Abstract
Cilia and flagella are dynamic organelles that are assembled and disassembled during cell differentiation, during stress, and during the cell cycle. Although intraflagellar transport (IFT) is well documented to be responsible for transport of ciliary/flagellar precursors from the cell body to the flagella, little is known about the molecular mechanisms for mobilizing the cell body-localized precursors to make them available for transport during organelle assembly or for disassembling the microtubule-based axoneme during shortening. Here, we show that Chlamydomonas kinesin-13 (CrKinesin-13), a member of the kinesin-13 family of microtubule depolymerizing kinesins best known for their roles in the cell cycle, functions in flagellar disassembly and flagellar assembly. Activation of a cell to generate new flagella induces rapid phosphorylation of CrKinesin-13, and activation of flagellar shortening induces the immediate transport of CrKinesin-13 via intraflagellar transport from the cell body into the flagella. Cells depleted of CrKinesin-13 by RNAi assemble flagella after cell division but are incapable of the rapid assembly of flagella that normally occurs after flagellar detachment. Furthermore, they are inhibited in flagellar shortening. Thus, CrKinesin-13 is dynamically regulated during flagellar assembly and disassembly in Chlamydomonas and functions in each.
Keywords: cilia, flagella, intraflagellar transport, Kinesin-13
Cilia and flagella play pivotal roles in cell motility and cell signaling during development and homeostasis (1–3). Even though we know much about the functions of these ubiquitous organelles, we are only just beginning to understand the molecular mechanisms that regulate their assembly and disassembly. After cell division is completed, the primary cilium is formed by assembly of ciliary components onto the distal end of the basal body, a modified centriole. Ciliary precursors in the cell body are delivered to the tip of the growing organelle by intraflagellar transport (IFT), a bidirectional transport system composed of protein complexes (called IFT particles) and their cargo (flagellar components such as tubulin and dynein arms). Anterograde movement from the cell body to the flagellar tip is driven by kinesin-2, whereas retrograde movement from the tip to the cell body depends on cytoplasmic dynein (4–6).
Regulation of the formation and removal of cilia and flagella is highly complex. During entry into the cell cycle, cilia, and flagella (which are functionally and structurally similar organelles whose names are used interchangeably) are resorbed (7–9). Many cells also modify their cilia in response to environmental or developmental cues (10, 11). Chlamydomonas, a unicellular biflagellated green alga, shortens it flagella in response to changes in the ionic properties of its medium and during zygote development. Recently, an aurora-like protein kinase (CALK) was shown to be required for flagellar resorption and rapidly phosphorylated during shortening induced by changes in the medium and after zygote formation (12). The resorption of primary cilia in mammalian cells that occurs during reentry into the cell cycle also requires an aurora kinase (9).
Cells also are capable of rapidly regenerating cilia after shortening or detachment. Studies on primary cilia of fibroblasts have shown that within 1 hour after growth factor-triggered loss of cilia, the fibroblasts regenerate the organelles (13). Early, classic studies on Chlamydomonas uncovered a cellular pool of flagellar precursors used for formation of new flagella. Chlamydomonas cells experimentally induced to detach their flagella by pH shock initiate flagellar regeneration immediately and grow two new flagella within 1–2 hours. Even in the absence of new protein synthesis, the cells can form two half-length flagella solely from a cell body pool of flagellar precursors (14). The mechanisms that regulate flagellar regeneration, including the accompanying up-regulation of transcription of flagellar genes, are likely to be complex (15–17). It is not known, for example, whether the tubulin dimers in the precursor pool are soluble or in the form of polymerized cytoplasmic microtubules. Wilson and Lefebvre reported that glycogen synthase kinase 3 (GSK3) was regulated during flagellar detachment and regeneration, and suggested that GSK3 might influence cargo loading in the IFT system (18).
In studies of the cellular and molecular events that underlie flagellar formation and removal, we observed that CrKinesin-13, the Chlamydomonas member of the kinesin-13 family of microtubule depolymerizing kinesins, is dynamically regulated during flagellar regeneration and during flagellar shortening. Within seconds after cells are triggered to generate new flagella, CrKinesin-13 in the cell body is posttranslationally modified, and modification is reversed as flagella reach nearly full-length. Cells depleted by RNAi of CrKinesin-13 fail to assemble flagella immediately after deflagellation and exhibit a ≈2 hours lag in initiation of assembly. During flagellar disassembly, CrKinesin-13, which lacks the neck domain implicated in diffusional translocation, is transported by IFT from the cell body into the flagella. Depletion of CrKinesin-13 inhibits flagellar shortening. Thus, in Chlamydomonas, Kinesin-13 functions both during flagellar assembly and during flagellar disassembly.
