Abstract
Alzheimer's disease is a neurodegenerative disorder characterized by the accumulation of amyloid plaques in the brain. This amyloid primarily contains amyloid-β (Aβ), a 39- to 43-aa peptide derived from the proteolytic cleavage of the endogenous amyloid precursor protein. The 42-residue-length Aβ peptide (Aβ1–42), the most abundant Aβ peptide found in plaques, has a much greater propensity to self-aggregate into fibrils than the other peptides and is believed to be more pathogenic. Synthetic human Aβ1–42 peptides self-aggregate into stable but poorly-ordered helical filaments. We determined their structure to ≈10-Å resolution by using cryoEM and the iterative real-space reconstruction method. This structure reveals 2 protofilaments winding around a hollow core. Previous hairpin-like NMR models for Aβ17–42 fit well in the cryoEM density map and reveal that the juxtaposed protofilaments are joined via the N terminus of the peptide from 1 protofilament connecting to the loop region of the peptide in the opposite protofilament. This model of mature Aβ1–42 fibrils is markedly different from previous cryoEM models of Aβ1–40 fibrils. In our model, the C terminus of Aβ forms the inside wall of the hollow core, which is supported by partial proteolysis analysis.
Keywords: Alzheimer's disease, iterative real-space reconstruction, protein misfolding, neurodegenerative disease, amyloid plaques
The deposition and accumulation of amyloid is a characteristic pathology for >20 different human diseases. These diseases develop with the misfolding and aggregation of normally-soluble peptides resulting in self-aggregation and accumulation of large insoluble fibrils. One of the most common forms of dementia, Alzheimer's disease, shows typical deposition of amyloid in the form of plaques in the extracellular spaces of the brain. The principal component of these plaques is the Aβ peptide, a 39- to 43-aa cleavage product of the amyloid precursor protein. The most abundant Aβ peptide found in plaques is the 42-residue Aβ1–42, which has a remarkable propensity to self-aggregate at high concentrations. Indeed, compared with Aβ1–40, Aβ1–42 demonstrates much more rapid fibril formation in vitro (1–3). In addition, recent studies in animal models demonstrate that Aβ1–42 is exclusively required for the formation of plaques. In these mice, overexpression of Aβ1–42 alone resulted in the development of plaque pathology, whereas Aβ1–40 overexpression did not (4). Therefore, studying differences between Aβ1–40 and Aβ1–42 and their aggregation products may be important in understanding pathological processes involved in fibril and plaque formation.
Because of their highly insoluble but noncrystalline nature, structural determination of amyloid fibrils is refractory to traditional methods such as X-ray crystallography and solution NMR spectroscopy. Early X-ray fiber diffraction experiments revealed a generic cross-β structure for nearly all amyloid fibrils, in which in-register β-strands run approximately perpendicular to the fibril axis and the β-sheets are parallel with the axis (5, 6). Recently, solid-state NMR (ss-NMR) spectroscopy has been used extensively to probe the atomic structures of Aβ1–40 and Aβ1–42 peptides in the fibrillar state (7–11), suggesting a hairpin-like arrangement of the peptides stacked within the fibril. More recently, the combination of quenched hydrogen/deuterium (H/D) exchange and solution NMR spectroscopy added yet another dimension to the ss-NMR models by identifying core regions of the fibril and revealing dynamic properties in solution (12–15). However, these models have been limited to local intermolecular and intramolecular interactions and have therefore been unable to describe the supramolecular architecture of the fibril. To directly determine the interprotofilament interactions in the fibrils, we have used cryoEM and single-particle reconstruction to solve the structure of Aβ1–42 fibrils.
Results
Aβ1–42 Fibrils.
