_120A08), respectively (Samson et al., 2004). The rpt5a-4 allele was isolated in the SALK collection (SIGnAL; http://signal.salk.edu/cgi-bin/tdnaexpress) as line S046321 in Col accession (Alonso et al., 2003). The rpt5b-1 allele was also isolated in the SALK collection as line S069366, whereas the rpt5b-2 allele was isolated in the SAIL collection (Sessions et al., 2002) as line SAIL681C04. All mutant seeds were provided by the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/). The pAL101 lines (harboring the CyclinA3-GUS reporter construct) have been described previously (Capron et al., 2003b).
For growth on plates, seeds were surface sterilized for 8 min in 95% ethanol with 0.1% Tween 20 and plated on Murashige and Skoog (MS) medium. For growth in both plates and soil, seeds were stratified for 2 d at 4°C and grown at 18 to 20°C, with 16 h light (fluorescent lights at ∼100 μmol photons m−2 s−1) and 8 h dark cycles. Immature flowers were emasculated and manually cross-pollinated for crosses, including allele transmission tests. For proteasome inhibition experiments, seeds were allowed to germinate for 4 d on MS plates then transferred to 24-well microtiter plates containing 1 mL of 0.5× MS salt mixture and 1% sucrose, pH 5.8, with 100 μM MG132 (Sigma-Aldrich). Seedlings were kept 36 h with gentle agitation in a growth chamber.
Genotyping of Mutants and Alleles by PCR
All primers are described in Supplemental Table 1 online.
Wild-type alleles and mutant alleles were genotyped with the following primers. For rpt5a-1 and rpt5a-2 alleles, wild-type RPT5a was genotyped with EAT-U and EAT-L, while mutant alleles were genotyped with EAT-U and TAG6. For rpt5a-3, wild-type RPT5a was scored with DYI-U and DYI-L and rpt5a-3 with DYI-L and TAG6. For rpt5a-4, wild-type allele was scored with S46321-U and S46321-L, while the mutant allele was genotyped with S46321-L and LBSalk2.
rpt5b-1 and rpt5b-2 alleles were genotyped with RPT5b-5′ and LBSalk2 or RPT5b-e6 and LB3Sail respectively while the wild type RPT5b was scored with RPT5b-5′ and RPT5b-e2, or RPT5b-e3f and RPT5b-e6.
Primers used for genotyping SNP between RPT5bCol and RPT5bWs alleles were designed using dCAPS Finder 2.0 (http://helix.wustl.edu/dcaps/dcaps.html) (Neff et al., 2002). For the SNP#1 located in the RPT5b promoter, a 266-bp fragment was amplified with primers JLV096 and JLV097 and digested with VspI. Only the fragment originating from the Ws allele was digested into 249- and 17-bp fragments. For the SNP#2 located in the RPT5b seventh intron, a 280-bp fragment was amplified with primers RPT5b-3369 and RPT5b-Hind3 and digested with HindIII. Only the fragment originating from the Col fragment was digested into 250- and 30-bp fragments.
For rpt5a-1, rpt5a-2, and rpt5a-3 mutant alleles, the linkage between the T-DNA insertion and the plant kanamycin resistance was checked by genotyping for the insertion 100 kanamycin-resistant plants in the progeny from two independent heterozygous mutants.
Histological Analysis
Pollen viability was assessed using Alexander staining (Alexander, 1969) and observed with a PL Fluotar X25 dry objective on a Leitz Diaplan microscope.
