Abstract
In plants, a diverse group of cell surface receptor-like protein kinases (RLKs) plays a fundamental role in sensing external signals to regulate gene expression. Roots explore the soil environment to optimize their growth via complex signaling cascades, mainly analyzed in Arabidopsis thaliana. However, legume roots have significant physiological differences, notably their capacity to establish symbiotic interactions. These major agricultural crops are affected by environmental stresses such as salinity. Here, we report the identification of a leucine-rich repeat RLK gene, Srlk, from the legume Medicago truncatula. Srlk is rapidly induced by salt stress in roots, and RNA interference (RNAi) assays specifically targeting Srlk yielded transgenic roots whose growth was less inhibited by the presence of salt in the medium. Promoter-β-glucuronidase fusions indicate that this gene is expressed in epidermal root tissues in response to salt stress. Two Srlk-TILLING mutants also failed to limit root growth in response to salt stress and accumulated fewer sodium ions than controls. Furthermore, early salt-regulated genes are downregulated in Srlk-RNAi roots and in the TILLING mutant lines when submitted to salt stress. We propose a role for Srlk in the regulation of the adaptation of M. truncatula roots to salt stress.
INTRODUCTION
Plants are affected by different environmental conditions, such as cold, drought, and soils with changing salt and nutrient concentrations. Salinity is one of the most important abiotic stresses for crop productivity, and the amount of land affected by salinity is increasing (Wang et al., 2003). Salt stress induces various complex biochemical, molecular, cellular, and physiological changes in plants (Wang et al., 2003; Tuteja, 2007; Munns and Tester, 2008). The fact that related plant genotypes showed large variations in their response to abiotic stresses suggests that activation of specific genes may lead to major changes in their adaptive responses to cope with unfavorable environmental conditions (de Lorenzo et al., 2007).
Because approximately one-third of the food required to feed the world's population depends on industrially produced nitrogen fertilizer, legumes are becoming a strategically important crop for grain and forage purposes (Smil, 1997; Graham and Vance, 2003). Indeed, these plants have the capacity to establish root symbioses with nitrogen-fixing bacteria commonly known as rhizobia, resulting in the formation of highly specialized root nodules where nitrogen fixation takes place (Limpens and Bisseling, 2003; Jones et al., 2007). Legume production is greatly constrained by numerous abiotic stresses, which can directly affect root growth and symbiotic interactions (Sharma and Lavanya, 2002; Morón et al., 2005). Understanding the different mechanisms by which these plants perceive and react to environmental stresses may lead to novel strategies for crop improvement.
In plants, a diverse group of cell surface receptor-like protein kinases (RLKs) play a fundamental role in sensing external signals and regulating gene expression responses at the cellular level (Stone and Walker, 1995; Lease et al., 1998). The multiplicity of external stimuli perceived by plants may be linked to the large number of RLK genes (at least 610 members in Arabidopsis thaliana; Torii, K.U., 2004). Members of this plant family are known to play roles in plant growth and development, plant defense responses against pathogens (Becraft, 2002; Shiu et al., 2004), and legume symbiotic interactions (Stacey et al., 2006). Nevertheless, few RLKs to date have been implicated in abiotic stress. The Arabidopsis LecRK2 receptor containing an extracellular lectin-like domain is salt responsive and regulated by ethylene signaling (He et al., 2004). The Arabidopsis cell wall–associated RLK gene WAK4 is regulated differentially by various biotic and abiotic factors and plays a vital role in cell elongation (Lally et al., 2001). More conclusively, using T-DNA insertion mutants and overexpressing and antisense plants, the RPK1 gene has been implicated in early abscisic acid (ABA) perception in Arabidopsis (Osakabe et al., 2005).
Receptor kinases are typically composed of an extracellular ligand binding domain, a transmembrane domain, and a cytoplasmic kinase domain. The extracellular domains of these receptors are quite divergent and enable them to respond selectively to different signals (Walker, 1994; Diévart and Clark, 2003; Torii, 2004; Johnson and Ingram, 2005). Based on the more than 20 structures of the extracellular domains, the plant RLK superfamily has been classified into various groups, such as leucine-rich repeats (LRRs), S domains (homologous to the S [self-incompatibility] locus glycoprotein), domains with epidermal growth factor repeats, and lectin domains (Torii and Clark, 2000; Shiu and Bleecker, 2001a). Among these, LRR-RLKs have been extensively studied in plants, although only a handful of receptors corresponding to mutants with clear phenotypes have been isolated. Thus, out of the 216 LRR-RLKs in Arabidopsis, only 10 have known functions, and only four have been studied in detail (Diévart and Clark, 2004, Osakabe et al., 2005). The abundance of plant LRR-RLKs may represent a plant-specific adaptation for the recognition of a wide variety of extracellular signals in these sessile organisms.
Using a suppressive subtractive hybridization (SSH) approach, we have previously identified several genes induced by salt stress in the model legume Medicago truncatula, including an LRR-RLK gene (named Srlk; de Lorenzo et al., 2007; Merchan et al., 2007). Here, we have evaluated the involvement of this gene in salt stress responses of legume roots and showed that this LRR-RLK plays a role in determining the sensitivity of legume roots to salt stress. Expression of the gene is rapidly and strongly increased in response to salt stress in roots and promoter-β-glucuronidase (GUS) fusions, suggesting that Srlk is activated in the root epidermis. Functional analysis of Srlk using RNA interference (RNAi) yielded roots whose growth was insensitive to the presence of salt in the medium, a phenotype similarly observed in two independent Srlk-mutant TILLING alleles. These plants accumulate fewer sodium ions than controls, and several early salt-regulated genes are downregulated after a salt stress. Taken together, these results suggest a role for Srlk in mediating early events in the response of M. truncatula Jemalong A17 roots to salt stress.
RESULTS
Srlk, a Novel LRR-RLK Induced by Salt Stress in M. truncatula
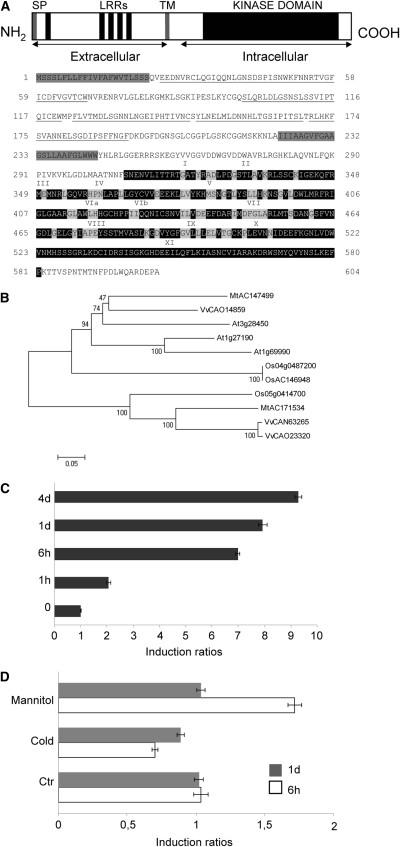
A partial cDNA of Srlk (for Salt-induced Receptor-Like Kinase) was previously isolated via an SSH approach for identifying genes involved in the reacquisition of root growth after salt stress in M. truncatula (Merchan et al., 2007). A sequence corresponding to Srlk was identified in The Institute for Genomic Research (TIGR) database, GenBank accession number TC101212, for tentative consensus sequence derived from several ESTs, corresponding to a transcript of ∼1.8 kb, and coding for a predicted protein of 604 amino acids. Analyses of structural properties of the Srlk-predicted protein using Pfam (Bateman et al., 2004) and SMART programs (Letunic et al., 2004) suggest that this protein encodes an LRR-RLK with four domains: an N-terminal hydrophobic signal peptide (1 to 22), extracellular LRRs (23 to 221), a transmembrane domain (from amino acid 222 to 244), and a cytoplasmic kinase domain (from amino acid 307 to 581) (Figure 1A). The Srlk extracellular region contains five predicted LRR domains, whereas the intracellular kinase domain contains the 11 conserved kinase subdomains (identified by roman numerals in Figure 1A). The previously described LRR-RLK extracellular domains (Shiu and Bleecker, 2001a) differ from those of Srlk, suggesting that this receptor recognizes a different signal.
