Abstract
The oncoproteins MDM2 and MDMX negatively regulate the activity and stability of the tumor suppressor protein p53—a cellular process initiated by MDM2 and/or MDMX binding to the N-terminal transactivation domain of p53. MDM2 and MDMX in many tumors confer p53 inactivation and tumor survival, and are important molecular targets for anticancer therapy. We screened a duodecimal peptide phage library against site-specifically biotinylated p53-binding domains of human MDM2 and MDMX chemically synthesized via native chemical ligation, and identified several peptide inhibitors of the p53-MDM2/MDMX interactions. The most potent inhibitor (TSFAEYWNLLSP), termed PMI, bound to MDM2 and MDMX at low nanomolar affinities—approximately 2 orders of magnitude stronger than the wild-type p53 peptide of the same length (ETFSDLWKLLPE). We solved the crystal structures of synthetic MDM2 and MDMX, both in complex with PMI, at 1.6 Å resolution. Comparative structural analysis identified an extensive, tightened intramolecular H-bonding network in bound PMI that contributed to its conformational stability, thus enhanced binding to the 2 oncogenic proteins. Importantly, the C-terminal residue Pro of PMI induced formation of a hydrophobic cleft in MDMX previously unseen in the structures of p53-bound MDM2 or MDMX. Our findings deciphered the structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX, shedding new light on structure-based rational design of different classes of p53 activators for potential therapeutic use.
p53 is best known as a tumor suppressor that transcriptionally regulates, in response to cellular stresses such as DNA damage or oncogene activation, the expression of various target genes that mediate cell-cycle arrest, DNA repair, senescence or apoptosis—all of these cellular responses are designed to prevent damaged cells from proliferating and passing mutations on to the next generation (1–3). In 50% of human cancers, p53 is defective due usually to somatic mutations or deletions primarily in its DNA-binding domain and, to a lesser extent, to posttranslational modifications such as phosphorylation, acetylation and methylation that affect p53 function and stability. Altered p53 fails to regulate growth arrest and cell death upon DNA damage, directly contributing to tumor development, malignant progression, poor prognosis and resistance to treatment (4). Conversely, restoring endogenous p53 activity can halt the growth of cancerous tumors in vivo by inducing apoptosis, senescence, and innate inflammatory responses (5–7).
As p53 mediates growth arrest and apoptosis, it is essential to keep its activity in check during normal development (2). Multiple mechanisms exist to negatively regulate p53 activity, among which the E3 ubiquitin ligase MDM2 and its homolog MDMX (also known as MDM4) play a central regulatory role in the developing embryo and in mature differentiated cells (8, 9). MDM2 consists of 491-aa residues, comprising an N-terminal p53-binding domain, a central domain preceded by nuclear export and localization signals essential for nuclear-cytoplasmic trafficking of MDM2, a zinc finger domain, and a C-terminal zinc-dependent RING finger domain that confers E3 ubiquitin ligase activity (10). Structurally related to MDM2, MDMX of 490-aa residues possesses domain structures arranged similarly to MDM2, except that MDMX lacks ubiquitin-ligase function (11, 12). Growing evidence supports that in unstressed cells MDM2 primarily controls p53 stability through ubiquitylation to target the tumor suppressor protein for constitutive degradation by the proteasome (13, 14), whereas MDMX mainly functions as a significant p53 transcriptional antagonist independently of MDM2 (15, 16). Under stress conditions, MDM2 and MDMX cooperate to activate p53 through mechanisms involving both MDM2 autodegradation (autoubiquitylation) and MDM2-depedent degradation of MDMX (17–20).
In many tumors, p53 is present in its wild-type form. The presence of wild-type p53 strongly correlates to amplification and/or over-expression of MDM2/MDMX, resulting directly in p53 suppression and malignant progression (8, 9). Inhibition of the p53-MDM2 interactions by MDM2 antagonists has been shown both in vitro and in vivo to reactivate the p53 pathway and selectively kill tumor cells in a p53-dependent manner. Acting synergistically in tumor cells, MDM2 and MDMX have become 2 of the most attractive molecular targets for anticancer therapy. Toward this end, much of the current efforts have been focused on combinatorial library search for and structure-based rational design of low molecular weight inhibitors that target the N-terminal p53-binding domains of MDM2 and MDMX (21). Successful examples include, but are not limited to, cis-imidazoline analogs termed Nutlins and, more recently, a spiro-oxindole-derived compound termed MI-219 (22, 23).
