Abstract
In planta secretion of fungal pathogen proteins, including effectors destined for the plant cell cytoplasm, is critical for disease progression. However, little is known about the endoplasmic reticulum (ER) secretion mechanisms used by these pathogens. To determine if normal ER function is crucial for fungal pathogenicity, Magnaporthe oryzae genes encoding proteins homologous to yeast Lhs1p and Kar2p, members of the heat shock protein 70 family in Saccharomyces cerevisiae, were cloned and characterized. Like their yeast counterparts, both LHS1 and KAR2 proteins localized in the ER and functioned in an unfolded protein response (UPR) similar to the yeast UPR. Mutants produced by disruption of LHS1 were viable but showed a defect in the translocation of proteins across the ER membrane and reduced activities of extracellular enzymes. The Δlhs1 mutant was severely impaired not only in conidiation, but also in both penetration and biotrophic invasion in susceptible rice (Oryza sativa) plants. This mutant also had defects in the induction of the Pi-ta resistance gene–mediated hypersensitive response and in the accumulation of fluorescently-labeled secreted effector proteins in biotrophic interfacial complexes. Our results suggest that proper processing of secreted proteins, including effectors, by chaperones in the ER is requisite for successful disease development and for determining host-pathogen compatibility via the gene-for-gene interaction.
INTRODUCTION
Rice blast is the most serious biotic threat to rice (Oryza sativa) production worldwide (Ou, 1985) and has also served as an excellent model for studying plant–pathogen interactions (Valent, 1990; Talbot, 2003; Ebbole, 2007). The availability of the full genomic sequences for both rice and the fungal pathogen Magnaporthe oryzae (Yu et al., 2002; Dean et al., 2005) has accelerated studies on underlying mechanisms of rice blast disease. So far, most studies have focused on stages of the disease cycle between germination of fungal conidia on the plant surface and mechanical penetration of that surface by the appressoria, which generate turgor pressure as high as 8 MPa (Howard et al., 1991; de Jong et al., 1997) to power entry by fungal penetration pegs (Talbot, 2003; Caracuel-Rios and Talbot, 2007; Ebbole, 2007). Once inside the rice cell lumen, the hemibiotrophic blast fungus undergoes a prolonged phase of biotrophic cell invasions, first growing in every invaded cell as filamentous hyphae that soon differentiate into bulbous invasive hyphae (IH) for further colonization (Heath et al., 1990; Kankanala et al., 2007). The IH are sealed in a plant membrane, the extra-invasive-hyphal membrane (EIHM), as they fill one invaded cell and then the next.
Recent studies of the in planta secretion of the blast avirulence (AVR) effectors, AVR-Pita and PWL proteins, have identified a pathogen-induced structure, the biotrophic interfacial complex (BIC), that develops within the EIHM compartment (Berruyer, R., Khang, C.H., Kankanala, P., Park, S.Y., Czymmek, K., Kang, S. and Valent, B., unpublished data). The BIC develops first within the EIHM at the filamentous hyphal tips and then moves beside the first differentiated IH cell. Effectors secreted by IH accumulate in BICs as IH continue to colonize the host cell. Only a small number of blast effectors, presumed to be secreted into the rice cytoplasm to control host defenses and cellular processes, have been identified, based on their avirulence activity resulting in resistance gene–mediated hypersensitive response (HR) (Flor, 1971; Sweigard et al., 1995; Bryan et al., 2000; Orbach et al., 2000; Farman et al., 2002; Böhnert et al., 2004; Qu et al., 2006; Ballini et al., 2008). Important challenges remaining include identifying additional blast effectors, understanding how effectors function to promote infection or to confer recognition and resistance, and understanding how effectors are secreted in planta.
The endoplasmic reticulum (ER) is a membraneous cellular structure unique to eukaryotes. Transport into the ER is the initial step in protein secretion and is a crucial step for the biosynthesis of secretory and membrane proteins (Zimmermann et al., 2006). A signal peptide at the N terminus of the preprotein directs its transport into the ER; protein translocation across the ER occurs co- or posttranslationally depending on the nature of the preprotein. In Saccharomyces cerevisiae, two members of the heat shock protein 70 (Hsp70) family, Kar2p and Lhs1p, together with the cochaperone Sec63p and the nucleotide exchange factor Sil1p, play key roles as molecular chaperones in protein import into the ER and in proper protein folding in the ER lumen (Brodsky et al., 1995; Craven et al., 1996; Tyson and Stirling, 2000). In yeast, Lhs1p regulates reciprocal ATPase activities coordinately with Kar2p and serves as a nucleotide exchange factor for Kar2p, like Sil1p (Steel et al., 2004). These genes encode components of the yeast unfolded protein response (UPR) machinery, and their transcription is induced by ER stressors, such as DTT (an inhibitor of disulfide bond formation) and tunicamycin (an inhibitor of N-glycosylation), or by the expression of heterologous proteins (Normington et al., 1989; Rose et al., 1989; Craven et al., 1996; Tyson and Stirling, 2000). Although neither SIL1 nor LHS1 is essential, deletion of both genes in yeast results in synthetic lethality (Tyson and Stirling, 2000). The deletion mutant Δlhs1 and the conditional mutants Δlhs1Δsil1, Δsec63, and Δkar2 accumulate numerous presecretory proteins in the cytoplasm as a result of defective transport into the ER (Young et al., 2001; Steel et al., 2004).
In addition to effectors (Tian et al., 2004; Bishop et al., 2005; Kamoun, 2006), many proteins secreted by plant pathogenic fungi have been implicated in pathogenesis. These include extracellular hydrolytic enzymes that help fungi breach the host surface and colonize plant cells. Small secretory proteins, such as hydrophobins (MPG1 and MHP1) and snodprot1 (MSP1), have been shown to be important for virulence in M. oryzae (Talbot et al., 1993, 1996; Kim et al., 2005; Jeong et al., 2007), and their secretion is critical for disease development (Kershaw et al., 2005). Despite the importance of secreted proteins during plant infection, the M. oryzae secretory system is poorly understood. Recently, a P-type ATPase (APT2) was determined to be essential for the release of several secretory proteins during the infection (Gilbert et al., 2006); however, to our knowledge, no other genes associated with protein secretion have been identified. In this study, we investigated whether two putative Hsp70 family proteins in M. oryzae, KAR2 and LHS1, function as chaperones for protein translocation and maturation in the ER and whether they participate in pathogenesis. The LHS1 gene is necessary for conidiation and for the ability to cause rice blast disease, supporting the model that proper secretion of certain proteins during development and pathogenesis is critical. Biotrophic tissue colonization, secretion into BICs, and induction of Pi-ta–mediated HR are especially sensitive to mutation of the LHS1 ER chaperone gene.
RESULTS
Two Putative Hsp70 Proteins in M. oryzae Are ER Localized
We searched for putative Hsp70 homologs in the M. oryzae genome using the InterProScan program incorporated in a bioinformatics portal system, Comparative Fungal Genomics Platform (Park et al., 2008) (Table 1). Thirteen proteins were identified to contain Hsp70-related domains (IPR001023, Hsp70; IPR013126, Hsp70; IPR012725, Chaperone DnaK; note that IPR001023 is the secondary accession number of IPR013126) (see Supplemental Figure 1 online). Among them, two proteins, MGG02503.5 (DK1) and MGG06648.5 (DK5), contain a well-characterized ER retention signal (IPR000886; ER-targeting sequence) at the C terminus (HDEL in DK1 and HEEL in DK5) (Munro and Pelham, 1987; Derkx and Madrid, 2001) and a probable signal peptide (0.998 and 1.000 signal peptide probability, respectively) at the N terminus as indicated by SIGNALP (Bendtsen et al., 2004). Based on both the neural network and the hidden Markov model, the predicted cleavage site for DK1 was between residues 26 and 27 (A-Q) and for DK5 was between residues 20 and 21 (A-I). Because the DK1 and DK5 proteins exhibited sequence similarity to the yeast ER Hsp70 proteins Kar2p (e-value 0, 71.2% identity) and Lhs1p (e-value 9.00e-51, 23.4% identity), respectively, they were designated as KAR2 and LHS1 in M. oryzae. The similarity between yeast Lhs1p and LHS1 is significantly lower than that between KAR2 and yeast Kar2p due to the presence of an apolipophorin III-like domain (IPR011000) and an extended C terminus after the Hsp70 domain in LHS1. However, no homologs of LHS1 in other fungi have the apolipophorin III-like domain, and there has been no report about the functionality of this domain in fungi yet.
Table 1.
