Abstract
Glucocorticoids are widely used to suppress inflammation and treat various immune-mediated diseases. Some glucocorticoid receptor (GR)-regulated genes mediate the therapeutic response, whereas others cause debilitating side effects. To discover selective modulators of the GR response, we developed a high-throughput, multiplexed system to monitor regulation of 4 promoters simultaneously. An initial screen of 1,040 natural products and Food and Drug Administration-approved drugs identified modulators that caused GR to regulate only a subset of its target promoters. Some compounds selectively inhibited GR-mediated gene activation without altering the repression of cytokine expression by GR. This approach will facilitate identification of genes and small molecules that augment beneficial effects of GR and diminish deleterious ones. Our results have important implications for the development of GR modulators and the identification of cross-talk pathways that control selective GR gene regulation.
Keywords: fluorescent protein, high-throughput screen, inhibitor, selective modulator, combinatorial therapy
Glucocorticoid receptor (GR) belongs to the nuclear receptor (NR) family of intracellular ligand-regulated transcription factors and is expressed ubiquitously in humans (1). Hormones such as cortisol activate GR, causing its nuclear translocation, interaction with coregulators, and binding to specific genomic sites to regulate transcription (2, 3). This mediates the broad systemic effects of GR signaling and underlies glucocorticoid treatment of diverse immune-mediated diseases such as asthma and rheumatoid arthritis. However, severe dose-limiting side effects occur, including osteoporosis, muscle wasting, and diabetes (4). No existing drugs induce only beneficial effects of GR.
The beneficial and harmful effects of glucocorticoids are due to selective activation or repression of particular genes by GR. This selectivity is based in part on tissue-specific factors and cross-talk pathways (5). For example, GR reduces the expression of certain inflammatory cytokines by inhibiting other transcription factors such as AP-1 and NF-κb (6). Conversely, GR increases expression of RANKL, a gene coregulated by the vitamin D receptor that activates bone resorption by osteoclasts (7). Controlling GR activity at certain tissues or promoters has profound therapeutic implications.
Efforts to achieve this goal have focused primarily on developing selective GR ligands that induce a subset of GR activities (8, 9). However, it is not yet possible to predict GR gene regulation based on ligand design, and it remains uncertain whether new ligands can produce therapeutically relevant transcriptional selectivity. Moreover, transcription-based screens for GR modulators generally measure GR activation at a single experimental promoter (10). This does not allow efficient identification of molecules that produce promoter-specific responses. Accordingly, although numerous GR agonists with different potencies and/or modes of delivery are in clinical use, dose equivalency generally results in similar clinical responses and side effects (11). Nonligand modulation of GR is an alternative strategy to achieve the desired transcriptional output, potentially enabling tissue or promoter-specific GR effects.
To address this problem, we developed a high-throughput system to measure GR activity simultaneously at 4 promoters. This permits discovery of genes or molecules that alter GR signaling in a promoter-specific fashion. We have applied this system to identify selective modulators of the GR response in an initial screen of 1,040 natural products and Food and Drug Administrtion (FDA)-approved compounds.
Results
Simultaneous Measurement of GR Response at 3 Promoters.
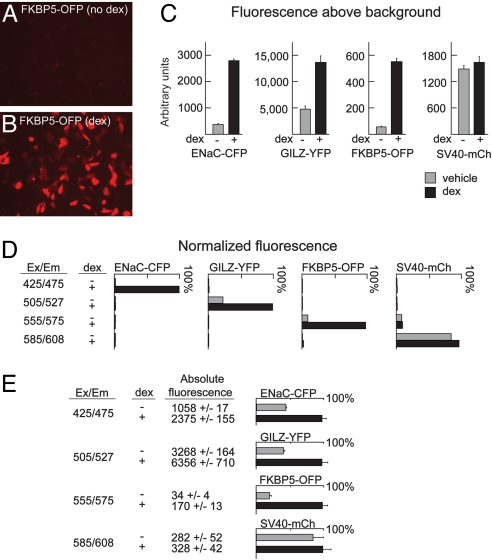
To measure GR regulation of multiple promoters simultaneously, we used fluorescent proteins (FPs) with different excitation/emission properties (12) in combination with regulatory elements for ENaC, GILZ, and FKBP5 (13, 14), 3 genes strongly induced by GR (15, 16) and believed to be clinically relevant GR targets (17–19). We linked GR-responsive regulatory regions for these genes to cDNAs encoding cerulean fluorescent protein (CFP), yellow fluorescent protein (YFP), and mOrange fluorescent protein (OFP). The FP-reporter constructs (pENaC-CFP, pGILZ-YFP, and pFKBP5-OFP) were transfected individually into A549 cells, a model of GR signaling. Fluorescence was induced in transfected cells by exposure to dexamethasone (dex), a synthetic GR ligand (Fig. 1 A and B).
Fig. 1.
