Abstract
Phenylamidine cationic groups linked by a furan ring (furamidine) and related compounds bind as monomers to AT sequences of DNA. An unsymmetric derivative (DB293) with one of the phenyl rings of furamidine replaced with a benzimidazole has been found by quantitative footprinting analyses to bind to GC-containing sites on DNA more strongly than to pure AT sequences. NMR structural analysis and surface plasmon resonance binding results clearly demonstrate that DB293 binds in the minor groove at specific GC-containing sequences of DNA in a highly cooperative manner as a stacked dimer. Neither the symmetric bisphenyl nor bisbenzimidazole analogs of DB293 bind significantly to the GC containing sequences. DB293 provides a paradigm for design of compounds for specific recognition of mixed DNA sequences and extends the boundaries for small molecule-DNA recognition.
Organic cations that bind in the DNA minor groove have biological activities that range from anti-opportunistic infection to anticancer properties (1–5). Such compounds have also provided a wealth of fundamental information about nucleic acid recognition properties, and they continue to be important models in the study of nucleic acid complexes (1, 2, 6–8). Netropsin (Fig. 1) was the first minor-groove-binding compound crystallized with a B-form DNA, and the structure of the complex provided clear suggestions about the molecular basis for AT base pair sequence-specific recognition (9). The structure also lead to proposals by the Dickerson and Lown groups for minor-groove binding netropsin analogs, lexitropsins, that could specifically recognize GC base pairs and could, thus, have extended sequence recognition capability (10–12). Initial efforts in the design of analogs of netropsin that could recognize GC base pairs, however, yielded compounds of limited specificity. A breakthrough in this area occurred with the discovery that the monocationic relative of netropsin, distamycin (Fig. 1), could bind into the minor groove of some AT sequences of DNA as a stacked, antiparallel dimer (7, 13, 14). Replacement of pyrrole groups in distamycin by imidazole provided lexitropsins with improved GC recognition specificity through dimer complexes, and current design efforts in the pyrrole-imidazole polyamide system have reached a high level of success (15–18). With recent developments by the Dervan group, AT and TA as well as GC and CG base pairs can now be effectively distinguished in DNA sequences by polyamides (15–18).
Figure 1.
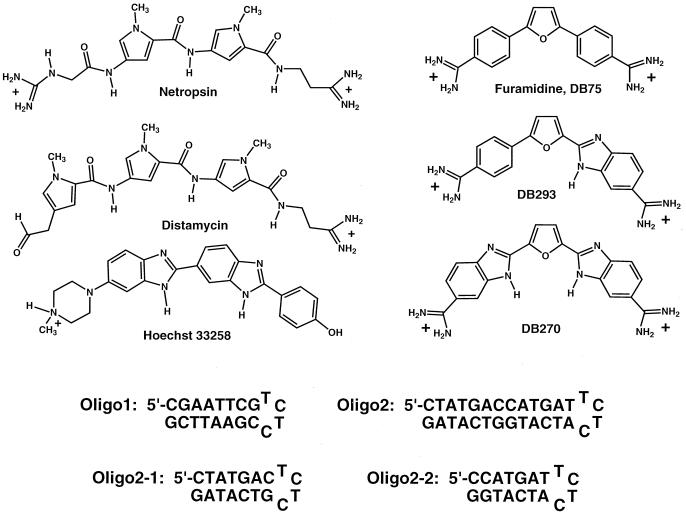
Structures for the minor-groove binding compounds netropsin, distamycin, Hoechst 33258, furamidine (DB75), DB270, and DB293; and sequences for oligo1, oligo2, oligo2-1, and oligo2-2.