Comparison of the M. oryzae Hsp70 Homologs with Those in Other Fungi
|
M. oryzae
|
G. zeae
|
A. nidulans
|
N. crassa
|
S. cerevisiae
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Name (Locus) | Locus | E-Value | Locus | E-Value | Locus | E-Value | Locus | Gene Name | E-Value |
| DK1 (MGG_02503.5) | FG09471.1 | 0 | AN2062.3 | 0 | NCU03982.2 | 0 | YJL034W | KAR2 | 0 |
| DK2 (MGG_02842.5) | FG08644.1 | 0 | AN4616.3 | 1.00E-167 | NCU00692.2 | 0 | YHR064C | SSZ1 | 1.00E-106 |
| DK3 (MGG_04191.5) | FG06154.1 | 0 | AN6010.3 | 0 | NCU08693.2 | 0 | YJR045C | SSC1 | 0 |
| DK4 (MGG_06065.5) | FG01950.1 | 0 | AN1047.3 | 0 | NCU05269.2 | 0 | YBR169C | SSE2 | 1.00E-177 |
| DK5 (MGG_06648.5) | FG10819.1 | 0 | AN0847.3 | 0 | NCU09485.2 | 0 | YKL073W | LHS1 | 9.00E-51 |
| DK6 (MGG_06958.5) | FG00838.1 | 0 | AN5129.3 | 0 | NCU09602.2 | 0 | YLL024C | SSA2 | 0 |
| DK7 (MGG_11513.5) | FG00921.1 | 1.00E-103 | AN10202.3 | 1.00E-117 | NCU02075.2 | 1.00E-124 | YDL229W | SSB1 | 1.00E-101 |
| DK8 (MGG_11517.5) | FG00838.1 | 1.00E-105 | AN11227.3 | 1.00E-114 | NCU02075.2 | 1.00E-131 | YNL209W | SSB2 | 1.00E-126 |
| DK9 (MGG_02548.5) | FG04419.1 | 8.00E-44 | AN6099.3 | 3.00E-42 | NCU01499.2 | 7.00E-44 | – | – | – |
| DK10 (MG03039.4) | FG08548.1 | 0 | AN0866.3 | 0 | NCU00573.2 | 0 | YLR369W | SSQ1 | 1.00E-09 |
| DK11 (MGG_07156.5) | FG04419.1 | 0 | AN0587.3 | 1.00E-64 | NCU01499.2 | 0 | – | – | – |
| DK12 (MGG_09390.5) | – | – | – | – | NCU06263.2 | 1.00E-09 | – | – | – |
| DK13 (MGG_09631.5) | FG11105.1 | 2.00E-44 | AN7370.3 | 2.00E-70 | NCU04396.2 | 7.00E-43 | YBL075C | SSA3 | 5.00E-07 |
The table was derived from the results of a BLAST matrix (http://cfgp.snu.ac.kr). M. oryzae, Magnaporthe oryzae; G. zeae, Gibberella zeae; A. nidulans, Aspergillus nidulans; N. crassa, Neurospora crassa; S. cerevisiae, Saccharomyces cerevisiae.
Two fusion constructs, ProTrpC-KAR2SP-GFP-HDEL and ProLHS1-LHS1SP-GFP-HEEL, were introduced into the wild-type strain KJ201 to visualize their cellular localization. The fluorescent signals were compared with those of transformants stained with an ER-Tracker dye (Cole et al., 2000; Derkx and Madrid, 2001). The conidia of transformants expressing the fusion proteins exhibited fluorescent signals around the nucleus and parts in the cytoplasm with a thread-like pattern, and the signals were colocalized with those by the ER-Tracker dye staining (Figure 1).
Figure 1.
Cellular Localization of KAR2 and LHS1 in M. oryzae.
Conidia of transformants of KJ201 expressing ProLHS1-LHS1SP-GFP-HEEL and ProTrpC-KAR2SP-GFP-HDEL were observed under an epifluorescence microscope. The cells were stained with the ER-Tracker dye Blue-White DPX and viewed using a 4',6-diamidino-2-phenylindole filter. The original blue color from Blue-White DPX staining changed to red to better visualize colocalization with GFP signal. DIC, differential interference contrast image. Bar = 10 μm.
Disruption of LHS1 Severely Reduced Pathogenicity
The M. oryzae LHS1 gene (locus MGG06648.5) in strain 70-15 (http://www.broad.mit.edu/annotation/fungi/magnaporthe) consists of a 3-kb open reading frame (ORF) without introns and encodes a protein of 999 amino acids. LHS1 in strain KJ201 is identical in sequence to that in strain 70-15. The LHS1 gene was deleted by targeted gene replacement via Agrobacterium tumefaciens–mediated transformation (ATMT) (Figure 2A). Three transformants, k5_44, k5_35, and k5_36, were confirmed by PCR to carry the desired mutation and were genetically purified via a single conidium. Deletion of LHS1 was confirmed by DNA gel blot hybridization analysis, and loss of the LHS1 transcript was confirmed by RT-PCR (Figures 2B and 2C). Because all three mutants were identical in terms of their growth rate, conidiation, and pathogenicity, one mutant, k5_44 (referred to hereafter as Δlhs1), was used in subsequent experiments. To confirm that the phenotype exhibited by the null mutant was due to the deletion of LHS1, we complemented the mutation by introducing an 11-kb SmaI fragment carrying LHS1 via protoplast-mediated fungal transformation (Figure 2B). One complemented transformant, k5cp_19 (cp19), which contained a single copy of LHS1, was selected for further analysis. RT-PCR using mycelial RNA from cultures grown in complete liquid medium confirmed the recovery of LHS1 transcripts in cp19 at a level comparable to that in wild-type strain KJ201 (Figure 2C).
Figure 2.
Generation of the Δlhs1 Mutant by Agrobacterium-Mediated Transformation and Complementation.
(A) Gene disruption strategy. Vertical bar, XhoI site; HPH, hygromycin B phosphotransferase marker gene cassette.
(B) Genomic DNA samples from wild type (lane 1), Δlhs1 (lane 2), ect32 (lane 3), and cp19 (lane 4) were digested with XhoI and probed with XhoI-digested LHS1 (5′-flanking 477 bp).
(C) RT-PCR to confirm the loss and recovery of LHS1 transcripts. Lane 1, the wild type; lane 2, Δlhs1; lane 3, cp19.
Conidial suspensions of equal concentration from the wild-type, Δlhs1, ect32 (an ectopic transformant), and cp19 were then spray inoculated onto 3-week-old plants of rice cultivar Nakdongbyeo, which is susceptible to the wild-type KJ201. The Δlhs1 mutant exhibited significantly reduced pathogenicity relative to that in the other strains. At 7 d after inoculation (DAI), rice plants infected with the wild-type, ect32, and cp19 showed typical blast symptoms; they were covered with expanded, sporulating (grayish-centered) lesions surrounded with a yellowish margin (Figure 3A). By contrast, plants infected with the Δlhs1 mutant showed a few smaller lesions with a white center surrounded by a dark-brown circle and yellowish margins. Reduced symptoms were also observed when the spore suspensions were infiltrated into rice leaves. The plants infected with the mutant developed limited lesions with white centers at 7 DAI, whereas those inoculated with the wild-type, ect32, and cp19 strains produced expanding lesions that appeared gray due to abundant conidiation (Figure 3B).
Figure 3.
Pathogenicity of the Δlhs1 Mutant.
(A) and (B) Three-week-old rice (compatible cultivar Nakdongbyeo) plants were observed 7 DAI by spraying with conidial suspensions (1 × 105 conidia/mL) (A) and by infiltration (2 × 104 conidia/mL) (B). Both assays were maintained under high humidity that induced sporulation of lesions (gray coloration).
(C) Sheath assay with Nakdongbyeo. Sheaths were observed 48 HAI with 2 × 104 conidia/mL. Bar = 50 μm.
(D) Expression of plant defense-related genes in response to infection by the wild type and the Δlhs1 mutant was analyzed by quantitative RT-PCR after spray inoculation of the leaves of Nakdongbyeo. The graph was generated with three replicates in a representative data set, and similar results were obtained in another independent biological repetition. The error bars indicate sd of three replicates.
To determine which step(s) of infection was defective due to the loss of LHS1, the abilities to adhere, germinate, form appressoria, penetrate, and grow infectiously in planta were compared among the four strains. When observed on a hydrophobic plastic cover slip, adhesion, conidial germination, and appressorium formation in the Δlhs1 mutant were comparable to those observed in the wild-type, ect32, and cp19 strains (see Supplemental Table 2 online; data not shown); however, when penetration of onion epidermal cells was observed (see Supplemental Figure 3 online), penetration by the mutant was only 34% of that by the other strains. In the leaf sheath assay at ∼36 h after inoculation (HAI), 43% of wild-type appressoria had formed IH in the first-invaded cell, while 19% of the mutant appressoria had formed IH (Table 2). By 48 HAI, most IH of the wild type, ect32, and cp19 filled the first invaded cell and grew into a second or third cell as expected (Figure 3C). By contrast, mutant IH rarely ramified beyond the first-invaded cell at 48 HAI. These results are consistent with those from rice infection assays (Figures 3A and 3B). In addition, expression of the rice defense-related genes PR1a and PBZ1 in response to Δlhs1 infection was less than that caused by infection with the wild-type KJ201 when analyzed by quantitative RT-PCR (Figure 3D). Taken together, the reduced pathogenicity of the Δlhs1 mutant resulted from defects in penetration and infectious growth in planta.
Table 2.
Quantitative Analysis of Compatible and Incompatible Interactions of the Δlhs1 Mutant
| Compatible Interactionsa
|
Incompatible Interactionsb
|
|||
|---|---|---|---|---|
| Wild Type | Δlhs1 | Wild Type | Δlhs1 | |
| Infect sites App. | 175 | 368 | ND | ND |
| App. sites with IH | 76 (43.4%) | 72 (19.5%) | 310 | 101 |
| IH sites with HR | ND | ND | 296 (95.5%) | 9 (8.9%) |
| IH sites with Flu. BIC | 74 (97.4%) | 4 (5.6%) | ND | ND |
Fungal strains were inoculated on the sheath of susceptible rice cultivar YT16 and observed 36 ± 2 HAI.