Male gametophyte development was analyzed using DAPI staining (Park et al., 1998), using UV epi-illumination with a Leitz Diaplan (Pl Fluotar ×50/0.7 oil objective) or propidium iodide DNA staining. For the latter, inflorescences were placed in ethanol:formaldehyde:propionic acid (70:10:5) for 24 h, washed, and stored at 4°C in 70% ethanol. After fixation, tissues were rehydrated in ethanol series and treated by RNaseA (Fermentas). Inflorescences were then stained with propidium iodide (Sigma-Aldrich) in 0.1 M arginine, pH 11 solution for 2 to 4 d (Motomayor et al., 2000). After washing with 0.1 M arginine, pH 8, anthers were gently dissected and observed in a drop of citifluor glycerol/PBS. The 568-nm laser line of an argon laser of a Leica TCS-NT confocal laser scanning microscope was used to illuminate and observe male gametophytes. Emission was collected with a band-pass filter at 590 nm, a Leica PL Fluotar ×40/1-0.5 oil objective, and the Leica LCS software. Development of the ovules and embryo sac was observed with DIC microscopy after clearing treatment (Grelon et al., 2001).
Histochemical GUS staining was performed as by Marrocco et al. (2003).
Plasmid Construction and Plant Transformation
A 4444-bp genomic RPT5a fragment (spanning 1895 bp of promoter and 2549 bp of gene sequence) was amplified using primers EAT-A and EAT-D from F22F7 BAC clone and cloned in pCambia 1200 (CAMBIA) as pC-RPT5a. For the Col genomic RPT5b complementation construct, the RPT5b locus was amplified as two fragments using primers JLV083 and JLV085, and JLV086 and JLV087, respectively. Both fragments were cloned as a 3875-bp fragment into pBIB-HYG binary vector (Becker, 1990) resulting in plasmid pBIB:RPT5bCol.
An 804-bp RPT5b promoter fragment was amplified with primers JLV094 and JLV077 as a HindIII-NcoI fragment on Col and Ws genomic DNA, respectively. Both fragments were inserted upstream the uidA coding sequence and cloned into pBIB-HYG as pJL299 and pJL300 (harboring the Col and Ws RPT5b promoters, respectively). A promoterless GUS (uidA) construct was also generated as pJL301.
All binary vectors were transformed into Arabidopsis using the floral dip method (Clough and Bent, 1998). pC-RPT5a and pBIB:RPT5bCol were transformed into rpt5a-1/RPT5a plants, whereas pJL299, pJL300, and pJL301 were transformed into wild-type Col and Ws plants.
All T1 plants were selected on MS plates supplemented with 15 mg/L hygromycin B.
Expression Analysis
Total RNA was extracted using TRIZOL-reagent (Invitrogen) from 100 mg of tissue (inflorescences). Contaminating DNA was removed by DNaseI treatment with RNase Free DNase set (Qiagen) using spin columns of the Rneasy plant mini kit (Qiagen).
RT-PCR was performed with RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas) on 1 μg RNA according to the supplier's instructions. Unless stated, RPT5a, RPT5b, and APT cDNAs were amplified with specific primers as follows: S46321-U and S46321-L (RPT5a), RPT5b-e5 and RPT5b-e10 (RPT5b), APT 5′ and APT 3′ (APT).
Transcript abundance was quantified using fluorescence-based real-time PCR. Total RNA was extracted and treated by DNaseI as described above. The RevertAid H Minus M-MuLV reverse transcriptase (Fermentas) was used to generate cDNA from 500 ng of RNA in a reaction volume of 20 μL. For each cDNA sample (diluted 1:3 in water), 5 μL was used as a template for quantitative PCR. Reactions were performed with Maxima SYBR Green qPCR Master Mix (Fermentas) using an Eppendorf Mastercycler RealPlex2 according to the manufacturer's instructions. Efficiency and specificity of each primer couple we used were checked. The RPT5b-specific primers used were RPT5b-e5 and RPT5b-e6. Expression levels were quantified with respect to APT expression levels (primers APT-5′ and APT-3′) and were averaged over at least two replicates from two independent biological samples.
RNA gel blots were performed according to standard procedures. Twenty micrograms of RNA extracted from inflorescences was loaded per sample. Genescreen membrane was hybridized with 32P-labeled exon3-exon6 cDNA fragment of RPT5a.