Measurement of activity from 4 promoters simultaneously with fluorescent reporters. (A and B) Fluorescence microscopy of A549 cells transfected with pFKBP5-OFP and treated with dex (100 nm) as indicated. (C) The peak fluorescence of A549 cells transfected with the indicated reporter constructs and treated with dex as indicated was measured by using a monochrometer-based plate reader. (D) Cells were transfected with each FP reporter construct and treated with dex as indicated. Fluorescence above background was measured sequentially with each of the 4 optimized excitation/emission pairs. Values above background measured with the optimal setting for each reporter with dex treatment were normalized to 100%. Note that minimal fluorescence was detected for each reporter at nonoptimal settings. (E) Fluorescence from cells in 96-well dishes transfected with all 4 FP reporters was sequentially measured by using the 4 excitation/emission pairs. Mean and standard deviations of absolute fluorescence measured from 8 wells treated with and without dex are graphed as percent activation.
We and others have previously used monochrometer-based fluorescence plate readers to quantify the expression of FPs (20, 21). We applied this system to measure dex-induced fluorescence from each of the GR-responsive reporters. We readily detected increases in peak fluorescence in A549 cells transfected with the pENaC-CFP, pFKBP5-OFP, and pGILZ-YFP reporters (Fig. 1C). In contrast, a constitutive SV40-mCherry (pSV40-mCh) FP reporter did not respond to dex (Fig. 1C).
For simultaneous measurement of 4 reporters, we defined 4 excitation/emission parameters in which each FP variant was strongly detected at a single setting (Fig. 1D). Only pSV40-mCh fluoresced at >1 setting: Fluorescence generated by pSV40-mCh with the excitation/emission pairing optimized for OFP (555 excite, 575 read; Fig. 1C) was a small, constant percentage of the signal measured at the optimized mCh setting (585 excite, 608 read, Fig. 1D). This did not significantly alter the value of the OFP reporter. By transfecting all 4 reporters and sequentially measuring with the 4 optimized emission/excitation settings, we simultaneously measured the changes in transcription associated with GR activation from 4 promoters (Fig. 1E). Although cotransfecting the reporters lowered the absolute fluorescent output for each, the low standard deviations of the GR-inducible reporters relative to the level of activation (Fig. 1E) implied that each would clearly indicate GR activity in screens. There was no evidence of fluorescence resonance energy transfer between the different reporters.
Application of a Multipromoter Screen to Identify GR Modulators.
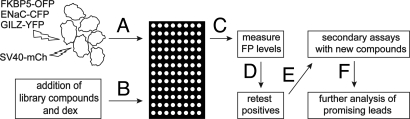
We used this system to screen a library of 1,040 natural products and FDA-approved drugs for modulators of GR signaling (22), as outlined in Fig. 2. The pENaC-CFP, pGILZ-YFP, and pFKBP5-OFP reporters indicated GR signaling, whereas pSV40-mCh, an internal control, excluded nonspecific transcription inhibitors and toxins. We screened the library in duplicate in saturating dex (100 nM). Z′, a function that reflects the ability to discriminate between control and experimental wells (23), was determined for each GR reporter. The Z′ values were: Z′(ENAC-CFP) = 0.68; Z′(GILZ-YFP) = 0.64; and Z′(FKBP5-OFP) = 0.66. These indicate that the screen should efficiently identify compounds that increase or decrease fluorescence from any of the 3 experimental promoters (23).
Fig. 2.
Design of multipromoter based screen for GR modulators. (A) Cells were transfected with the indicated fluorescent protein reporter plasmids and plated into 96-well dishes the next day. (B) Two hours later, dex (100 nM) and library compounds (2.5 μM) were added to the cells via liquid-handling robot. (C) The next day, fluorescence for each reporter was measured with a plate reader. (D) Hits were selected based on predefined criteria (see Materials and Methods) and retested in the screening assay. (E) Fresh compounds of repeat hits were obtained and retested in FP assays and dose–response studies. (F) Selected compounds were analyzed in further detail.
In the primary screen, 66 compounds significantly modulated one or more of the experimental reporters in duplicate. Primary hits included both general and promoter-specific inhibitors and potentiators of GR activity. Fourteen were known NR ligands, comprising 7 glucocorticoids, 2 antiandrogens, 3 estrogens, and 2 progesterones. Of the hits not known to be NR ligands, repeat FP assays with library compounds followed by standard luciferase assays using newly acquired compounds yielded 6 compounds that dose-dependently altered GR signaling (Table S1 and Table 1). Luciferase assays were used here and elsewhere because they are a standard method to quantify transcription and thus provide validation for the FP-based screening method. Further analyses of the validated compounds are described below.
Table 1.
Effects of lead compounds on FP reporters
| Compound | ENaC-CFP | GILZ-YFP | FKBP5-OFP |
|---|---|---|---|
| Acl | ↓ ↓ ↓* | ↓ ↓ ↓ | ↓ ↓ ↓ |
| Ciclo | ↓ † | ↓ | No effect |
| Forskolin | No effect | ↑‡ | No effect |
| Hpg | No effect | ↓ ↓ ↓ | ↓ ↓ ↓ |
| Mitoxantrone | ↓ ↓ ↓ | ↓ ↓ ↓ | ↓ ↓ ↓ |
| Para | ↓ †§ | ↓ | No effect§ |
| Rosolic acid | ↓ | No effect | ↓ |
*More than 2 SD less than the mean.
†Indicates 0.8 to 2 SD less than the mean.
‡Indicates 0.8 to 2 SD greater than the mean.
§Potential interference from compound fluorescence.
Anthracyclines Prevent GR-Mediated Gene Activation.