Results
Posttranslational Modification of Kinesin-13 in Chlamydomonas.
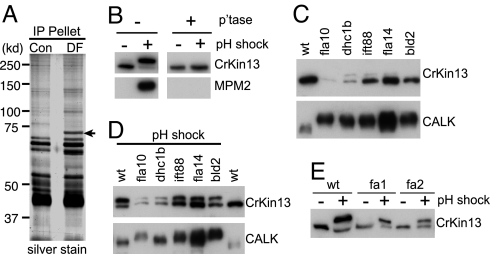
Based on the known roles of “mitotic kinases” in cilia and flagella (9, 12, 19, 20), we searched for additional mitosis-linked proteins that appeared after pH shock-induced flagellar detachment/regeneration using the MPM2 antibody, which recognizes a phospho-epitope (pS/T -P) present on a set of proteins, including microtubule-associated proteins MAP-1, MAP-2, and NIMA kinase, that regulate microtubule dynamics during the cell cycle (21). Immunoblotting and immunoprecipitation with the MPM2 antibody (Fig. 1A), MALDI-TOF mass spectrometry, and BLAST analysis identified a 70 kDa, MPM2-binding protein that appeared after pH shock as a 647 aa-containing kinesin-like protein with a highly conserved 343 aa kinesin motor domain in the middle of the protein (Fig. S1). Analysis by BLAST and alignment with other kinesin family members showed that the protein, which we have named CrKinesin-13, is a member of the microtubule depolymerizing kinesin-13 family. Kinesin-13 family members in other organisms are well established as key regulators of mitotic microtubules and enforce the proper length of the spindle and regulate chromosome segregation by catalyzing a 100-fold increase in the rate of depolymerization of microtubules (22–25).
Fig. 1.
Posttranslational modification of CrKinesin-13 (A) MPM2 antibody immunoprecipitates a 70-kDa protein from pH-shocked cells. Cell lysates from control (con) cells and pH-shocked cells (DF) were incubated with MPM2 antibody, and the immunoprecipitates were analyzed by immunoblotting (Fig. S1) and silver staining. (B) CrKinesin-13 is phosphorylated during pH shock, and the phosphorylated CrKinesin-13 is the antigen recognized by the MPM2 antibody. Cell lysates from control and deflagellated cells were treated with or without phosphatase followed by immunoblot analysis with anti-CrKinesin-13 and mAb-MPM2. (C) CrKinesin-13 properties in mutants defective in flagellar assembly. The indicated mutant and wild-type (21gr) cells were analyzed by immunoblotting using the CrKinesin-13 antibody. (D) CrKinesin-13 is modified during the pathways activated by pH shock. The flagellar mutants were subjected to pH shock along with wild-type cells, frozen within 1 minute, and subsequently analyzed by SDS/PAGE and immunobloting. (E) The fa1 and fa2 mutants, which are defective in flagellar detachment, undergo pH shock-induced modification of CrKinesin-13. Immunoblot analysis of 21gr, fa1, and fa2 mutant cells was carried out before and after pH shock.
CrKinesin-13 contains all of the conserved domains of the kinesin-13 family members except the neck domain, (SI Text and Fig. S2). Kinesin-13 family members recently reported to be present in the flagella of the unicellular pathogens Leishmania and Giardia also are missing the neck domain (26, 27). Because the neck domain is implicated in movement of kinesin-13s along microtubules in mitosis (23), cells that possess neckless kinesin-13 family members must use other mechanisms for transporting kinesin-13.
As shown in Fig. 1B (upper-left panel), immunoblotting with an anti-CrKinesin-13 antibody that we generated (Fig. S3) showed that the lower isoform of CrKinesin-13 was converted to the upper form when cells were exposed to a pH shock. Incubation of lysates with a protein phosphatase converted the upper isoform to the lower isoform (Fig. 1B, upper-right panel), indicating that the upper band was a phosphorylated form of the protein. Immunoblotting with the MPM2 antibody confirmed the anti-CrKinesin-13 results (Fig. 1B, lower panels).
