Abstract
Heroin remains a major drug of abuse and is preferred by addicts over morphine. Like morphine, heroin has high affinity and selectivity for μ-receptors, but its residual analgesia in exon 1 MOR-1 knockout mice that do not respond to morphine suggests a different mechanism of action. MOR-1 splice variants lacking exon 1 have been observed in mice, humans, and rats, raising the possibility that they might be responsible for the residual heroin and morphine-6β-glucuronide (M6G) analgesia in the exon 1 knockout mice. To test this possibility, we disrupted exon 11 of MOR-1, which eliminates all of the variants that do not contain exon 1. Morphine and methadone analgesia in the exon 11 knockout mouse was normal, but the analgesic actions of heroin, M6G, and fentanyl were markedly diminished in the radiant heat tail-flick and hot-plate assays. Similarly, the ability of M6G to inhibit gastrointestinal transit was greatly diminished in these exon 11 knockout mice, whereas the ability of morphine was unchanged. These findings identify receptors selectively involved with heroin and M6G actions and confirm the relevance of the exon 11-associated variants and raise important issues regarding the importance of atypical truncated G-protein-coupled receptors.
Keywords: knockout, analgesia, opiate receptor
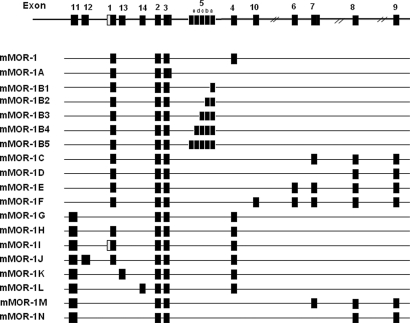
Clinicians have long appreciated analgesic differences among the μ-opioid-analgesics, with some patients responding better to one drug than another (1), results that have been recapitulated in animal models (2, 3). Heroin remains a major drug of abuse and is preferred by addicts over morphine (4, 5). Why addicts and patients should be able to distinguish between 2 μ-agonists that presumably act through only μ-opioid-receptors is not clear. Although some investigators have favored pharmacokinetic factors, several studies raised the possibility of pharmacodynamic causes (2, 6). This inability to reconcile the diverse pharmacology of μ-opioids with their binding selectivity led to the proposal of multiple μ-opioid-receptors (7), a concept initially proposed based on binding and pharmacological studies in animal models and subsequently confirmed with the identification of a large number of splice variants of the cloned μ-opioid-receptor MOR-1 in mice, rats, and humans (Fig. 1A) (8–15).
Fig. 1.
Schematic of MOR-1 splicing in the mouse.
MOR-1 is essential for morphine actions, as shown by the complete loss of morphine activity in a series of MOR-1 knockout mice (16–19). However, detailed studies on one exon 1 knockout mouse revealed persistent heroin and morphine-6β-glucuronide (M6G) analgesia despite the absence of morphine actions (19). It has been suggested that heroin could act through δ-opioid-receptors (20, 21), raising the possibility that this residual activity reflected cross-labeling of δ- or possibly even κ-receptors. However, this seems unlikely because the residual M6G activity is not reversed by δ or κ antagonists. Alternatively, the drugs could be acting through MOR-1 variants lacking exon 1, such as those we have previously reported in mice (Fig. 1) (8) and, most recently, in humans (22). These variants are associated with exon 11, which is located ≈30 kb upstream of exon 1 in the mouse and is under the control of its own exon 11 promoter (8, 23). This possibility was supported by the observation that exon 1 knockout animals that still responded to heroin continued to express mRNA transcripts from the MOR-1 gene (19). To explore the role of these exon 11-associated variants in heroin and M6G actions, we generated an exon 11 knockout mouse and now report that selective disruption of exon 11 leads to a unique phenotype for μ-opioids showing a marked reduction of heroin, M6G, and fentanyl actions with little effect on the actions of morphine or methadone.
Results
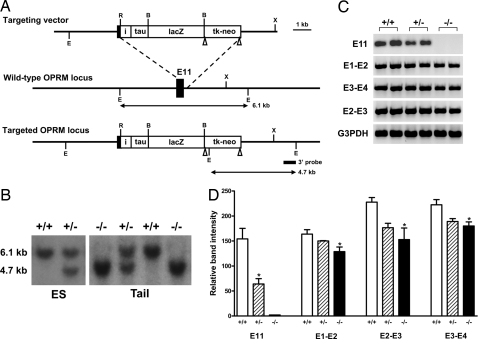
Exon 11 was targeted in 129/Sv embryonic stem (ES) cells by homologous recombination by using a targeting vector in which exon 11 and its adjacent intron region were replaced by an IRES/τ/LacZ-loxP/tk-neo/loxP cassette (Fig. 2A). Transfection of the targeting vector into ES cells yielded 4 positive clones from the 192 neo-resistant clones selected, as determined by Southern blot analysis of EcoRV-digested ES cell DNA by using a 3′ probe (Fig. 2B). Two targeted ES clones were used to generate germ-line-transmitting chimeras. Homozygous mice then were produced through heterozygous mating. Heterozygous mating produced 26.2% wild-type mice, 49.2% heterozygotes, and 24.6% homozygotes (n = 309), indicating that mice lacking exon 11 developed normally. Homozygous mice were healthy and fertile with no obvious morphological abnormalities. RT-PCR confirmed the loss of exon 11, revealing a loss of ≈58% of the mRNA transcripts containing exon 11 in the heterozygotes compared with wild-type animals and its complete absence in the homozygotes (Fig. 2C).