Interestingly, the polyamide system is the only one of the well known minor-groove binding motifs that has yielded conclusive evidence for formation of the stacked-dimer recognition unit, although recent evidence indicates that some monocationic cyanine dyes can form an array of stacked dimers in the DNA minor groove (19). No dications have been found to form the stacked-dimer recognition motif. Netropsin, the first minor-groove-binding agent to be characterized in detail and a dicationic relative of the monocation distamycin (Fig. 1), for example, does not form a dimer recognition unit. In a recent crystal structure of a 2:1 netropsin–DNA complex, the two netropsin molecules in the complex bind in the minor groove as tandem monomers instead of the side-by-side stacked dimer observed with distamycin (20). Repulsion of the two charged groups of netropsin as well as of other minor-groove agents, such as the furan derivatives shown in Fig. 1, has been postulated to prevent stacked-dimer formation. There are, however, also many monocationic minor-groove agents, such as Hoechst 33258 and analogs, that apparently do not form stacked-dimer DNA recognition motifs (21), although under some conditions Hoechst 33258 can form 2:1 complexes with DNA (22). These results indicate that the electrostatic and stereochemical requirements for minor-groove recognition of DNA by dimers are very restrictive and suggest that stacked dimer formation by dications is unlikely.
We are conducting a series of systematic studies on how compound structure and substituents affect the DNA recognition properties of unfused aromatic cations such as furamidine (Fig. 1) and analogs (see, for example, refs. 3–5, 23, and 24). As part of these studies, we made the very surprising finding that the unsymmetric dication DB293 (Fig. 1) binds to specific sequences of DNA that contain GC base pairs very strongly, and we propose that this is through a stacked-dimer motif. We report here the initial evidence for dimer formation, the initial DNA recognition properties, and a characterization of the binding of DB293 to GC containing sequences. This dimer recognition mode offers new possibilities for development of agents for recognition of mixed base pair sequences of DNA and requires reevaluation of ideas about the limits of DNA recognition by the stacked-dimer motif.
Materials and Methods
DNase I Footprinting.
Plasmid DNA restriction fragments were prepared, and DNase I footprinting experiments were conducted as described (25, 26).
Thermal Melting (Tm).
Tm experiments were conducted with a Cary 4 spectrophotometer (Varian) interfaced to a microcomputer. A thermistor fixed into a reference cuvette was used to monitor the temperature. The oligomers were added to 1 ml of buffer (0.01 M Mes/0.001 M EDTA) in 1-cm path length reduced volume quartz cells, and the concentration was determined by measuring the absorbance at 260 nm. Experiments were generally conducted at a concentration of ≈3 × 10−6 M hairpin oligomer. Tm experiments for the complexes were conducted as a function of ratio.
Immobilization of DNA and Surface Plasmon Resonance (SPR) Binding Studies.
5′-biotin-labeled DNA hairpins were purchased with HPLC purification (Midland Certified Reagent, Midland, TX). Samples of the DNA in Mes 10 buffer (0.01 M Mes and 0.001M EDTA, with 0.1 M NaCl) at 50 nM concentration were applied to a BIAcore (Piscataway, NJ) SA (streptavidin) chip by direct flow at 5 μl/min in a BIAcore 2000 SPR instrument (27). Nearly the same amount of all oligomers were immobilized on the SA chip. Steady state binding analysis was performed with multiple injections of different compound concentrations over the immobilized DNA surface at a flow rate of 10 or 20 μl/min and 25°C. Binding results from the SPR experiments were fit with either a single site model (K2 = 0) or with a two site model:
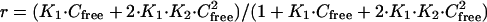 |
1 |
where r represents the moles of bound compound per mole of DNA hairpin duplex, K1 and K2 are macroscopic binding constants, and Cfree is the free compound molar concentration in equilibrium with the complex (28). The free compound is fixed by the concentration in the flow solution.
NMR.
All NMR spectra were acquired with a Varian Unity Plus 600-MHz spectrometer as described (28). Typical conditions for the collection of spectra in D2O: one-dimensional (1D) spectra were collected with a spectral width of 6,000 Hz and 32 K data points, 0.6-ml sample in a 5-mm NMR tube. Two-dimensional experiments were obtained with a spectral width of 6,000 Hz in both dimensions, with 2,048 complex data points in the time 2 (t2) dimension and 512 points in the time 1 (t1) dimension.
Results
DNase I Footprinting Analysis of Binding Specificity.