Peptides, because of their large interacting surfaces, offer the prospect of enhanced potency, high specificity and low toxicity. However, most of the peptidic and peptidomimetic inhibitors examined to date bind MDM2 at affinities ranging from high nanomolar to low micromolar concentrations, and none is nearly as effective as Nutlins and MI-219 in tumor killing in vitro (21). Further, because the structural basis for MDMX inhibition is much less understood than that for MDM2 inhibition, antagonists designed for MDM2 are, in general, significantly less inhibitory toward MDMX. Potent peptide inhibitors against MDM2 and/or MDMX are needed as important cellular probes of the p53 pathway in cancer biology and as useful templates for structure-based rational design of different classes of p53 activators for potential therapeutic use. Here, we report identification and functional and structural characterizations of a high-affinity peptide inhibitor, termed PMI (p53-MDM2/MDMX inhibitor), of p53 interactions with both MDM2 and MDMX.
Results
PMI—a Potent Inhibitor of the p53-MDM2/MDMX Interactions Selected from a Phage Displayed Peptide Library.
The p53-binding domains of MDM2 and MDMX (25–109MDM2 and 24–108MDMX, referred to thereafter as synMDM2 and synMDMX) and their site-specifically biotinylated forms were chemically synthesized using native chemical ligation (SI Text). Using biotin-synMDM2 and biotin-synMDMX as bait, we screened a duodecimal peptide library displayed on M13 phage. Shown in Fig. S1 are the amino acid sequences from 15 binding clones obtained after 4 rounds of selection. Two consensus sequences emerged for both MDM2 and MDMX: LTFEHYWAQLTS, also termed pDI (24), and PMI–TSFAEYWNLLSP. The 3 most critical residues involved in p53-MDM2/MDMX recognition, i.e., Phe-19, Trp-23 and Leu-26 (p53 numbering), were all present in the phage-selected consensus sequences. pDI was recently identified from the same Ph.D.-12TM phage library, using GST-tagged recombinant MDM2 and MDMX immobilized on glutathione-agarose beads (24). However, because of low solubility and relatively weak activity of pDI, this work focused only on PMI, which is highly soluble in aqueous solution and of higher affinity for MDM2 and MDMX than pDI. Two p53 peptides, (15–29)p53 (SQETFSDLWKLLPEN) and (17–28)p53 (ETFSDLWKLLPE), were used for comparison.
We quantified direct interactions between synMDM2/synMDMX and the 3 peptide inhibitors, using isothermal titration calorimetry (ITC), and the results are tabulated in Table S1 (see Fig. S2 for additional data). PMI bound to synMDM2 and synMDMX with Kd values of 3.3 and 8.9 nM, respectively, ≈20-fold stronger than (15–29)p53 for either protein. Compared with that of (17–28)p53—the wild type sequence of the same length, the binding affinity of PMI increased by a factor of 89 for MDM2 and of 43 for MDMX. In both cases, favorable enthalpy changes overcame unfavorable entropy changes, contributing to a dramatically enhanced binding of PMI to synMDM2 and synMDMX.
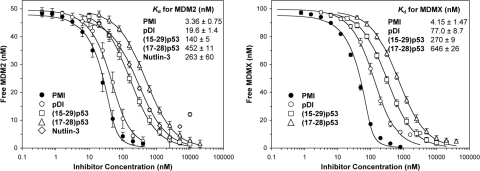
To verify the ITC results, we devised a surface plasmon resonance (SPR)-based competition assay, in which (15–29)p53 was immobilized on a CM5 sensor chip for kinetic analysis of a fixed concentration of synMDM2 (50 nM) or synMDMX (100 nM) preincubated with varying concentrations of PMI, (15–29)p53, or (17–28)p53. Nutlin-3—a racemic mixture of Nutlin-3a and Nutlin-3b (23), was used as a control. As shown in Fig. 1, all 4 inhibitors competed, in a dose-dependent manner, with immobilized (15–29)p53 for synMDM2 or synMDMX binding. Nonlinear regression analyses yielded for PMI a Kd value of 3.4 nM for synMDM2 and of 4.2 nM for synMDMX. By contrast, (15–29)p53 bound to synMDM2 and synMDMX at affinities of 140 nM and 270 nM, respectively. Although Nutlin-3 showed little binding to MDMX, its Kd value of 263 nM for MDM2 was largely in line with the published result (IC50: 90 nM for Nutlin-3a and 13.6 μM for Nutlin-3b) (23). Importantly, compared with (17–28)p53, PMI bound to synMDM2 135-fold stronger and to synMDMX 156-fold tighter. Our data suggest that PMI, with low nanomolar affinities for both MDM2 and MDMX, is one of the strongest peptidic inhibitors of the p53-MDM2/MDMX interactions ever reported.
Fig. 1.
Quantification of the interactions of synMDM2 (50 nM) and synMDMX (100 nM) with varying concentrations of PMI, pDI, (15–29)p53, (17–28)p53, or Nutlin-3 by SPR-based competition assays. Each curve is the mean of 4 independent measurements at 25 °C in 10 mM Hepes, 150 mM NaCl, 0.005% surfactant P20, pH 7.4. Nutlin-3 was too weak for synMDMX and is not reported. Solubility of both Nutlin-3 and pDI decreased at the highest concentrations used, attributing to an upward curvature of the inhibition curves for synMDM2-Nutlin-3 and synMDMX-pDI.