Figure 1.
An LRR-RLK Is Induced by Salt Stress in M. truncatula Jemalong A17.
(A) Predicted amino acid sequence of Srlk. Hydrophobic regions corresponding to the signal peptide sequence and the transmembrane region (gray boxes), the LRRs (amino acids underlined), and the kinase domain (black) are indicated. Roman numerals indicate the 11 characteristic subdomains of protein kinases. Letters shaded in gray indicate amino acids highly conserved among protein kinases.
(B) Phylogenetic relationship among Srlk homologs. Phylogram of deduced full-length protein sequences of Srlk homologs constructed with the MEGA4 software (Tamura et al., 2007). The bootstrapping value (out of 10,000 samples) for each node, obtained with the same software, is shown. Species are as follows: Mt, Medicago truncatula; At, Arabidopsis thaliana; Vv, Vitis vinifera, and Os, Oryza sativa.
(C) Srlk expression levels in response to salinity treatments. M. truncatula Jemalong A17 roots (4-d grown) were transferred to 150 mM NaCl for different times. Real time RT-PCR analysis of Srlk expression in roots at 0, 1 h, 6 h, 1 d, and 4 d of treatments is shown.
(D) Srlk expression levels in response to mannitol (300 mM) and a cold (4°C) stress during 6 and 24 h. For (C) and (D), histogram represents quantification of specific PCR amplification products for Srlk gene normalized against the constitutive control actin11. Numbers on the x axis indicate fold induction of gene expression in treated compared with untreated samples. A representative example out of two biological experiments is shown, and error bars represent sd between three technical replicates.
To identify Srlk homologous proteins, a phylogenetic tree was constructed using the deduced protein sequences of these genes, the MEGA4 program, and the Arabidopsis proteins At MSrlkl1 (for Medicago Smrlk1-like protein, accession number At3g28450), At MSrlk2 (accession number At1g27190), and At MSrlk3 (accession number At1g69990), three rice homologs (accessions numbers Os05g0414700, AC146948.2, and Os04g0487200), two Vitis vinifera homologs (Vv/CAN63265.1 and Vv/CAO23320.1), and one Medicago homolog (Mt/AC171534.4) as shown in Figure 1B. The closest homolog to Mt Srlk was an Arabidopsis protein (At MSrkl1 for Medicago Salt Receptor kinase-like 1) that exhibits 62% identity. No function has been assigned to this gene in any species.
Next, we examined expression of the Slrk gene in response to salt and other abiotic stresses. The ATLAS of gene expression for M. truncatula recently developed based on Affymetrix chips (http://bioinfo.noble.org/gene-atlas/; Benedito et al., 2008) revealed that the Srlk gene is mainly expressed in root tissues (see Supplemental Figure 1A online). We determined Srlk expression levels in M. truncatula Jemalong A17 roots submitted to salt (150 mM NaCl), mannitol (300 mM), and cold stress (4°C) at different time points (0, 1 h, 6 h, 1 d, and 4 d for salt stress, and 6 h and 1 d for mannitol and cold stress treatments). Srlk expression levels were already induced at 1 h after salinity treatment and reached their highest level from within 6 h and maintained up to 4 d of salt stress (Figure 1C). In mannitol and cold stress conditions, the Srlk expression was not significantly induced (Figure 1D). The Zpt2-1 and CorA1 genes (Merchan et al., 2007) were used as positive controls of stress treatments (see Supplemental Figure 1B online). Thus, we have identified a LRR-RLK in M. truncatula Jemalong A17 that is induced early in response to salt stress.
Functional Characterization of Salt Signaling Pathways in M. truncatula Jemalong A17 Roots
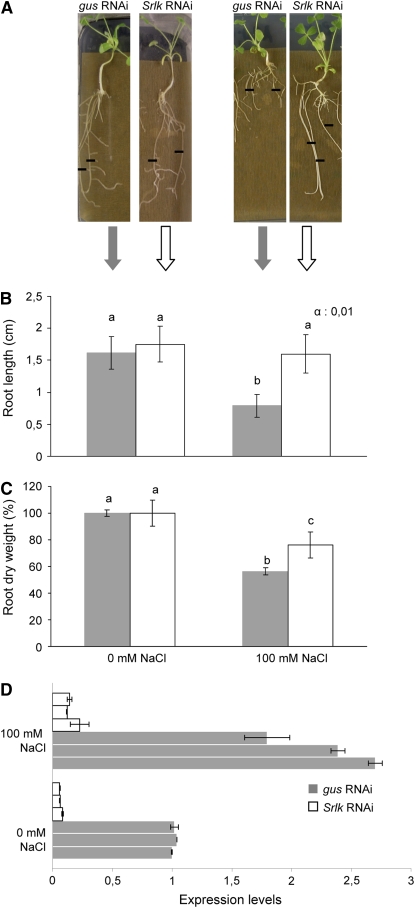
To investigate the putative role of Srlk in response to salt stress, an RNAi approach was used in M. truncatula Jemalong A17 roots using composite plants (Boisson-Dernier et al., 2001). RNAi knockdown of candidate genes is an efficient way to suppress gene expression (Smith et al., 2000; González-Rizzo et al., 2006). A 203-bp fragment of Srlk with maximal RNAi specificity was used for RNAi constructs by selecting a region with <21-bp-long stretches of full complementarity with any other sequence of M. truncatula. We compared transgenic Agrobacterium rhizogenes–transformed roots carrying an Srlk-RNAi construct with a gus-RNAi control to rule out any indirect effect induced by the activation of the silencing machinery. Two weeks after A. rhizogenes infection in the appropriate medium, similarly grown composite plants were transferred to a medium containing 100 mM NaCl or control medium without salt and incubated for six more days (Figure 2). Root length for each plant was determined from the moment of transfer into these media up to the 6-d period. A significant difference in root growth was detected in the population of Srlk-RNAi lines compared with control roots under salt stress conditions (Figures 2A and 2B). By contrast, a similar behavior was observed in the absence of salt stress. Measurements of n > 100 independent transgenic roots per construct (from three biological replicates) were examined to evaluate the salinity-dependent effect on root growth (Student's t test, P < 0.001). The phenotype observed in Srlk-RNAi transgenic roots was further confirmed using root dry weight measurements (Figure 2C).
Figure 2.
Functional Analysis of the Srlk Gene Using RNAi.
(A) Representative images of gus-RNAi and Srlk-RNAi A. rhizogenes–transformed M. truncatula roots 6 d after transfer to control medium (left) or 100 mM NaCl (right). Black lines indicate the position of root tips at the moment of transfer to the salt or control medium. Similar root lengths were observed for both plants in control conditions.