CrKinesin-13 modification upon pH shock is unrelated to flagellar detachment per se. Previous work from several laboratories has established that exposure of Chlamydomonas cells to treatments (such as pH shock) that activate signals for deflagellation also activate pathways for flagellar regeneration (15, 17). To determine whether the pH shock-induced posttranslational modification of CrKinesin-13 was related to deflagellation per se or to the flagellar regeneration that occurs immediately after deflagellation, we studied CrKinesin-13 in several mutants defective in genes essential for flagellar assembly or flagellar detachment. The mutants were a fla10 null mutant (flagella-less; defective in anterograde IFT motor kinesin2) (28); dhc1b (flagella-less; defective in retrograde IFT motor cytoplasmic dynein 1b) (29); fla14 (short, stumpy flagella; defective in the retrograde motor cytoplasmic dynein light chain) (30, 31); ift88 (flagella-less; defective in IFT particle protein IFT88) (32); bld2 (flagella-less; defective in epsilon tubulin essential for basal body assembly) (33); and fa1 and fa2 (flagella of normal length; defective in the FA1 and FA2 proteins required for pH shock-induced flagellar detachment) (19).
Similar to wild-type cells, the phosphorylated form of CrKinesin-13 was essentially undetectable in all of the mutants under standard culture conditions (Fig. 1C, upper panel). (We should note that, unlike CrKinesin-13, the aurora-like protein kinase CALK was phosphorylated in all of the flagella-less mutants before pH shock (Fig. 1C, lower panel) (12), indicating that CALK phosphorylation and CrKinesin-13 phosphorylation were decoupled in these cells.) When we subjected the mutants to pH shock, CrKinesin-13 was phosphorylated in each (Figs. 1D and 1E). Thus, these results indicated that the pH shock-induced posttranslational modification of the kinesin was unrelated to flagellar detachment per se.
CrKinesin-13 and Flagellar Regeneration.
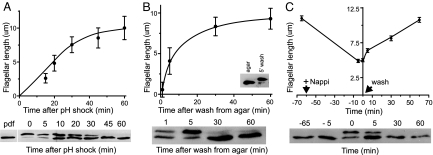
Analysis of CrKinesin-13 in cells regenerating their flagella implicated the protein in flagellar growth. We examined flagellar regeneration in cells induced to detach their flagella by pH shock (Fig. 2A); in agar-grown cells, which are aflagellate but can be activated to regenerate flagella by transfer to liquid medium (Fig. 2B); and in cells previously treated (see below) to bring their flagella to half length and then returned to standard culture conditions to allow re-growth of the organelles to full length (Fig. 2C). In cells regenerating flagella in all three conditions, only the lower isoform of CrKinesin-13 was present before regeneration was induced. Importantly, the upper form appeared immediately after regeneration was triggered. In addition, when the flagella began to approach full length, the CrKinesin-13 in all 3 sets of regenerating cells returned to the lower form. These experiments strongly linked cellular regulation of CrKinesin-13 to flagellar regeneration.
Fig. 2.
Modification of CrKinesin-13. (A) CrKinesin-13 is modified during flagellar regeneration. 21gr cells were triggered to grow new flagella by pH shock (error bars, SD); (B) by transfer from agar plates into liquid medium (error bars, SEM); and (C) by washing into fresh medium cells whose flagella had shortened approximately to half length during incubation in 20 mM NaPPi (error bars, SEM). A flagellum on at least 50 cells was measured for each time point. In (A), pdf is a sample taken just before deflagellation; the T = 0 sample was taken immediately after deflagellation. (B, inset) Cells scraped from agar directly into sample buffer, and a sample of the same cells 5 minutes after washing into liquid medium; all of the CrKinesin-13 was in the unphosphorylated form in the agar-grown cells.
CrKinesin-13 Is Dispensable for Flagellar Detachment but Functions During Flagellar Assembly.