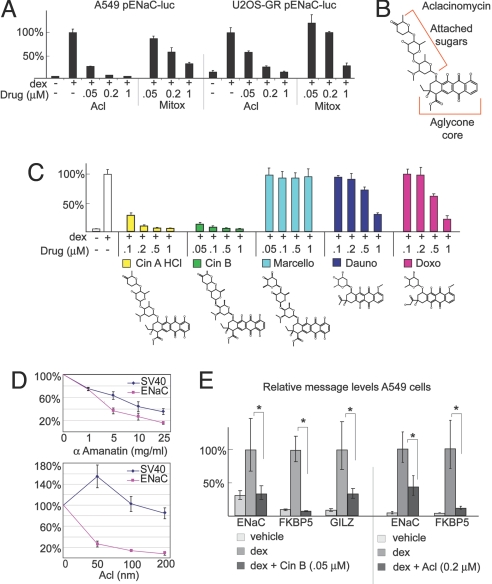
Aclacinomycin (Acl), an anthracycline antineoplastic, and mitoxantrone, a related anthracenedione, were nonselective inhibitors of GR (Table 1); these activities were validated in A549 cells with luciferase assays (Fig. 3A). Both also inhibited GR in U2OS-GR cells (Fig. 3A), a bone sarcoma cell line model of GR signaling (15).
Fig. 3.
Anthracyclines inhibit GR-mediated transcriptional activation. (A) Luciferase assays were performed with lysates from A549 and U2OS-GR cells transfected with pENaC-luc and pSV40-rl after treatment with different concentrations of Acl and Mitox for 18 h. Mean and standard deviation of 4 or more replicates are shown. The absolute level of the control reporter was only modestly altered by treatment with Acl or Mitox (S3). (B) Structure of Acl. (C) Luciferase assays were performed with lysates from A549 cells transfected with pENaC-luc and pSV40-rl after treatment with increasing concentrations of drugs as shown for 18 h. Cin, cinerubin; Marcello, marcellomycin; Doxo, doxorubicin; Dauno, daunorbicin. (D) Luciferase assays were performed on A549 cells transfected with pENaC-luc and pSV40-rl and treated with dex and increasing concentrations of either α-amanatin or Acl. Absolute activities of the dex-responsive pENaC-luc reporter and the dex-unresponsive pSV40-RL reporter with dex treatment alone were each normalized to 100%. Increases in pSV40-luc activity with low doses of Acl were observed consistently. (E) A549 cells were treated for 4 h (Cin B) and 24 h (Acl), as indicated. Relative mRNA levels of the indicated genes were determined in triplicate or quadruplicate by using qPCR. The mean relative mRNA level for each GR target gene with dex treatment was normalized to equal 100%. Results are shown from a representative experiment; qualitatively similar data were obtained in replicate experiments. *, P ≤ 0.05.
To investigate structure activity relationships, we tested the effects of additional anthracyclines on GR signaling. Anthracyclines contain a tetracyclic aglycone core attached to at least 1 sugar residue (e.g., Acl; Fig. 3B) and intercalate GC sequences within DNA. Cinerubin (Cin) B and Cin A HCl (NSC 267634), which both contain a trisaccharide chain attached to the aglycone moiety and are very similar to Acl (Fig. 3C, compare with Fig. 3B), potently inhibited GR activity (Fig. 3C) in luciferase assays. In contrast, marcellomycin, an anthracycline with a slightly divergent trisaccharide chain, had no effect on GR activity (Fig. 3C). Doxorubicin and daunorubicin, which are monosaccharide anthracyclines, inhibited GR, although they were >10-fold less potent than Acl and Cin B. Taken together, these results show that the sugar moieties attached to the aglycone core mediate the effects on GR transcriptional regulation.
To rule out nonspecific effects on RNA polymerase II (Pol II), we compared the effects of α-amanitin, a direct inhibitor of Pol II, to Acl (Fig. 3D). Both drugs were tested on a GR-dependent and a GR-independent reporter (Fig. 3D). As expected, α-amanitin similarly reduced the activity of both reporters. In contrast, treatment with Acl dramatically reduced GR activation of pENaC-luc compared with the constitutive reporter, pSV40-rl. Thus, Acl does not inhibit GR solely through nonspecific blockade of Pol II.
To test whether the plasmid-based results predict regulation of native promoters, we used quantitative PCR (qPCR) to measure the effects of 2 of the most potent anthracylines, Cin B and Acl, on endogenous GR target genes. Expression of ribosomal protein L19 (RPL19), which does not respond to GR, was the reference standard (Fig. S1). Both drugs inhibited GR at the endogenous promoters (Fig. 3E). These anthracyclines thus are potent inhibitors of GR-mediated activation of genes within native chromatin.
Selective GR Modulators.