(B) Quantification of mean root growth of independent transgenic roots transformed with the constructs mentioned in (A) and grown 6 d on media without salt (left graph) or supplemented with 100 mM NaCl (right graph). A representative example out of three biological experiments is shown (n > 30 per construct and condition per experiment). The different letters indicate mean values significantly different between gus-RNAi and Srlk-RNAi roots (Student's t test, P < 0.001).
(C) Quantification of root dry weights of Srlk-RNAi or gus-RNAi plants under salt stress or control conditions after 15 d. The different letters indicate mean values significantly different using the Kruskal and Wallis statistical method (n = 20). For (B) and (C), error bars indicate the interval of confidence (α = 0.01).
(D) Real-time RT-PCR analysis of expression levels of Srlk in pools from three independent Srlk-RNAi (white bars) and three gus-RNAi transgenic lines (gray bars) treated or not with salt. Values were normalized against the actin gene, and error bars are sd.
As several LRR-RLKs are involved in symbiosis (Endre et al., 2002; Searle et al., 2003; Torii, 2004; Schnabel et al., 2005; Stacey et al., 2006), we also analyzed a potential role of Srlk in nodulation, particularly under salt stress conditions. Nodulation capacity under control and salt stress conditions of several independent transgenic root lines expressing gus-RNAi and Srlk-RNAi constructs was analyzed. After 3 weeks of growth in control medium, these composite plants were transferred to a nitrogen-deprived medium without or with salt (100 mM NaCl) and subsequently inoculated with the Sinorhizobium meliloti 2011 wild-type strain. We determined the total number of nodules per plant 21 d after inoculation, and no obvious differences were observed. A similar inhibitory effect of salt stress on the symbiotic interaction was detected both in gus-RNAi and Srlk-RNAi composite plants (see Supplemental Figure 2 online).
The effect of the RNAi construct on Srlk expression in transgenic roots was analyzed. A strong and significant decrease of Srlk expression was observed both under normal or salt stress conditions (Figure 2D) in Srlk-RNAi roots compared with gus-RNAi controls. These results confirmed the downregulation of Srlk gene expression by RNAi in these transgenic roots.
Therefore, repression of Srlk expression in M. truncatula Jemalong A17 plants prevents the inhibition of root growth by salt, suggesting that Srlk-RNAi roots may be less sensitive to this environmental constraint.
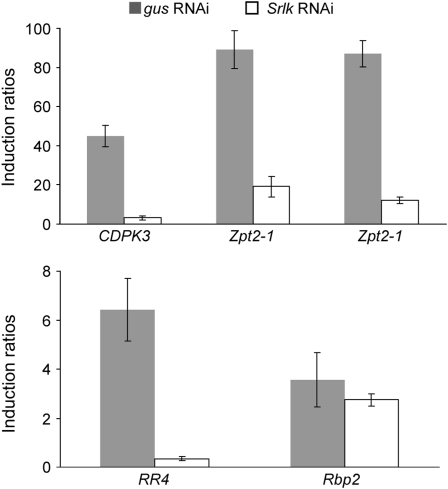
Several Salt-Responsive Genes Are Downregulated in Srlk-RNAi Roots under Salt Stress
Our previous work (de Lorenzo et al., 2007; Merchan et al., 2007) identified several early markers of salt stress responses for M. truncatula Jemalong A17 roots. Five genes induced by short-term salinity treatments were selected to investigate their behavior in Srlk-RNAi roots. A calcium-dependent protein kinase (CDPK3) gene was induced at 1 and 6 h after salt stress (Gargantini et al., 2006), whereas a cytokinin-related response regulator (RR4; Merchan et al., 2007) was induced within 1 h for up to 4 d. In addition, two transcription factors (Zpt2-1 and Zpt2-2; de Lorenzo et al., 2007; Merchan et al., 2007) and an RNA binding protein (Rbp2) gene showed increased levels from 1 h of salt treatment (see Supplemental Figure 3 online). Expression of these five genes in Srlk-RNAi and gus-RNAi transgenic roots treated with or without salt for 6 h (see Methods) was analyzed using real-time RT-PCR. Four (CDPK3, RR4, Zpt2-1, and Zpt2-2) were significantly downregulated in Srlk-RNAi plants under salt stress (Figure 3), whereas the Rbp2 gene was less affected. Expression levels are presented as induction ratios between salt and control conditions.
Figure 3.
Expression of Salt-Regulated Genes in Srlk-RNAi Plants under Salt Stress.
The effect of Srlk suppression on the expression of early induced salt-responsive genes was determined in transgenic root apexes treated with 150 mM NaCl during 6 h. RNA samples from three independent Srlk-RNAi transgenic lines and their corresponding controls were analyzed by real-time RT-PCR. Histograms represent the induction ratios between salt and control conditions of the expression levels of each gene in Srlk-RNAi and in gus-RNAi roots. The salt-regulated genes studied were CDPK3, RR4, Zpt2-1, Zpt2-2, and Rbp2. Values were normalized against the actin gene.
These results strongly suggest that Srlk-RNAi roots are less sensitive to the external addition of salt and that the genes tested (except for Rbp2) may be involved in a salt-responsive signaling pathway activated by this RLK.
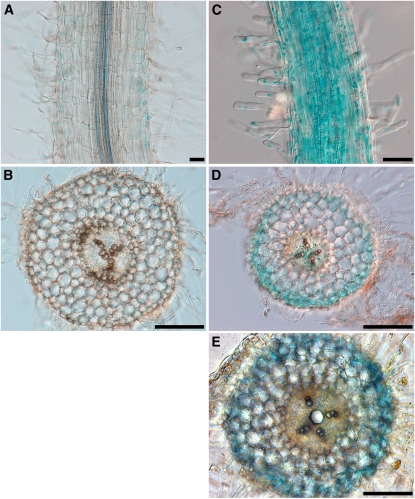
Spatial Expression of Srlk in M. truncatula Roots
The temporal and spatial expression patterns of Srlk in M. truncatula roots and its regulation by salt stress were investigated using a 2.2-kb Srlk promoter:GUS fusion. Two weeks after A. rhizogenes infection, the histological GUS activity of the resulting transgenic root was determined. The Srlk promoter was only weakly active in the root epidermis under control conditions
Based on results obtained for Srlk expression levels at different time points (Figure 1C), we evaluated the effect of salt stress conditions on GUS gene expression directed by the Srlk promoter. After 6 h of NaCl treatment, we observed an increase in GUS expression in the root epidermis (Figure 4). Whole-mount longitudinal views and transversal sections of transgenic roots in the absence (Figures 4A and 4B) or presence of salt revealed strong staining in epidermal cells and root hairs (Figures 4C to 4E). In half of the observed roots, an additional staining of vascular tissues could be observed after a NaCl treatment (Figure 4D).
Figure 4.
Spatial Expression Pattern of the Srlk Promoter.
Histochemical localization of GUS activity in M. truncatula transgenic roots expressing the Srlk promoter:GUS fusion. Plants in (C) to (E) were treated for 6 h with salt (150 mM NaCl) before GUS staining. Bars = 250 μm in (A) and (C) and 500 μm in (B), (D), and (E).
(A) and (B) Whole-mount staining (A) of untreated roots allowed weak detection of blue staining in the epidermis that is barely detected in transverse sections (B).
(C) Whole-mount staining of NaCl-treated roots revealed strong staining of the epidermis and root hairs.
(D) and (E) Transverse sections of the transgenic roots after NaCl treatment showing strong blue staining, representative of GUS activity, in the root epidermis. In half of the cases, weak staining was observed in vascular tissues (E). GUS staining data are representative of at least 25 independent transgenic roots.