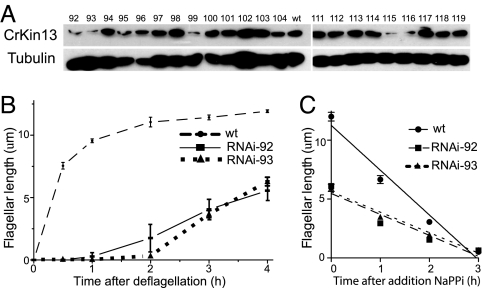
To examine the function of kinesin-13 in flagellar regeneration, we used RNAi methods to deplete CrKinesin-13, which is encoded by a single copy gene (34, 35) (see SI Text). Several transformants (92, 93, 99, 115, and 116) showed reduced levels of CrKinesin-13 protein by immunoblotting (Fig. 3A). Examination by phase contrast microscopy (not shown) indicated that the RNAi transformants also possessed shorter flagella, whereas clones with normal expression levels of CrKinesin-13 did not. Four of the transformants (92, 93, 99, and 115) that were examined in more detail possessed flagella that ranged in average length from 5.6 to 7.5 μm, whereas the average length of wild-type cells is ≈12 μm (see SI Text). Recovering cells with a short flagella phenotype is relatively uncommon in Chlamydomonas mutant screens (36). Cultures of 99 and 115 sometimes contained many undivided or non-flagellated cells. All four transformants behaved similarly in the experiments to be described below, but transformants 92 and 93 exhibited consistent growth properties and high proportions of flagellated cells (>90%) and were used for the experiments reported here.
Fig. 3.
Effect of CrKinesin-13 depletion on flagellar shortening. (A) Immunoblots of CrKinesin-13 RNAi transformants. (B) Flagellar regeneration in 21gr wild-type cells (predeflagellation length, 12.6 μm), RNAi transformant 92 (predeflagellation length, 6.5 μm), and RNAi transformant 93 (predeflagellation length, 8.0 μm). Curves for 21gr and RNAi-92 represent results from three independent experiments and for RNAi-93 from one experiment. Error bars indicate SEM. (C) Flagellar shortening induced by NaPPi in 21gr cells and RNAi transformants 92 and 93. The length of one flagellum on at least 50 cells was measured for each time point. Error bars show SEM.
When we subjected the RNAi transformants to pH shock, they detached their flagella similarly to wild-type cells, indicating that Crkinesin-13 was not essential in the pathway for signaling of flagellar detachment or for detachment per se. On the other hand, the CrKinesin-13-depleted cells behaved much differently than wild-type cells after deflagellation. As expected, the wild-type cells began assembling new flagella within minutes after pH shock-induced flagellar detachment, and by 1.5–2 hours their flagella had reached nearly their predeflagellation length (12.6 μm; Fig. 3B). In contrast, the CrKinesin-13-depleted RNAi transformants 92 and 93 failed to initiate flagellar regeneration immediately after pH shock-induced detachment (Fig. 3B). It was only the ability of the cells to respond rapidly to flagellar detachment that was blocked, however, and after a 1.5–2 hour lag, the RNAi cells initiated flagellar regeneration (albeit at a rate lower than wild-type cells), and their flagella reached nearly their predeflagellation length (6–8 μm) ≈4 hours after deflagellation. The short flagella phenotype of the RNAi cells along with the lag in initiation of flagellar regeneration after detachment indicated that CrKinesin-13 functioned during flagellar growth.
CrKinesin-13 Is a Cell Body Protein and Moves into Flagella During Flagellar Disassembly.
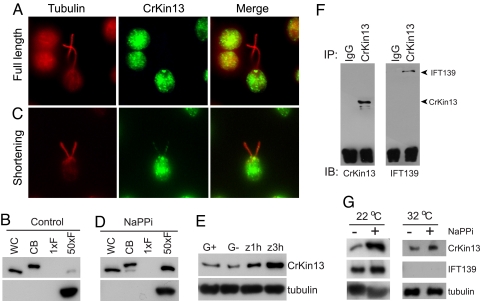
In other protists, kinesin-13 family members are present in flagella and function in length control (26, 27). We investigated the cellular localization of CrKinesin-13 by use of immunofluorescence microscopy and cell fractionation. Immunofluorescence microscopy showed that CrKinesin-13 was predominantly localized to the basal body region of the cells, and little if any was detected in the flagella (Fig. 4A). Confirming the IF results, CrKinesin-13 was present in the cell body fraction (in the phosphorylated form, as we used pH shock to deflagellate the cells), and none was detected in the flagella isolated from the same number of cells (Fig. 4B). When we loaded 50 cell equivalents of flagella (the organelles account for ≈2% of total cellular protein), which allowed us to analyze approximately equal amounts of protein in each lane, faint staining of CrKinesin-13 could be detected. Thus, these results indicated that under standard culture conditions the protein was almost exclusively localized in the cell body.
Fig. 4.