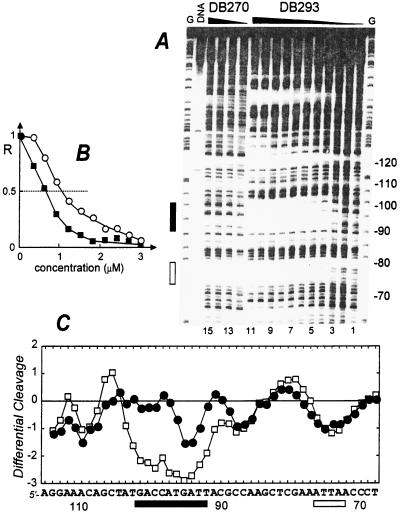
As part of a continuing effort to characterize the DNA recognition properties of furan derivatives related to those in Fig. 1, we have conducted quantitative DNase I footprinting studies with the compounds of Fig. 1 and several different DNA sequences. Space prevents presentation of all results here, but example results from a 265-bp restriction fragment are shown in Fig. 2 for illustration. Footprints with the symmetric compounds furamidine and the bisbenzimidazole DB270 are as expected for AT-specific minor-groove binding agents and agree with observations on other furan derivatives and standard minor-groove binding compounds (1, 26). With the unsymmetric compound DB293 (Fig. 1), however, the footprinting results present a number of surprises in the form of strong footprints in unexpected GC-rich regions (Fig. 2). In the 90- to 100-base region of the 265-mer pBS fragment in Fig. 2, for example, DB293 gave a very strong footprint whereas DB270 and furamidine gave negligible footprints. The most surprising feature of the footprint in this sequence region is its GC content relative to the AT rich sequences in which footprints are usually observed with minor-groove agents (1). Qualitative analysis of the footprinting data reveals that the C50 value, the drug concentration required for half-maximal footprinting, in the 90–100 region is significantly lower than at the neighboring ATTA site, indicating that DB293 prefers the site including GC base pairs. The differential cleavage plots show that both DB270 and DB293 bind similarly to sites composed exclusively of AT base pairs (Fig. 2).
Figure 2.
Quantitative DNase I footprinting titration experiment with DB293 on the 265-bp DNA fragment. The EcoRI-PvuII restriction fragment from plasmid pBS was 3′-end-labeled at the EcoRI site with [α-32P]dATP in the presence of AMV reverse transcriptase. (A) The products of the DNase I digestion were resolved on an 8% polyacrylamide gel containing 8M urea. Drug concentrations are (lanes 1–11) 0, 0.3, 0.6, 0.9, 1.2, 1.5, 1.8, 2.1, 2.4, 2.7, and 3.0 μM for DB293 and (lanes 12–15) 0, 1, 2, and 5 μM for DB270. Tracks labeled G represent dimethyl sulfate-piperidine markers specific for guanines. The track labeled DNA contained no drug and no enzyme. Numbers at the right side of the gel refer to the numbering scheme of the fragment. The rectangles on the right side refer to the positions of an AT-rich (open box) and a GC-rich (filled box) binding site for DB293. (B) Footprinting plots for the binding of DB293 to the AT site 5′-AATTAA (open circles) and the GC-rich site 5′-ACCATG (filled squares). The relative band intensity R corresponds to the ratio Ic/Io, where Ic is the intensity of the band at the ligand concentration c and Io is the intensity of the same band in the absence of DB293. The differential cleavage plots in C compare the susceptibility of the DNA to cutting by DNase I in the presence of 5 μM DB270 (filled circles) or 1.5 μM DB293 (open squares). Deviation of points toward the lettered sequence (negative values) corresponds to a ligand-protected site and deviation away (positive values) represents enhanced cleavage. The vertical scale is in units of ln(fa) − ln(fc), where fa is the fractional cleavage at any bond in the presence of the drug and fc is the fractional cleavage of the same bond in the control. The results are displayed on a logarithmic scale for the sake of convenience. The rectangles below the sequence show the positions of the AT binding site (open box) and the GC-rich site (filled box). Footprinting reactions, separation of cleavage products, and data analysis were carried out as described (10).
Thermal Melting Analysis of Binding.