For the mRNA in situ hybridization, antisense probes were synthesized in vitro and labeled with DIG-UTP (Roche) using a gel-purified PCR product including the T7 RNA polymerase binding site as template. A specific RPT5a probe was amplified with primers EAT3194 and JLV100 and the RPT5b probe with primers S69366e3 and JLV111. Tissue fixation, embedding, sectioning, and in situ hybridization were performed as described by Nikovics et al. (2006).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: At3g05530 (RPT5a) and At1g09100 (RPT5b), both in Col accession. Sequence for the RPT5b locus in Ws accession has been deposited with the GenBank/EMBL data libraries under accession number EU980096. The identification numbers for the mutants described here are FLAG_218D02 (rpt5a-1), FLAG_130B03 (rpt5a-2), FLAG_120A08 (rpt5a-3), S046321 (rpt5a-4), S069366 (rpt5b-1), and SAIL681C04 (rpt5b-2). rpt5a-4 has been previously described as rpt5 (Huang et al., 2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Single-Locus T-DNA Insertions in rpt5a-1, rpt5a-2, and rpt5a-3 Mutants.
Supplemental Figure 2. Male Gametophyte Development in rpt5a-2, rpt5a-3, and rpt5a-4 Mutants.
Supplemental Figure 3. Splicing at RPT5b Intron 7 Is Not Significantly Affected between Col and Ws Accessions.
Supplemental Figure 4. rpt5b Mutants Characterization.
Supplemental Figure 5. rpt5b Mutants Display Wild-Type Phenotype under High Sugar and Low Nutrient Conditions.
Supplemental Figure 6. Pollen Mitosis I Occurs Normally in rpt5a/RPT5a rpt5b/RPT5b Plants.
Supplemental Figure 7. Ovule Development in rpt5a/RPT5a rpt5b/RPT5b Plants.
Supplemental Figure 8. RPT5a and RPT5b Are Expressed Ubiquitously, with RPT5a Being More Strongly Expressed.
Supplemental Figure 9. The RPT5bWs Promoter Is Upregulated through the Proteasome Feedback Loop in Sporophytes and Gametophytes.
Supplemental Figure 10. RPT5bCol and RPT5bWs Promoters React Similarly to the Proteasome Feedback Signal in Both Col and Ws Sporophytes.
Supplemental Figure 11. rpt5a-2/rpt5a-2 and rpt5a-3/rpt5a-3 Sporophytic Phenotypes Are Rescued in a RPT5bCol/RPT5bCol Background.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Table 2. RPT5bCol and RPT5bWs Promoters Both React Similarly to the Proteasome Feedback Signal in Female Gametophytes.
Supplementary Material
Acknowledgments
We thank Mathilde Grelon for her very helpful suggestions on both the project and the manuscript. We thank Alain Lecharny and Veronique Brunaud for bioinformatic analysis, Christine Horlow for the pollen DNA propidium iodide staining protocol, and Halima Morin and Jocelyne Kronenberger for their sound advice on mRNA in situ hybridization. We also thank Rebecca Stevens and Martine Miquel for their advice on improving the manuscript and Carole Caranta for support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jean-Luc Gallois (jlgallois@avignon.inra.fr).
Online version contains Web-only data.