Transport of CrKinesin-13 into flagella is triggered when the flagellar shortening pathway is activated and transport requires IFT. (A) Immunofluorescence of control cells under standard culture conditions. Staining with anti-CrKinesin-13 was detected in the basal body region, and little if any was detected in the flagella. (B) Immunoblot of whole cells (WC), cell bodies (CB), and flagella (F) with anti-CrKinesin-13. CrKinesin-13 was predominantly localized to the cell body (note that the pH shock used for deflagellation caused CrKinesin-13 phosphorylation). (1xF: One cell equivalent of flagella; 50xF: 50 cell equivalents of flagella.) Staining with alpha tubulin antibody documented equal loading. (C) Immunofluorescence of cells undergoing flagellar disassembly. CrKinesin-13 staining was detected along the length of the flagella and at the tip. (D) CrKinesin-13 in the flagella increased within 5 minutes after cells were placed in NaPPi. (E) Immunoblots of flagella from gametes and the zygotes that formed at 1 hour and 3 hours after gametes were mixed. CrKinesin-13 in the flagella increased during zygotic flagellar disassembly. G+, mt+ gametes; G−, mt− gametes; Z, zygotes. Alpha tubulin was used as loading control. (F) Co-immunoprecipitation of CrKinesin-13 and IFT particles. The flagellar membrane/matrix fractions of flagella isolated 5 minutes after transfer of cells to NaPPi were used for immunoprecipitation with anti-CrKinesin-13 or preimmune IgG. The immunoprecipitates were analyzed with anti-CrKinesin-13 and anti-IFT139 antibodies. (G) Failure of CrKinesin-13 to be transported into flagella in the fla10–1 mutant at the non-permissive temperature after induction of flagellar disassembly by NaPPi. fla10–1 Cells were incubated at 22 °C or 32 °C for 1 hour followed by incubation in 20 mM NaPPi for 5 minutes at the same temperatures, and the flagella were isolated and analyzed by immunoblotting. Alpha tubulin was used as a loading control and an anti-IFT139 antibody was used to detect IFT particles.
In contrast, in the flagella of zygotes undergoing flagellar shortening (37), we detected faint CrKinesin-13 immunofluorescence (data not shown). In addition, when we examined cells induced to undergo flagellar shortening by incubation in NaPPi, an agent long known to activate flagellar shortening in Chlamydomonas (31), we detected CrKinesin-13 staining in the flagella in a punctate distribution along their length (Fig. 4C).
Immunoblot analysis of cell bodies and flagella isolated from cells undergoing shortening during both zygote maturation and NaPPi treatment confirmed the immunofluorescence data. The amount of CrKinesin-13 in the flagella undergoing NaPPi-induced shortening increased several-fold compared with flagella from control cells (Fig. 4D). Similarly, immunoblots of flagella isolated from zygotes as they underwent flagellar shortening around 2 hours after mt+ and mt− gametes were mixed together showed a several-fold increase in CrKinesin-13 (Fig. 4E).
IFT Is Essential for Transport of This “Neckless” Kinesin-13 into Flagella During Shortening.
The absence of the neck domain in CrKinesin-13 raised the possibility that IFT might be essential for transport of CrKinesin-13 into flagella. The punctate flagellar staining of CrKinesin-13, which is similar to the distribution of IFT particles in flagella (38), was consistent with this idea. To test directly for a relationship between CrKinesin-13 and IFT, we immunoprecipitated CrKinesin-13 from shortening flagella and immunoblotted the immunoprecipitate with an antibody against IFT particle protein IFT139. As shown in Fig. 4F, the IFT particle protein was present in the immunoprecipitates prepared using anti-CrKinesin-13 serum and was absent in those prepared using preimmune serum. These results indicated that CrKinesin-13 interacted with IFT particles.
We also took advantage of the conditional kinesin-2 mutant, fla10–1 to determine whether IFT was required for CrKinesin-13 transport (39). At the permissive temperature (22 °C), IFT is functional, but when transferred to the non-permissive temperature (32 °C), IFT gradually diminishes; and after ≈1 hours, IFT becomes undetectable microscopically and the amounts of IFT particle proteins are substantially reduced in flagella. The flagella also gradually begin to shorten, albeit at rates (1–1.5 μm/h) that are 3–4-fold lower than in wild-type cells undergoing NaPPi-induced shortening (4–5 μm/h) (12, 31, 40–42).