PMI Binds to MDM2 and MDMX in a Canonical Mode Previously Described for p53 Peptides.
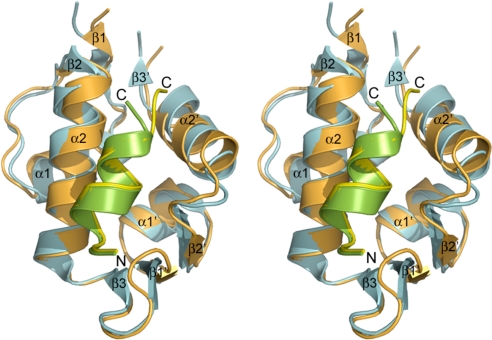
The overall structures of human synMDM2 and synMDMX are similar, as evidenced by a root mean square deviation (rmsd) of 1.2 Å between superimposed Cα atoms. synMDM2 and synMDMX share the basic structural elements and conserved global fold reported for human MDM2 (23, 25–28) and zebra fish MDMX (29). Both molecules are characterized by structural repetition of 2 assemblies of a βαβαβ topology, which are related by an approximate dyad axis of pseudosymmetry (Fig. 2). Superposition of synMDM2 and synMDMX points toward a large structural variation in the C-terminal half of the molecules, whereas the N-terminal halves are nearly identical. Except for the loop regions, the largest conformational changes are located in the α2′ helix, stemming from a change of His-96 in MDM2 to Pro-95 in MDMX. Interestingly, a crystallographic 2-fold axis generates a dimer of the PMI-synMDM2 complex (Fig. S3), although no evidence for dimerization has been found using native gel electrophoresis and size exclusion chromatography.
Fig. 2.
Stereoview of superimposed structures of synMDM2-PMI (orange/green) and synMDMX-PMI (blue/yellow) in a ribbon diagram.
The N-terminal transactivation domain of p53 encompasses T18F19S20D21L22W23K24L25L26 minimally required for effective MDM2 binding (30, 31). The side chains of Phe-19, Trp-23 and Leu-26, involved in p53 transactivation (32–34), dock, in an amphipathic α-helix, inside a hydrophobic cavity of the oncoprotein (25). Not surprisingly, PMI retains the functionally conserved hydrophobic triad, Phe-3/Trp-7/Leu-10. Structural analysis indicates that the Phe-3-binding pockets are nearly identical in MDM2 and in MDMX. However, the Trp-7-binding pockets slightly differ in geometry (Leu-54 and Ile-99 in MDM2 versus Met-53 and Leu-98 in MDMX). Nevertheless, the Phe/Trp dyads of PMI and p53 appear indistinguishable in MDM2/MDMX binding, as indicated by calculations of buried surface area (BSA) and binding energy (Table S2). In contrast, the Leu-10-binding pocket in MDM2 (lined by Leu-54, Val-93, His-96, Ile-99, Tyr-100) differs from that in MDMX (lined by Met-53, Val-92, Pro-95, Leu-98, Tyr-99), mainly because of the shift of the α2′ helix caused by the His-96 to Pro-95 change. Because Leu-10 of PMI is buried in the peptide-protein complexes to a different extent from Leu-26 of p53, the energetic contribution of that position may be context-dependent.
In addition to the hydrophobic interactions, 3 intermolecular H-bonds contribute to PMI binding to MDM2 and MDMX. The same number of intermolecular H-bonds is also present in the p53-MDM2 complex between Phe-19 N, Trp-23 Nε1, and Asn-29 OXT of the peptide and Gln-72 Oε1, Leu-54 O, and Tyr-100 Oη of the protein, respectively (25). However, the H-bonding patterns involving Tyr-100 in PMI-synMDM2 and Tyr-99 in PMI-synMDMX differ. Tyr-100 Oη donates an H-bond to Leu-10 O of PMI, whereas Tyr-99 Oη forms an H-bond with Ser-11 N of PMI, reflecting structural differences in the C-terminal region of PMI and in the vicinity of Tyr-100/Tyr-99 between different protein-peptide complexes.
PMI Differs from p53.