As the root apex determines root growth in the soil and is known to be particularly sensitive to a variety of environmental stimuli (Dinneny et al., 2008; Gruber et al., 2009), we evaluated Srlk expression in this zone. Under salt stress, GUS expression driven from the Srlk promoter was also strongly detected in the basal meristematic region of the lateral root tip and vascular tissues in contrast with untreated root apexes (see Supplemental Figure 4 online). Transgenic roots carrying a GUS transgene without promoter did not show any GUS activity (see Supplemental Figure 4A online).
Expression of the Srlk promoter was predominantly localized in the root epidermis and is induced in this tissue as well as in the root apex, two regions potentially linked to the perception of soil environmental conditions.
Two Mutants Carrying TILLING-Derived Alleles of Srlk Are Affected in Salt Stress Responses
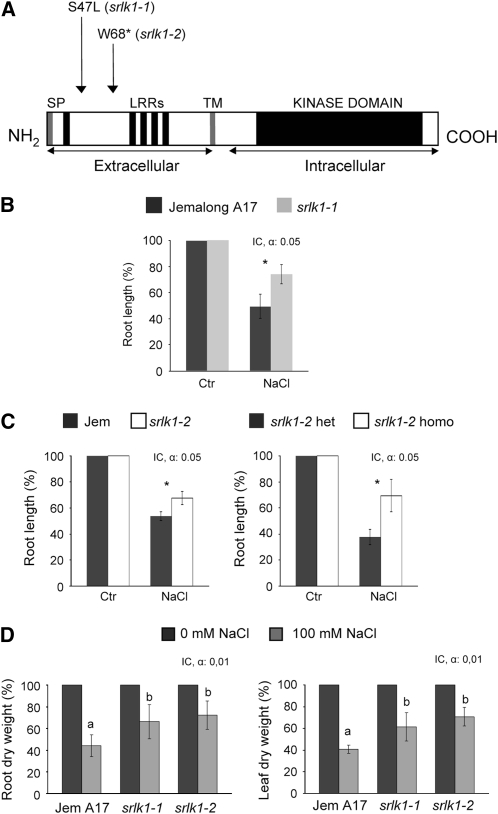
We next characterized two alleles of the Srlk genes through TILLING (for targeting-induced local lesions in genomes) in M. truncatula (Comai and Henikoff, 2006). Mutations predicted by CODDLE (for codons optimized to discover deleterious lesions) as potentially deleterious for gene function in the LRR region of the receptor were screened and two lines carrying different alleles of the Srlk gene were characterized. One corresponded to an S-to-L substitution at position S47 in the N-terminal LRR region (srlk1-1), while the second line had a single G-to-A base change in the N-terminal LRR sequence, creating a stop codon (srlk1-2). Both mutations are indicated in Figure 5A. We have identified heterozygous and homozygous mutants for the null and the S-to-L allele, respectively.
Figure 5.
Phenotypic Analysis of TILLING Srlk Mutants.
(A) Schematic representation of Srlk, as shown in Figure 1, indicating the positions of the changed amino acids in the TILLING lines.
(B) and (C) Quantification of root length after salt stress treatments (100 mM NaCl) of different TILLING Srlk-mutants and control plants (M. truncatula Jemalong A17 or heterozygous siblings). Plants were scored at 6 d after germination in an in vitro system to monitor root growth under salt stress or control medium. The relative root length at 100 mM NaCl was calculated as percentage of corresponding untreated plants (homozygous, segregating heterozygous, or wild-type Jemalong A17). In (B), a representative example of three biological replicates is shown. Asterisks indicate mean values statistically different between pairwise plants at P < 0.05. Error bars indicated the interval of confidence (α = 0.05), and the Kruskal and Wallis test has been used (n = 20 per plant line).
(D) Five-day-old plants of the two Srlk mutants and Jemalong A17 were grown in the growth chamber in the presence of 0 and 100 mM of NaCl. Relative dry weights of roots (left) and leaves (right) from M. truncatula Jemalong A17 and srlk1-1 and srlk1-2 TILLING homozygous mutants after 12 d of growth in “i” medium submitted to 100 mM NaCl are shown. Results are represented as percentage of control without salt. IC, interval of confidence. Error bars indicate the interval of confidence at the indicated probability. The Krustal and Wallis test has been used (P < 0.01 and n < 20).
These Srlk-mutants were assayed for their root growth under control and salt stress conditions (100 mM NaCl). After germination, root length was measured after 6 d in each medium, and the effect of salt stress was measured as the percentage of root length in control conditions (percentage of control). This allowed comparison of root growth between different alleles independently of the initial growth for each plant. Figure 5B shows a comparison of root growth for Jemalong A17 plants and srlk1-1 homozygous lines under salt stress conditions. A significantly enhanced root length in the srlk1-1 lines under salt stress was observed (n = 20, Kruskal and Wallis test, P < 0.05). The srlk1-2 homozygous mutants were compared with Jemalong A17 and also segregating siblings heterozygous for the Srlk mutation. A statistically significant difference in root length was observed in both cases (Figure 5C).
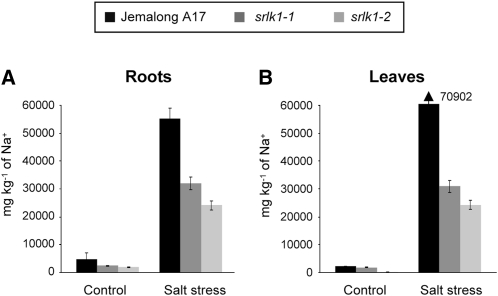
A role of Srlk in the response of M. truncatula to salt stress was further examined by measuring root and leaf dry weights in these plants. Significant differences were found for both parameters between Jemalong A17 and the Srlk mutants under salt stress conditions (Figure 5D; see Supplemental Figure 5 online). The accumulation of sodium was analyzed in roots and leaves of mutant and control plants after exposure to 100 mM NaCl during 14 d. Roots and leaves of srlk1-1 and srlk1-2 plants had reduced levels of Na+ (up to 50 and 65% reduction) than control Jemalong A17 plants (Figures 6A and 6B, roots and leaves, respectively). These results suggest that the Srlk mutants are less sensitive to salt stress due to a differential accumulation of sodium inside plant tissues.
Figure 6.
Sodium Accumulation in Different Tissues of Srlk Mutants.
Sodium concentrations were determined in roots (A) and leaves (B) of plants treated twice with 100 mM NaCl during 7 d (a total of 14 d) and in untreated plants using inducted coupled plasma optical spectrophotometry. Results are expressed in mg kg−1 (n = 20 per plant line), and two biological replicate experiments were performed. Error bars indicate the interval of confidence (α = 0.01).
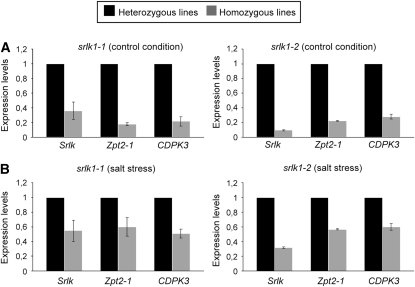
Gene expression was analyzed in these mutant plants in comparison to levels found in segregating heterozygous siblings to avoid any origin-driven variation with Jemalong A17 plants. Roots of srlk1-1 and srlk1-2 had reduced levels of Srlk transcripts, respectively (Figure 7A, 35 and 9% for the S-to-L and null mutants, respectively). The S-to-L substitution did not drastically affect accumulation of Srlk, whereas a strong reduction was observed in homozygous null alleles. The presence of a 5′ stop codon mutation may change its mRNA stability. Under salt stress and control conditions, the expression levels of the two genes tested, Zpt2-1 and CDPK3 (selected due to their differential expression levels between gus-RNAi and Srlk-RNAi), were significantly downregulated in homozygous lines, when compared with heterozygous lines (Figures 7A and 7B, respectively). The same results were obtained when the aerial parts of these lines were analyzed (see Supplemental Figure 6 online).