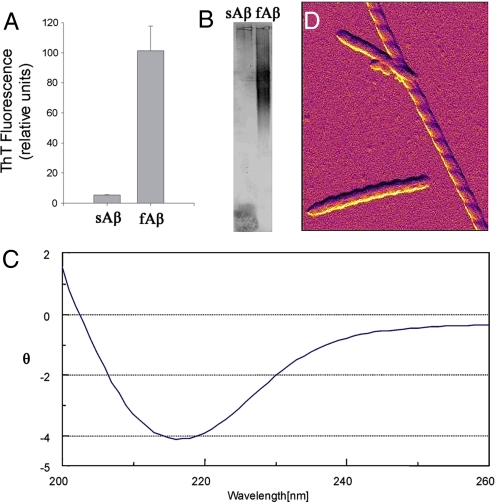
Soluble human synthetic Aβ1–42 (sAb, 100 μM) incubated at 37 °C for 4 weeks (see Materials and Methods) developed biochemical characteristics of fibrils (fAb), including thioflavin T fluorescence (Fig. 1A), high-molecular weight aggregates detected by agarose gel electrophoresis (Fig. 1B), and a typical CD spectrum for β-pleated sheet structure (Fig. 1C). Direct visualization using atomic force microscopy (AFM) revealed a highly-abundant population of long straight forms with pronounced left-handed helical twist and fixed width (Fig. 1D).
Fig. 1.
Biochemical characterization of Aβ1–42 fibrils. Human synthetic Aβ1–42 was incubated at 37 °C for 1 month. (A–C) Compared with sAβ, fAβ demonstrated increased thioflavin-T fluorescence (A), large molecular weight aggregates on agarose gel electrophoresis (B), and a CD spectrum characteristic of β-pleated sheet structure (at 20 μM) (C). θ = residue ellipticity (deg·cm2·dmol−1) × 10−3. (D) Amplitude AFM image of fAβ1–42 using the AC mode shows fibrils with left-handed helical twist (the field of view in the vertical direction spans 5,000 Å).
Single-Particle Analysis and Classification.
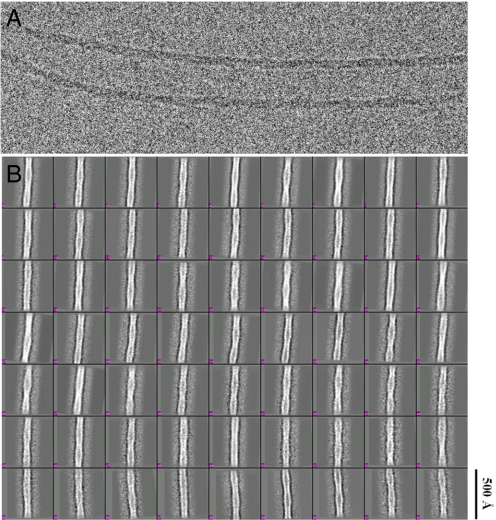
Electron micrographs of fibrils embedded in vitreous ice (Fig. 2A) were analyzed by using the single-particle method (16) as outlined in Fig. S1. Variations of fibril periodicity (helical pitch) were evident in the images, even along the length of a single fibril. Long, straight fibril segments were boxed out from the raw micrographs and then computationally segmented into short square boxes with 90% overlap between consecutive boxes. Each isolated box was treated as a single particle. The boxes were iteratively aligned and classified by multivariate statistical analysis (MSA). Class averages (Fig. 2B) further confirmed the length variation of the helical pitch that was apparent in the raw micrographs. In agreement with the findings of others (8, 17), the side profiles of all of the fibrils were most consistent with 2 helical strands comprising the filament. Therefore, a 2-start helix was assumed in the 3D reconstruction step. Class averages made from boxes containing bent fibrils or fibrils without evident helical twist were excluded from the final reconstruction.
Fig. 2.
CryoEM image of Aβ1–42 fibrils. (A) Fibrils embedded in vitreous ice show long unbranched fibrils with distinct helical twist and fixed width. (B) Samples show structural heterogeneity revealed by class averages of the raw particles calculated by MSA using EMAN software. The box size is 288 in pixels or 521 Å in length. The scale bar is the same for A and B.
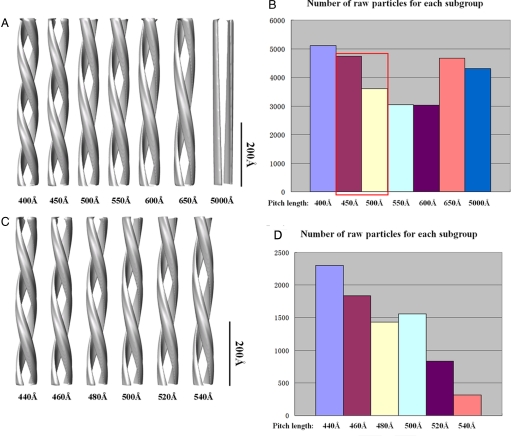
Having established that the structural heterogeneity of the particles was mainly caused by variations in helical pitch, several smooth double-stranded helical models with different pitch lengths were created, and a multireference classification for all of the raw particles against the projections of these models was performed (Fig. S1). A 2-step coarse-to-fine classification strategy was used to split the whole heterogeneous dataset into several homogenous subclasses according to their pitch lengths (Fig. 3).