Fig. 2.
Gene targeting exon 11 in the OPRM gene. (A) Schematic of the targeting strategy. The exon 11 coding region and its adjacent intron region are replaced with the IRES/τ/LacZ/loxP-pgk/neo-loxP cassette in 129/SV ES cells through homologous recombination with the targeting vector. Exon 11 is indicated by a black box: i, the internal ribosome entry sequence (IRES); tk, the herpes simplex virus thymidine kinase gene; neo, the neomycin resistance gene. The expected EcoRV-digested fragment lengths for wild-type and targeted alleles are indicated by arrows. 3′ probe is indicated by a short line. Restriction sites: E, EcoRV; R, EcoRI; B, BamHI; X, XbaI. LoxP sites are indicated by triangles. (B) Southern blot analysis of genomic DNA from ES cells and mouse tail. DNA is cut with EcoRV and hybridized with the 3′ probe. The 6.1 kb upper band represents the wild-type allele and the 4.7 kb lower band is the targeted allele. Genotypes: +/+, wild-type; +/−, heterozygous; and −/−, homozygous. (C) RT-PCR. RT is performed by using SuperScript II reverse transcriptase (Invitrogen) with total RNA from mouse brain. The first-strand cDNA is then used as template in PCRs to amplify the corresponding fragments with appropriate primers. (D) Semiquantification of PCR products. The PCR products are analyzed in 1.5% agarose gel, stained with ethidium bromide, and photographed with FluorChem 8000 Image system (Alpha Innotech). The relative band intensities from the gel are quantified with AlphaEase FC software and normalized with the intensities from G3PDH bands. Comparisons are made by using ANOVA, which demonstrated a significant difference among the groups (P <0.04) with post hoc Tukey tests confirming that the knockout groups are significantly lower (P <0.05) than wild-type groups, except for E11 where only the wild-type and heterozygous groups are compared by using a Student's t test (P <0.02) because there is no observable product in the knockout animals.
The gene encoding MOR-1 has 2 independent promoters. One is located immediately upstream of exon 1, whereas the second is associated with exon 11 and is located ≈30 kb upstream of exon 1. To determine the relative contribution of the exon 11-generated variants to total MOR-1 expression, we compared mRNA levels in wild-type and knockout animals with 3 sets of primers spanning exons 1 and 2, 2 and 3, and 3 and 4 by using RT-PCR (Fig. 2D). The products from the exon 1 and 2 primers revealed a 22% decrease in mRNA levels, whereas the exon 2 and 3 primers showed a decrease of 33% and the exon 3 and 4 primers were lowered by 19%. Thus, the exon 11 promoter appears to be responsible for approximately a quarter of MOR-1 expression at the mRNA level.
We observed a similar loss at the protein level in the exon 11 knockout animals, assessed with receptor binding (Table 1). Disruption of exon 11 did not significantly alter the affinity of the μ-agonist [3H][d-Ala2, MePhe4,Gly(ol)5]enkephalin (DAMGO) in brain tissue, but it lowered the binding site density (Bmax) by 21% in the knockout mice, which was significantly less than the wild-type (P < 0.001) and the heterozygotes (P < 0.05), with an intermediate decrease in the heterozygotes (8%) that was not significantly different from the wild-type mice. The selectivity profile of [3H]DAMGO binding in the knockout and heterozygous mice was essentially the same as in wild-type animals (Table 2). [3H]DAMGO binding in the knockout animals retained its high selectivity for a variety of μ-ligands, binding the δ-agonist [d-Pen2,d-Pen5]enkephalin (DPDPE) and the κ1 agonist U50,488H poorly. The affinity and levels of δ-or κ1 receptor binding were unaffected in the knockout mouse (Table 1).
Table 1.
Opioid binding in wild-type, heterozygous, and exon 11 knockout mice: Receptor saturation studies
| Radioligand | Wild-type (+/+) |
Heterozygote (+/−) |
Homozygote (−/−) (knockout) |
|||
|---|---|---|---|---|---|---|
| Kd, nM | Bmax, fmol/mg protein | Kd, nM) | Bmax, fmol/mg protein | Kd, nM | Bmax, fmol/mg protein) | |
| Mu: [3H]DAMGO | 0.33 ± 0.05 | 57.1 ± 1.3* | 0.31 ± 0.05 | 52.6 ± 2.6* | 0.30 ± 0.05 | 45.3 ± 1.3* |
| Delta: [3H]DPDPE | 0.36 ± 0.03 | 19.5 ± 1.4 | 0.31 ± 0.03 | 18.3 ± 2.0 | 0.34 ± 0.01 | 20.5 ± 1.5 |
| Kappa1: [3H]U50,488H | 0.67 ± 0.12 | 16.1 ± 1.3 | 0.67 ± 0.14 | 14.6 ± 1.2 | 0.84 ± 0.26 | 16.3 ± 2.3 |
Brain tissue was obtained from wild-type, heterozygous, and homozygous mice, and saturation analysis was performed with agonists selective for μ ([3H]DAMGO), δ ([3H]DPDPE) and κ1 ([3H]U50,488H) binding sites as previously described (27). Results are the means ± SEM of at least 3 independent determinations.