To investigate the complexes of these compounds at higher resolution with GC-rich sequences, a hairpin duplex model containing the 93- to 104-base sequence region from the 265-mer pBS restriction fragment was synthesized (oligo2 in Fig. 1). Oligo1 (Fig. 1) with the AATT sequence that has been used in the analysis of a large number of minor-groove agents provides a reference. Tm determinations of oligo2 on titration with DB293 gave up to a 30°C increase in Tm and did not level off until a ratio of 4:1 DB293:hairpin duplex had been reached. The high ratio of DB293 to oligomer duplex is surprising for a duplex of only 13 base pairs, and, to again simplify the system to better understand the nature of the complex, we divided oligo2 into two similar hairpin duplexes, oligo2-1 and oligo2-2 (Fig. 1). As an illustration of the results obtained, derivative Tm curves of DB270 and DB293 complexes with oligo2-1 are shown in Fig. 5, published as supplemental data on the PNAS web site, www.pnas.org. The DB293 complex has a biphasic melting curve at a 1:1 ratio, with a high temperature phase and a low temperature phase near the Tm of the free hairpin duplex. At a 2:1 ratio, the low temperature phase disappears, and only the high temperature transition is present. Melting curves of DB270 and furamidine complexes with oligo2-1 have single transitions at 1:1 and 2:1 ratios with melting temperatures below the DB293 value (Fig. 5). As with the footprinting experiments, these results illustrate the significant differences in DNA interactions between the symmetric compounds relative to the unsymmetric DB293. In addition, the Tm ratio results suggest that the unusual DNA recognition properties of DB293 are attributable to formation of 2:1 complexes with oligo2-1 and -2-2, and a 4:1 complex with oligo2. Such dimer complexes could also explain the unexpected footprinting behavior of DB293; however, based on the +2 charge of DB293, only tandem dimer complexes would be expected, and the oligomers used do not appear of adequate length or sequence for such tandem complexes.
Surface Plasmon Resonance (SPR) Analysis of Binding.
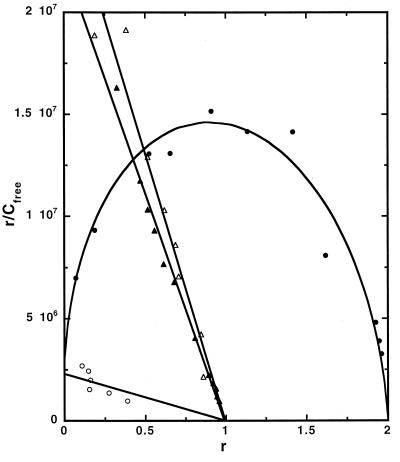
To quantitatively evaluate the interactions of the furan derivatives with specific DNA sequences, SPR was used with 5′-biotin-labeled oligo2-1 and -2-2 (Fig. 1). Oligo1 was immobilized as a reference sequence, and one flow cell was left as an unmodified control. The results for DB293 binding to oligo2-1 and -2-2 are quite different from results with the symmetric compounds and from the results obtained with oligo1 and DB293 (Fig. 3). Sensorgrams for DB270 and DB293 binding to oligo1 and -2-1 are shown in Fig. 6, published as supplemental data, and provide additional evidence for the interaction differences with these compounds. The sensorgrams increase with compound concentration in a similar manner for DB270 and DB293 with oligo1, as expected for the single AATT site. The results yield linear Scatchard plots (Fig. 3) for the two compounds, indicating the expected single type of strong binding site with similar equilibrium constants (K) in the range 2.3–2.6 × 107. Quite different results are observed with the oligo2-1 sensorgrams (Fig. 6). Rapid increases in response as a function of DB293 concentration are observed, and the final level reached is twice as large as with oligo1. The increases with DB270, on the contrary, are much smaller than observed with oligo1. Binding of DB270 to oligo2-1 is over a factor of 10 weaker than its binding to oligo1 (Fig. 3). Surprisingly, binding of DB293 with oligo2-1 is highly cooperative (K2 > K1) and saturates at two molecules of DB293 per oligo2-1 hairpin duplex. Fitting of the binding results to a two-site model to determine the macroscopic binding constants gave a K1 of 2.8 × 106 for initial binding and a K2 of 7.3 × 107 for binding of the second molecule (Fig. 3). Similar results are obtained for binding of DB293 to oligo2-2. The similarity of binding constants for DB270 and the first molecule of DB293 that binds to oligo2-1 suggests that these are similar processes. The striking difference occurs when the second molecule of DB293 binds cooperatively with a K2 that is >25× larger than for binding of DB270 and the first molecule of DB293 (K1) to the oligomers. The Hill coefficient for DB293 binding to oligo1 is between 0.9 and 1 whereas it is between 1.9 and 2 for binding to oligo2-1 and -2-2, in agreement with highly cooperative binding of two molecules to the latter oligomers. The combined binding constant for dimer formation (K1 × K2) is very large, >2 × 1014 M−1. Cooperative binding of stacked distamycin dimers and related polyamides to sequences with GC base pairs has also been observed and may be a common feature for stacked dimers in GC containing complexes (7, 30). These results indicate that the unusual footprinting pattern observed with DB293 is attributable to formation of a very strong and cooperative 2:1 complex in specific, mixed DNA sequences. The close analogs, furamidine and DB270, do not bind strongly or footprint at these DNA sequences. Because all three furan compounds are dications, it is clearly structure, and not charge, that prevents the symmetric derivatives from forming the dimer complex.