Fig. 3.
Quantitative classification of raw particle images according to pitch length. A 2-step coarse-to-fine strategy was used. (A) In the first step, 6 models were built with pitch lengths varying from 400 to 650 Å, plus 1 model with very large pitch length (5,000 Å) to extract undesirable particles with no apparent twist. (B) Multireference sorting was performed against the projections of these 7 models for the raw particles to distribute into 7 subgroups. (C) In the second step, another 6 models were built with pitch lengths varying from 440 to 540 Å. (D) A multireference sorting was performed against the projections of these 6 models for the central 2 bins in step 1 (A, red) to further distribute into 6 subgroups.
3D Reconstruction.
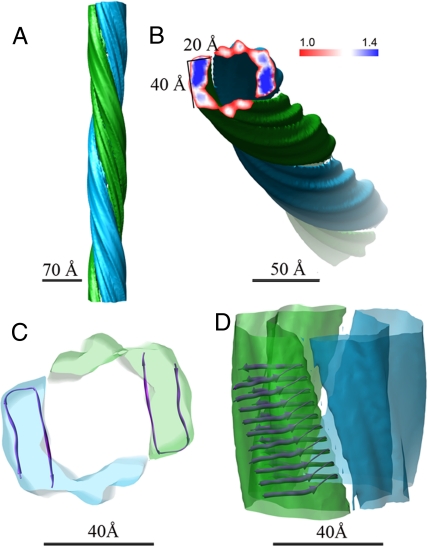
Using the iterative helical real-space reconstruction (IHRSR) algorithm (16), we generated a 3D model (Fig. 4A) from the image data in one of the homogenous subclasses with a pitch length of 460 Å. In this algorithm, we assumed that the specimen possesses a helical symmetry, which is characterized by a pair of parameters: azimuthal angle Δθ and axial rise Δz, between adjacent subunits. The pitch length is defined by (360/Δθ) × Δz. Starting with different permutations of (Δθ, Δz), it is apparent that our dataset does not yield a unique solution of (Δθ, Δz) (Fig. S2 A and B). Such ambiguity is strong independent evidence for the smoothness of the Aβ1–42 fibrils along the protofilament axis. Nevertheless, in different runs, all of the final models converged to a consistent helical pitch (Fig. S2C) close to that used to classify the data initially (Fig. 3 C and D). The consistent pitch length observed here suggests that the fibril can be adequately described as a smooth, axially-continuous helix at current resolution that is ≈10 Å (Fig. S3). In fact, the global protofilament packing and, importantly, the density profile in the cross-section of the 3D reconstruction were unaffected by imposing different helical parameters (Fig. S2D).
Fig. 4.
The cryoEM structure of Aβ1–42 amyloid fibrils. (A) Shown is the final reconstruction assuming a smooth continuous helix by fixing the axial rise to 1 pixel. (B) The cross-section of the reconstruction clearly defined the boundaries of 2 protofilaments. The dimension of each large density region in the cross-section is ≈40 Å × 20 Å. (C) The atomic model (Protein Data Bank ID code 2BEG) fit into the cryoEM density map (top view). (D) Ten Aβ1–42 molecules are shown stacked in the fibrillar state into 1 protofilament of the EM density map (side view).
Interpretation of the Density Map.
The reconstructed map revealed 2 smooth protofilaments winding around a hollow core, assuming either a discrete subunit repeat or a continuous helix (Fig. 4A). The cross-section of the final structure (Fig. 4B) resolved the boundaries of the 2 protofilaments perpendicular to the fibril axis, with a large high-density region and a flanking small lower-density region for each protofilament. The fibril was assumed to have a left-handed twist based on the AFM images (Fig. 1B), consistent with the reported handedness for other amyloid fibrils (18–20).