*The Bmax values for μ sites varied significantly in the various groups of animals (P < 0.0005; ANOVA), with the knockout animals significantly lower than wild-type animals (P < 0.001) and heterozygous animals (P < 0.05).
Table 2.
Opioid binding in wild-type, heterozygous, and exon 11 knockout mice: Mu receptor competition studies
| Ligand |
Ki, nM |
||
|---|---|---|---|
| Wild type (+/+) | Heterozygote (+/−) | Homozygote (−/−) (knockout) | |
| Morphine | 0.51 ± 0.01 | 0.53 ± 0.06 | 0.62 ± 0.16 |
| M6G | 2.41 ± 0.41 | 2.72 ± 0.17 | 1.98 ± 0.21 |
| 6-Acetylmorphine | 4.61 ± 1.46 | 3.46 ± 0.34 | 2.57 ± 0.17 |
| Methadone | 1.36 ± 0.46 | 0.91 ± 0.35 | 0.96 ± 0.39 |
| Fentanyl | 0.82 ± 0.12 | 1.15 ± 0.39 | 0.57 ± 0.18 |
| Naloxone | 0.99 ± 0.25 | 0.90 ± 0.11 | 0.43 ± 0.03 |
| DPDPE | 257.7 ± 39.6 | 151.7 ± 47.9 | 158.7 ± 36.7 |
| U50,488H | >1,000 | >1,000 | >1,000 |
Competition studies were carried out in brain membranes from wild-type, heterozygous, and knockout mice with the μ agonist ([3H]DAMGO), as previously described (27). IC50 values were determined and converted to Ki values. Results are the means ± SEM of at least 3 independent determinations.
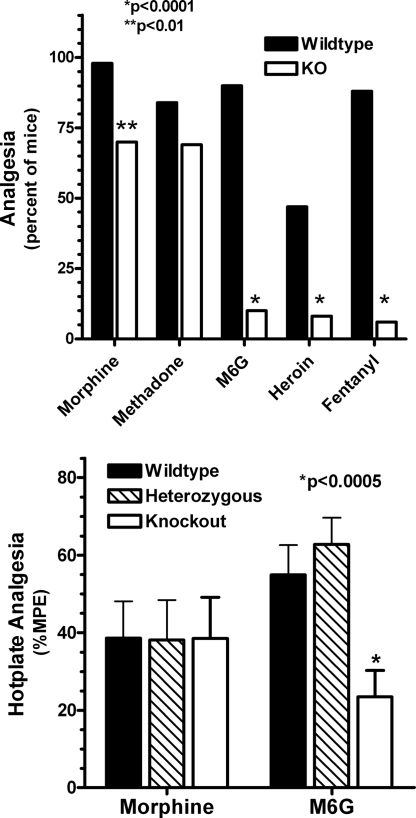
The pharmacological consequences of disrupting exon 11 differed markedly from those observed in earlier studies by using an exon 1 knockout animal (19). In contrast to the exon 1 knockouts in which morphine was totally inactive, disruption of exon 11 had little effect on analgesia in the radiant heat tail-flick assay for fixed doses of either morphine or methadone (Fig. 3Upper). However, we observed a decreased response for heroin, M6G, and fentanyl in these same knockout animals (Fig. 3 Upper). Full dose–response curves confirmed these observations (Table 3). Disruption of exon 11 had no significant effect on the analgesic ED50 value for morphine or methadone. In contrast, the ED50 values for heroin, M6G, and fentanyl were significantly increased. Inspection of these dose–response curves revealed a parallel shift to the right with no decrease in the maximal response. It was interesting that the heterozygotes showed no decrease in analgesic sensitivity, suggesting the existence of spare receptors. Similar results were observed in a second analgesic test, the hot-plate assay (Fig. 3 Lower). As in the tail-flick assay, disruption of exon 11 had no effect on morphine actions while significantly lowering the activity of M6G.
Fig. 3.
Analgesic characterization of exon 11 knockout mice. (Upper) Radiant heat tail-flick assay. Groups of mice (n = 16–24) receive morphine (4 mg/kg, s.c.), methadone (2 mg/kg, s.c.), M6G (2 mg/kg, s.c.), heroin (1 mg/kg, s.c.), or fentanyl (0.5 mg/kg, s.c.) and test in the tail-flick assay at 15 min (heroin) or 30 min (morphine, methadone, M6G, and fentanyl). Responses in wild-type and knockout animals are compared for each drug separately by using the Fisher exact test. (Lower) Hot-plate assay. After determining baseline latencies for all animals to the first pain response (licking or jumping), groups of mice (n = 10–15) receive either morphine (2.5 mg/kg, s.c.) or M6G (1.5 mg/kg) and are tested for analgesia on a 52 °C hot plate 15 min later. The latencies are determined and converted to %MPE [(latency − baseline)/(maximal latency − baseline)]. ANOVA is used to compare morphine-treated groups and to compare M6G-treated groups.