To test for a role of IFT in transport of CrKinesin-13, we preincubated fla10–1 cells at the permissive and non-permissive temperatures for 1 hours, added NaPPi for 5 minutes to stimulate flagellar disassembly, and then determined the amounts of CrKinesin-13 in isolated flagella by immunoblotting. As expected, activation of flagellar disassembly at 22 °C resulted in a several-fold increase of CrKinesin-13 in flagella compared with non-NaPPi treated cells (Fig. 4G). Also as expected, the IFT particles were reduced substantially in flagella isolated from the fla10–1 cells treated with NaPPi at the non-permissive temperature (Fig. 4G). Importantly, however, although the amount of CrKinesin-13 in the flagella increased slightly upon NaPPi treatment at the non-permissive temperature [which was consistent with a small NaPPi-induced increase in shortening rate at 32 °C (37)], the increase was substantially less than that at the permissive temperature (Fig. 4G). Thus, our results indicated that the transport of CrKinesin-13 into flagella during flagellar shortening depended on IFT.
CrKinesin-13 Functions During Flagellar Shortening.
Finally, we investigated the function of CrKinesin-13 in flagellar shortening using the RNAi cells described above. As shown in Fig. 3C, wt cells shortened their flagella rapidly (4.2 μm/h) when incubated in NaPPi. On the other hand, although the RNAi cells underwent flagellar shortening, the rate of shortening (2 μm/h) was less than 50% of that of wild-type cells and was closer to the basal rate of flagellar disassembly (41). Thus, in addition to functioning during rapid re-growth of flagella, CrKinesin-13 also functions during the disassembly of the axoneme that occurs during shortening.
Discussion
Our major findings are the following: (i) CrKinesin-13 was phosphorylated when flagellar growth was initiated and dephosphorylated as the flagella reached full length. (ii) The protein was transported by IFT into flagella during flagellar shortening. (iii) CrKinesin-13-depleted cells exhibited a nearly 2-h lag in initiation of flagellar regeneration. (iv) Flagellar shortening was inhibited in CrKinesin-13-depleted cells.
CrKinesin-13 Posttranslational Modifications Track Flagellar Length During Regeneration.
Our results that three independent methods for inducing flagellar growth in cells all led to CrKinesin-13 phosphorylation, which was followed by dephosphorylation as the flagella reached nearly full length, strongly linked the protein to flagellar assembly. CrKinesin-13 thus joins a short list of proteins whose posttranslational modifications parallel flagellar length during regeneration. Previous work showed that after pH shock-induced flagellar detachment, the ratio of tyrosine-phosphorylated glycogen synthase kinase 3 (GSK3) to non-phosphorylated GSK3 increased ≈1.4-fold within 5 minutes after deflagellation and then decreased to predeflagellation levels at 30 minutes (18). Given the implication of GSK3 in flagellar length control, it will be interesting to determine if the protein kinase is functionally linked to CrKinesin-13.
Moreover, the phosphorylation and activation of Chlamydomonas aurora-like kinase CALK during deflagellation and flagellar regeneration (Fig. 1) (12), suggest that CrKinesin-13 could be a substrate for CALK. The action of aurora protein kinases on mammalian kinesin-13 regulates its localization and microtubule depolymerization activity (43), which are essential for mitotic spindle assembly and chromosome segregation (23, 24). We found, however, that although CALK was phosphorylated in cells defective in flagellar assembly, CrKinesin-13 was not modified until the mutants were subjected to pH shock (Fig. 1). This evidence does not rule out a possible role for CALK in phosphorylating CrKinesin-13, but it does indicate that the relationship, if any, between CALK and CrKinesin-13 is complex.
CrKinesin-13 Is Translocated by IFT Into Flagella During Shortening.
Although our results showed that CrKinesin-13 is a cell body protein, when flagellar shortening was triggered by environmental or developmental cues, it was rapidly transported into flagella by IFT (Fig. 4). Given that many kinesin-13 family members require the neck domain for diffusional translocation along microtubules, it might have been expected that this “neckless” CrKinesin-13 would fail to translocate into flagella on its own. Even though it possesses a neck domain, Kif2a, another member of the kinesin-13 family, is transported to the spindle pole by a cytoplasmic dynein motor complex (44). Thus, both kinesin and dynein motors have now been shown to mediate the translocation of kinesin-13 family members.
CrKinesin-13 Functions During Flagellar Shortening and During Flagellar Growth.