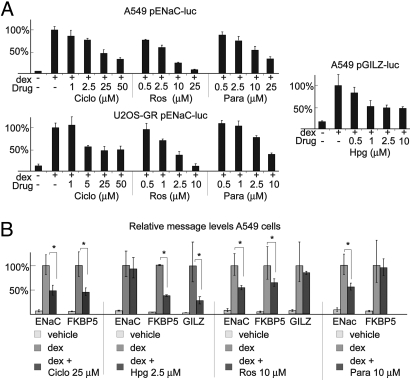
Ciclopirox olamine (Ciclo), rosolic acid, and pararosaniline (Para), not previously known to modulate GR signaling, were identified in the primary screen as selective inhibitors of the GR response, as was hydroxyprogesterone caproate (Hpg), a progesterone-like steroid hormone (Table 1). These compounds also exhibited dose-dependent effects on GR activity in luciferase assays (Fig. 4A and Fig. S2), suggesting drug-like activities for each. We next tested whether the effects on the FP reporters predicted regulation of endogenous genes. qPCR showed that all 4 compounds affected the expression of endogenous GR target genes (Fig. 4B). Ciclo was a general inhibitor of the GR response, whereas, similar to the primary screen, Hpg, rosolic acid, and Para selectively modulated endogenous GR target genes (Fig. 4B). Specifically, Hpg had no impact on induction of ENaC but reduced induction of GILZ and FKBP5, whereas rosolic acid inhibited GR induction of ENaC and FKBP5 but failed to inhibit GILZ (Fig. 4B). Para also selectively inhibited the induction of endogenous ENaC transcription (Fig. 4B). Taken together, these data show that selective modulation of GR signaling by compounds in the multiplexed plasmid screen can predict selective effects on endogenous gene transcription.
Fig. 4.
Hydroxyprogesterone caproate, rosolic acid, and Para are selective GR modulators. (A) Luciferase assays were performed on A549 and U2OS-GR cells transfected with pENaC-luc and pSV40-rl and treated with dex and various drugs as indicated. The relative activity of pENaC-luc with dex treatment was normalized to 100%. Ros, rosolic acid. (B) Relative mRNA levels of the indicated genes were determined by using qPCR. Cells were treated for 24 h as indicated. *, P ≤ 0.05.
Nonligand Modulation of GR Signaling.
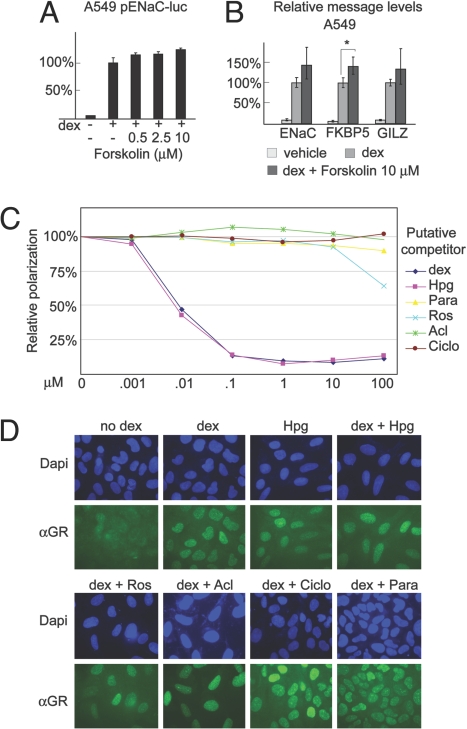
Forskolin, previously known to be a nonligand potentiator of GR signaling (24), increased GR activity in our primary screen (Table 1) and luciferase assays (Fig. 5A). We used qPCR to establish that forskolin also significantly increased the dex-induced expression of the endogenous FKBP5 locus (Fig. 5B). Thus, our screening system identified nonligand compounds that augment GR activity at endogenous genes.
Fig. 5.
Nonligand modulation of GR. (A) Luciferase assays were performed by using A549 cells that were transfected with pENaC-luc and pSV40-rl and treated with dex and forskolin, as indicated. The relative activity of pENaC-luc with dex treatment was normalized to 100%. (B) Relative mRNA levels of the indicated target genes were determined by using qPCR. Cells were treated as indicated for 24 h. *, P ≤ 0.05. (C) Fluorescence polarization assays were performed by using the indicated compounds as potential competitors to GR binding of a fluorescent control ligand. The value in the absence of competitor drug was normalized to 100%. Results are an average of 2 samples; the standard deviation between paired samples was <7% for all data points. (D) U2OS-GR cells were treated with Hpg (2.5 μM), Ros (10 μM), Acl (200 nM), Ciclo (25 μM), or Para (10 μM) for 24 h and then treated with dex or ethanol for 1 h before fixation. GR distribution was visualized by using indirect immunofluorescence. Drug doses were: Hpg (2.5 μM), Ros (10 μM), Acl (200 nM), Ciclo (25 μM), and Para (10 μM).
To verify that the screen also identified nonligand inhibitors of the GR response, we tested whether several inhibitory compounds interacted with the ligand-binding domain (LBD) of GR in a fluorescence polarization assay. As expected, the steroid hormone Hpg competed for hormone binding to the GR LBD; however, none of the other compounds interacted with the GR LBD in this assay. To corroborate these experiments, we examined whether the compounds inhibited nuclear localization of GR in U2OS-GR cells. None of the compounds prevented dex-mediated nuclear localization of GR (Fig. 5C), and Hpg induced nuclear translocation of GR in the absence of dex.
We also tested whether the compounds altered the protein levels of GR in A549 cells by using Western blot analysis. Ciclo and Para reduced GR protein levels, but Cin B, rosolic acid, and Hpg did not induce substantial alteration in GR protein levels after 24 h (Fig. S3). Together, these data indicate that Cin B and rosolic acid act neither as GR ligands, nor through altering GR protein expression; however, reduction in GR protein levels may contribute to the effects of Para and Ciclo on GR-mediated gene activation.