Table 3.
Opioid analgesia in wild-type and exon 11 knockout mice
| Drug | ED50, mg/kg (s.c.) |
Fold shift | P | ||
|---|---|---|---|---|---|
| Wild type (+/+) | Heterozygote (+/−) | Homozygote (−/−) | |||
| Morphine | 1.58 ± 0.17 | 1.30 ± 0.20 | 2.58 ± 0.52 | 1.6 | 0.07 |
| Methadone | 1.53 ± 0.06 | 1.56 ± 0.08 | 1.8 ± 0.14 | 1.2 | 0.11 |
| M6G | 0.92 ± 0.12 | 0.97 ± 0.25 | 19.3 ± 4.2 | 21 | 0.0005 |
| Heroin | 0.58 ± 0.11 | 0.48 ± 0.05 | 3.2 ± 0.8 | 5.5 | 0.0005 |
| Fentanyl | 0.024 ± 0.002 | 0.020 ± 0.001 | 0.23 ± 0.09 | 9.7 | 0.024 |
Analgesia was assessed in cumulative dose–response studies with at least 8 animals/groups using the radiant heat tail-flick assay and was defined quantally as a doubling or greater of the baseline latency with a maximal latency of 10 sec to minimize tissue damage, as previously described (28). ED50 values were determined using probit analysis. Results are the means ± SEM of the ED50 values determined from independent dose–response studies for morphine (n = 5), methadone (n = 5), heroin (n = 7), M6G (n = 7), and fentanyl (n = 4). Differences among groups for each drug were determined by ANOVA.
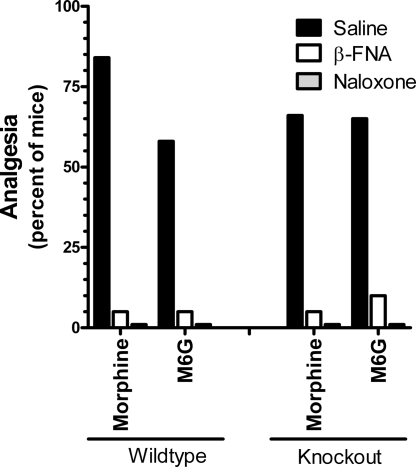
Naloxone (5 mg/kg, s.c.) eliminated the analgesic responses of both drugs in wild-type and knockout mice, documenting the opioid nature of the response (Fig. 4). Morphine and M6G analgesia in the wild-type mice was mediated through μ-receptors, as illustrated by the antagonism of analgesia by the mu-selective antagonist β-funaltrexamine (β-FNA) (Fig. 4) and the inability of the δ-antagonist naltrindole and the κ1 antagonist to significantly lower the response of morphine, confirming results from the literature. Similarly, β-FNA antagonized morphine analgesia in the exon 11 knockout animals, indicating that morphine was still acting through μ-receptors in the knockout mice. The ability of higher doses of heroin, M6G, and fentanyl to elicit a full analgesic response in the exon 11 knockout mice implied that the higher doses were acting through a secondary receptor system. The reversal of the analgesia produced by the higher M6G doses in these knockout animals by β-FNA indicated that this secondary system also involved μ-opioid-receptors (Fig. 4).
Fig. 4.
Effects of naloxone and the μ-selective antagonist β-FNA on morphine and M6G analgesia. Groups of mice (n = 15–25) receive morphine (4.5 mg/kg) or M6G (3 mg/kg for wild type; 24 mg/kg for knockouts) 24 h after administration of β-funaltrexamine (40 mg/kg) or 15 min after naloxone (5 mg/kg, sc), and are tested in the radiant heat tail-flick assay. Responses in saline and antagonist-treated animals are compared for each opioid analgesic separately by using the Fisher exact test.
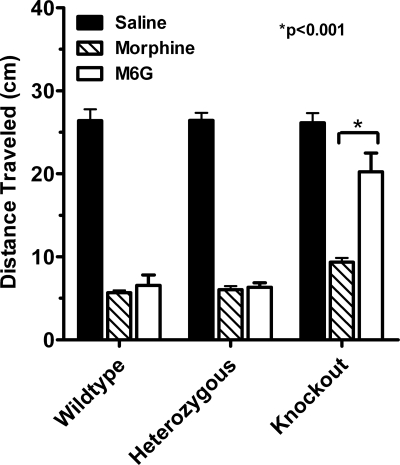
Phenotypic differences due to the disruption of exon 11 were not limited to analgesia. Opiates slow gastrointestinal transit, an important factor contributing to constipation. In wild-type mice, both morphine and M6G inhibited gastrointestinal transit, decreasing the distance traveled by a charcoal meal by about 75% (Fig. 5). Disruption of exon 11 did not appreciably alter the ability of morphine to inhibit gastrointestinal transit. In contrast, the inhibition produced by M6G was greatly diminished in the knockout mice. As with analgesia, M6G retained full activity in the heterozygotes, implying the existence of spare receptors.