Our studies of CrKinesin-13-depleted cells demonstrated that the protein functions during both flagellar assembly and flagellar disassembly (Fig. 3). Even though the CrKinesin-13-depleted cells failed to initiate flagellar growth for nearly 2 hours, they retained the ability to regenerate flagella and by 4 hours had grown flagella of predetachment length. These results suggested that the signaling pathway for regeneration was intact, and that mobilization of the pool of flagellar precursors required CrKinesin-13.
Moreover, the rate of shortening in the CrKinesin-13-depleted cells was reduced more than 2-fold compared with that in wild-type cells (Fig. 3C). The reduced rate of shortening in the knockdown cells was similar to the basal rate of shortening observed in fla10–1 cells at the restrictive temperature (37, 40). This result, coupled with the short-flagella rather than a long-flagella phenotype in the knockdown cells, suggests that the basal rate of microtubule depolymerization in Chlamydomonas flagella of steady-state length might be independent of CrKinesin-13.
Our results that CrKinesin-13 is posttranslationally modified in the cell body during regeneration and functions during regeneration raise the possibility that microtubule dynamics in the cell body might be just as important for flagellar growth and length control as microtubule dynamics in the flagellum.
Materials and Methods
Strains, Culture Conditions, and Special Chemicals.
Chlamydomonas reinhardtii strains and growth conditions, methods for inducing flagellar disassembly, and special chemicals were essentially as described previously (12).
Cell Culture, Cell Fractionation, and Flagellar Length Measurements.
For pH shock treatment, the cultures were treated with 0.2 M HAc to reduce the pH to 4.5 rapidly and after 30 seconds the cultures were brought to pH 7.2 with 0.2 M KOH. To induce flagellar shortening, cultures were incubated in medium containing 20 mM NaPPi. Flagella and cell bodies were isolated as previously described (37). Cell samples for flagellar length measurements were fixed in a final 1% glutaraldehyde solution and imaged by differential interference contrast with a 40× lens on a Zeiss Axio Observer Z1 microscope. Flagellar length was measured by using ImageJ software (National Institutes of Health) and the length was calibrated with a micrometer. Flagellar length results were graphed using GraphPad Prism version 5.0a for Mac OS S.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis and Immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS/PAGE) and immunoblotting were performed essentially as described previously (12). Antibodies used were mouse anti-alpha-tubulin (1:5000, Sigma), mouse MPM2 (1:2000, DakoCytomation), mouse anti-IFT139 (1:5000), and rabbit anti-CALK (1:2000). IFT antibodies were gifts from Dennis Deiner and Joel Rosenbaum (Yale University) and Douglas Cole (University of Idaho). Phosphatase treatment of the cell lysate was essentially as described previously (12).
Immunofluorescence Microscopy.
We followed a protocol provided by Susan Dutcher (33) for cell extraction and labeling with the antibodies. A Zeiss Axiovert 200M inverted Imaging Microscope equipped with a CCD camera (Zeiss AxioCam) and an AxioVision image analysis system were used to acquire images. The images were processed in AxioVision and/or Adobe Photoshop. Primary antibodies used were anti–CrKinesin-13 (1: 50), anti-acetylated alpha tubulin (1:200, Sigma), anti-IFT139 (1:50). Secondary antibodies used were Alexa fluor 488 goat anti-rabbit IgG, and Texas Red goat anti-mouse IgG (all 1:400; Molecular Probes).
Supplementary Material
Acknowledgments.
We thank Dennis Deiner (Yale University, New Haven, CT), Joel Rosenbaum (Yale University, New Haven, CT), and Douglas Cole (University of Idaho, Moscow, ID) for providing IFT antibodies, and George Witman (University of Massachusetts, Amherst, MA), Susan Dutcher (Washington University, St. Louis), and Masafumi Hirono (University of Tokyo, Tokyo) for providing Chlamydomonas strains. We are grateful to Fred Grinnell for discussions and for helpful comments on the manuscript and to the Kazusa DNA Research Institute for providing cDNA clones. We thank Chen Yue for assistance in the mass spectrometry analysis. This work was supported in part by Grant GM25661 from the National Institutes of Health (to W.J.S.), and by the National Natural Science Foundation of China (Grants 30671090, 30830057, and 30771084), National Basic Research Program of China [also called 973 program (Grant 2007CB914401) (to J. P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808671106/DCSupplemental.