Activity of Selected GR Modulators in Cell-Based Assays of GR Function.
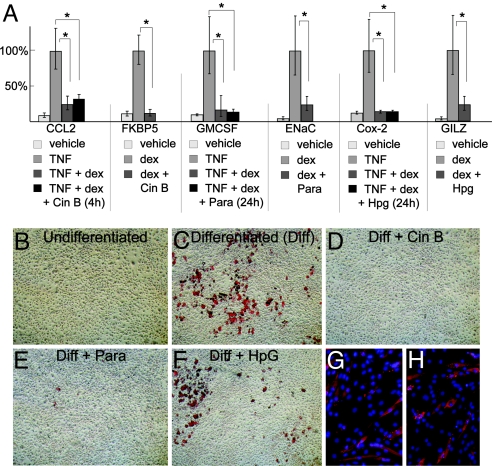
This screen was initially designed to discover GR modulators based on changes in activity of 3 promoters that are activated by GR but it left uncertain whether leads would alter GR physiology. GR is a well-known suppressor of TNF-α activity, an important aspect of the antiinflammatory effects of glucocorticoids (6). Thus, we tested whether Cin B, Hpg, and Para would alter the repression of TNF-α target genes by GR. Cin B did not alter GR repression of CCL2 expression with TNF-α treatment. Para and Hpg also had little or no effect on the GR-mediated repression of several TNF-α-regulated genes (Fig. 5 and Fig. S4). These data indicate that Cin B, Hpg, and Para selectively block activation of GR target genes, with little effect on the repression of TNF-α activity.
GR signaling regulates cell fate and energy homeostasis. We therefore tested whether several of the compounds altered GR-mediated adipogenesis of 3T3 L1 cells. Cin B and Para both inhibited GR-induced fat differentiation, whereas Hpg had no effect (Fig. 6 B–F). Inhibition of differentiation was not associated with gross alterations in cell viability or morphology (compare Fig. 6 A with D and E). Moreover, Cin B had almost no effect on myogenic differentiation of C2C12 myoblasts (Fig. 6 G and H), a GR-independent process. Thus, we have identified chemical modulators of the GR response with selective effects on GR-regulated promoters and GR physiology.
Fig. 6.
Impact of selected GR modulators on GR-mediated cytokine repression and adipogenesis. (A) Relative mRNA levels of the indicated target genes were determined by using qPCR. Cells were treated for 4 h (Cin B) or 24 h (Para and Hp). Doses used were TNF-α (2.5 ng/mL); Cin B (50 nM), Para (10 μM), and Hpg (2.5 μM). *, P ≤ 0.05. (B–F) Oil red staining of 3T3-L1 adipocytes. Cells were grown to confluence and then treated with 250 nM dex, 0.5 μM 3- isobutyl-1-methylzanthine, and .83 μM insulin for 48 h, followed by 5–7 days of additional culture as described (38). Various compounds as indicated were added to the culture media concomitant with the 48 h of dex treatment. (G and H) C2C12 cells were placed in differentiation medium (G) and differentiation medium plus 50 nm Cin B (H) for 72 h. Myosin heavy chain expression was visualized with indirect immunofluorescence.
Discussion
We have developed an assay in which simultaneous measurement of 4 promoter-FP reporters quantifies selective promoter activity. In a high-throughput screen, we identified compounds that modulate GR signaling at endogenous target genes, but neither interact with the LBD of GR, nor affect nuclear localization of the receptor. Importantly, we discovered several nonligand compounds that “tuned” GR responses, i.e., those that caused GR to regulate only a subset of its normal spectrum of target genes and cellular responses. This work thus has important implications for drug discovery and genetic studies to elucidate factors and signaling pathways that selectively regulate GR function.
Power of a MultiPromoter Readout.
Transcriptional regulation is usually measured in high throughput with a single experimental promoter driving an enzyme-based reporter or FP (25–27). Laser-based systems and high-content microscopy can be used to measure multiple cellular processes but have not been widely applied for multiplexed measurement of transcription (28). The ease of measurement, cost, and low variability of FP expression offer advantages over screening systems that directly measure mRNA levels (29). Our system can also use mixtures of cell types harboring distinct FP reporters, thereby distinguishing cell selective modulators. Thus, our system is an important, low-cost alternative to current systems that use a single reporter or directly measure mRNA levels.
Most importantly, the multiplexed system identified compounds that differentially modulated GR activity at a subset of 3 promoters. Although the screen was not designed to identify them, several of the compounds inhibited GR activation but not the repression of inflammatory cytokines. By choosing promoters to include both beneficial and harmful targets, it should be possible to identify more efficiently small molecules that tune receptor output to a desired pattern through effects on relevant cross-talk pathways.
In the screen, there were 66 primary hits, of which 14 were steroid-like NR ligands. Six additional hits were validated as nonsteroidal GR modulators. Thus, the overall screening efficiency was 30%. However, the percentage of nonsteroidal hits that validated as dose-dependent GR regulators was only 11% (6 of 52). The false positive rate was in part due to edge effects, which have been described (30). In the future, specific library and plate configurations can be used to reduce variability.
Inhibitors of GR.