Fig. 5.
Effects of disruption of exon 11 on gastrointestinal transit. Groups of mice (n = 6–8) receive morphine (4.5 mg/kg, s.c.) or M6G (4.5 mg/kg) 15 min before a 0.5-ml charcoal meal (2.5% gum tragacanth in 10% activated charcoal in water), and are killed 30 min later, and the distance the charcoal traveled is measured. Results are analyzed by using 2-way ANOVA (P < 0.001). The response in the morphine groups are significantly lower than saline in all 3 groups of mice (P < 0.001 in all groups). The response in the M6G group is significantly lower than saline in wild-type (P < 0.001), heterozygous (P < 0.001), and knockout (P < 0.01) mice. The response in the morphine group is not significantly different from that of M6G in saline or heterozygous groups, but is significantly different in the knockout group (P < 0.001).
Discussion
Multiple μ-opioid-receptors, originally proposed more than 25 years ago (7), have now been confirmed with the cloning of an array of MOR-1 splice variants (Fig. 1). Early studies established the existence of 3′-splicing, with a series of exon 1 variants differing only in their amino acid sequence at the tip of the intracellular C terminus. A second set of 5′ splice variants associated with exon 11, located about 30 kb upstream of exon 1 and under the control of a physically distinct promoter, illustrates the complex nature of this gene (8). The current study establishes both the expression and relevance of the exon 11-associated splice variants. These exon 11-associated variants represent a significant contribution to total Oprm gene expression, corresponding to approximately a quarter of the total expression of MOR-1 and its variants at both the mRNA and protein levels in the whole brain. Previously, we observed distinct regional distributions for the exon 11 variants that differed from that of MOR-1 itself (8), implying functional significance. However, the phenotype of the exon 11 knockout mice provides the first documentation of the functional role of this set of MOR-1 variants.
A number of studies have illustrated pharmacological differences between morphine and heroin (2), whereas others clearly established the role of MOR-1 in their actions by using antisense (24, 25) and knockout approaches (16–19). However, the mechanisms responsible for heroin and M6G actions were not clear. The current studies demonstrate the importance of the exon 11-associated variants in mediating heroin, M6G, and fentanyl analgesia and provide the first association of specific MOR-1 splice variants with these drug actions. The exon 11 knockout mouse illustrates a unique pharmacological phenotype and how alternative splicing of the μ-opioid-receptor gene encoding MOR-1 may help explain the behavioral differences among μ-opioids, including the ability of addicts to discriminate between heroin and morphine (5).
However, the role of the exon 11-associated variants in heroin and M6G actions raises another interesting issue at the molecular level. The exon 11-associated variants that yield full, 7-transmembrane-domain receptors (MOR-1H, MOR-1J, and MOR-1K) contain exon 1. Because heroin and M6G are still active in the exon 1 knockout mice, these variants are not likely to play a major role in the actions of these drugs. Of the 5 remaining exon 11-associated variants that lack exon 1, three (MOR-1G, MOR-1M, and MOR-1N) have been demonstrated in the brain by Western blot and RT-PCR. However, these 3 exon 11-associated variants are structurally unique. They contain only the 6 transmembrane domains encoded by exons 2 and 3. Although there always remains the possibility that additional exon 11 variants lacking exon 1, and which possess a full complement of 7 transmembrane domains, remain to be identified, the evidence currently available suggests that the truncated proteins are pharmacologically active. This possibility is further supported by the continued expression of exon 11 variants in the Pintar exon 1 knockout mouse.
In conclusion, MOR-1 knockout animals have yielded many insights into the actions of opioid analgesics. The disruption of exon 11 generated an analgesic phenotype unlike that seen with the exon 1 knockout and establishes a role for the exon 11-associated variants in heroin action. In addition to providing potential insights into the clinical effects of these agents, the observations also open questions into the potential functional significance of truncated splice variants that may extrapolate to other G-protein coupled receptors, many of which also express truncated forms.
Experimental Procedures
Construction of the E11 Knockin/Knockout (KI/KO) Targeting Vector.