Figure 7.
Expression of Salt-Regulated Genes in Srlk Mutants.
Effect of Srlk mutation on the expression of early induced salt-responsive genes was determined in roots treated with 150 mM NaCl during 6 h. RNA samples from srlk1-1 and srlk1-2 homozygous lines and their corresponding segregating heterozygous siblings were analyzed by real-time RT-PCR. Histograms represent the ratio of expression levels of each gene in homozygous mutant lines in relation to their heterozygous sibling (black columns). The genes studied were Zpt2-1, CDPK3, and Srlk in control (A) and salt stress conditions (B). Values were normalized against the actin gene.
These results demonstrate that two different mutant alleles of Srlk are affected in their responses to salt stress at both phenotypic (root growth) and molecular levels in a similar manner to the RNAi composite plants. These results indicate a major role of Srlk in the regulation of early salt stress responses of M. truncatula roots.
DISCUSSION
To cope with abiotic stresses, plants have developed various strategies that usually involve three steps: recognition of the stress condition, signal transduction, and activation of gene expression leading to adaptive or protective physiological responses, such as metabolite accumulation (Rathinasabapathi, 2000; Denby and Gehring, 2005; Valliyodan and Nguyen, 2006). Membrane-located receptor kinases play important roles in many plant signal transduction pathways (Tichtinsky et al., 2003). In this study, we report the characterization and functional analysis of a novel LRR-RLK gene, Srlk, involved in the regulation of early M. truncatula root responses to salt stress.
RLKs constitute one of the largest gene families in plant genomes (Shiu and Bleecker, 2001a, 2001b). The basic structure of RLKs can be adapted through diverse extracellular domains to recognize an extremely diverse array of ligands (Johnson and Ingram, 2005). The SRLK putative extracellular domain contains five LRRs, placing this receptor into the LRR-RLK family. The Srlk gene shows high sequence similarity with the kinase domain of three Arabidopsis genes (At3g28450, At1g27190, and At1g69990) but contains a different extracellular LRR domain.
Many receptor kinases have been shown to be regulated by abiotic stresses, and, interestingly, the closest Arabidopsis homolog of Mt Srlk is apparently induced by salt stress (Zimmermann et al., 2004; GENEVESTIGATOR, https://www.genevestigator.ethz.ch/). However very few LRR-RLK genes have known functions in stress responses. In Arabidopsis, the cell wall–associated RLK gene WAK4 is regulated differentially by various biotic and abiotic factors and plays a vital role in cell elongation (Lally et al., 2001), and RPK1, an LRR-RLK upregulated by ABA, dehydration, high salt, and low temperature (Hong et al., 1997), regulates ABA early signaling (Osakabe et al., 2005). The involvement of a novel LRR-RLK in regulating a salt signaling pathway in M. truncatula further demonstrates potential roles of these genes in abiotic stress responses, notably in the agriculturally important legumes.
Recent genetic and biochemical studies have revealed mechanisms that underlie ligand recognition, dimerization, activation, regulation of activity, and signaling specificity of some LRR-RLK receptors (Diévart and Clark, 2003). The srlk1-1 allele carrying an S-to-L transition within an LRR domain-coding region suggests that this missense allele compromises its function. Mutations that affect the β-sheet sequence of LRRs of d61-2 in rice (Oryza sativa; Yamamuro et al., 2000), bri1-9 in Arabidopsis (Noguchi et al., 1999), and the FLS2 receptor, fls2-24 (Gómez-Gómez et al., 1999; Gómez-Gómez and Boller, 2000), interfere with receptor activation, suggesting that this region may be involved in brassinosteroid hormone or flagellin recognition, respectively. In addition, the extracellular domains of the LRR-RLKs play a role in receptor dimerization in different plants (Koka et al., 2000; Shah et al., 2001). The srlk1-2 allele contains a stop codon in the N-terminal LRR sequence and encodes a severely truncated protein. Interestingly, Srlk expression was barely detectable in root and leaves of M. truncatula srlk1-2 mutants even under salt stress. This null-mutation may affect Srlk mRNA stability and, eventually, initiate a non-sense-mediated RNA decay that (Conti and Izaurralde, 2005) as the other inactive Srlk1-1 allele did not show any significant reduction in its own transcript levels.
High salinity causes an imbalance in sodium and can lead to toxic accumulation of this ion in the cytosol and negatively impact the acquisition and homeostasis of essential nutrients, such as K+ and Ca2+ (Manchanda and Garg, 2008). This imbalance can be compensated by the coordinated action of various pumps, ions, Ca2+ sensors, and its downstream interacting partners. The roles of various ion pumps/channels in salinity tolerance have been described (Zhu, 2002; Mahajan et al., 2006). Among them, the SOS pathway results in the exclusion of excess Na+ ions of the cell via a plasma membrane Na+/H+ antiporter to maintain cellular ion homeostasis (Mahajan and Tuteja, 2005, Martínez-Atienza et al., 2007; Manchanda and Garg, 2008). We could not reliably identify an SOS1 homolog in M. truncatula to establish any link between Srlk and SOS pathways. These diverse mechanisms ultimately lead to the efflux of excess Na+ ions, to the reduction of Na+ entry into the cell, to Na+ sequestration in the vacuole, and/or in the closure of stomata to prevent water loss and reduce water (and Na+) uptake (Zhu, 2003; Munns and Tester, 2008). Differential sodium ion accumulation in plant tissues of Srlk mutants suggests that they limit Na+ entry into the cell or have reduced stomatal conductance (hydroactive closure; Mahajan and Tuteja, 2005). As the phenotype was initially observed in RNAi composite plants, where the transgene is only present in root tissues and its downregulation blocks early salt stress molecular responses in roots, we think that a change in stomatal conductance is unlikely. Nonetheless, the Srlk kinase seems to control a salt avoidance response in the model legume M. truncatula.
A signal transduction pathway mediated by Srlk is linked to the activation of a Zpt2-1 Transcription Factor and a CDPK gene. CDPKs regulate plant adaptation responses to biotic and abiotic stresses by sensing Ca2+ and phosphorylating downstream components (Ludwig et al., 2004; Zhu et al., 2007). For example, in Arabidopsis, the roots of plant affected in CPK4 and CPK11 function are insensitive to salt (Zhu et al., 2007). CPK11 was found to interact and phosphorylate in vitro a zinc-finger transcripton factor At Di19 (Milla et al., 2006). In addition, the SOS3 gene is a Ca2+ binding protein that senses the change in Ca2+ concentration and activates SOS2 protein kinase activity in a calcium-dependent manner. This complex was found to phosphorylate SOS1, a plasma membrane Na+/H+ antiporter, to maintain cellular ion homeostasis (Mahajan and Tuteja, 2005; Martínez-Atienza et al., 2007; Manchanda and Garg, 2008).