References
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44 117–122. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J.D., Boavida, L.C., Carneiro, J., Haury, M., and Feijo, J.A. (2003). Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 133 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj, N., Zwickl, P., Seemuller, E., Baumeister, W., and Goldberg, A.L. (2003). ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 11 69–78. [DOI] [PubMed] [Google Scholar]
- Boavida, L.C., Becker, J.D., and Feijo, J.A. (2005a). The making of gametes in higher plants. Int. J. Dev. Biol. 49 595–614. [DOI] [PubMed] [Google Scholar]
- Boavida, L.C., Vieira, A.M., Becker, J.D., and Feijo, J.A. (2005b). Gametophyte interaction and sexual reproduction: How plants make a zygote. Int. J. Dev. Biol. 49 615–632. [DOI] [PubMed] [Google Scholar]
- Bonhomme, S., Horlow, C., Vezon, D., de Laissardiere, S., Guyon, A., Ferault, M., Marchand, M., Bechtold, N., and Pelletier, G. (1998). T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol. Gen. Genet. 260 444–452. [DOI] [PubMed] [Google Scholar]
- Brukhin, V., Gheyselinck, J., Gagliardini, V., Genschik, P., and Grossniklaus, U. (2005). The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17 2723–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron, A., Okresz, L., and Genschik, P. (2003a). First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 8 83–89. [DOI] [PubMed] [Google Scholar]
- Capron, A., Serralbo, O., Fulop, K., Frugier, F., Parmentier, Y., Dong, A., Lecureuil, A., Guerche, P., Kondorosi, E., Scheres, B., and Genschik, P. (2003b). The Arabidopsis anaphase-promoting complex or cyclosome: Molecular and genetic characterization of the APC2 subunit. Plant Cell 15 2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J.M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–671. [DOI] [PubMed] [Google Scholar]
- Cho, Y.H., Yoo, S.D., and Sheen, J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127 579–589. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Doelling, J.H., Phillips, A.R., Soyler-Ogretim, G., Wise, J., Chandler, J., Callis, J., Otegui, M.S., and Vierstra, R.D. (2007). The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol. 145 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R.J., Willers, I., and Marques, A.J. (2007). Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function. Biochim. Biophys. Acta 1773 1599–1604. [DOI] [PubMed] [Google Scholar]
- Dreher, K., and Callis, J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Doelling, J.H., Rubin, D.M., and Vierstra, R.D. (1999). Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteasome. Plant J. 18 529–539. [DOI] [PubMed] [Google Scholar]
- Fu, H., Reis, N., Lee, Y., Glickman, M.H., and Vierstra, R.D. (2001). Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik, P., Criqui, M.C., Parmentier, Y., Derevier, A., and Fleck, J. (1998). Cell cycle-dependent proteolysis in plants. Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. Plant Cell 10 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H., Rubin, D.M., Fried, V.A., and Finley, D. (1998). The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67 425–479. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Pi, L., Liang, W., Xu, B., Wang, H., Cai, R., and Huang, H. (2006). The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 18 2479–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J., Oh, S.A., Brownfield, L., Hong, S.H., Ryu, H., Hwang, I., Twell, D., and Nam, H.G. (2008). Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455 1134–1137. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Kurepa, J., and Smalle, J.A. (2007). Structure, function and regulation of plant proteasomes. Biochimie 90 324–335. [DOI] [PubMed] [Google Scholar]
- Kurepa, J., Toh, E.A., and Smalle, J.A. (2008). 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53 102–114. [DOI] [PubMed] [Google Scholar]
- Kwee, H.S., and Sundaresan, V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. Plant J. 36 853–866. [DOI] [PubMed] [Google Scholar]
- Lam, Y.A., Lawson, T.G., Velayutham, M., Zweier, J.L., and Pickart, C.M. (2002). A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416 763–767. [DOI] [PubMed] [Google Scholar]
- Liu, J., et al. (2008). Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20 1538–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhaupt, G., Schnall, R., Karpov, V., Vetter, I., and Feldmann, H. (1999). Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450 27–34. [DOI] [PubMed] [Google Scholar]
- Marrocco, K., Lecureuil, A., Nicolas, P., and Guerche, P. (2003). The Arabidopsis SKP1-like genes present a spectrum of expression profiles. Plant Mol. Biol. 52 715–727. [DOI] [PubMed] [Google Scholar]