A 1.1-kb τ fragment was amplified by PCR with a sense primer containing a NcoI site (5′-GAA CCA CCA TGG CTG AGC CCC GCC AGG AGT TCG ACG-3′) and an antisense primer containing a BamHI site (5′-GAT GGG ATC CCC GGA CAC GAT CTC CGC CCC GTG GTC GGT CTT GG-3′) using the first-cDNA synthesized from bovine brain mRNA (Clontech) as the template. A 0.6-kb EcoRI/NcoI IRES fragment cut from a IRES/τ/LacZ/LTNL cassette (a kind gift from Peter Mombaerts, The Rockefeller University), and the 1.1-kb NcoI/BamHI τ fragment were subcloned into EcoRI/BamHI site of pcDNA3(−) to construct IRES/τ/pcDNA3(−). A 3.1-kb LacZ fragment cut from pMC1871 vector (Amersham Pharmacia) was inserted into the BamHI site of IRES/τ/pcDNA3(−) to make IRES/τ/LacZ/pcDNA3(−), in which the τ was in frame with LacZ. A 3.7-kb KpnI/XhoI fragment containing 5′ of E11 digested from pSP1 that was amplified from a ES-129svJ-derived genomic BAC clone (GenomeSystems) was subcloned into KpnI/XhoI sites of pBluescript SKII vector containing a unique polyliner (PacI-AscI-FseI-NheI-AflII) to make 5′-arm/SK. The IRES/τ/LacZ cassette cut from IRES/τ/LacZ/pcDNA3(−) with XhoI/AflII was subcloned into XhoI/AflII sites of 5′-arm/SK to make 5′-arm/IRES/τ/LacZ/SK. A 2-kb XbaI loxP-pgk/neo-loxP fragment cut from loxP-pgk/neo-loxP/pGEM7 (a kind gift from Qin Wen Yang, The Rockefeller University) was then subcloned into NheI site of 5′-arm/IRES/τ/LacZ/SK to make 5′-arm/IRES/τ/LacZ/loxP-pgk/neo-loxP/SK. Finally, a 2-kb FseI/AscI genomic fragment containing 3′ of E11 amplified by PCR with a sense primer containing a FseI site (5′-GAT CTT GGC CGG CCT CTT GTT CTC GTG GCT GTA TTG GGC-3′) and an antisense primer containing a AscI site (5′-GAT CTT GGC GCG CCT TTA GAG CTG GTT AAA TCC CAT GCT CTA G-3′) by using genomic DNA from 129/Sv ES cells, was subcloned into FseI/AscI sites of 5′-arm/IRES/τ/LacZ/loxP-pgk/neo-loxP/SK to make the complete targeting construct (Fig. 1A). All of the PCR products and cloning junctions were verified by sequencing with appropriate primers.
Generation of E11 KI/KO Mice.
Forty micrograms of the targeting vector was linearized with AscI, and electroporated into 1 × 106 ES cells at 800 V and 3 μF with a Bio-Rad GenePulser. G418-resistant colonies were picked after 7 to 8 days selection with 150 μg/ml G418. Genomic DNA purified from G418-resistant cells was digested with EcoRV and screened by Southern blot analysis with a 0.7-kb external 3′ probe. Four positives out of 196 G418-resistant clones were identified. Southern blot analysis with several restriction enzymes by using a 3′ probe and a tk-neo probe further verified single-copy integration of the targeting vector in the correct locus. Two positive ES clones were injected into C57BL/6 blastocysts to produce chimeras, which then were bred with C57BL/6. Offspring of the chimeras were genotyped by Southern blot analysis of tail DNAs by using the 3′ probe.
RT-PCR.
Total RNA was extracted from mouse brain with the guanidinium thiocyanate/phenol/chloroform extraction method (26), treated with DNase I by using TURBO DNA-free reagents (Ambion) to remove potentially contaminating genomic DNA, and reversed transcribed with random hexamer or oligo(dT) and SuperScript II reverse transcriptase (Invitrogen) as previously described (9). The reverse transcriptase product was treated with RNase H to remove RNA complementary to the first-strand cDNA. The first-strand cDNA was then used as a template in PCRc to amplify a 173-bp exon 11 fragment with a sense primer (5′-GTCCTTGAGAATGGAGAGGATCAGCAAAGC-3′) and an antisense primer (5′-GGTAACTCTTCCCCTCTTGATTTCCATC 3′), a 402-bp fragment for exons 1 and 2 with a sense primer (5′-GGCTCCTGGCTCAACTTGTCCCAC-3′) and an antisense primer (5′-GGTGCAGAGGGTGAAGATACTGGTGAAC-3′), a 561-bp fragment for exons 2 and 3 with a sense primer (5′-GTGTTAACTACCTGATGGGAACGTGGC-3′) and an antisense primer (5′-GTGGTTTCTGGAATCGTGATCAGTGC-3′), and a 449-bp fragment for exons 3 and 4 with a sense primer (5′-GCACTGATCACGATTCCAGAAACCAC-3′) and an antisense primer (5′-CTGACGTGTAGCGTGAATAGCCAGAGAGG-3′). RNA loading was estimated by a parallel PCR with a pair of glyceraldehyde-3-phosphate dehydrogenase (G3PDH) (Clontech). PCRs were carried out by using Platinum TaqDNA polymerase (Invitrogen) for 28–45 cycles after 2 min at 94 °C, each cycle consisting of a 20-sec denaturing step at 94 °C, a 20-sec annealing step at 65 °C, and a 45- to 90-sec extension at 72 °C. The PCR products were analyzed in 1.5% agarose gel, stained with ethidium bromide and photographed with FluorChem 8000 Image system (Alpha Innotech). The relative band intensities from the gel were quantified with AlphaEase FC software and normalized with the intensities from G3PDH bands. To quantify the amount of transcripts from different targets with a wide range of abundance, various PCR cycle numbers and extension times were adjusted to amplify cDNA at linear phase. For lower abundant exon 11, we used 45 cycles/45 sec extension. For medium abundant exons 1/2, 2/3, and 3/4, we amplified for 35 cycles/45 sec. For higher abundant G3PDH transcript, we only used 28 cycles/90 sec extension.