The size of the large high-density region was ≈40 × 20 Å (Fig. 4B). This size is in good agreement with that of a protofilament (≈45 × 25 Å) in previous cryoEM images of 35MoxAβ1–42 fibrils (21) and with the protofilament dimensions deduced from AFM images (22–24). This large-density region is able to accommodate 2 β-sheets, fitting well with a recent hairpin-like NMR model for Aβ17–42 (21) (Protein Data Bank ID code 2BEG) (Fig. 4C). The small lower-density region of each protofilament is able to accommodate the flexible N terminus (residue 1–16), which is absent in the NMR model. In our fitting, we assumed the axial packing of the peptides adopts the generic cross-β structure in which β-strands run approximately perpendicular to the fibril axis with an equal spacing of 4.8 Å (Fig. 4D). The only remaining degree of freedom in this packing scheme is whether to assume that the peptides stack in register (i.e., with the hydrogen bonds between the same residues in the strands above and below), or whether they are shifted by 1 or more residues. It can be calculated that if there were even a 1-residue shift, this would give an azimuthal angle of 10.8° at the radius of the fibril, and, with an axial rise of 4.8 Å, lead to a short pitch of 160 Å, which has not been observed in our analysis. Therefore, our model assumes an in-register stacking of the peptides along the protofilaments, as is the case with previous models (5, 6).
When fitting the NMR model into the cryoEM density map, it is unclear whether the C- or N-terminal portions of the hairpin (residues 17–42) face the external surface of the fibril. To resolve this ambiguity, a partial proteolysis assay was used to determine whether specific proteolytic sites in monomer Aβ were protected in the fibrillar structure. A variety of proteases (trypsin, chymotrypsin, pepsin, thermolysin, and MMP-9) was incubated with either monomeric Aβ1–42 or purified Aβ1–42 fibrils. Cleavage sites were deduced from proteolytic fragments detected by MALDI-TOF mass spectroscopy. In general, for monomer Aβ, proteolytic cleavage sites were found throughout the peptide; however, for fibrillar Aβ, cleavage sites were restricted to the N-terminal arm of the hairpin loop (Fig. 5), suggesting that the C terminus was protected in the fibril structure. Based on these data, we hypothesize that the N- terminal arm of the hairpin loop forms the external surface of the fibril and is thus susceptible to proteolysis (Fig. 6). This orientation of Aβ is consistent with recent quenched H/D exchange NMR data showing that the C-terminal residues Lys-28–Ala-42 are well protected from solvent access compared with the N-terminal arm of the hairpin loop (14, 25). Moreover, the C-terminal residues were more protected in Aβ1–42 fibrils compared with Aβ1–40 fibrils, consistent with differences in the suprastructure of the fibrils.
Fig. 5.
Summary of Aβ proteolysis analysis. Various proteases (trypsin, thermolysin, chymotrypsin, pepsin, and MMP-9) were incubated with freshly-solubilized Aβ1–42 peptides or purified Aβ1–42 fibrils. Shown above the Aβ1–42 peptide sequence are monomer Aβ cleavage sites for each protease (color-coded arrows); below the sequence are color-coded arrows representing fAβ1–42 cleavage sites. Cleavage sites were deduced from peptide fragments liberated after proteolysis as detected by MALDI-TOF mass spectrometry.
Fig. 6.
Model for interprotofilament interactions in Aβ1–42 amyloid fibrils. Based on this hybrid model, the N terminus of the peptide from one protofilament interacts with the loop region of the peptide from the opposite protofilament.