Fungal strains were inoculated on the sheath of the rice cultivar Yashiro-mochi and observed 48 HAI after staining with Trypan blue.
App., appressorium; Flu. BIC, fluorescent biotrophic interfacial complex; ND, not determined.
The M. oryzae Δlhs1 Mutant Exhibits Defects in Asexual Development
Because conidia of M. oryzae are the primary source of inoculum, asexual development is critical to sustain the polycyclic disease cycle. The Δlhs1 mutant produced significantly fewer conidia compared with the wild type (0.3 to 4% of the wild type) (Figure 4A). The conidiation of cp19 was comparable to that of the wild type, suggesting that deletion of LHS1 caused the conidiation defect. Transcription of the class II hydrophobin gene MHP1, which is specifically induced during conidiation and in planta colonization (Kim et al., 2005), was also noticeably reduced in the Δlhs1 mutant (Figure 4B).
Figure 4.
Conidiation of the Δlhs1 Mutant and MHP1 Expression.
(A) Conidiation was compared among the wild type, the Δlhs1 mutant, and cp19 (an ectopic transformant) after growth on oatmeal agar medium (OMA) for 13 d. The asterisk denotes 0.4 ± 0.1. The values are the mean ± sd of three replicates.
(B) Transcription of MHP1 during conidiation in the Δlhs1 mutant was compared with that in the wild-type strain by quantitative RT-PCR. The profiles were normalized using CYP1 (a gene encoding cyclophilin as endogenous control) transcripts and calibrated against the vegetative growth profile. Both strains were sampled at day 10 (conidiating mycelia on OMA) and at day 3 (mycelia in complete liquid medium). The quantitative RT-PCR reactions were repeated twice independently with three replicates, and a representative set of data is presented. The error bars are sd of three replicates.
(C) Conidia development was observed under a light microscope 16 (top) and 24 h (bottom) after induction of conidiation under cover slips. Schematic drawings of conidiophores and conidia from each culture at 24 HAI were prepared based on microscopy observations. Bar = 100 μm.
To determine which stage(s) of conidial development was defective in the mutant, we observed newly developing conidia. After 16 h, one or two conidia were generated on the conidiophores of the wild type and cp19, while the mutant developed a significantly reduced number of conidiophores and conidium formation at the terminus was barely initiated. Three or more conidia were formed sympodially on the conidiophores of the wild type and cp19, while the mutant formed one, or rarely two, conidia after 24 h (Figure 4C). Therefore, the strikingly reduced conidiogenesis in the mutant resulted from a combination of decreased frequency of conidiophore formation and reduced generation of conidia.
The Δlhs1 Mutant Accumulates Less Pigment in Medium Supplemented with MnCl2 and Is Sensitive to CuSO4
To analyze additional changes in the Δlhs1 mutant, the growth rate of the mutant was compared with that of the wild type under stress-inducing conditions. The growth rate of the mutant was slightly reduced (90%) compared with that of the wild type on complete medium (CMA) (see Supplemental Figures 4 and 5 online). On CMA supplemented with Congo Red, methyl viologen, hydrogen peroxide, 3-amino-1,2,4 triazole, calcium chloride, sodium chloride, p-coumaric acid, and SDS, the growth rates of the mutant and the wild type were not significantly different from those on CMA (see Supplemental Table 3 online). By contrast, when CMA was amended with 10 mM MnCl2, the wild type produced a reddish brown exudate from the center of the colony, but the mutant produced only lightly brown or colorless exudate (see Supplemental Figure 4 online). However, the growth rates of the mutant and the wild type in the presence of 10 mM MnCl2 were similar to those of CMA. Meanwhile, although the growth rates of both the mutant and the wild type on CMA supplemented with 4 mM CuSO4 were reduced relative to those on CMA, the degree of reduction in the mutant was more severe (73% for the mutant versus 37% for the wild type), indicating that the mutant was more sensitive to CuSO4. Both strains showed reduced aerial mycelia (see Supplemental Figure 5 online).
LHS1 Expression Is Induced by ER Stress, and UPR Target Genes Are Upregulated in the Δlhs1 Mutant
To characterize if (and how) expression of LHS1 and related genes was affected by the Δlhs1 mutation, quantitative RT-PCR was performed using RNA extracted from cultures of the wild type and the mutant at various developmental stages and under stress conditions. In response to heat (42°C for 5, 10, 20, and 40 min) and cold (4°C for 40 min) shocks, levels of KAR2 and LHS1 transcripts did not change markedly from the control. However, KAR2 transcription was reduced after 80 min of a cold or heat shock (see Supplemental Figure 6 online). We then tested whether LHS1 and KAR2 transcription was altered during the recovery from heat shock. After the induction of heat tolerance by incubating cultures at 37°C for 1 h, the cultures were incubated at 50°C for 20 min and then transferred to a 25°C incubator for 15, 30, 60, 120, or 240 min. Transcription of KAR2 and LHS1 was measured at 25°C at several time points. No remarkable induction was observed for either gene, and level of LHS1 transcripts was reduced by half after 120 min (see Supplemental Figure 7 online).
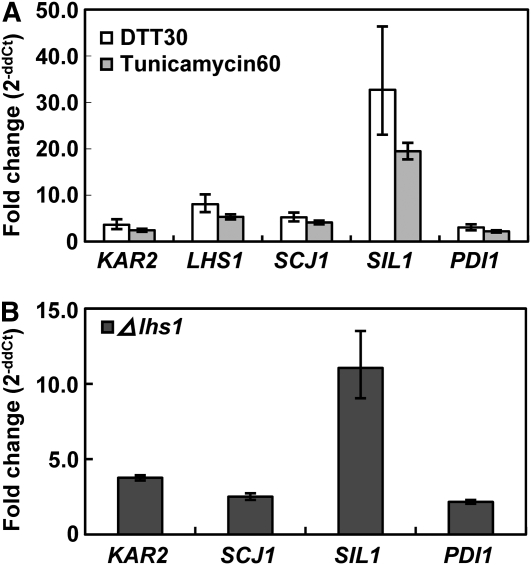
By contrast, when ER stress was induced by treatments with 10 mM DTT or 10 μg/mL tunicamycin, expression of both genes was significantly elevated (Figure 5A). Both agents have been shown to induce the transcription of genes associated with posttranslational modification in yeast and several filamentous fungi (van Gemeren et al., 1997; Kasuya et al., 1999; Travers et al., 2000; Arvas et al., 2006). Next, the homologs of Scj1p (an Hsp40 family protein), Sil1p (a nucleotide exchange factor), and Pdi1p (protein disulfide isomerase1), which are found in the ER of S. cerevisiae, were identified in the Magnaporthe genome database using BLASTP. The transcriptional regulation of these homolog genes was investigated under the same conditions. As a control, TUB, which encodes β-tubulin, was used. Upon DTT or tunicamycin treatment, transcription of SIL1, PDI1, and SCJ1 was induced more than twofold with degree of induction for SIL1 being highest (32-fold by DTT and 20-fold by tunicamycin) (Figure 5A). During vegetative growth and conidiation, transcription of KAR2, SIL1, PDI1, and SCJ1 was significantly increased (more than twofold) in the Δlhs1 mutant compared with that in the wild type (Figure 5B).
Figure 5.
Expression Profiles of UPR Pathway Genes under ER Stress and in the Δlhs1 Mutant Background.
(A) Cultures started with conidial suspension (1 × 106 conidia/flask) were grown on complete medium for 2 d, and then 10 mM DTT and 10 μg/mL tunicamycin were individually applied to cultures. DTT30 indicates samples from the wild type treated with DTT for 30 min, and Tunicamycin60 denotes samples from the wild type treated with tunicamycin for 60 min. Expression data were normalized using the β-tubulin gene (TUB) and calibrated against the profile of mycelia grown without treatment. The graph was generated with three replicates in a representative data set, and similar results were obtained from at least two independent biological repetitions. The error bars represent sd of three replicates.
(B) UPR gene expression in the Δlhs1 mutant and the wild type during conidiation was quantified using quantitative RT-PCR. Both cultures were conidiated on OMA for 10 d before RNA extraction. CYP1 was used for normalization, and the values were calculated by 2−ddCT methods with quantitative RT-PCR data. Two biological repetitions with three replicates were assayed, and a representative set of data is presented. The error bars represent sd of three replicates.
Overexpression of SIL1 Complements All Defects in the Δlhs1 Mutant
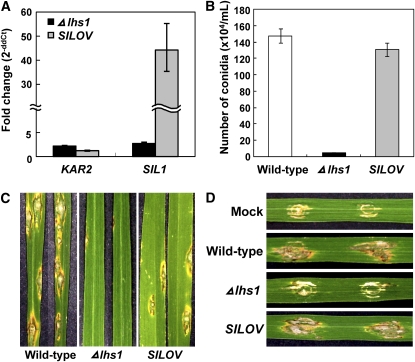
We examined whether SIL1 can substitute for LHS1 in the Δlhs1 mutant since both Lhs1p and Sil1p in S. cerevisiae serve as nucleotide exchange factor for Kar2p (Steel et al., 2004) and overexpression of SIL1 suppressed Δlhs1 mutant phenotypes (Tyson and Stirling, 2000). Overexpression of SIL1, the homolog of yeast Sil1p, using the Aspergillus nidulans TrpC promoter resulted in ∼44-fold and 16-fold increases compared with the wild type and the mutant, respectively (Figure 6A). SIL1 overexpression in a transformant carrying this construct (named SILOV) rescued all defects in pathogenicity, conidiation, and growth on CuSO4 medium (Figures 6B to 6D; see Supplemental Figure 5 online), and KAR2 expression was restored to the wild-type level (Figure 6A).