Among salt-responsive transcription factors, TFIIIA zinc finger proteins play an important role in plant responses to salt stress in M. truncatula roots (de Lorenzo et al., 2007; Merchan et al., 2007). SSH approaches using salt-treated roots have revealed several salt-regulated genes in M. truncatula (Gargantini et al., 2006; de Lorenzo et al., 2007; Merchan et al., 2007). Similarly, the RPK1 mutant and antisense RPK1 transgenic plants showed reduced expression levels of various ABA-inducible genes (Osakabe et al., 2005). Srlk may be a new candidate receptor to function in an early step of a salt-dependent signal transduction pathway leading to expression of downstream genes, such as Zpt2-1, Zpt2-2, RR4, and CDPK. Promoter-GUS fusion analyses suggest that the Srlk is expressed in epidermal tissues, as are the HAK-type K+ transporters and the At KC1 and AKT1 (Arabidopsis Shaker K+ channel genes) (Lagarde et al., 1996; Su et al., 2002; Pilot et al., 2003). These results may link the Srlk receptor with perception of high salinity and activation of a signaling pathway leading to plant avoidance of deleterious effects of salt in internal tissues.
Further research should focus on identifying the extracellular ligand/s recognized by Srlk to activate this pathway. This may provide tools to reduce the negative impact of high salinity in agriculture, particularly in legume crops.
METHODS
Plant Material and Agrobacterium rhizogenes Transformation
Medicago truncatula genotype Jemalong A17 seeds were sterilized as described by de Lorenzo et al. (2007). After washing with sterilized water, seeds were sown on 1% agar plates and stored for 2 d at 4°C before being incubated overnight at 24°C in the dark to ensure uniform germination. Germinated seedlings were transferred to square plates containing appropriate medium for treatment (see below) and grown vertically in chambers at 24°C under long-day conditions (16 h light/8 h dark). Different constructs were introduced into A. rhizogenes ARqua1 (Smr-derivative strain of A4T) and used for transformation of M. truncatula Jemalong A17 as described by Boisson-Dernier et al. (2001).
M. truncatula TILLING Srlk-Mutants
M. truncatula TILLING mutants of Srlk were obtained from the TILLING platform (J. Clarke and D. Baker, John Innes Centre Genome Laboratory, Norwich, UK; http://jicgenomelab.co.uk/services/mutation-detection/tilling.html). Genomic DNA was isolated from each line according to Sambrook et al. (1989), and the mutation of each line was confirmed by PCR, sequencing, and digestion of the amplified fragment. In the same way, the segregation of the mutation was checked to identify heterozygous and homozygous siblings of the same progeny.
Phenotypes (root growth and dry weights) were scored on low-nitrogen medium containing or not 100 mM NaCl as described previously (de Lorenzo et al., 2007). Root length under salt stress was expressed as percentage of root growth without salt using homozygous lines for the mutation in Srlk and wild-type Jemalong A17 or segregating heterozygous siblings as control. Significant differences were tested at 5% probability by the Kruskal and Wallis statistical method (Georgin and Gouet, 2000).
Salinity Treatments
The salt sensitivity of M. truncatula Jemalong A17 was determined as previously described (de Lorenzo et al., 2007). NaCl at 150 mM severely inhibits root growth, whereas at 100 mM NaCl, primary root elongation was inhibited ∼50% when compared with control without salt. For this reason, the phenotype of Srlk-RNAi transgenic roots and TILLING Srlk-mutants was evaluated at 100 mM NaCl.
Root responses to salt stress in composite plants were tested on growth papers (Mega International) placed on low-nitrogen medium supplemented or not with 100 mM NaCl (Merchan et al., 2007). The position of root tips was marked at the time of transfer, and root growth from this point was measured after 6 d. Three biological experiments were performed, and at least 100 independent transgenic roots per construct and per condition were analyzed. In a second set of experiments, root dry weights of composite plants were analyzed at 15 d after the start of salt treatment. Roots of each plant were harvested and dried at 60°C for 48 h, and dry weight under salt stress was represented as percentage of untreated roots dry weight.
Nodulation assays were performed as described by de Lorenzo et al. (2007). Two biological replicate experiments were performed, and a minimum of 60 independent transgenic roots per construct and per condition were analyzed. In both experiments, the statistical method used for evaluation of samples was the Student's t test (P < 0.001).
Salt stress treatment for determination of sodium concentrations in different plant tissues (root and leaf) was performed on 10-d-old plants grown and then exposed twice to a 100 mM NaCl treatment during 7 d (for a total of 14 d).
Determination of Sodium in Plant Tissues
Samples of different plant tissues were dried for 48 h at 60°C, homogenized with a mortar, and filtered. Sodium element was determined by inducted coupled plasma optical spectrophotometry after treatment in a microwave system (Murillo et al., 1999). A sample of untreated plants was also processed in the same way and used as a control. A BCR Certified Reference Material was used as quality control of the analytical procedures. Two biological replicate experiments were performed, and 20 independent plant tissues were analyzed.
Gene Expression Analysis
Total RNAs were extracted from frozen roots using the RNeasy plant mini kit (Qiagen). First-strand cDNA was synthesized from 1.5 μg of total RNA, using the SUPERSCRIPT II first-strand synthesis system (Invitrogen). One-tenth of the cDNAs was used as a template in 10-μL PCR reactions. PCR was performed with a Light Cycler apparatus and the LC FastStart DNA Master SYBR Green IR (Roche Diagnostics) in a standard PCR reaction according to the manufacturer's instructions. A negative control without cDNA template was always included for each primer combination. Technical replicates in three independent syntheses of cDNA (derived from the same RNA sample) and two independent biological replicate experiments were performed. Parallel reactions to amplify actin11 were used to normalize the amount of template cDNA (see Supplemental Table 1 online for gene-specific primers).
Gene Constructions
An Srlk promoter-GUS fusion was constructed. A 2.2-kb genomic DNA fragment lying upstream of the Srlk translation initiation codon was amplified using Srlk promoter primers (see Supplemental Table 1 online) and Pfu DNA polymerase (Promega). The PCR product was sequenced and cloned between the EcoRI and BamHI polylinker sites of the binary vector pPR97 (Szabados et al., 1995). The resulting chimeric construct was transformed into M. truncatula Jemalong A17 roots as described (Boisson-Dernier et al., 2001). A histochemical assay of GUS activity (overnight) was performed on roots of M. truncatula Jemalong A17 composite plants as described by Pichon et al. (1992). Briefly, GUS expression was visualized after soaking the tissues in staining buffer and washing several times in 70% ethanol. To improve the contrast between stained and nonreactive tissues, the samples were briefly cleared with sodium hypochlorite (Boivin et al., 1990). Using this method, endogenous GUS activity was not observed in root tissues from untransformed M. truncatula Jemalong A17 plants. Transversal root sections (100 μm) were done on material embedded in 6% agarose using a Leica VT1200S vibratome. Observations were performed using a Nikon AZ100 macroscope equipped with a Nikon DS-Ri1 digital camera. GUS staining data were representative of at least 25 independent transgenic roots.
To construct Srlk-RNAi and gus-RNAi plasmids, specific fragments of Srlk and uidA genes were cloned into a pFRN destination vector (González-Rizzo et al., 2006). Primers used were Srlk-LC and UidA (GUS) (see Supplemental Table 1 online for primer sequences). Efficiency of silencing was verified using real-time RT-PCR on various individual clones.
Gene Expression in Response to Salt Stress
For gene expression studies in M. truncatula plants, 15 germinated seedlings were placed in flasks with 30 mL of low-nitrogen liquid medium (Blondon, 1964) and grown in a shaking incubator (125 rpm) at 24°C under long-day conditions (16 h light/8 h dark). After 4 d, seedlings were treated with 150 mM NaCl for various incubation times (0, 1 h, 6 h, 1 d, and 4 d) under the same growth conditions. Plants were similarly treated with mannitol (300 mM) or exposed to cold stress (4°C) during 6 h and 1 day. Roots were collected at the indicated time points and immediately frozen in liquid nitrogen for RNA extraction.