References
- 1.Pan J, Wang Q, Snell WJ. Cilium-generated signaling and cilia-related disorders. Lab Invest. 2005;85:452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- 2.Afzelius BA. Cilia-related diseases. J Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fliegauf M, Benzing T, Omran H. When cilia go bad: Cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 5.Cole DG. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic. 2003;4:435–442. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- 6.Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 7.Tucker RW, Scher CD, Stiles CD. Centriole deciliation associated with the early response of 3T3 cells to growth factors but not to SV40. Cell. 1979;18:1065–1072. doi: 10.1016/0092-8674(79)90219-8. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- 9.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley MW, Ochiai CK, Corwin JT. Maturation of kinocilia in amphibian hair cells: Growth and shortening related to kinociliary bulb formation. Hear Res. 1992;59:108–115. [PubMed] [Google Scholar]
- 11.Bloodgood RA. Resorption of organelles containing microtubules. Cytobios. 1974;9:142–161. [PubMed] [Google Scholar]
- 12.Pan J, Wang Q, Snell WJ. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell. 2004;6:445–451. doi: 10.1016/s1534-5807(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 13.Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- 14.Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre PA, Rosenbaum JL. Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Annu Rev Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- 16.Tam LW, Wilson NF, Lefebvre PA. A CDK-related kinase regulates the length and assembly of flagella in Chlamydomonas. J Cell Biol. 2007;176:819–829. doi: 10.1083/jcb.200610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheshire JL, Evans JH, Keller LR. Ca2+ signaling in the Chlamydomonas flagellar regeneration system: Cellular and molecular responses. J Cell Sci. 1994;107:2491–2498. doi: 10.1242/jcs.107.9.2491. [DOI] [PubMed] [Google Scholar]
- 18.Wilson NF, Lefebvre PA. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot Cell. 2004;3:1307–1319. doi: 10.1128/EC.3.5.1307-1319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quarmby LM. Cellular deflagellation. Int Rev Cytol. 2004;233:47–91. doi: 10.1016/S0074-7696(04)33002-0. [DOI] [PubMed] [Google Scholar]
- 20.Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol Biol Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westendorf JM, Rao PN, Gerace L. Cloning of cDNAs for M-phase phosphoproteins recognized by the MPM2 monoclonal antibody and determination of the phosphorylated epitope. Proc Natl Acad Sci USA. 1994;91:714–718. doi: 10.1073/pnas.91.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walczak CE. The Kin I kinesins are microtubule end-stimulated ATPases. Mol Cell. 2003;11:286–288. doi: 10.1016/s1097-2765(03)00067-4. [DOI] [PubMed] [Google Scholar]
- 23.Moore A, Wordeman L. The mechanism, function and regulation of depolymerizing kinesins during mitosis. Trends Cell Biol. 2004;14:537–546. doi: 10.1016/j.tcb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Moores CA, Milligan RA. The role of the kinesin-13 neck in microtubule depolymerization. J Cell Sci. 2006;119:3905–3913. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- 25.Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pages M, Bastien P. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol. 2007;17:778–782. doi: 10.1016/j.cub.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 27.Dawson SC, Sagolla MS, Mancuso JJ, Woessner DJ, House SA, et al. Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell. 2007;6:2354–2364. doi: 10.1128/EC.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Kinesin-II is not essential for mitosis and cell growth in Chlamydomonas. Cell Motil Cytoskeleton. 2002;52:195–201. doi: 10.1002/cm.10051. [DOI] [PubMed] [Google Scholar]
- 29.Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefebvre PA, Nordstrom SA, Moulder JE, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. IV Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J Cell Biol. 1978;78:8–27. doi: 10.1083/jcb.78.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. Mol Biol Cell. 2002;13:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson DN, Simmons MP, Reddy AS. Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics. 2006;7:18. doi: 10.1186/1471-2164-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tam LW, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan J, Snell WJ. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev Cell. 2005;9:431–438. doi: 10.1016/j.devcel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walther Z, Vashishtha M, Hall JL. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall WF, Qin H, Rodrigo Brenni M, Rosenbaum JL. Flagellar length control system: Testing a simple model based on intraflagellar transport and turnover. Mol Biol Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker JD, Quarmby LM. Chlamydomonas fla mutants reveal a link between deflagellation and intraflagellar transport. BMC Cell Biol. 2003;4:11. doi: 10.1186/1471-2121-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 44.Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.