Anthracyclines were identified as inhibitors of the GR response. The most potent of these, Acl and Cin B, both contain a trisaccharide chain attached to the anthracycline aglycone core that mediates DNA intercalation and is critical for the cytotoxic properties of this drug class (31). Although anthracyclines inhibit the activity of RNA polymerases (32), the effects we observed cannot be ascribed to nonspecific inhibition of transcription (Fig. 3D). The impact of the trisaccharide chain on GR inhibition also indicates that Acl and Cin B do not block the GR response simply through competing for DNA binding via the aglycone core, a mechanism previously reported for inhibition of SP-1 by other anthracyclines (33, 34). Rather, intercalation of the anthracycline aglycone core with DNA may allow specific interactions between the attached sugars and DNA-associated proteins such as transcriptional cofactors and chromatin remodeling complexes that regulate subsets of the Pol II transcriptome.
Selective Modulators of the GR Response.
Progestins can bind GR and mediate transcriptional effects (35). We identified hydroxyprogesterone caproate as a selective regulator of GR in the primary screen, and at endogenous genes. We also identified 7 known glucocorticoids as selectively regulating GR in the primary screen (Table S2). Thus, the multipromoter assay can be harnessed to identify selective ligands that specify a desired transcriptional output.
Rosolic acid and Para, which share a common structure of a central carbon bound to 3 phenyl groups, also selectively modulated the GR response in the primary screen and at endogenous genes. Para was previously reported to inhibit androgen receptor (AR) signaling (29), and 2 additional compounds with structural similarity to Para and rosolic acid also inhibited GR in luciferase assays (Fig. S5). It will be of interest to test whether the compounds identified here selectively regulate other nuclear receptors and to determine their mechanisms of action. It does not appear that their predominant mode of action is via interactions with the LBD of GR or through preventing the nuclear localization of GR, suggesting that GR cross-talk pathways are mediating the effects.
Conclusion
Selective modulation of GR and other nuclear receptors is a major therapeutic goal. Most efforts to achieve selective modulation have been directed at the LBD of each receptor, which represents only a single facet of the nuclear receptor's activity and does not directly address receptor interactions with cross-talk pathways (36, 37). In contrast, the multiplexed reporter screen we have described here is a powerful approach for identifying ligands that specify a desired transcriptional output or for identifying and targeting therapeutically relevant receptor cross-talk pathways. Subsequent analysis using qPCR or microarrays can rapidly determine whether hits have bona fide impact on receptor signaling. For putative GR modulators, there are also numerous cell-based assays, such as repression of cytokine expression and modulation of cell phenotype, to test physiologic function. Our application of such assays here suggests that GR signaling can be altered in a potentially beneficial fashion through combinations of traditional ligands and nonligand modulators. We anticipate that broad interrogation of NR cross-talk pathways in compound screens will lead to improved therapies in which physiologic responses are tuned via combinations of classical receptor ligands and heterologous modulators.
Materials and Methods
Plasmids.
mCherry and mOrange, cerulean, pENaC-luc, pFKBP5-luc and pGILZ-luc were gifts from R. Tsien (University of California, San Diego), D. Piston (Vanderbilt University, Nashville), C. Thomas (University of Iowa, Iowa City), T. Scammell (University of South Alabama, Mobile), and K. Yamamoto (University of California, San Francisco), respectively. pSV40-RL is from Promega. Luciferase coding regions in pENaC-luc, pFKBP5-luc, pGILZ-luc, and pSV40-RL were replaced with various FPs by using standard techniques. When necessary, site-directed mutagenesis used the QuikChange kit and protocol (Promega).
Cell Culture.
A549 (ATCC), U2OS-GR (gift of Keith Yamamoto's laboratory), and C2C12 (ATCC) cells were grown in high-glucose DMEM supplemented with glutamine, penicillin, streptomycin, and 5% or 10% FBS (HyClone). Dex treatment was at 100 nm unless otherwise indicated. Culture and differentiation of 3T3-L1 cells (ATCC) was as described (38). C2C12 cells were cultured in DMEM supplemented with 2% heat-inactivated horse serum (GIBCO) to induce differentiation.
Chemicals.
α-Amanitin, 3-isobutyl-1-methylzanthine, Hpg, mitoxantrone, dex, DMSO, forskolin, rosolic acid, Para, and Ciclo were from Sigma. TNF-α was from Roche. Acl, doxorubicin, daunorubicin, marcellomycin, Cin B, and Cin A HCL (NSC 267694) were from the National Cancer Institute/DTP Open Chemical Repository (http://dtp.nci.nih.gov).
Transfections.
Approximately 3 × 105 cells were plated in 6-well dishes. The next day, 500 μL of Lipofectamine 2000 (Invitrogen)/plasmid DNA (4 μg total) complex was added to 1.5 mL of DMEM with pen/strep and 5% FBS (A549 cells) or to 1.5 mL of Optimem (U2OS cells). After 3–4 h of incubation, media were replaced with standard growth media.
Luciferase Assays.
Luciferase assays used the Promega Dual-Luciferase Reporter system and protocol. Briefly, cells were transfected with firefly luciferase plasmids and pSV40-RL at a ratio of 10:1. The next day, cells were split into 96-well dishes and treated with dex and various drugs. After 12–18 h, cells were harvested in 50 μL of PLB. Luminescence was detected from 10 μL of lysate by using an Ultra Evolution plate reader (Tecan).