In RNAi-transformed roots, 2-week-old composite plants were placed in sterile plastic pots containing perlite/sand (3:1, v/v) as mixed substrate. Plants were grown in control low-nitrogen solution until the root tips emerged out of each pot ∼2 cm. Then, the emerging root tips were immersed in salt or control solution for 6 h and immediately frozen in liquid nitrogen for RNA extraction. A minimum of 10 independent transgenic roots per construction and condition were pooled to perform quantitative RT-PCR analyses, and measurements of five pools of transgenic roots per construct and conditions were examined.
For analysis of salt-regulated gene expression in TILLING mutants, these plants were grown in agar plates and then treated for 6 h with 150 mM NaCl. A minimum of 10 independent transgenic roots per line and condition were pooled to perform quantitative RT-PCR analyses.
Sequencing and Data Analysis
DNA was sequenced with the BigDye terminator system (GenPak), and nucleotide sequences were determined in an automatic laser sequencer 373A (Applied Biosystems).
Srlk sequence was used to query the TIGR Medicago Gene Index (http://www.tigr.org/) and National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/BLAST/) using the BlastN and BlastX sequence comparison algorithms. Conserved protein domains were examined using the Conserved Domains at NCBI (http://www.ncbi.nih.gov/Structure/cdd/cdd.shtml) in conjunction with the Pfam Protein Families database (http://pfam.wustl.edu/) and SMART programs (http://smart.embl-heidelberg.de/). Related LRR-RLKs from Arabidopsis and others plant species were identified by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) algorithms. Sequence alignment was made using the ClustalW program (http://www.ebi.ac.uk/clustalw/). The phylogenetic tree was produced using the neighbor-joining method (Saitou and Nei, 1987) with midpoint rooting. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to each branch (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. There were a total of 522 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TC101212 (for Srlk gene), At3g28450 (for At MSrlkl1), At1g27190 (for At MSrlkl2), At1g69990 (for At MSrlkl3), Os05g0414700 (for Os MSrlkl1), AC146948.2 (for Os MSrlkl2), Os04g0487200 (for Os MSrlkl3), Vv/CAN63265.1 (for Vv MSrlkl1), Vv/CAO23320.1 (for Vv MSrlkl2), Mt/AC171534.4 (for MtSrlkl), TC77176 (for Zpt2-1), TC86494 (for Zpt2-2), TC76706 (for CorA1), TC69270 (for CDPK3), TC64310 (for RR4), TC87010 (for Rbp2), and TC85697 (for actin11).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Levels of Srlk in Different Plant Tissues According to the Affymetrix Gene ATLAS of M. truncatula (http://bioinfo.noble.org/gene-atlas/) and of Zpt2-1 and CorA1 in Response to Mannitol and a Cold Stress, Respectively.
Supplemental Figure 2. Evaluation of Nodulation Capacity of Transgenic gus-RNAi and Srlk-RNAi M. truncatula Roots.
Supplemental Figure 3. Expression Levels of Different Salt-Related Genes in Response to Short-Term Salinity Treatments.
Supplemental Figure 4. Expression of the Srlk Promoter in Lateral Root Apexes with or without Salt and a Control Carrying the Vector without Promoter.
Supplemental Figure 5. Effect of a Long-Term NaCl Treatment on M. truncatula Jemalong A17 and Two Mutants of Srlk.
Supplemental Figure 6. Expression of Salt-Regulated Genes in Srlk Mutants in Salt Stress Condition.
Supplemental Table 1. Sequences of Oligonucleotides Used for PCR and Quantitative RT-PCR.
Supplemental Data Set 1. Text File of Alignment of Srlk Homologs in Several Plant Species.
Supplementary Material
Acknowledgments
L.D.L. and F.M. were supported by an F.P.U. (University Professor Training grant) and postdoctoral grant from the Spanish Department of Education and Science, respectively. We wish to acknowledge the contributions of Christine Lesignor and Françoise Moussy at Institut National de la Recherche Agronomique, Dijon, France, for generating the TILLING resource and David Baker, Lee McPherson, Bethany McCullagh, and Richard Gorman for the Srlk TILLING at the John Innes Centre. We also thank Liliane Troussard for sequencing, the Imaging and Cell Biology platform of the IFR87 “La plante et son environnement” (FR-W2251, supported by Action de Soutien à la Technologie et la Recherche en Essonne, France) for microscopy support, and Florian Frugier for useful comments on the manuscript. The support of the “Grain legumes” FP6 EEC project (FOOD-CT-2004-506223) is also acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Martín Crespi (crespi@isv.cnrs-gif.fr).
Online version contains Web-only data.
References
- Bateman, A., et al. (2004). The Pfam protein families database. 32: D138–D141. [DOI] [PMC free article] [PubMed]
- Becraft, P.W. (2002). Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18 163–192. [DOI] [PubMed] [Google Scholar]
- Benedito, V., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55 504–513. [DOI] [PubMed] [Google Scholar]
- Blondon, F. (1964). Contribution à l'étude du développement de graminées fourragères: Ray-grass et dactyle. Rev. Gen. Bot. 71 293–381. [Google Scholar]
- Boisson-Dernier, A., Chabaud, M., García, F., Becard, G., Rosenberg, C., and Barker, D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700. [DOI] [PubMed] [Google Scholar]
- Boivin, C., Camut, S., Malpica, C.A., Ruchet, G., and Rosenberg, C. (1990). Rhizobium meliloti genes encoding catabolism of trigonel-line are induced under symbiotic conditions. Plant Cell 2 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L., and Henikoff, S. (2006). TILLING: Practical single-nucleotide mutation discovery. Plant J. 45 684–694. [DOI] [PubMed] [Google Scholar]
- Conti, E., and Izaurralde, E. (2005). Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17 316–325. [DOI] [PubMed] [Google Scholar]
- de Lorenzo, L., Merchan, M., Blanchet, S., Megías, M., Frugier, F., Crespi, M., and Sousa, C. (2007). Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol. 145 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby, K., and Gehring, C. (2005). Engineering drought and salinity tolerance in plants: Lessons from genome-wide expression profiling in Arabidopsis. Trends Plant Sci. 23 547–552. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., Long, T.A., Wang, J.Y., Jung, J.W., Mace, D., Pointer, S., Barron, C., Brady, S.M., Schiefelbein, J., and Benfey, P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320 942–945. [DOI] [PubMed] [Google Scholar]
- Diévart, A., and Clark, S.E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6 507–516. [DOI] [PubMed] [Google Scholar]
- Diévart, A., and Clark, S.E. (2004). LRR-containing receptors regulating plant developmental and defence. Development 131 251–261. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Gargantini, P., González-Rizzo, S., Chinchilla, D., Raices, M., Giammaria, V., Ulloa, R.M., Frugier, F., and Crespi, M. (2006). A CDPK isoform participates in the regulation of nodule number in Medicago truncatula. Plant J. 48 843–856. [DOI] [PubMed] [Google Scholar]
- Georgin, P., and Gouet, M. (2000). Statistiques avec Excell. (Paris: Eyrolles).