Figure 6.
Phenotypic Restoration in the Δlhs1 Mutant by SIL1 Overexpression.
(A) Quantitative RT-PCR was performed to quantify transcripts from KAR2- and SIL1-specific primers, and CYP1 transcript was used for normalization. RNA samples were extracted from cultures of the wild type, the Δlhs1 mutant, and SILOV grown in the liquid complete medium. Expression data from the Δlhs1 mutant and SILOV were calibrated using data from the wild type. Values were calculated from three replicates in a representative data set, and similar results were shown in another independent biological repetition. The error bars represent sd of three replicates.
(B) The number of conidia produced by individual strains on OMA was counted 12 DAI.
(C) and (D) Conidial suspensions (5 × 104 conidia/mL for spray inoculation and 2 × 104 conidia/mL for infiltration assay) of the individual strains were inoculated on rice cultivar Nakdongbyeo. The photos were taken 7 DAI.
LHS1 Is Required for Protein Translocation into the ER
To assess protein translocation in the Δlhs1 mutant, the ProTrpC-KAR2SP-GFP-HDEL construct (Figure 1) was introduced into the mutant. Protein gel blot analysis with anti-GFP antibody was used to investigate whether this precursor protein would be properly processed in a transformant of KJ201 carrying ProTrpC-KAR2SP-GFP-HDEL and the Δlhs1 mutant carrying the same construct. If the cytosolic precursor is properly processed by protein translocation into the ER, its molecular weight would be reduced by cleavage of the signal peptide. In the Δlhs1 mutant, the nonprocessed precursor form (30 kD) accumulated, while a 27-kD protein was detected in KJ201 (Figure 7).
Figure 7.
Defects in Protein Translocation Caused by the Δlhs1 Mutation.
Protein gel blot analysis was performed to indirectly assess protein translocation followed by removal of the signal peptide. Proteins extracted from the wild-type KJ201 (lane 1), KJ201 carrying ProTrpC-KAR2SP-GFP-HDEL (lane 2), the Δlhs1 mutant carrying ProTrpC-KAR2SP-GFP-HDEL (lane 3), and the Δlhs1 mutant (lane 4) were separated using an SDS-PAGE gel. Separated proteins were transferred to nitrocellulose membrane and probed with monoclonal anti-GFP N-terminal (top panel) and mouse anti-actin monoclonal antibody as primary antibody and then goat anti-mouse IgA+IgG+IgM (H+L) as secondary antibody (bottom panel). The scheme of the expected protein product and the signal peptide with corresponding molecular mass is depicted. [See online article for color version of this figure.]
The Δlhs1 Mutation Reduces the Activities of Extracellular Enzymes in Liquid Culture
We compared the activities of several extracellular enzymes in culture filtrates from the Δlhs1 mutant and the wild type. Six enzymes, including xylanase, xylosidase, arabinosidase, glucanase, polygalacturonase, and laccase, were assayed. The enzymatic activities of xylosidase, arabinosidase, glucanase, and laccase were significantly reduced in the Δlhs1 mutant (41 to 67% of the wild-type activity), and those of xylanase and polygalacturonase were slightly reduced in the mutant (85 to 90% of the wild-type activity) (Figure 8).
Figure 8.
The Δlhs1 Mutant Appears Defective in the Secretion of Extracellular Enzymes.
Enzymatic activities of xylanase, polygalacturonase, glucanase, xylosidase, arabinosidase, and laccase were assayed using culture filtrates from the wild type and the Δlhs1 mutant grown in modified HMT medium supplemented with 1% rice cell wall. Relative enzymatic activities (%) in the Δlhs1 mutant compared with those of the wild type were calculated after normalization with the amount of proteins in each extract. The values were calculated from the results with a mixture of two replicates in a representative data set, and similar results were shown in a separate biological repetition. Ara, arabinosidase; Xylo, xylosidase; Lac, laccase; Xyl, xylanase; PGa, polygalacturonase; Glu, glucanase.
The Δlhs1 Mutant Negatively Affects AVR-Pita Function and Secretion
To determine whether the deletion of LHS1 affects the secretion of effector proteins, we investigated the effect of this mutation on the AVR-Pita- and Pi-ta–mediated gene-for-gene interaction (Bryan et al., 2000; Jia et al., 2000; Orbach et al., 2000). Rice cultivar Yashiro-mochi containing the cognate resistance protein Pi-ta displayed an extensive HR to the AVR-Pita–containing wild-type KJ201 (Figure 9A), which often included macroscopically visible, uniformly dark-brown Type 1 resistance spots (Valent et al. 1991). By contrast, the Δlhs1 mutant no longer induced the extensive Type 1 HR spots on Yashiro-mochi. As in the compatible interactions on Nakdongbyeo and YT16 (Figures 3A, 6B, and 9A), the mutant forms rare, restricted lesions on Yashiro-mochi compared with those formed by the fully compatible pathogen O-135. At the cellular level, hypersensitive death of invaded Yashiro-mochi sheath epidermal cells was assessed by staining with Trypan blue. The HR occurred in 95.5% of the rice cells invaded by the wild type, but only in 8.9% of the cells invaded by the Δlhs1 mutant (Table 2). Therefore, as observed at both the macroscopic and microscopic levels, mutation of the LHS1 ER chaperone greatly impaired Pi-ta–mediated resistance, presumably due to a defect in secretion of the AVR-Pita protein.
Figure 9.
Mutation of LHS1 Impacted AVR-Pita Function and Secretion.
(A) Whole-plant assays with cultivars Yashiro-mochi (Pi-ta) and YT16 (pi-ta−) inoculated with wild-type KJ201 (AVR-Pita), mutant Δlhs1 (AVR-Pita), and a positive control for infection, strain O-135 (avr-pita−). Incompatible KJ201 generally failed to form visible symptoms except for rare, dark-brown, nonsporulating spots due to extended HR on Yashiro-mochi, while it generated typical expanding lesions on compatible cultivar YT16. The Δlhs1 mutant formed rare, small, white lesions on Yashiro-mochi, similar to those it produced on susceptible cultivar YT16. Photographed at 7 DAI.
(B) In Yashiro-mochi rice sheaths at 48 HAI, the wild-type strain had induced hypersensitive death in 96% of first-invaded cells containing IH (left), but the mutant had induced HR in only 9% of the rare rice cells containing IH (right). Tissues were stained with Trypan blue to highlight the fungus and rice HR. Typical images are shown. Bars = 20 μm.
(C) In susceptible YT16 sheath cells, GFP expressed by wild-type KJ201 using the AVR-Pita promoter and signal peptide (APitaSP:GFP) was secreted and accumulated in the BIC (arrowheads) (left). As shown in Table 2, GFP expressed by the Δlhs1 mutant (APitaSP:GFP) was generally not observed in BICs (right). Photos taken at 36 ± 2 HAI (top, differential interference contrast image; bottom, GFP filter with 4-s exposure time). Both pictures were taken with the same magnification. Bars = 10 μm. Schematic drawings of development of BICs in the first cells were prepared based on microscopy observations.
To directly examine secretion of AVR-Pita by the Δlhs1 mutant, we compared in planta secretion into BICs by KJ201 and Δlhs1 transformants expressing GFP fused to the AVR-Pita signal peptide (APitaSP:GFP) under control of the native AVR-Pita promoter. For wild-type strains, such fluorescent effector reporters are secreted into BICs at filamentous hyphal tips, and the fluorescence remains in the BICs when they assume their final locations beside the first IH cell (Berruyer, R., Khang, C.H., Kankanala, P., Park, S.Y., Czymmek, K., Kang, S. and Valent, B., unpublished data). We focused on primary BICs beside IH in first-invaded cells (Figure 9C) to quantify infection sites that secreted APitaSP:GFP into BICs. Considering the infection sites with wild-type IH, 97.4% had fluorescence in BICs, but only 5.6% of the infection sites with mutant IH had fluorescence in BICs (Table 2). In those cells where BIC fluorescence was observed in the mutant, this fluorescence was much weaker than in the wild type. We concluded that the LHS1 ER chaperone is required for proper secretion of the AVR-Pita effector protein in planta.
DISCUSSION
Secreted effector proteins play key roles in pathogenesis (Nimchuk et al., 2003; Kamoun, 2006). Unlike the well-characterized mechanisms underlying effector protein secretion in many bacterial plant pathogens (Alfano and Collmer, 2004; Ghosh, 2004), little is known about how eukaryotic pathogens secrete effectors (Caracuel-Rios and Talbot, 2007; Kamoun, 2007). Most eukaryotic effectors have an N-terminal signal peptide for translocation into the ER, implicating classical ER-mediated secretion of effectors outside the pathogen (Zimmermann et al., 2006). However, experimental data supporting this hypothesis were limited.