- McCormick, S. (2004). Control of male gametophyte development. Plant Cell 16(Suppl): S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiners, S., Heyken, D., Weller, A., Ludwig, A., Stangl, K., Kloetzel, P.M., and Kruger, E. (2003). Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J. Biol. Chem. 278 21517–21525. [DOI] [PubMed] [Google Scholar]
- Monte, E., Al-Sady, B., Leivar, P., and Quail, P.H. (2007). Out of the dark: How the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J. Exp. Bot. 58 3125–3133. [DOI] [PubMed] [Google Scholar]
- Motomayor, J., Vezon, D., Bajon, C., Sauvanet, A., Grandjean, O., Marchand, M., Bechtold, N., Pelletier, G., and Horlow, C. (2000). Switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sex. Plant Reprod. 12 209–218. [Google Scholar]
- Neff, M.M., Turk, E., and Kalishman, M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18 613–615. [DOI] [PubMed] [Google Scholar]
- Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., and Laufs, P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat, G.C., Yu, H.J., Ngo, Q.A., Rajani, S., Mayalagu, S., Johnson, C.S., Capron, A., Xie, L.F., Ye, D., and Sundaresan, V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614. [DOI] [PubMed] [Google Scholar]
- Park, S.K., Howden, R., and Twell, D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125 3789–3799. [DOI] [PubMed] [Google Scholar]
- Perez-Perez, J.M., Serralbo, O., Vanstraelen, M., Gonzalez, C., Criqui, M.C., Genschik, P., Kondorosi, E., and Scheres, B. (2008). Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C). Plant J. 53 78–89. [DOI] [PubMed] [Google Scholar]
- Pina, C., Pinto, F., Feijo, J.A., and Becker, J.D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines, J. (2006). Mitosis: A matter of getting rid of the right protein at the right time. Trends Cell Biol. 16 55–63. [DOI] [PubMed] [Google Scholar]
- Rubin, D.M., Glickman, M.H., Larsen, C.N., Dhruvakumar, S., and Finley, D. (1998). Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17 4909–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, D.M., van Nocker, S., Glickman, M., Coux, O., Wefes, I., Sadis, S., Fu, H., Goldberg, A., Vierstra, R., and Finley, D. (1997). ATPase and ubiquitin-binding proteins of the yeast proteasome. Mol. Biol. Rep. 24 17–26. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Duchene, S., De Oliveira, Y., Caboche, M., Lecharny, A., and Aubourg, S. (2004). FLAGdb++: a database for the functional analysis of the Arabidopsis genome. Nucleic Acids Res. 32 D347–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, X., Hu, Z., Lu, H., Wang, X., Baluska, F., Samaj, J., and Lin, J. (2006). Roles of the ubiquitin/proteasome pathway in pollen tube growth with emphasis on MG132-induced alterations in ultrastructure, cytoskeleton, and cell wall components. Plant Physiol. 141 1578–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Babiychuk, E., Kushnir, S., Durski, A., and Vierstra, R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., Kurepa, J., Yang, P., Emborg, T.J., Babiychuk, E., Kushnir, S., and Vierstra, R.D. (2003). The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Smith, D.M., Chang, S.C., Park, S., Finley, D., Cheng, Y., and Goldberg, A.L. (2007). Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza, A., Scoccianti, V., Crinelli, R., Calzoni, G.L., and Magnani, M. (2001). Inhibition of proteasome activity strongly affects kiwifruit pollen germination. Involvement of the ubiquitin/proteasome pathway as a major regulator. Plant Physiol. 126 1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.S., and Raes, J. (2004). Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 38 615–643. [DOI] [PubMed] [Google Scholar]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 661–665. [DOI] [PubMed] [Google Scholar]
- Ueda, M., Matsui, K., Ishiguro, S., Sano, R., Wada, T., Paponov, I., Palme, K., and Okada, K. (2004). The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131 2101–2111. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
- Walz, J., Erdmann, A., Kania, M., Typke, D., Koster, A.J., and Baumeister, W. (1998). 26S proteasome structure revealed by three-dimensional electron microscopy. J. Struct. Biol. 121 19–29. [DOI] [PubMed] [Google Scholar]
- Wilkinson, J.E., Twell, D., and Lindsey, K. (1997). Activities of CaMV 35S and nos promoters in pollen: Implications for field release of transgenic plants. J. Exp. Bot. 48 265–275. [Google Scholar]
- Xie, Y., and Varshavsky, A. (2001). RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. USA 98 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari, R., and Drews, G.N. (2004). Female gametophyte development. Plant Cell 16(Suppl): S133–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Fu, H., Walker, J., Papa, C.M., Smalle, J., Ju, Y.M., and Vierstra, R.D. (2004). Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 279 6401–6413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.