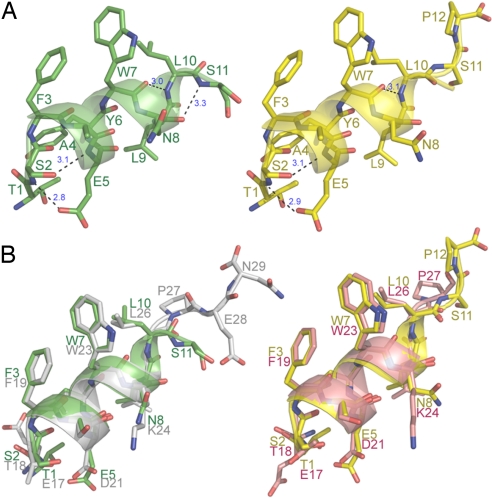
A secondary-structure analysis, using the Kabsch–Sander algorithm (35), reveals that the α-helix of PMI is more extended than that of p53. In PMI, the regular α-helix starts at Phe-3 and ends at Asn-8 (6 residues), followed by a helical turn comprising Leu-9 and Leu-10 in MDM2 and only Leu-9 in MDMX. By contrast, the regular α-helix in p53, starting at Phe-19 and ending at Trp-23, is shortened by 1 residue. Two (i, i + 3) >C O… HN< H-bonds involving Trp-7-Leu-10 (3.0 Å) and Asn-8-Ser-11 (3.3 Å) in PMI-synMDM2, or the Trp-7-Leu-10 H-bond (3.1 Å) in PMI-synMDMX, are missing in the helical turn region of p53 (Fig. 3). Further, Ser-2 of PMI participates in a more extensive and stronger H-bonding network than does Thr-18 of p53. In addition to Ser-2 O-Glu-5 N and Ser-2 O-Tyr-6 N—two backbone H-bonds also found for Thr-18 of p53, PMI possesses 2 additional main chain-side chain H-bonds, i.e., Ser-2 N-Glu-5 Oε1 (2.8 Å in MDM2 and 2.9 Å in MDMX) and Ser-2 Oγ-Glu-5 N (3.1 Å in both MDM2 and MDMX) (Fig. 3). Optimally aligned, these two H-bonds likely contribute to the conformational stability of PMI in the MDM2/MDMX complex. By contrast, the topologically equivalent main chain-side chain H-bonds involving Thr-18 of p53, i.e., Thr-18 N-Asp-21 Oδ2 (3.5 Å) and Thr-18 Oγ1-Asp-21 N (3.6 Å) (25), appear too long to be energetically significant. It is worth noting that a side chain-side chain H-bond was thought to exist between Thr-18 Oγ1 and Asp-21 Oδ2 as part of the Thr-18-Asp-21 H-bonding network important for p53 stability (25). Similar interactions also exist between Ser-2 Oγ and Glu-5 Oε1 in PMI-MDM2 and PMI-MDMX.
O… HN< H-bonds involving Trp-7-Leu-10 (3.0 Å) and Asn-8-Ser-11 (3.3 Å) in PMI-synMDM2, or the Trp-7-Leu-10 H-bond (3.1 Å) in PMI-synMDMX, are missing in the helical turn region of p53 (Fig. 3). Further, Ser-2 of PMI participates in a more extensive and stronger H-bonding network than does Thr-18 of p53. In addition to Ser-2 O-Glu-5 N and Ser-2 O-Tyr-6 N—two backbone H-bonds also found for Thr-18 of p53, PMI possesses 2 additional main chain-side chain H-bonds, i.e., Ser-2 N-Glu-5 Oε1 (2.8 Å in MDM2 and 2.9 Å in MDMX) and Ser-2 Oγ-Glu-5 N (3.1 Å in both MDM2 and MDMX) (Fig. 3). Optimally aligned, these two H-bonds likely contribute to the conformational stability of PMI in the MDM2/MDMX complex. By contrast, the topologically equivalent main chain-side chain H-bonds involving Thr-18 of p53, i.e., Thr-18 N-Asp-21 Oδ2 (3.5 Å) and Thr-18 Oγ1-Asp-21 N (3.6 Å) (25), appear too long to be energetically significant. It is worth noting that a side chain-side chain H-bond was thought to exist between Thr-18 Oγ1 and Asp-21 Oδ2 as part of the Thr-18-Asp-21 H-bonding network important for p53 stability (25). Similar interactions also exist between Ser-2 Oγ and Glu-5 Oε1 in PMI-MDM2 and PMI-MDMX.
Fig. 3.
Structures of PMI and p53 in bound state. (A) PMI bound to MDM2 (Left, green) and to MDMX (Right, yellow). In PMI, Ser-2 to Ala-4 make 3 regular (i, i + 4) >C O···HN< H-bonds in the α-helix. Ser-2 and residues from Tyr-6 to Asn-8 (in the MDM2 complex) or Tyr-6 to Trp-7 (in the MDMX complex) also make 4 or 3 (i, i + 3) >C
O···HN< H-bonds in the α-helix. Ser-2 and residues from Tyr-6 to Asn-8 (in the MDM2 complex) or Tyr-6 to Trp-7 (in the MDMX complex) also make 4 or 3 (i, i + 3) >C O···HN< H-bonds. In addition, Ser-2 also forms 2 main chain-side chain, and 1 side chain-side chain H-bonds with Glu-5. However, only the (energetically significant) H-bonds unique to PMI are shown in dashes. (B) Superposition of PMI (green) and p53 bound to MDM2 (Left), and of PMI (yellow) and p53 bound to MDMX (Right). Residues 17–29 of p53 (gray) bound to human MDM2 (PDB entry 1YCR) and 17–27 of p53 (pink) bound to zebra fish MDMX (PDB entry 2Z5T) are shown.