Receptor Binding Assays.
Membranes were prepared from mouse brain and assays conducted as described previously (27). [3H]DAMGO, or [3H]DPDPE, or [3H]U69,593 binding assays were performed at 25 °C for 60 min in 50 mM potassium phosphate buffer, pH 7.4, containing 5 mM magnesium sulfate. Specific binding was defined as the difference between total binding and nonspecific binding, determined in the presence of 10 μM levallorphan. Protein concentrations were determined as described previously by using BSA as the standard (28). Kd, Bmax, and Ki values were calculated by nonlinear regression analysis (GraphPad Prism).
Tail-Flick and Hot-Plate Assays.
All behavioral studies were performed with mice generated from heterozygous × heterozygous matings to provide wild-type, heterozygous, and homozygous animals with comparable genetic backgrounds. In the radiant heat tail-flick assay, analgesia was determined quantally as a doubling or greater of baseline tail-flick latencies. A maximal latency of 10 sec was used to minimize tissue damage. Cumulative dose-response curves were determined by starting all mice at the lowest opioid dose and escalating the dose. Mice that showed analgesia were removed from further testing and carried forward as analgesic. ED50 values for each replication was calculated by using probit analysis. Results are the mean ± SEM of 4 to 7 independent ED50 determinations, each with groups of at least 6 mice, as indicated. ED50 values among the groups were compared by ANOVA followed by Tukey's post hoc test.
To examine the effects of opioid antagonists, groups of mice received the indicated antagonist 15 min before the agonist, with the exception of β-funaltrexamine, which was administered 24 h before the agonist.
The hot-plate assay was performed at 52 °C. The time(s) elapsing to the first pain response (licking or jumping) was scored. A maximal latency of 30 sec was used to minimize any tissue damage. Results were determined as %MPE ([latency after drug − baseline latency]/[30 − baseline latency]).
Gastrointestinal Transit.
Gastrointestinal transit was assessed as previously described (29). Groups of mice (n = 6–8) were treated with the indicated drugs 15 min before administering a 0.5-ml charcoal meal (2.5% gum tragacanth in 10% activated charcoal in water) by oral gavage. The mice were killed 30 min later and the distance the charcoal traveled was measured. Results are presented as the means ± SEM for each group.
Statistics.
Analgesia was assessed quantally and analyzed by using the Fisher exact test. Other comparisons were made by using either Student's t test or analysis of variance, depending on the comparison.
Acknowledgments.
This work was supported, in part, by research grants from the National Institute on Drug Abuse to G.W.P. (DA02165, DA06241, DA07242) and to Y.-X.P. (DA13997); a Senior Scientist Award (K05DA00220) and a grant from the National Genetics Foundation to G.W.P.; and a Core Grant from the National Cancer Institute to Memorial Sloan–Kettering Cancer Center (CA08748).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Payne R, Pasternak GW. In: Principles of Drug Therapy in Neurology. Johnston MV, Macdonald RL, Young AB, editors. Philadelphia: Davis; 1992. pp. 268–301. [Google Scholar]
- 2.Rossi GC, Brown GP, Leventhal L, Yang K, Pasternak GW. Novel receptor mechanisms for heroin and morphine-6β-glucuronide analgesia. Neurosci Lett. 1996;216:1–4. doi: 10.1016/0304-3940(96)12976-1. [DOI] [PubMed] [Google Scholar]
- 3.Walker JR, King M, Izzo E, Koob GF, Pasternak GW. Antagonism of heroin and morphine self-administration in rats by the morphine-6β-glucuronide antagonist 3-O-methylnaltrexone. Eur J Pharmacol. 1999;383:115–119. doi: 10.1016/s0014-2999(99)00633-0. [DOI] [PubMed] [Google Scholar]
- 4.Fraser HF, Van Horn GD, Martin WR, Wolbach AB, Isbell H. Methods for evaluating addiction liability. (A) “Attitude” of opiate addicts toward opiate-like drugs. (B) A short-term “direct” addiction test. J Pharmacol Exp Ther. 1961;133:371–387. [PubMed] [Google Scholar]
- 5.Martin WR, Fraser HF. A comparative study of physiological and subjective effects of heroin and morphine administered intravenously in postaddicts. J Pharmacol Exp Ther. 1961;133:388–399. [PubMed] [Google Scholar]
- 6.Lange DG, Roerig SC, Fujimoto JM. Absence of cross-tolerance to heroin in morphine-tolerant mice. Science. 1980;208:72–74. doi: 10.1126/science.7361110. [DOI] [PubMed] [Google Scholar]