For rice blast disease, we have demonstrated that the ER chaperone LHS1 and proper ER function play a role in effector secretion. The Δlhs1 mutant was highly impaired in secretion of AVR-Pita:GFP fusion proteins into BICs, complex interfacial aggregations of lamellar membranes, and vesicles that accumulate secreted effectors in planta (Berruyer, R., Khang, C.H., Kankanala, P., Park, S.Y., Czymmek, K., Kang, S. and Valent, B., unpublished data). Additionally, the mutant exhibited dramatically reduced pathogenicity toward two rice cultivars lacking major gene resistance (Figures 3A to 3C and 9A) and dramatically reduced avirulence toward a rice cultivar containing resistance gene Pi-ta (Figure 9). At the macroscopic level, both the compatible and incompatible interactions were reduced to low infection levels characterized by small, sparse lesions (Figures 3 and 9). At the cellular level in leaf sheath assays, vigorous colonization by IH in the compatible interaction and relatively uniform HR blocking IH growth in the incompatible interaction were replaced by limited growth of Δlhs1 IH at some infection sites (Figures 3B, 9B, and 9C). A mutant with impaired abilities to induce HR in resistant plants and pathogenicity in susceptible plants resembles hrp− mutants in bacterial plant pathogens, which are defective in the specialized Type III system for secreting effectors inside host cells. However, our results suggest that a highly conserved component of the eukaryotic secretion system is involved in effector secretion in rice blast disease.
Disease processes that occur in planta were more affected by loss of LHS1 than processes that occur on the outer plant surface. Conidial germination and appressorium formation were both normal in the mutant. By contrast, the mutant was defective in appressorial penetration, in plant colonization, in inducing HR, and in sporulation. The Δlhs1 mutant produced very few conidia compared with the wild-type strain (Figure 4) due to infrequent conidiophore formation and slow conidial development. In the field, each fully compatible blast lesion produces thousands of conidia that reinitiate the disease cycle (Ou, 1985). Considering that rice blast is a polycyclic disease, reduction in conidial development would be highly detrimental to development of disease epidemics. Taken together, our results confirm that secreted proteins play a significant role in diverse aspects of disease development and resistance reactions. With such diverse aspects of pathogenicity affected, it is somewhat surprising that the mutant was consistently capable of forming some small lesions on rice. However, the reduced secretion exhibited by the mutant was apparently sufficient to form the limited disease symptoms.
In this study, we investigated the role of the well-conserved Hsp70 family of chaperones, which are involved in protein transport into the ER and proper protein folding (Conesa et al., 2001; Ni and Lee, 2007), in pathogenesis and development. In yeast, Kar2p and Lhs1p (homologs of human GRP78/Bip and GRP170/ORP150, respectively) play critical roles in protein translocation and protein folding in the ER (Normington et al., 1989; Rose et al., 1989; Baxter et al., 1996; Craven et al., 1996; Hamilton and Flynn, 1996; Saris et al., 1997; Tyson and Stirling, 2000; Young et al., 2001). We identified proteins homologous to Kar2p (KAR2) and Lhs1p (LHS1) in M. oryzae based on sequence homology and functionally characterized them. Both proteins had a signal peptide at the N terminus and an ER retention signal at the C terminus (see Supplemental Figure 1 online). When the signal peptides from KAR2 and LHS1 were fused to a GFP reporter and introduced to M. oryzae, green fluorescence colocalized with ER (Figure 1), suggesting that these proteins function in the ER. Efforts to obtain null mutants for KAR2 have been unsuccessful even after screening several hundred transformants from three independent transformations (data not shown), suggesting that KAR2 may be an essential gene in M. oryzae, similar to its homolog in S. cerevisiae and Aspergillus niger (Rose et al., 1989; Punt et al., 1998). However, null mutants generated via targeted gene disruption of LHS1 provided insight into the roles of ER chaperones in pathogenicity and development of M. oryzae.
Although sequence identity between LHS1 and yeast Lhs1p (23%) is very limited compared with that between KAR2 and yeast Kar2p (71%) and LHS1 contains extra domain(s) that are absent in Lhs1p, the function, regulation of expression of LHS1, and its functional relationships with other known ER chaperone genes appear conserved. The ER stress-induced UPR includes transcriptional upregulation of chaperone genes, attenuation of protein synthesis, and retro-translocation of misfolded proteins to the cytosol for degradation. This evolutionarily well-conserved pathway plays key roles in overcoming stresses that negatively affect ER function and cause accumulation of unfolded protein (Marciniak and Ron, 2006; Ni and Lee, 2007). During UPR, many genes encoding secreted proteins are downregulated in yeast and Arabidopsis thaliana (Martinez and Chrispeels, 2003; Pakula et al., 2003; Al-Sheikh et al., 2004).
By contrast, expression of genes involved in protein translocation and folding, ER-activated degradation, and protein secretion are induced (Travers et al., 2000; Martinez and Chrispeels, 2003). In A. niger, a genome-wide transcriptional analysis of genes differentially regulated during UPR stress revealed that genes homologous to M. oryzae LHS1 and KAR2 were significantly upregulated (Guillemette et al., 2007). In M. oryzae, genes homologous to previously known ER chaperone genes, such as KAR2, SCJ1, SIL1, PDI1, and LHS1, were concurrently induced in response to ER stresses caused by DTT and tunicamycin (Figure 5), which is consistent with findings in other organisms (Travers et al., 2000; Martinez and Chrispeels, 2003; Guillemette et al., 2007). The first four genes were also expressed at higher levels in the Δlhs1 mutant than in the wild type, suggesting that UPR is constitutively induced in the mutant.
Expression of SIL1 in the Δlhs1 mutant was highly upregulated, probably to compensate for the absence of LHS1. Both proteins may function as nucleotide exchange factors for KAR2, similar to their homologs in S. cerevisiae (Steel et al., 2004). However, the magnitude of induction seemed insufficient to ensure normal conidial development and pathogenicity. When SIL1 was overexpressed in the Δlhs1 mutant using a strong constitutive promoter (A. nidulans TrpC promoter), mutant phenotypes in asexual development and pathogenicity were restored (Figure 6). At the molecular level the transcription of KAR2 was reduced nearly to the wild-type level, supporting that UPR was alleviated by overexpression of SIL1.
Two additional lines of evidence indicate that LHS1 is required for proper secretion of some proteins, thus rendering the Δlhs1 mutant hypovirulent compared with wild-type strains. First, the Δlhs1 mutant could not efficiently remove the signal peptide from GFP fused to the KAR2 signal peptide (Figure 7). This result implies that such effectors as well as certain secretory proteins are not or are partially translocated into the ER and that they are blocked or they proceed improperly through the secretory pathway. The yeast lhs1 mutant displayed defects in the translocation of many types of proteins, including secreted proteins (e.g., prepro-α-factor), ER proteins (e.g., Kar2p and PDI), and vacuolar proteins (e.g., proteinase A and carboxy peptidase) (Baxter et al., 1996; Craven et al., 1996; Tyson and Stirling, 2000). Second, a subset of extracellular enzymes displayed reduced activities (41 to 90% of the wild-type level) in the culture filtrate of the Δlhs1 mutant (Figure 8), suggesting that these secreted proteins need LHS1 for their proper maturation and secretion. However, considering that not all enzymes exhibited reduced activity and that there was no complete blocking of the enzymes, proteins other than LHS1 also carry out such functions. The SIL1 protein, which can complement the loss of LHS1 when overexpressed, is a candidate. It also remains possible that there exists more than one pathway to secrete proteins in M. oryzae. The MgAPT2 gene, which encodes a Golgi-localized protein required for exocytosis and shows defects in a subset of extracellular enzyme secretion (Gilbert et al., 2006), may participate in an alternative pathway.
In M. oryzae, a large number of genes involved in the prepenetration stages of infection have been identified (Talbot, 2003; Caracuel-Rios and Talbot, 2007; Ebbole, 2007; Jeon et al., 2007). By contrast, only a few genes have been shown to be involved in the postpenetration ramification stages, including biotrophic growth in the first cell and neighboring cells and lesion expansion. The integral membrane P-type ATPase, encoded by the MgAPT2 gene, has been shown to be a Golgi-localized protein and to function in host cell colonization and exocytosis (Gilbert et al., 2006). Similar to Δlhs1, the ΔMgapt2 mutant failed to induce plant defenses during incompatible interactions, presumably due to unsuccessful delivery of effector proteins. However, unlike Δlhs1, ΔMgapt2 mutants exhibited normal sporulation. Since Golgi-localized secretion functions lie downstream from ER-localized functions, it remains reasonable that effector secretion pathways might diverge after the ER translocation step.
In summary, successful fungal growth in planta would require proper functioning of the secretory machinery and chaperones in the ER so that effectors counteract and/or manipulate host defense responses. In the Δlhs1 mutant, where UPR is already induced, the ER system was probably overloaded, resulting in reduced effector protein secretion. This restricts ramification of the pathogen after penetration of the host plant. The contribution of LHS1 to protein translocation and secretion of proteins, including effectors, revealed the importance of the ER chaperones for successful disease development by the rice blast fungus. Our results suggest that many other structural and regulatory components in the LHS1-mediated secretory pathway will be important for disease development. Some of the proteins that are secreted through this machinery will also play important roles in pathogenesis. Thus, characterization of both types of genes may lead to novel strategies for controlling rice blast disease.