O···HN< H-bonds. In addition, Ser-2 also forms 2 main chain-side chain, and 1 side chain-side chain H-bonds with Glu-5. However, only the (energetically significant) H-bonds unique to PMI are shown in dashes. (B) Superposition of PMI (green) and p53 bound to MDM2 (Left), and of PMI (yellow) and p53 bound to MDMX (Right). Residues 17–29 of p53 (gray) bound to human MDM2 (PDB entry 1YCR) and 17–27 of p53 (pink) bound to zebra fish MDMX (PDB entry 2Z5T) are shown.
Tyr-6 of PMI or Leu-22 of p53 makes van der Waals contacts with Val-93 of MDM2 or Val-92 of MDMX. As reported in refs. 24 and 36, Tyr is strongly selected by phage display over a Leu residue at the same position. This selection probably can be rationalized by the tyrosyl side chain capable of making additional hydrophobic, π-cation, and electrostatic interactions with residues inside the PMI-binding pocket. Tyr-6 forms cation-π interactions with Lys-94 of MDM2 or possibly Lys-93 of MDMX. Further, the buried surface area of Tyr-6 of PMI is significantly larger than that of Leu-22 of p53 (Table S2). Finally, the hydroxyl group of Tyr-6 participates in an elaborate, water-mediated H-bonding network comprising the side chain(s) of Gln-72 and Lys-94 of MDM2 or Gln-71 of MDMX (Fig. S4).
synMDM2-PMI Differs from synMDMX-PMI.
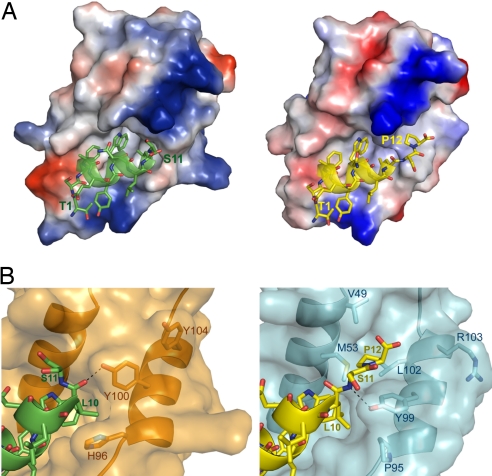
The most profound structural difference between the 2 complexes centers on the 2 C-terminal residues (Ser-11 and Pro-12) of PMI in the vicinity of Tyr-100/Tyr-99 (Fig. 4). In the synMDM2-PMI complex, Pro-12 of PMI is fully disordered (refer to the electron density map, Fig. S5), and the side chain of Ser-11 does not contribute to MDM2 binding. This finding is consistent with the previous observation that C-terminal residues flanking Leu-26 of p53 (equivalent to Leu-10 of PMI) do not make direct contact with MDM2 (25). The aromatic ring of a protruding Tyr-100, H-bonded to the carbonyl O of Leu-10 of PMI, appears to “squeeze out” Ser-11 and Pro-12 to point away from the protein. By contrast, structurally ordered Ser-11 and Pro-12 of PMI fit snugly in the synMDMX-PMI complex, where Tyr-99 recesses to form an H-bond with Ser-11 N of the peptide.
Fig. 4.
Structural differences in PMI binding between MDM2 and MDMX. (A) Electrostatic potential distribution (negative in red, positive in blue, and neutral in white) at the molecular surfaces of MDM2 (Left) and MDMX (Right). PMI is shown in a ribbon and stick diagram. (B) Close-up view of the binding pockets for Leu-10 and Pro-12 of PMI, where only the residues lining the binding pockets are shown in sticks. The side chain of Tyr-100 of MDM2 is H-bonded to Leu-10 O of PMI (Left), whereas Tyr-99 Oη of MDMX forms an H-bond with Ser-11 N of PMI (Right).
The α2′ helix of MDMX moves outward in relation to the α2′ helix of MDM2, coinciding with the Cα atoms of His-96 of MDM2 and Pro-95 of MDMX moving apart by 2.4 Å. This change propagates to Tyr-99 of MDMX. A resultant conformational adjustment surrounding Tyr-99, characterized by a shift of its Cα atom by 1.5 Å, creates for Pro-12 a new binding cleft lined by Val-49, Met-53, Tyr-99 and Leu-102 (a BSA of Pro-12: 99 Å2). Notably, Leu-10 of PMI is largely shielded from the bulk solvent by His-96 and Tyr-100 of MDM2 (a BSA of Leu-10: 122 Å2). However, it becomes less buried in the complex with MDMX (a BSA of Leu-10: 84 Å2) as a result of the significant conformational change in the α2′ helix region. Pro-12 interacting with its newly formed hydrophobic cleft in synMDMX-PMI likely compensates, at least in part, for the lost binding energy by a partially exposed Leu-10.