- 7.Wolozin BL, Pasternak GW. Classification of multiple morphine and enkephalin binding sites in the central nervous system. Proc Natl Acad Sci USA. 1981;78:6181–6185. doi: 10.1073/pnas.78.10.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y-X, et al. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci USA. 2001;98:14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Y-X, et al. Identification and characterization of three new alternatively spliced mu opioid receptor isoforms. Mol Pharmacol. 1999;56:396–403. doi: 10.1124/mol.56.2.396. [DOI] [PubMed] [Google Scholar]
- 10.Pan Y-X, et al. Identification of four novel exon 5 splice variants of the mouse mu-opioid receptor gene: Functional consequences of C-terminal splicing. Mol Pharmacol. 2005;68:866–875. doi: 10.1124/mol.105.011858. [DOI] [PubMed] [Google Scholar]
- 11.Zimprich A, Bacher B, Höllt V. Cloning and expression of an isoform of the rmu-opioid receptor (rmuOR1B) Regul Pept. 1994;54:347–348. [Google Scholar]
- 12.Bare LA, Mansson E, Yang D. Expression of two variants of the human μ opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett. 1994;354:213–216. doi: 10.1016/0014-5793(94)01129-x. [DOI] [PubMed] [Google Scholar]
- 13.Pan L, et al. Identification and characterization of six new alternatively spliced variants of the human mu opioid receptor gene, Oprm. Neuroscience. 2005;133:209–220. doi: 10.1016/j.neuroscience.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 14.Pan Y-X, et al. Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochem Biophys Res Commun. 2003;301:1057–1061. doi: 10.1016/s0006-291x(03)00089-5. [DOI] [PubMed] [Google Scholar]
- 15.Pasternak DA, et al. Identification of three new alternatively spliced variants of the rat mu opioid receptor gene: Dissociation of affinity and efficacy. J Neurochem. 2004;91:881–890. doi: 10.1111/j.1471-4159.2004.02767.x. [DOI] [PubMed] [Google Scholar]
- 16.Matthes HWD, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the μ-opioid-receptor gene. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 17.Sora I, et al. Opiate receptor knockout mice define μ receptor roles in endogenous nociceptive responses and morphine-induced analgesia. Proc Natl Acad Sci USA. 1997;94:1544–1549. doi: 10.1073/pnas.94.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh HH, et al. μ opioid receptor knockout in mice: Effects on ligand-induced analgesia and morphine lethality. Mol Brain Res. 1998;54:321–326. doi: 10.1016/s0169-328x(97)00353-7. [DOI] [PubMed] [Google Scholar]
- 19.Schuller AG, et al. Retention of heroin and morphine-6beta-glucuronide analgesia in a new line of mice lacking exon 1 of MOR-1. Nat Neurosci. 1999;2:151–156. doi: 10.1038/5706. [DOI] [PubMed] [Google Scholar]
- 20.Holmes BB, Rady JJ, Fujimoto JM. Heroin acts on delta opioid receptors in the brain of streptozocin-induced diabetic rats. Proc Soc Exp Biol Med. 1998;218:334–340. doi: 10.3181/00379727-218-44301. [DOI] [PubMed] [Google Scholar]
- 21.Rady JJ, Aksu F, Fujimoto JM. The heroin metabolite, 6-monoacetylmorphine, activates delta opioid receptors to produce antinociception in Swiss-Webster mice. J Pharmacol Exp Ther. 1994;268:1222–1231. [PubMed] [Google Scholar]
- 22.Xu J, Xu M, Pasternak GW, Pan Y-X. Isolation and characterization of new exon 11-associated N-terminal splice variants of the human mu opioid receptor gene, OPRM1. J Neurochem. 2009;108:962–972. doi: 10.1111/j.1471-4159.2008.05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan Y-X. Identification and characterization of a novel promoter of the mouse mu opioid receptor gene (Oprm) that generates eight splice variants. Gene. 2002;295:97–108. doi: 10.1016/s0378-1119(02)00825-9. [DOI] [PubMed] [Google Scholar]
- 24.Rossi G, Pan Y-X, Cheng J, Pasternak GW. Blockade of morphine analgesia by an antisense oligodeoxynucleotide against the mu receptor. Life Sci. 1994;54:L375–L379. doi: 10.1016/0024-3205(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 25.Rossi GC, Pan Y-X, Brown GP, Pasternak GW. Antisense mapping the MOR-1 opioid receptor: Evidence for alternative splicing and a novel morphine-6β-glucuronide receptor. FEBS Lett. 1995;369:192–196. doi: 10.1016/0014-5793(95)00757-z. [DOI] [PubMed] [Google Scholar]
- 26.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 27.Bolan EA, Pasternak GW, Pan Y-X. Functional analysis of MOR-1 splice variants of the mu opioid receptor gene, Oprm. Synapse. 2004;51:11–18. doi: 10.1002/syn.10277. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y-X, et al. Cloning and functional characterization of a kappa3-related opioid receptor. Mol Pharmacol. 1995;47:1180–1188. [PubMed] [Google Scholar]
- 29.Bolan EA, Tallarida RJ, Pasternak GW. Synergy between mu opioid ligands: Evidence for functional interactions among mu opioid receptor subtypes. J Pharmacol Exp Ther. 2002;303:557–562. doi: 10.1124/jpet.102.035881. [DOI] [PubMed] [Google Scholar]