METHODS
Fungal Strains and Culture Conditions
Magnaporthe oryzae strain KJ201 was used as the wild-type strain throughout this research. This strain and its transformants were cultured on OMA (5% oatmeal [w/v] and 2.5% agar powder) at 25°C under continuous fluorescent light and were cultured in liquid complete medium (0.6% yeast extract, 0.6% casamino acid, and 1% sucrose) at 25°C for 3 d with agitation (150 rpm) for genomic DNA extraction. For RNA extraction, conidial suspensions of individual strains were cultured in liquid complete medium as described by Talbot et al. (1993) for 2 to 3 d and were subsequently subjected to heat shock, cold shock, or ER stress treatments, such as DTT (10 mM) and tunicamycin (10 μg/mL). To observe vegetative mycelial growth under stress, 4 mM CuSO4 or 10 mM MnCl2 was individually added to CM agar medium (Talbot et al., 1993). A mycelial disc (7 mm in diameter) of each strain was inoculated upside down on these stress media, and the growth rate was assessed by measuring culture diameters every 4th day until the 12th day. Rice (Oryza sativa) pathogen O-135 was used as a control for normal disease development on cultivar Yashiro-mochi (Valent et al., 1991)
Nucleic Acid Manipulation and Quantitative RT-PCR
Most molecular biology related techniques, including preparation of plasmid DNA, restriction enzyme digestion, cloning, and DNA gel blot hybridization analysis, were performed as previously described (Sambrook and Russell, 2001). Genomic DNA and total RNA was extracted as described by Kim et al. (2005). For the PCR screening of the transformants, genomic DNA was extracted by a quick and safe (QS) DNA extraction method (Chi et al., 2009).
To quantify levels of transcripts, quantitative RT-PCR was performed as described before (Yi and Lee, 2008). Briefly, 5 μg of RNA was reverse transcribed into cDNA using the SuperScript first-strand synthesis system (Invitrogen) according to the manufacturer's instructions. Reactions were performed in 10 μL volume containing 150 nM of each primer, 25 ng of cDNA, and 5 μL of SYBR Green PCR Master Mix (Applied Biosystems) using 7500 Real-Time PCR system with instrument default cycle conditions (Applied Biosystems). To analyze relative abundance of transcripts with the 2−ΔΔCt method (Livak and Schmittgen, 2001), average threshold cycle (Ct) was normalized either by β-tubulin or cyclophilin for each condition as 2−ΔCt. Fold changes were calculated as 2−ΔΔCt. The primer pairs used for quantitative RT-PCR are listed in Supplemental Table 1 online. The PCR reactions were repeated at least twice independently with three replicates, and a representative set of data was presented.
Cellular Localization of LHS1 and KAR2 in M. oryzae
A 1.2-kb genomic fragment including the putative promoter and the predicted signal peptide of LHS1 gene was amplified with primers DK5fu1F and DK5fu_SigR (see Supplemental Table 1 online). The eGFP ORF (0.7 kb) was amplified with primers eGFP_M and *HEELgfpR using pSK2707 as the template. The resulting PCR fragments were fused by double-joint PCR (Yu et al., 2004) using primers DK5fu1F and *HEELgfpR, and the resulting 1.9-kb fragment was cloned to XL-TOPO vector (Invitrogen). The Aspergillus nidulans TrpC promoter, amplified with primers pTrpC_F and pTrpC_R (0.3 kb) from pBCATPH (Yun, 1998), the KAR2 ORF without the stop codon, amplified with primers trpDK1fuF and DK1fu1R from KJ201 genomic DNA, and eGFP ORF, amplified with primers eGFP_M and eGFP_P1 from pSK2707. A 0.4-kb fragment carrying the TrpC promoter and the signal peptide region of KAR2 was amplified with primers pTrpC_F and DK1fu_sigR, and the eGFP ORF, amplified with primers eGFP_M and *HDELgfpR from pSK2707, were fused by double-joint PCR with primers pTrpC_F and *HDELgfpR, and the resulting 1.1-kb fragment was cloned to XL-TOPO vector.
The constructs encoding fusion proteins ProLHS1-LHS1SP-GFP-HEEL and ProTrpC-KAR2SP-GFP-HDEL were introduced to the wild type by cotransformation with pII99 vector (Lee et al., 2003) carrying a gene conferring resistance to geneticin. Fluorescence signals in the conidia of transformants were observed after staining with the ER-Tracker Blue-White DPX (Molecular Probes) using a fluorescence microscope (Zeiss Axio Imager A1; Cal Zeiss) with GFP and 4',6-diamidino-2-phenylindole filters.
Disruption of LHS1 and Complementation
Based on the sequences at the locus MGG_06648.5 in the M. grisea genome (http://www.broad.mit.edu/annotation/fungi/magnaporthe/), the 5′ 1.5-kb and 3′ 1.3-kb flanking regions of the gene were amplified using primers nH_kdk5F and f_kdk5_5r and primers nXb_kdk5R and f_kdk5_3f, respectively, from genomic DNA of KJ201. The primers f_kdk5_5r and f_kdk5_3f contain extra sequences that are homologous to the ends of the HPH marker cassette. The 2.1-kb HPH marker cassette was amplified from pBCATPH (Seo et al., 2007) using primers HygB_F and HygB_R. These three amplicons were fused by double-joint PCR, and the resulting mutant allele was amplified using primers H3_kdk5_Fw and Xb_kdk5_Rv from the diluted fusion PCR product as previously described (Yu et al., 2004). The mutant allele was cloned into pCR-TOPO 2.1 vector (Invitrogen), and the HindIII-XbaI fragment was cloned into pSK1834 ATMT binary vector (Khang et al., 2005). The resulting clone was introduced to Agrobacterium tumefaciens strain AGL-1, and ATMT of KJ201 was performed as previously described (Rho et al., 2001). Transformants were selected on medium supplemented with 200 ppm hygromycin twice and screened by PCR. Transformants carrying the expected null mutation and ectopic transformants were genetically purified via single conidium isolation. Deletion of the LHS1 gene was confirmed by DNA gel blot hybridization analysis, and the loss of LHS1 transcripts was also confirmed by RT-PCR.
For complementation, a fosmid clone of KJ201 carrying the LHS1 gene, FOSKJ_A_11_L10, was isolated, and the 11-kb SmaI fragment in this fosmid clone was subcloned to pBluescript SK+. The resulting plasmid was named as 11_L10pBlu_20 and used for cotransforming Δlhs1 protoplast with pII99 vector (Lee et al., 2003) as previously described (Sweigard et al., 1992). Resulting transformants were selected on TB3 agar medium amended with 800 ppm geneticin. After genetic purification by single conidium isolation, the presence of a single-copy insertion of LHS1 was checked by DNA gel blot hybridization analysis, and RT-PCR using mycelial RNA extracted from cultures grown in complete medium confirmed the recovery of LHS1 transcript in the transformants.
Assays of Conidial Germination, Appressorium Formation, and Conidiation, and Pathogenicity
A disk of culture was placed on OMA and cultured at 25°C under constant fluorescent light for 12 d with four to five replicates. Conidia were collected from these cultures, resuspended in 5 mL of sterile distilled water, and counted using a hemacytometer with 20 μL of conidial suspension. Observation of the conidiation process was performed as previously reported (Lau and Hamer, 1998). Conidial suspensions (2 × 104 conidia/mL) were used for assaying conidial germination and appressorium formation. Thirty microliters of each suspension was dropped onto microscope cover slips (Marienfeld) with three replicates. After 12 h of incubation in a moistened box at 25°C, conidial germination and appressorium formation were observed with a light microscope.
Conidia harvested from 13-d-old OMA culture using sterile distilled water were adjusted to105 conidia/mL with 250 ppm Tween 20 before inoculation of plants. Five milliliters of the conidial suspension was sprayed on each pot with rice plants at the 3- to 4-leaf stage. Inoculated plants were placed in a dark dew chamber overnight and moved to a growth chamber. The results were observed 7 DAI. For the infiltration assay, 100 to 200 μL of 2 × 104 conidia/mL conidial suspension was injected into rice leaves using a syringe without a needle. One-month-old rice sheaths of cultivars Nakdongbyeo, Yashiro-mochi, or YT16 and conidia from 13-d-old OMA cultures (conc. 1 to 2 × 104 conidia/mL) were used for sheath assays. At 48 HAI, sheaths were trimmed to produce epidermal layers three to four cells thick from above the midvein. Trimmed sheath tissue sections were mounted on glass slides. The specimens were observed using either an Zeiss Axio Imager A1 fluorescence microscope with ×40 objective or a Zeiss Axioplan 2 IE MOT microscope with a C-Apochromat ×63 (numerical aperture of 1.2) water immersion objective (all from Carl Zeiss). Fluorescence was observed with an X-Cite 120 mercury lamp source (EXFO Life Sciences) and a GFP-specific filter (excitation 480 ± 20 nm, emission 510 ± 20 nm, filter set 41020; Chroma Tech.). To assess the incompatible interactions, rice cultivar Yashiro-mochi was used with either Nakdongbyeo or YT16 as compatible interaction controls, and other protocols were applied as described above for spray inoculation and sheath assay. All pathogenicity assays were repeated at least four times with similar results. For observing cell death, the sheath samples were stained with Trypan blue for 1 h and destained in lactophenol for 1 h. The onion assay to observe infectious growth and autofluorescence was performed as described (Kim et al., 2005) without staining.