Figure 3.
Scatchard plots of the results for binding of DB293 and DB270 to oligo1 and oligo2-1 along with best fit binding curves are shown: closed and open triangles are for DB293 and DB270 binding to oligo1; and closed and open circles are for DB293 and DB270 binding to oligo2-1. Because of the weak binding of DB270 to oligo2-1, the results were fit with the assumption of a single DB270 binding to the duplex (13). Sensorgrams with the data for this plot are shown in Fig. 6, published as supplemental data.
NMR Analysis of the Dimer Complex with Oligo2-1.
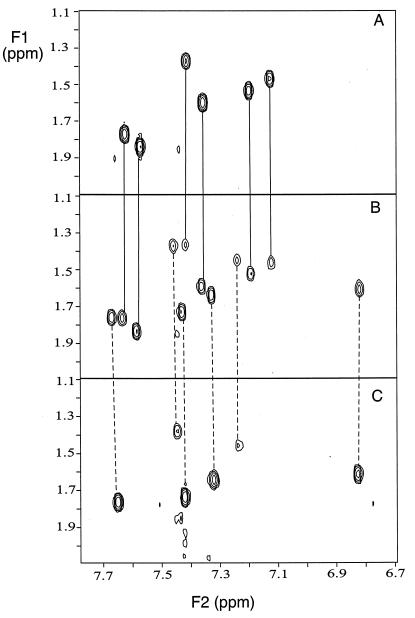
In 1D and 2D proton NMR titrations of the oligo2-1 duplex with DB293, only two DNA species are detected at a 1:1 molar ratio. The two species are clearly illustrated with 2D COSY spectra in Fig. 4 for the aromatic-to-thymine methyl NMR spectral region, and other 2D spectra provide similar examples. The 2:1 complex and the free DNA are the only species observed in the 1:1 ratio COSY spectrum (Fig. 4), in agreement with the high cooperativity observed in the binding experiments. In contrast, two sets of cross peaks are detected for DB293 in the oligo2-1 complex in both COSY and NOESY spectra, as expected for two distinct bound molecules in slow exchange. NOESY cross peaks between the two DB293 molecules and from DB293 to DNA minor-groove protons clearly show that the compound binds in the minor groove as an antiparallel dimer and makes contact with both DNA strands. Strong crosspeaks from the two bound DB293 molecules to DNA base pairs in the region from T4⋅A15 to C7⋅G12 are observed, and these interactions place the dimer in the ATGA sequence that is common to both oligo2-1 and -2-2. Analysis of the NMR results is continuing.
Figure 4.
Two-dimensional COSY spectra of the TH6-TCH3 spectral region are shown for free DNA (A); a 1:1 ratio sample of DB293 to oligo2-1 (B); and a 2:1 ratio (C). Signals for the free DNA and for the 2:1 complex in the 1:1 ratio sample are indicated by connecting lines to the top and bottom spectra.