Discussion
Inhibition of p53 interactions with the oncogenic proteins MDM2 and MDMX is of important therapeutic value in cancer treatment. However, potent inhibitors active at low nanomolar concentrations against both MDM2 and MDMX are nonexistent. Using phage display coupled with chemical protein synthesis via native chemical ligation (37, 38), we identified PMI—the strongest peptide ligand ever reported for MDM2/MDMX. Further, high-resolution crystal structures of synMDM2 and synMDMX in complex with PMI have been determined, unveiling not only the structural differences between MDM2 and MDMX but also the molecular determinants for high-affinity peptide inhibition of the 2 oncogenic proteins.
PMI binds MDM2 and MDMX similarly to p53 peptides. However, the improvement in binding affinity of PMI over (17–28)p53 by 2 orders of magnitude was surprising, partly because both peptides contained the same hydrophobic triad, Phe/Trp/Leu, known to be most critical for p53 transactivation and for MDM2 interactions (32–34, 39). Structural analyses of the PMI complexes and of p53-bound MDM2/MDMX structures suggest that an extensive, tightened intramolecular H-bonding network found in PMI likely plays an important role in stabilizing its more extended α-helical conformation in the complex, thus contributing to high-affinity PMI binding to the oncoproteins. Central to the N-terminal H-bonding network in PMI is Ser-2, which donates and accepts up to 5 H-bonds as judged by their geometry. Not surprisingly, mutation of Ser-2 to Ala reduced the binding affinity of PMI for MDM2 by 1 order of magnitude, underscoring the importance of intramolecular hydrogen bonding in PMI-MDM2/MDMX interactions. It has been shown that Thr-18, highly conserved in p53 and important for MDM2 binding, can be substituted only by Ser (31). Consistent with the structural findings, the results from ITC measurements showed that the free energy of association gained by PMI over (17–28)p53 for MDM2/MDMX came entirely from a favorable enthalpy change counteracted by an unfavorable entropy change. A greater entropy loss for PMI (Table S1) was indicative of a more stable peptide conformation in the complex than (17–28)p53.
An enhanced α-helicity of PMI and the tightening of its intramolecular H-bonding network seen in the MDM2 and MDMX complexes likely result, at least in part, from the selection of Ser-11 (as opposed to Pro-27 in p53). Zondlo et al. recently showed that mutation of the highly conserved Pro-27 in (12–30)p53 to Ser increased its binding affinity for MDM2 by 50-fold (a decrease in Kd from 229 to 4.7 nM) (40). The authors attributed the improvement in MDM2 binding to increased α-helicity of the peptide in the complex—a thesis largely confirmed in a molecular dynamic simulation study by Dastidar et al. (41).
One of the most interesting structural findings entails the C-terminal residue Pro-12 of PMI interacting with a hydrophobic cleft unique to MDMX. Pro-12 is the third most buried residue at the PMI-MDMX interface with the second highest solvation energy effect (Table S2). By contrast, residues flanking Leu-26 of p53 do not make energetically meaningful contact with either human MDM2 or zebra fish MDMX (25, 29). In fact, deletion of Ser-11 and Pro-12 in PMI, while exhibiting little effect on MDM2 binding, weakened MDMX binding by 1 order of magnitude (Kd jumped from 3.6 to 29 nM), indicative of the importance of Pro-12 in binding MDMX but not MDM2. As shown in the known structures of MDM2 and zebra fish MDMX (23, 25–29), the geometry of the binding pocket for the C-terminal residues of p53 hinges on the conformation of Tyr-100 of MDM2 or Tyr-99 of MDMX. The orientation of Tyr-100 or Tyr-99 seen in different MDM2 and MDMX complex structures (Fig. 5) is clearly determined not only by the intrinsic properties of the α2′ helices but also by the chemical nature of the C-terminal residues of the ligand, supporting the notion that mutual conformational modulation of a protein-peptide complex leads to higher affinity (41). The hydrophobic binding cleft in MDMX induced by Pro-12 of PMI obviously affords an excellent opportunity to fine-tune binding affinity and specificity for MDM2/MDMX inhibitors.
Fig. 5.
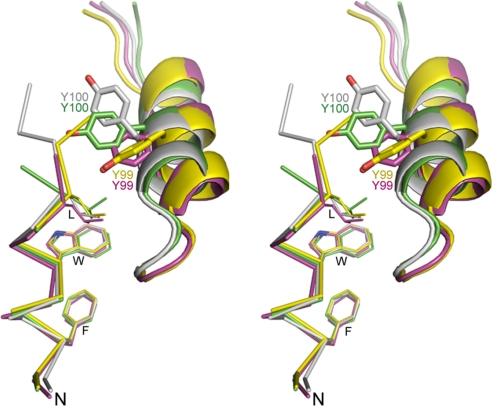
Conformational changes of Tyr-100/Tyr-99 seen in different peptide-protein complexes: PMI-synMDM2, green; p53-MDM2, gray (PDB entry 1YCR); PMI-synMDMX, yellow; and p53-MDMX, pink (PDB entry 1Z5T).