Overexpression of SIL1
The ORF of SIL1 (MGG_10843.5) plus the 1.1-kb 3′ untranslated region was fused to a strong promoter, the A. nidulans TrpC promoter, by fusion PCR. After cloning the amplicon into pCR-XL-TOPO vector (Invitrogen), the sequences of the insert were confirmed by sequencing using an ABI 3700 DNA sequencer (PE Applied Biosystems) at The National Instrumentation Center for Environmental Management (Seoul National University, Seoul, Korea). The Δlhs1 mutant was cotransformed with pII99 vector (Lee et al., 2003) and a sequence-confirmed clone via protoplast-mediated fungal transformation, and resulting transformants were selected on a geneticin-containing medium and verified by DNA gel blot hybridization analysis and quantitative RT-PCR as described above.
Protein Translocation Assay
Fungal strains were grown in liquid complete medium (Talbot et al., 1993) for 2 d and transferred to new medium after pulverization of mycelia. After 24 h subculture, the mycelia from each strain were collected. Proteins were extracted from individual cultures according to the method described previously by Bruno et al. (2004). Total proteins (∼20 μg) were separated on an SDS-12% polyacrylamide gel and transferred to nitrocellulose membrane (Bio-Rad Laboratories). The blot was prepared in duplicate and probed with monoclonal anti-Green Fluorescent Protein, N-terminal (Sigma-Aldrich) as primary antibody (1:500) and goat anti-mouse IgA+IgG+IgM (H+L) (Pierce Biotechnology) as secondary antibody (1:20,000). As controls, mouse anti-actin monoclonal antibody (Chemicon International) (1:1,000) and goat anti-mouse IgA+IgG+IgM (H+L) (1:100,000) were used.
Assays of Secreted Enzyme Activities
Mycelia of the wild type and the Δlhs1 mutant were stationary cultured for 16 d in the HMT medium (van Hoof et al., 1991), substituted with rice cell wall as the major carbon source. Activities of xylanase, polygalacturonases, and glucanase were assayed using the DNS method (Miller, 1959) with 0.1% xylan and 0.1% polygalacturonic acid as substrates. Activities of β-xylosidase and α-arabinosidase (Chaplin and Kennedy, 1986) and laccase were assessed using 1 mM 4-nitrophenyl β-d-xylopyranoside, 4-nitrophenyl α-l-arabinofuranoside and 2,2′- azino-bis-3-ethylbenzothiazoline-6-sulfonic acid as respective substrates.
Analysis of AVR-Pita Fusion Protein Secretion into the BIC
The AVR-Pita promoter and signal peptide fused to eGFP construct (Berruyer, R., Khang, C.H., Kankanala, P., Park, S.Y., Czymmek, K., Kang, S. and Valent, B., unpublished data) was used to cotransform KJ201 and the Δlhs1 mutant with the pII99 vector via protoplast-mediated fungal transformation. Transformants were screened by PCR and DNA gel blot hybridization analysis. Several independently isolated transformants were inoculated on the sheath of YT16 and observed under a fluorescent microscope 36 ± 2 h after inoculation.
Accession Numbers
The GenBank accession numbers for the M. oryzae sequences described in this article are as follows: LHS1 (MGG06648, ABS87374), KAR2 (MGG02503, ABS87375), SIL1 (MGG10843, EU156060), SCJ1 (MGG07502, EU156061), MHP1 (MGG01173, AF126872), MSP1 (MGG05344, XP359969.1), and CYP1 (MGG10447, AF293848). The sequences for PDI1 (MGG05753, XM360379), TUB (MGG00604, XM368640), and other homologs in Supplemental Figure 1 online, DK2 (MGG02842, XM366766), DK3 (MGG04191, XM361717), DK4 (MGG06065, XM369399), DK6 (MGG06958, XM370461), DK7 (MGG11513, XM001410873), DK8 (MGG11517, XM001410877), DK9 (MGG02548, XM366472), DK10 (MGG03039, XM366963), DK11 (MGG07156, XM367231), DK12 (MGG09390, XM364536), and DK13 (MGG09631, XM364786), were taken from the M. grisea 70-15 genome database. PBZ1 (D38170), PR1a (AJ278436), and ACT (Os03g50890) are genes from O. sativa.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of the Hsp70 Homologs in M. oryzae.
Supplemental Figure 2. Complementation of the Yeast Mutant Δlhs1 with LHS1.
Supplemental Figure 3. Penetration and Infectious Growth of the Δlhs1 Mutant and Its Complementation Transformant (cp19) on Onion Cells.
Supplemental Figure 4. Growth of the Δlhs1 Mutant on Manganese-Containing Medium.
Supplemental Figure 5. Growth of the Δlhs1 Mutant on Copper Sulfate–Containing Medium.
Supplemental Figure 6. Changes in KAR2 and LHS1 Expression after Heat and Cold Shocks.
Supplemental Figure 7. Changes in KAR2 and LHS1 Expression during the Recovery from a Heat Shock.
Supplemental Table 1. Primers Used in This Study.
Supplemental Table 2. Conidial Germination and Appressorium Formation of the Δlhs1 Mutant and Its Complements.
Supplemental Table 3. Growth Rate of Δlhs1 under Various Stress Conditions.
Supplementary Material
Acknowledgments
We thank Joong-Hoon Ahn (Konkuk University) for insightful suggestions and for providing materials used for the extracellular enzyme assay and Soonok Kim and Yangseon Kim for helpful discussions. This research was supported by a grant from the Crop Functional Genomics Center (CG1141) of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology, by a grant from the Biogreen21 project (20080401-034-044-008-01-00) funded by Rural Development Administration, and by a Korea Science and Engineering Foundation grant funded by the Korean government (Ministry of Education, Science, and Technology) (R11-2008-062-03001-0) to Y.-H.L. This work was also supported by USDA-National Research Initiative grants 2006-35319-17296 to B.V. and 2005-35319-15310 to S.K. M.Y. is grateful for the graduate fellowships by the Ministry of Education through the Brain Korea 21 Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yong-Hwan Lee (yonglee@snu.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alfano, J.R., and Collmer, A. (2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42 385–414. [DOI] [PubMed] [Google Scholar]
- Al-Sheikh, H., Watson, A.J., Lacey, G.A., Punt, P.J., MacKenzie, D.A., Jeenes, D.J., Pakula, T., Penttila, M., Alcocer, M.J., and Archer, D.B. (2004). Endoplasmic reticulum stress leads to the selective transcriptional downregulation of the glucoamylase gene in Aspergillus niger. Mol. Microbiol. 53 1731–1742. [DOI] [PubMed] [Google Scholar]
- Arvas, M., Pakula, T., Lanthaler, K., Saloheimo, M., Valkonen, M., Suortti, T., Robson, G., and Penttila, M. (2006). Common features and interesting differences in transcriptional responses to secretion stress in the fungi Trichoderma reesei and Saccharomyces cerevisiae. BMC Genomics 7 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini, E., Morel, J.B., Droc, G., Price, A., Courtois, B., Notteghem, J.L., and Tharreau, D. (2008). A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 21 859–868. [DOI] [PubMed] [Google Scholar]
- Baxter, B.K., James, P., Evans, T., and Craig, E.A. (1996). SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol. Cell. Biol. 16 6444–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. [DOI] [PubMed] [Google Scholar]
- Bishop, J.G., Ripoll, D.R., Bashir, S., Damasceno, C.M.B., Seeds, J.D., and Rose, J.K.C. (2005). Selection on glycine beta-1,3-endoglucanase genes differentially inhibited by a phytophthora glucanase inhibitor protein. Genetics 169 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnert, H.U., Fudal, I., Dioh, W., Tharreau, D., Notteghem, J.L., and Lebrun, M.H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., Goeckeler, J., and Schekman, R. (1995). BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92 9643–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, K.S., Tenjo, F., Li, L., Hamer, J.E., and Xu, J.R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, G.T., Wu, K.S., Farrall, L., Jia, Y., Hershey, H.P., McAdams, S.A., Faulk, K.N., Donaldson, G.K., Tarchini, R., and Valent, B. (2000). A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuel-Rios, Z., and Talbot, N.J. (2007). Cellular differentiation and host invasion by the rice blast fungus Magnaporthe grisea. Curr. Opin. Microbiol. 10 1–7. [DOI] [PubMed] [Google Scholar]
- Chaplin, M.F., and Kennedy, J.F. (1986). Carbohydrate Analysis: A Practical Approach. (Washington DC: IRL Press).