GR Binding.
Compound binding to the GR LBD was assessed by using fluorescence polarization competition (P2816; Invitrogen). Polarization with no competitor was normalized to 100%.
Compound Screen.
A549 cells were transfected with an equal mixture of pENaC-CFP, pGILZ-YFP, and pFKBP5-OFP (1.3 μg of each) or with pSV40-ChFP (4 μg). The next day, cells were mixed (≈1.8 × 104 of the triple-transfected cells; ≈2 × 103 cells transfected with pSV40-ChFP alone) and replated into 96-well plates (3603; Costar). Two hours later, library compounds (2.5 μM) and dex (100 nM) were added by using a Biomek robot. The next day, cells were fixed for 4 min in 4% paraformaldehyde. Fluorescence was measured for each reporter by using a Safire plate reader (Tecan). Hits were scored based on the mean and SD of the experimental wells on each plate. Criteria for a hit were if 1 reporter was >1.5 times the SD from the mean, or if 2 reporters were >0.8 times the SD from the mean. Wells were generally excluded if SV40-ChFP was >2 times the SD from the mean in a single plate, or 0.8 times the SD in duplicate plates. Compounds with intrinsic fluorescence were not scored for reporters with overlapping fluorescence.
Quantitative PCR.
Total RNA isolation used TRIzol and Pure Link (Invitrogen). Random-primed cDNA was prepared from 1 μg of total RNA by using MMLV-RT (Promega). Resultant cDNA (50 ng) was used per 50-μL reaction containing 1.25 units of TaqDNA polymerase (Invitrogen), 1.5 mM MgCl2, 300 nM concentrations of each primer (sequences available on request), 0.15 mM dNTP mix, and 0.2× SYBR green dye (Molecular Probes) in 1× PCR buffer (Qiagen). Real-time PCR used an Applied Biosystems 7700 machine and was analyzed by using the δδCt method (Applied Biosystems Prism 7700 Users Bulletin No. 2). RPL19 expression was used for data normalization. Relative message levels between samples were compared by using nonparametric rank-sum tests. Data from representative experiments are shown; qualitatively similar data were obtained in replicate experiments.
Immunohistochemistry.
Cells were fixed in ice-cold methanol (αGR) or 4% PFA [myosin heavy chain (MyHC)] and washed three times in PBS, incubated with a polyclonal GR antibody (1 μg/mL; 3579; Abcam) or MF20 (MyHC), washed in PBS, and incubated with secondary antibody (Invitrogen). Oil-red staining was performed as described (38).
Supplementary Material
Acknowledgments.
We thank Jeremy Jones, Sebastian Meijsing, Stefan Taubert, Abigail Kroch, and other members of M.I.D.'s and Keith R. Yamamoto's laboratories for stimulating discussions and critical comments on the manuscript and David Pearce, David Erle, and Monica and Nicholas Gerber for their critiques. A.N.G. was supported by National Institutes of Health Grant K08 HL077159, a SABRe award (Sandler Foundation), and an American Lung Association of California research grant. M.I.D. was supported by National Institutes of Health Grants R01 NS50284-01A1 (National Institute of Neurological Disorders and Stroke) and R01CA131226-01 (National Cancer Institute).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812308106/DCSupplemental.
References
- 1.McMaster A, Ray DW. Modelling the glucocorticoid receptor and producing therapeutic agents with anti-inflammatory effects but reduced side-effects. Exp Physiol. 2007;92:299–309. doi: 10.1113/expphysiol.2006.036194. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto KR, Darimont BD, Wagner RL, Iniguez-Lluhi JA. Building transcriptional regulatory complexes: Signals and surfaces. Cold Spring Harb Symp Quant Biol. 1998;63:587–598. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- 3.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schacke H, Docke W-D, Asadullah K. Mechanisms involved in the side effects of glucocorticoids. Pharmacol Ther. 2002;96:23–43. doi: 10.1016/s0163-7258(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 5.Kassel O, Herrlich P. Crosstalk between the glucocorticoid receptor and other transcription factors: Molecular aspects. Mol Cell Endocrinol. 2007;275:13–29. doi: 10.1016/j.mce.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Smoak KA, Cidlowski JA. Mechanisms of glucocorticoid receptor signaling during inflammation. Mech Ageing Dev. 2004;125:697–706. doi: 10.1016/j.mad.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-{kappa}B ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26:6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosen J, Miner JN. The search for safer glucocorticoid receptor ligands. Endocr Rev. 2005;26:452–464. doi: 10.1210/er.2005-0002. [DOI] [PubMed] [Google Scholar]
- 9.Cole TJ, Mollard R. Selective glucocorticoid receptor ligands. Med Chem. 2007;3:494–506. doi: 10.2174/157340607781745474. [DOI] [PubMed] [Google Scholar]
- 10.Fan F, Wood KV. Bioluminescent assays for high-throughput screening. ASSAY Drug Dev Technol. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 11.Adams N, Lasserson TJ, Cates CJ, Jones PW. Fluticasone versus beclomethasone or budesonide for chronic asthma in adults and children. Cochrane Database Syst Rev. 2007:CD002310. doi: 10.1002/14651858.CD002310.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 13.Mick VE, et al. The {{alpha}}-subunit of the epithelial sodium channel is an aldosterone-induced transcript in mammalian collecting ducts, and this transcriptional response is mediated via distinct cis-elements in the 5′-flanking region of the gene. Mol Endocrinol. 2001;15:575–588. doi: 10.1210/mend.15.4.0620. [DOI] [PubMed] [Google Scholar]