Finally, it is worth noting that PMI, despite its high binding affinity for MDM2/MDMX, is much less active than Nutlin-3 in the killing of p53+/+ HCT116 cells (Fig. S6). The attenuated cytotoxic activity in vitro may reflect a low intracellular concentration of PMI, likely resulting from a combination of proteolytic degradation, inefficient cellular uptake, and endosomal sequestration (42). For these reasons, peptide inhibitors as potential therapeutic agents are generally considered “undruggable” compared with traditional low molecular weight compounds. Importantly, however, the discovery of small molecule drugs depends on high-resolution crystal structures of MDM2/MDMX complexed with high-affinity peptide ligands (22, 23). The information obtained from high-affinity peptide inhibition of MDM2/MDMX is equally valuable for the design of miniature proteins that potently activate the p53 pathway and effectively kill p53+/+ tumor cells in vitro (24, 43, 44). Our structural work reported here should aid in silico library screening and structure-based rational design of different classes of p53 activators for anticancer therapy.
Shortly before we submitted this manuscript, the crystal structure of human (23–111)MDMX in complex with (15–29)p53 at 1.9 Å resolution was reported (45). The complex structure is very similar to zebra fish MDMX- (15–29)p53 used for comparison in our article. Superposition of human MDMX- (15–29)p53 and synMDMX-PMI yielded a RMSD of 0.8 Å. As observed in zebra fish MDMX- (15–29)p53 (29), the C-terminal residues of p53 (Pro-27 and Glu-28) are loosely bound to human MDMX with a single stabilizing H-bond between Tyr-99 Oη and Pro-27 O. The hydrophobic cleft for Pro-12 of PMI seen in synMDMX-PMI is not formed in the human MDMX- (15–29)p53 complex.
Materials and Methods
Phage Display.
Ph.D.-12—a combinatorial library of random peptide 12-mers fused, via a short spacer GlyGlyGlySer, to the N terminus of a minor coat protein (pIII) of M13 phage—was purchased from New England Biolabs, Inc. The basic procedures for library screening are as follows: (i) incubate input phage (10 μL) with 400 μL of 10 nM biotin-synMDM2/biotin-synMDMX for 60 min before adding phage-target solution to 50 μL of streptavidin-agarose resin (Pierce) for affinity capturing; (ii) wash unbound phage; (iii) elute bound phage with 1 mM (15–29)p53; (iv) amplify the eluate and collect phage for the next round of panning; (v) repeat steps 1–4; (vi) sequence selected binding clones according to the procedures recommended by the manufacturer.
Surface Plasmon Resonance (SPR) Spectroscopy.
Competition binding kinetics was carried out at 25 °C on a Biacore T100 SPR instrument, using a (15–29)p53-immobilized CM5 sensor chip (17 RUs for MDM2 and 36 RUs for MDMX). The buffer was 10 mM Hepes, 150 mM NaCl, 0.005% surfactant P20, pH 7.4. 50 nM synMDM2 or 100 nM synMDMX was incubated at room temperature for 30 min with varying concentrations of inhibitor, and injected at a flow rate of 20 μL/min for 2 min, followed by 4 min dissociation. The concentration of unbound synMDM2 or synMDMX in solution was deduced, based on p53-association RU values, from a calibration curve established by RU measurements of different concentrations of synMDM2 or synMDMX injected alone. Nonlinear regression analysis was performed using GraphPad Prism 4 to give rise to Kd values. Protein and peptide solutions were quantified by UV absorbance measurements at 280 nm, using molar extinction coefficients calculated from an algorithm published in ref. 46.
Structure Determination and Refinement.
Conditions for crystallization and data collection are described in SI Text. The structures of both complexes were solved by molecular replacement, using Phaser (47) and search models based on the previously solved and refined structures of (17–125)MDM2- (15–29)p53 (PDB entry 1YCR) and zebra fish (15–129)MDMX- (15–29)p53 (PDB entry 2Z5T) (25, 29). The structures were refined to 1.65 Å for synMDM2-PMI (R = 0.155; Rfree = 0.194) and 1.63 Å for synMDMX-PMI (R = 0.154; Rfree = 0.168) with the program Refmac (48), and rebuilt using the program COOT (49). Parameters for data collection and results of refinement are summarized in Table S3. The atomic coordinates of synMDM2-PMI (3EQS) and synMDMX-PMI (3EQY) have been deposited in the Protein Data Bank.
Supplementary Material
Acknowledgments.
This work was supported by American Cancer Society Research Scholar Grant CDD112858, National Institutes of Health Grants AI056264 and AI061482 (to W.L.), and the Intramural Research Program of the National Institutes of Health (S.G.T.).