- Chi, M.H., Park, S.Y., Kim, S., and Lee, Y.H. (2009). A quick and safe method for fungal DNA extraction. Plant Pathol. J. 25 108–111. [Google Scholar]
- Cole, L., Davies, D., Hyde, G.J., and Ashford, A.E. (2000). ER-Tracker dye and BODIPY-brefeldin A differentiate the endoplasmic reticulum and golgi bodies from the tubular-vacuole system in living hyphae of Pisolithus tinctorius. J. Microsc. 197 239–249. [DOI] [PubMed] [Google Scholar]
- Conesa, A., Punt, P.J., van Luijk, N., and van den Hondel, C.A. (2001). The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33 155–171. [DOI] [PubMed] [Google Scholar]
- Craven, R.A., Egerton, M., and Stirling, C.J. (1996). A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 15 2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434 980–986. [DOI] [PubMed] [Google Scholar]
- de Jong, J.C., McCormack, B.J., Smirnoff, N., and Talbot, N.J. (1997). Glycerol generates turgor in rice blast. Nature 389 244–245. [Google Scholar]
- Derkx, P.M., and Madrid, S.M. (2001). The foldase CYPB is a component of the secretory pathway of Aspergillus niger and contains the endoplasmic reticulum retention signal HEEL. Mol. Genet. Genomics 266 537–545. [DOI] [PubMed] [Google Scholar]
- Ebbole, D.J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45 437–456. [DOI] [PubMed] [Google Scholar]
- Farman, M.L., Eto, Y., Nakao, T., Tosa, Y., Nakayashiki, H., Mayama, S., and Leong, S.A. (2002). Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol. Plant Microbe Interact. 15 6–16. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9 275–276. [Google Scholar]
- Ghosh, P. (2004). Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68 771–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, M.J., Thornton, C.R., Wakley, G.E., and Talbot, N.J. (2006). A P-type ATPase required for rice blast disease and induction of host resistance. Nature 440 535–539. [DOI] [PubMed] [Google Scholar]
- Guillemette, T., van Peij, N.N., Goosen, T., Lanthaler, K., Robson, G.D., van den Hondel, C.A., Stam, H., and Archer, D.B. (2007). Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics 8 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, T.G., and Flynn, G.C. (1996). Cer1p, a novel Hsp70-related protein required for posttranslational endoplasmic reticulum translocation in yeast. J. Biol. Chem. 271 30610–30613. [DOI] [PubMed] [Google Scholar]
- Heath, M.C., Valent, B., Howard, R.J., and Chumley, F.G. (1990). Interactions of two strains of Magnaporthe grisea with rice, goosegrass, and weeping lovegrass. Can. J. Bot. 68 1627–1637. [Google Scholar]
- Howard, R.J., Ferrari, M.A., Roach, D.H., and Money, N.P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 88 11281–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J., et al. (2007). Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 39 561–565. [DOI] [PubMed] [Google Scholar]
- Jeong, J.S., Mitchell, T.K., and Dean, R.A. (2007). The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 273 157–165. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2007). Groovy times: Filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 10 358–365. [DOI] [PubMed] [Google Scholar]
- Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuya, T., Nakajima, H., and Kitamoto, K. (1999). Cloning and characterization of the bipA gene encoding ER chaperone BiP from Aspergillus oryzae. J. Biosci. Bioeng. 88 472–478. [DOI] [PubMed] [Google Scholar]
- Kershaw, M.J., Thornton, C.R., Wakley, G.E., and Talbot, N.J. (2005). Four conserved intramolecular disulphide linkages are required for secretion and cell wall localization of a hydrophobin during fungal morphogenesis. Mol. Microbiol. 56 117–125. [DOI] [PubMed] [Google Scholar]
- Khang, C.H., Park, S.Y., Lee, Y.H., and Kang, S. (2005). A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum. Fungal Genet. Biol. 42 483–492. [DOI] [PubMed] [Google Scholar]
- Kim, S., Ahn, I.P., Rho, H.S., and Lee, Y.H. (2005). MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 57 1224–1237. [DOI] [PubMed] [Google Scholar]
- Lau, G.W., and Hamer, J.E. (1998). Acropetal: A genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet. Biol. 24 228–239. [DOI] [PubMed] [Google Scholar]
- Lee, J., Lee, T., Lee, Y.W., Yun, S.H., and Turgeon, B.G. (2003). Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50 145–152. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Marciniak, S.J., and Ron, D. (2006). Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 86 1133–1149. [DOI] [PubMed] [Google Scholar]
- Martinez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G.L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31 426–428. [Google Scholar]
- Munro, S., and Pelham, H.R. (1987). A C-terminal signal prevents secretion of luminal ER proteins. Cell 48 899–907. [DOI] [PubMed] [Google Scholar]
- Ni, M., and Lee, A.S. (2007). ER chaperones in mammalian development and human diseases. FEBS Lett. 581 3641–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt III, B.F., and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Normington, K., Kohno, K., Kozutsumi, Y., Gething, M.J., and Sambrook, J. (1989). S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 57 1223–1236. [DOI] [PubMed] [Google Scholar]
- Orbach, M.J., Farrall, L., Sweigard, J.A., Chumley, F.G., and Valent, B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, S.H. (1985). Rice Diseases. (Kew, UK: Commonwealth Mycological Institute).
- Pakula, T.M., Laxell, M., Huuskonen, A., Uusitalo, J., Saloheimo, M., and Penttila, M. (2003). The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. Evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem. 278 45011–45020. [DOI] [PubMed] [Google Scholar]
- Park, J., et al. (2008). CFGP: A web-based, comparative fungal genomics platform. Nucleic Acids Res. 36 D562–D571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt, P.J., van Gemeren, I.A., Drint-Kuijvenhoven, J., Hessing, J.G., van Muijlwijk-Harteveld, G.M., Beijersbergen, A., Verrips, C.T., and van den Hondel, C.A. (1998). Analysis of the role of the gene bipA, encoding the major endoplasmic reticulum chaperone protein in the secretion of homologous and heterologous proteins in black Aspergilli. Appl. Microbiol. Biotechnol. 50 447–454. [DOI] [PubMed] [Google Scholar]
- Qu, S., Liu, G., Zhou, B., Bellizzi, M., Zeng, L., Dai, L., Han, B., and Wang, G.L. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho, H.S., Kang, S., and Lee, Y.H. (2001). Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea. Mol. Cells 12 407–411. [PubMed] [Google Scholar]
- Rose, M.D., Misra, L.M., and Vogel, J.P. (1989). KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57 1211–1221. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Saris, N., Holkeri, H., Craven, R.A., Stirling, C.J., and Makarow, M. (1997). The Hsp70 homologue Lhs1p is involved in a novel function of the yeast endoplasmic reticulum, refolding and stabilization of heat-denatured protein aggregates. J. Cell Biol. 137 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, B.W., Kim, H.K., Lee, Y.W., and Yun, S.H. (2007). Functional analysis of a histidine auxotrophic mutation in Gibberella zeae. Plant Pathol. J. 23 51–56. [Google Scholar]
- Steel, G.J., Fullerton, D.M., Tyson, J.R., and Stirling, C.J. (2004). Coordinated activation of Hsp70 chaperones. Science 303 98–101. [DOI] [PubMed] [Google Scholar]
- Sweigard, J.A., Carroll, A.M., Kang, S., Farrall, L., Chumley, F.G., and Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard, J.A., Chumley, F.G., and Valent, B. (1992). Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 232 183–190. [PubMed] [Google Scholar]
- Talbot, N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57 177–202. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J., Ebbole, D.J., and Hamer, J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N.J., Kershaw, M.J., Wakley, G.E., De Vries, O., Wessels, J., and Hamer, J.E. (1996). MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M.Y., Huitema, E., da Cunha, L., Torto-Alalibo, T., and Kamoun, S. (2004). A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 279 26370–26377. [DOI] [PubMed] [Google Scholar]
- Travers, K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101 249–258. [DOI] [PubMed] [Google Scholar]
- Tyson, J.R., and Stirling, C.J. (2000). LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 19 6440–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, B. (1990). Rice blast as a model system for plant pathology. Phytopathology 80 33–36. [Google Scholar]
- Valent, B., Farrall, L., and Chumley, F.G. (1991). Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gemeren, I.A., Punt, P.J., Drint-Kuyvenhoven, A., Broekhuijsen, M.P., van't Hoog, A., Beijersbergen, A., Verrips, C.T., and van den Hondel, C.A. (1997). The ER chaperone encoding bipA gene of black Aspergilli is induced by heat shock and unfolded proteins. Gene 198 43–52. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., Leykam, J., Schaeffer, H.J., and Walton, J.D. (1991). A single beta 1,3-glucanase secreted by the maize pathogen Cochliobolus carbonum acts by an exolytic mechanism. Physiol. Mol. Plant Pathol. 39 259–267. [Google Scholar]
- Yi, M., and Lee, Y.H. (2008). Identification of genes encoding heat shock protein 40 family and the functional characterization of two Hsp40s, MHF16 and MHF21, in Magnaporthe oryzae. Plant Pathol. J. 24 131–142. [Google Scholar]
- Young, B.P., Craven, R.A., Reid, P.J., Willer, M., and Stirling, C.J. (2001). Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 20 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Yu, J.H., Hamari, Z., Han, K.H., Seo, J.A., Reyes-Dominguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41 973–981. [DOI] [PubMed] [Google Scholar]
- Yun, S.H. (1998). Molecular Genetics and Manipulation of Pathogenicity and Mating Determinants in Mycoshpaerella zeae-maydis and Cochliobolus heterostrophus. PhD dissertation (Ithaca, NY: Cornell University).
- Zimmermann, R., Muller, L., and Wullich, B. (2006). Protein transport into the endoplasmic reticulum: Mechanisms and pathologies. Trends Mol. Med. 12 567–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.