Discussion
From the results presented in this report, it is clear that all three furan derivatives of Fig. 1 bind to the AATT sequence in oligo1 as classical minor-groove monomer complexes. The results, where comparable, are equivalent to those in the literature (4, 5, 23, 24) and indicate that the oligomer hairpin models and surface attachment in SPR experiments do not strongly affect the compound–DNA complexes. The agreement between footprinting and SPR results for relative binding of DB293 and DB270 also supports use of the hairpin model systems. The results clearly demonstrate that DB293 forms an antiparallel, stacked dimer in complex with some DNA sites that contain GC base pairs (such as oligo2-1), and this accounts for the unusual footprints observed with that compound (Fig. 2). Symmetric compounds that are closely related to DB293, such as DB270 and furamidine, do not form the dimer species in a DNA complex and do not bind significantly to or footprint in DNA sequences that do not have classical AT minor-groove binding sites. The dimer complex provides a motif for understanding and design of compounds that can recognize DNA sequences containing both AT and GC base pairs. What molecular features of DB293 allow it to form a stacked-dimer complex whereas so many other minor-groove-binding agents, including many other benzimidazole derivatives, do not? Favorable stacking of DB293 to give a dimer in the context of the anionic DNA minor groove can contribute to formation of the 2:1 complex, but many other compounds, such as DB270 and Hoechst 33258, stack much more strongly in solution than DB293 but do not form the dimer motif. It appears that the reasons for cooperative formation of the DB293 dimer complex are encoded in the interactions between a specific DNA sequence and the specific orientation of chemical groups in the dimer. Our results show that the binding of aromatic dications to mixed DNA sequences is exquisitely sensitive to compound structure and DNA sequence. The concept that dications do not form dimers for recognition of the DNA minor groove must be modified to incorporate stacked complexes of a new class of DNA recognition units based on the phenyl-furan-benzimidazole core of DB293. Synthetic expansion of the DB293 parent structure is in progress and should provide new agents for control of gene expression at the DNA level. The new compounds should also be of significant benefit in defining the sequence recognition limits for the DB293 dimer motif. Another question that naturally arises out of these results is, What other types of cooperative interactions of small organic cations are possible to generate new nucleic acid recognition motifs for RNA as well as DNA? The results with the furan derivatives suggest that a wider search using combinatorial methods is likely to produce additional DNA recognition motifs.
Supplementary Material
Acknowledgments
We thank Farial Tanious for valuable assistance with Tm experiments on oligo2 and other aspects of this research. This research was supported by National Institutes of Health research grant AI-33363 (to W.D.W. and D.W.B.) and from the Institut National de la Santé et de la Recherche Médicale, Association pour la Recherche sur le Cancer, and the Ligue Nationale Frangaise Contre le Cancer (Comité du Nord) (to C.B.). The BIAcore 2000 was purchased with support from the Georgia Research Alliance, and the NMR spectrometers were purchased with support from the National Science Foundation and the Georgia Research Alliance.
Abbreviations
- Tm
thermal melting
- SPR
surface plasmon resonance
- 1D
one-dimensional
References
- 1.Bailly C. In: Advances in DNA Sequence-Specific Agents. Hurley L H, editor. Vol. 3. Greenwich, CT: Jai; 1998. pp. 97–156. [Google Scholar]
- 2.Mountzouris J A, Hurley L H. In: Bioorganic Chemistry: Nucleic Acids. Hecht S M, editor. New York: Oxford Univ. Press; 1996. pp. 288–323. [Google Scholar]