- 14.Hubler TR, Scammell JG. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones. 2004;9:243–252. doi: 10.1379/CSC-32R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogatsky I, et al. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA. 2003;100:13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JC, et al. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA. 2004;101:15603–15608. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodruff PG, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayroldi E, et al. GILZ mediates the antiproliferative activity of glucocorticoids by negative regulation of Ras signaling. J Clin Invest. 2007;117:1605–1615. doi: 10.1172/JCI30724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HC, et al. Oxidative stress disrupts glucocorticoid hormone-dependent transcription of the amiloride-sensitive epithelial sodium channel alpha-subunit in lung epithelial cells through ERK-dependent and thioredoxin-sensitive pathways. J Biol Chem. 2000;275:8600–8609. doi: 10.1074/jbc.275.12.8600. [DOI] [PubMed] [Google Scholar]
- 20.Richards B, et al. Stable expression of Anthozoa fluorescent proteins in mammalian cells. Cytometry. 2002;48:106–112. doi: 10.1002/cyto.10117. [DOI] [PubMed] [Google Scholar]
- 21.Pollitt SK, et al. A rapid cellular FRET assay of polyglutamine aggregation identifies a novel inhibitor. Neuron. 2003;40:685–694. doi: 10.1016/s0896-6273(03)00697-4. [DOI] [PubMed] [Google Scholar]
- 22.Abbott A. Neurologists strike gold in drug screen effort. Nature. 2002;417:109. doi: 10.1038/417109a. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Liu JL, Papachristou DN, Patel YC. Glucocorticoids activate somatostatin gene transcription through co-operative interaction with the cyclic AMP signalling pathway. Biochem J. 1994;301(Pt 3):863–869. doi: 10.1042/bj3010863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Necela BM, Cidlowski JA. Development of a flow cytometric assay to study glucocorticoid receptor-mediated gene activation in living cells. Steroids. 2003;68:341–350. doi: 10.1016/s0039-128x(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T, et al. Screening of novel nuclear receptor agonists by a convenient reporter gene assay system using green fluorescent protein derivatives. Phytomedicine. 2006;13:401–411. doi: 10.1016/j.phymed.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Sonneveld E, Jansen HJ, Riteco JAC, Brouwer A, van der Burg B. Development of androgen- and estrogen-responsive bioassays, members of a panel of human cell line-based highly selective steroid-responsive bioassays. Toxicol Sci. 2005;83:136–148. doi: 10.1093/toxsci/kfi005. [DOI] [PubMed] [Google Scholar]
- 28.Auld DS, et al. Fluorescent protein-based cellular assays analyzed by laser-scanning microplate cytometry in 1,536-well plate format. Methods Enzymol. 2006;414:566–589. doi: 10.1016/S0076-6879(06)14029-X. [DOI] [PubMed] [Google Scholar]
- 29.Hieronymus H, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J Biomol Screen. 2003;8:566–570. doi: 10.1177/1087057103256465. [DOI] [PubMed] [Google Scholar]
- 31.Binaschi M, et al. Anthracyclines: Selected new developments. Curr Med Chem Anticancer Agents. 2001;1:113–130. doi: 10.2174/1568011013354723. [DOI] [PubMed] [Google Scholar]
- 32.Long BH, Willis CE, Prestayko AW, Crooke ST. Effect of anthracycline analogues on the appearance of newly synthesized total RNA and messenger RNA in the cytoplasm of erythroleukemia cells. Mol Pharmacol. 1982;22:152–157. [PubMed] [Google Scholar]
- 33.Punchihewa C, De Alba A, Sidell N, Yang D. XR5944: A potent inhibitor of estrogen receptors. Mol Cancer Ther. 2007;6:213–219. doi: 10.1158/1535-7163.MCT-06-0392. [DOI] [PubMed] [Google Scholar]
- 34.Martin B, Vaquero A, Priebe W, Portugal J. Bisanthracycline WP631 inhibits basal and Sp1-activated transcription initiation in vitro. Nucleic Acids Res. 1999;27:3402–3409. doi: 10.1093/nar/27.17.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas CP, Liu KZ, Vats HS. Medroxyprogesterone acetate binds the glucocorticoid receptor to stimulate {alpha}-ENaC and sgk1 expression in renal collecting duct epithelia. Am J Physiol. 2006;290:F306–F312. doi: 10.1152/ajprenal.00062.2005. [DOI] [PubMed] [Google Scholar]
- 36.Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: Therapeutic opportunities and assay designs. Assay Drug Dev Technol. 2003;1:843–852. doi: 10.1089/154065803772613471. [DOI] [PubMed] [Google Scholar]
- 37.Kremoser C, Albers M, Burris TP, Deuschle U, Koegl M. Panning for SNuRMs: Using cofactor profiling for the rational discovery of selective nuclear receptor modulators. Drug Discov Today. 2007;12:860–869. doi: 10.1016/j.drudis.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 38.Wang JC, et al. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes Dev. 2006;20:689–699. doi: 10.1101/gad.1400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.