- Gómez-Gómez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez, L., Felix, G., and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18 277–284. [DOI] [PubMed] [Google Scholar]
- González-Rizzo, S., Crespi, M., and Frugier, F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, P.H., and Vance, C.P. (2003). Legumes: Importance and constraints to greater use. Plant Physiol. 131 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, V., Blanchet, S., Diet, A., Zahaf, O., Boualem, A., Kakar, K., Alunni, B., Udvardi, M., Frugier, F., and Crespi, M. (2009). Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol. Genet. Genomics 281 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.-J., Zhang, Z.-G., Yan, D.-Q., Zhang, J.-S., and Chen, S.-Y. (2004). A salt-responsive receptor-like kinase gene regulated by the ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor. Appl. Genet. 109 377–383. [DOI] [PubMed] [Google Scholar]
- Hong, S.W., Jon, J.H., Kwak, J.M., and Nam, H.G. (1997). Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 113 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K.L., and Ingram, G.C. (2005). Sending the right signals: Regulating receptor kinase activity. Curr. Opin. Plant Biol. 8 648–656. [DOI] [PubMed] [Google Scholar]
- Jones, K.M., Kobayashi, H., Davies, B.W., Taga, M.E., and Walker, G.C. (2007). How rhizobial symbionts invade plants: The Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka, C.V., Cerny, R.E., Gardner, R.G., Noguchi, T., Fujioka, S., Takatsuto, S., Yoshida, S., and Clouse, S.D. (2000). A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and responses. Plant Physiol. 122 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde, D., Basset, M., Leoetit, M., Conejero, G., Gaymard, F., Astruc, S., and Grignon, C. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9 195–203. [DOI] [PubMed] [Google Scholar]
- Lally, D., Ingmire, P., Tong, H.Y., and He, Z.H. (2001). Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13 1317–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, K., Ingham, E., and Walker, J.C. (1998). Challenges in understanding RLK function. Curr. Opin. Plant Biol. 1 388–392. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. (2004). SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32 D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens, E., and Bisseling, T. (2003). Signaling in symbiosis. Curr. Opin. Plant Biol. 6 343–350. [DOI] [PubMed] [Google Scholar]
- Ludwig, A.A., Romeis, T., Jones, J.D.G. (2004). CDPK-mediated signalling pathways: specificity and crosstalk. J. Exp. Bot. 55 181–188. [DOI] [PubMed] [Google Scholar]
- Mahajan, S., Sopoy, S.K., and Tuteja, N. (2006). CBL-CIPK paradigm: Role in calcium and stress signaling in plants. Proc. Indian Nat. Sci. Acad. 72 63–78. [Google Scholar]
- Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 444 139–158. [DOI] [PubMed] [Google Scholar]
- Manchanda, G., and Garg, N. (2008). Salinity and its effects on the functional biology of legumes. Acta Physiol. Plant. 30 595–618. [Google Scholar]
- Martínez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J.K., Pardo, J.M., and Quintero, F.J. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchan, F., de Lorenzo, L., González-Rizzo, S., Niebel, A., Megías, M., Frugier, F., Sousa, C., and Crespi, M. (2007). Analysis of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J. 51 1–17. [DOI] [PubMed] [Google Scholar]
- Milla, M.A.R., Uno, Y., Chang, I.F., Townsend, J., Maher, E.A., Quilici, D., and Cushman, J.C. (2006). A novel yeast two-hybrid approach to identify CDPK substrates: Characterization of the interaction between CPK11 and AtDi19, a nuclear zinc finger protein. FEBS Lett. 580 904–911. [DOI] [PubMed] [Google Scholar]
- Morón, B., Soria, M.E., Ault, J., Verroios, G., Noreen, S., Rodríguez, D., Gil, A.M., Thomas-Oates, J.E., Megías, M., and Sousa, C. (2005). Acidic ph induces the production of novel nodulation factors by Rhizobium tropici CIAT899. Production of NOD factors by R. tropici at acid ph. Chem. Biol. 12 1029–1040. [DOI] [PubMed] [Google Scholar]
- Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 651–681. [DOI] [PubMed] [Google Scholar]
- Murillo, J.M., Marañón, T., Cabrera, F., and López, R. (1999). Accumulation of heavy metals in sunflower and sorghum plants affected by the Guadiamar spill. Sci. Total Environ. 242 281–292. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe, Y., Maruyama, K., Seki, M., Satou, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2005). Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon, M., Journet, E., Dedieu, A., de Billy, F., Truchet, G., and Barker, D. (1992). Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot, G., Gaymard, F., Mouline, K., Chérel, I., and Sentenac, H. (2003). Regulated expression of Arabidopsis Shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 51 773–787. [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi, B. (2000). Metabolic engineering for stress tolerance: Installing osmoprotectant synthesis pathways. Ann. Bot. (Lond.) 86 709–716. [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schnabel, E., Journet, E.-P., de Carvalho-Niebel, F., Duc, G., and Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58 809–822. [DOI] [PubMed] [Google Scholar]
- Searle, I.R., Men, A.E., Laniya, T., Buzas, D., Iturbe-Ormaetxe, I., Carroll, B.J., and Gresshoff, P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299 109–112. [DOI] [PubMed] [Google Scholar]
- Shah, K., Gazella, T.W., Jr., van Erp, H., Hecht, V., and de Vries, S.C. (2001). Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J. Mol. Biol. 309 641–655. [DOI] [PubMed] [Google Scholar]
- Sharma, K.K., and Lavanya, M. (2002). Recent developments in transgenics for abiotic stress in legumes of the semi-arid tropics. JIR-CAS Working Rep. 23 61–73. [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. a). Plant receptor-like kinase gene family: Diversity, function, and signalling. Sci. STKE 113 re22. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001. b). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., Karlowski, W.M., Pan, R., Tzeng, Y., Mayer, K.F.X., and Li, W. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smil, V. (1997). Global population and the nitrogen cycle. Sci. Am. 277 76–81. [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Gene expression: Total silencing by intron-spliced hairpin RNAs. Nature 407 319–320. [DOI] [PubMed] [Google Scholar]
- Stacey, G., Libault, M., Brechenmacher, L., Wan, J., and May, G.D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9 110–121. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., and Walker, J.C. (1995). Plant protein kinase families and signal transduction. Plant Physiol. 108 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., Golldack, D., Zhao, C., and Bohnert, H.J. (2002). The expression of HAK-type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol. 129 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados, L., Charrier, B., Kondorosi, A., de Bruijn, F., and Ratet, P. (1995). New plant promoter and enhancer testing vectors. Mol. Breed. 1 419–423. [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tichtinsky, G., Vanoosthuyse, V., Cock, J.M., and Gaude, T. (2003). Making inroads into plant receptor kinase signalling pathways. Trends Plant Sci. 8 231–237. [DOI] [PubMed] [Google Scholar]
- Torii, K.U. (2004). Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathways. Int. Rev. Cytol. 234 1–46. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., and Clark, S.E. (2000). Receptor-like kinases in plant development. In Advances in Botanical Research: Plant Protein Kinases, M. Kreis, and J.C. Walker, eds (London: Academic Press), pp. 270–298.
- Tuteja, N. (2007). Mechanisms of high salinity tolerance in plants. Methods Enzymol. 428 419–438. [DOI] [PubMed] [Google Scholar]
- Valliyodan, B., and Nguyen, H. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 9 189–195. [DOI] [PubMed] [Google Scholar]
- Walker, J.C. (1994). Structure and function of the receptor-like protein kinases of higher plants. Plant Mol. Biol. 26 1599–1609. [DOI] [PubMed] [Google Scholar]
- Wang, W., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 218 1–14. [DOI] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6 441–445. [DOI] [PubMed] [Google Scholar]
- Zhu, S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.