- 3.Del Poeta M, Schell W, Dykstra C C, Jones S, Tidwell R R, Czarny A, Bajic M, Kumar A, Boykin D W. Antimicrob Agents Chemother. 1998;42:2495–2510. doi: 10.1128/aac.42.10.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopkins K, Kumar A, Bajic M, Wilson W D, Bender B K, McCurdy D R, Hall J E, Tidwell R R, Boykin D W. J Med Chem. 1998;41:3872–3878. doi: 10.1021/jm980230c. [DOI] [PubMed] [Google Scholar]
- 5.Neidle S, Kelland L R, Trent J O, Simpson I J, Boykin D W, Kumar A, Wilson W D. Bioorg Med Chem. 1997;7:1403–1408. [Google Scholar]
- 6.Zimmer C, Wahnert U. Prog Biophys Mol Biol. 1986;47:31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]
- 7.Geierstanger B H, Wemmer D E. Annu Rev Biophys Biomol Struct. 1995;24:463–493. doi: 10.1146/annurev.bb.24.060195.002335. [DOI] [PubMed] [Google Scholar]
- 8.Wilson W D. In: Nucleic Acids in Chemistry and Biology. Blackburn G M, Gait M J, editors. Oxford: IRL; 1996. pp. 329–374. [Google Scholar]
- 9.Kopka M L, Yoon C, Goodsell D, Pjura P, Dickerson R E. Proc Natl Acad Sci USA. 1985;82:1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lown J W, Krowicki K, Bhat U, Skorobogaty A, Ward B, Dabrowiak J C. Biochemistry. 1986;25:7408–7416. doi: 10.1021/bi00371a024. [DOI] [PubMed] [Google Scholar]
- 11.Kopka M L, Larsen T A. In: Nucleic Acid Targeted Drug Design. Probst C L, Perun T J, editors. New York: Dekker; 1992. pp. 303–374. [Google Scholar]
- 12.Kopka M L, Goodsell D, Han G, Chiu T, Lown J W, Dickerson R E. Structure (London) 1997;5:1033–1046. doi: 10.1016/s0969-2126(97)00255-4. [DOI] [PubMed] [Google Scholar]
- 13.Pelton J G, Wemmer D E. Proc Natl Acad Sci USA. 1989;86:5723–5727. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelton J G, Wemmer D E. J Am Chem Soc. 1990;112:1393–1399. [Google Scholar]
- 15.Kielkopf C L, Baird E E, Dervan P B, Rees D C. Nat Struct Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- 16.White S, Szewezyk J W, Turner J M, Baird E E, Dervan P B. Nature (London) 1998;39(1):468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 17.Swalley S E, Baird E E, Dervan P B. J Am Chem Soc. 1999;121:1113–1120. [Google Scholar]
- 18.Herman D M, Turner J M, Baird E E, Dervan P B. J Am Chem Soc. 1999;121:1121–1127. [Google Scholar]
- 19.Seifert J L, Connor R E, Kushon S A, Wang M, Armitage B A. J Am Chem Soc. 1999;121:2987–2995. [Google Scholar]
- 20.Chen X, Mitra S N, Rao S T, Sekar K, Sundaralingam M. Nucleic Acids Res. 1998;26:5464–5547. doi: 10.1093/nar/26.23.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haq I, Ladbury J E, Chowdhry B Z, Jenkins T C, Chaires J B. J Mol Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- 22.Browne K A, He G-X, Bruice T C. J Am Chem Soc. 1993;115:7072–7079. [Google Scholar]
- 23.Trent J O, Clark G, Kumar A, Wilson W D, Boykin D W, Hall J E, Tidwell R R, Blagburn B, Neidle S. J Med Chem. 1996;39:4554–4562. doi: 10.1021/jm9604484. [DOI] [PubMed] [Google Scholar]
- 24.Wilson W D, Tanious F A, Ding D, Kumar A, Boykin D W, Colson P, Houssier C, Bailly C. J Am Chem Soc. 1998;120:10310–10321. [Google Scholar]
- 25.Bailly C, Hamy F, Waring M J. Biochemistry. 1996;35:1150–1157. doi: 10.1021/bi951696p. [DOI] [PubMed] [Google Scholar]
- 26.Bailly C, Dassonneville L, Carrasco C, Lucas D, Kumar A, Boykin D W, Wilson W D. Anti-Cancer Drug Des. 1999;14:47–60. [PubMed] [Google Scholar]
- 27.BIAcore. BIAcore BIApplications Handbook. Piscataway, NJ: BIAcore; 1998. [Google Scholar]
- 28.Connors K A. Binding Constants. New York: Wiley; 1987. [Google Scholar]
- 29.Ding D, Gryaznov S M, Wilson W D. Biochemistry. 1998;37:12082–12093. doi: 10.1021/bi980711y. [DOI] [PubMed] [Google Scholar]
- 30.Trauger J W, Baird E E, Mrksich M, Dervan P B. J Am Chem Soc. 1996;118:6160–6166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.