Note Added in Proof.
Several new MDMX structures have been determined in complexes with a single-domain antibody (PDB ID code 2VYR) (50), peptidomimetic inhibitors (PDB ID codes 3FE7 and 3FEA) (51), and pDI (PDB ID code 3FDO).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3EQS and 3EQY).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900947106/DCSupplemental.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lu X. Live or let die: The cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4.Kirsch DG, Kastan MB. Tumor-suppressor p53: Implications for tumor development and prognosis. J Clin Oncol. 1998;16:3158–3168. doi: 10.1200/JCO.1998.16.9.3158. [DOI] [PubMed] [Google Scholar]
- 5.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 7.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Toledo F, Wahl GM. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 9.Marine JC, Dyer MA, Jochemsen AG. MDMX: From bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 10.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 11.Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20:1001–1007. doi: 10.1128/mcb.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shvarts A, et al. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 13.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 14.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 15.Toledo F, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4–p53 regulatory network. Cancer Cell. 2006;9:273–285. doi: 10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Francoz S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Graaf P, et al. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315–38324. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- 20.Kawai H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 21.Murray JK, Gellman SH. Targeting protein–protein interactions: Lessons from p53/MDM2. Biopolymers. 2007;88:657–686. doi: 10.1002/bip.20741. [DOI] [PubMed] [Google Scholar]
- 22.Shangary S, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 24.Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 25.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai K, Schubert C, Kahne D. Crystallographic analysis of an 8-mer p53 peptide analogue complexed with MDM2. J Am Chem Soc. 2006;128:11000–11001. doi: 10.1021/ja063102j. [DOI] [PubMed] [Google Scholar]
- 27.Grasberger BL, et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005;48:909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 28.Fasan R, et al. Structure-activity studies in a family of beta-hairpin protein epitope mimetic inhibitors of the p53-HDM2 protein–protein interaction. Chembiochem. 2006;7:515–526. doi: 10.1002/cbic.200500452. [DOI] [PubMed] [Google Scholar]
- 29.Popowicz GM, et al. Molecular Basis for the Inhibition of p53 by Mdmx. Cell Cycle. 2007:6. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- 30.Lai Z, Auger KR, Manubay CM, Copeland RA. Thermodynamics of p53 binding to hdm2(1–126): Effects of phosphorylation and p53 peptide length. Arch Biochem Biophys. 2000;381:278–284. doi: 10.1006/abbi.2000.1998. [DOI] [PubMed] [Google Scholar]
- 31.Schon O, et al. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 32.Bottger A, et al. Molecular characterization of the hdm2–p53 interaction. J Mol Biol. 1997;269:744–756. doi: 10.1006/jmbi.1997.1078. [DOI] [PubMed] [Google Scholar]
- 33.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 34.Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2—fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 35.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 36.Bottger V, et al. Identification of novel mdm2 binding peptides by phage display. Oncogene. 1996;13:2141–2147. [PubMed] [Google Scholar]
- 37.Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 38.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 39.Massova I, Kollman PA. Computational alanine scanning to probe protein–protein interactions: A novel approach to evaluate binding free energies. J Am Chem Soc. 1999;121:8133–8143. [Google Scholar]
- 40.Zondlo SC, Lee AE, Zondlo NJ. Determinants of specificity of MDM2 for the activation domains of p53 and p65: proline27 disrupts the MDM2-binding motif of p53. Biochemistry. 2006;45:11945–11957. doi: 10.1021/bi060309g. [DOI] [PubMed] [Google Scholar]
- 41.Dastidar SG, Lane DP, Verma CS. Multiple peptide conformations give rise to similar binding affinities: Molecular simulations of p53-MDM2. J Am Chem Soc. 2008;130:13514–13515. doi: 10.1021/ja804289g. [DOI] [PubMed] [Google Scholar]
- 42.Vives E, Schmidt J, Pelegrin A. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta. 2008;1786(2):126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Li C, et al. Turning a scorpion toxin into an antitumor miniprotein. J Am Chem Soc. 2008;130:13546–13548. doi: 10.1021/ja8042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kritzer JA, et al. Miniature protein inhibitors of the p53-hDM2 interaction. Chembiochem. 2006;7:29–31. doi: 10.1002/cbic.200500324. [DOI] [PubMed] [Google Scholar]
- 45.Popowicz GM, Czarna A, Holak TA. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- 46.Pace CN, et al. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Yu GW, Vaysburd M, Allen MD, Settanni G, Fersht AR. Structure of human Mdm4 N-terminal domain bound to a single-domain antibody. J Mol Biol. 2009;385:1578–1589. doi: 10.1016/j.jmb.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 51.Kallen J, et al. Crystal structures of human MdmX (HdmX) in complex with p53 peptide-analogues reveal surprising conformational changes. J Biol Chem. 2009 doi: 10.1074/jbc.M809096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.