Discussion
Amyloid fibrils with classic biochemical and morphological appearance formed spontaneously after the prolonged incubation of synthetic human Aβ1–42 at acid pH under quiescent conditions. Our studies produced a 10-Å cryoEM density map that demonstrated 2 winding protofilaments surrounding a hollow core (Fig. 4). At this resolution, secondary structural elements (such as α-helices or β-sheets) within the fibril or protofilaments cannot be resolved. Our reconstruction and the derived model show the density along the protofilament to be smooth with no feature contrast variation out to our map resolution. In our model, we expect that the contrast variation along the protofilament will become apparent only at 4.8 Å, which is the spacing between 2 β-strands, as shown in Fig. 4D. In our analysis, we grouped the images with the same pitch length for the reconstruction. It is possible to have different combinations of axial rise and azimuthal angle that result in the same pitch length. Indeed we found that this was the case. Therefore, we cannot derive a unique axial rise and azimuthal angle in our analysis. Previously-modeled structures using X-ray or NMR approaches, however, can be fit within our density map. Indeed, the hairpin model of Aβ17–42 recently determined by NMR spectroscopy (21) fits very well within the larger high-density structural subunit of the protofilament (Fig. 4C), and the adjacent lower density structure accommodates the flexible N-terminal residues 1–16. The fitting of the NMR model to our cryoEM map (Fig. 4) demonstrates the importance of such a hybrid approach to characterize a complex assembly like the amyloid fibril.
This hybrid model reveals the molecular interactions that bind the twin helical protofilaments together. We propose that the N terminus of the Aβ peptide (stacked in 1 protofilament) associates with the hairpin loop of another Aβ peptide stacked in the opposite protofilament (Fig. 6). Considering the strongly-acidic environment in which the fibril was formed, such a contact is most likely to occur between hydrophobic residues. This head-to-tail interaction between 2 flexible regions of the peptides lends itself to producing variability in the pitch lengths, because there is no strong long-range constraint in the axial direction, and tiny deviations of the azimuthal angle can cumulatively result in large changes in pitch length. These contacts can also explain the generic polymorphism of Aβ fibrils, in that several protofilaments can be loosely assembled into a bundle or unwrap to become a flat ribbon. It is highly possible that the unstructured flexible N termini may play an important role in protofilament assembly. Another important implication of this structure is that this interaction may provide an attractive target to disrupt the fibrillar suprastructure. Small molecules that can disrupt the interaction between these flexible regions may be developed to first break down the mature fibrils into their protofilaments so that other “β-sheet breaker” drugs can have more access to the fibril's surface and can work more efficiently.
Previous NMR studies of Aβ1–40 and Aβ1–42 fibrils have suggested that juxtaposed protofilaments are assembled via direct packing of corrugated surfaces of β-sheets from different protofilaments (11, 25). However, our cryoEM structural model suggests that this is highly unlikely because the dimensions of the core region in the cross-section of our map can only accommodate 2 β-sheets (Fig. 4B).
The hollow core in the center of the fibril revealed by the cross-section is in good agreement with many previous experimental results. For example, Serpell and Smith (26) examined the Aβ11–25 fibrils by using cryoEM and calculated a density map from the equatorial layer line of a single fibril image. The cross-section of their map revealed a hollow core and a high density region at ≈20-Å radius. In addition, Serpell et al. (27, 28) averaged multiple cross-sectional images of ex vivo amyloid fibrils by using a thin-section EM technique and found that all of the fibrils had an electron-lucent core surrounded by several protofilaments. Finally, a reconstruction of amyloid fibrils formed from the SH3 peptide also showed 2 pairs of protofilaments twisting around a hollow core (29). Altogether, these findings strongly suggest that cross-β structure only exists within the protofilament, whereas the supramolecular assembly of the protofilaments is made by other types of interactions.
The partial proteolysis experiments suggest that the C terminus of the Aβ peptide forms the wall of the central pore of the fibril. Proteolytic sites in the C terminus that were accessible in the soluble form of Aβ were inaccessible in the fibril form, suggesting that these sites were shielded by the fibrils' supramolecular structure (Fig. 6). Because many of the residues in the C terminus are hydrophobic, it is likely that these residues may be buried within internal interactions to avoid exposure to the central pore, which is likely filled with solvent.
In comparing our cryoEM map of Aβ1–42 fibril with previous cryoEM maps of Aβ1–40 fibril (20, 30), there are some similarities but many striking differences. Both structures show that mature fibrils are assemblies of pairs of protofilaments. Moreover, both models derived from their respective maps suggest that protofilaments are joined by the interactions between flexible regions. However, our map of Aβ1–42 fibrils reveals a hollow core between the 2 protofilaments, whereas in the map of Aβ1–40 fibrils the density extends to and across the central axis of the fibril. In addition, the cross-over distances (equal to half the pitch, for this 2-stranded structure) for the majority of our Aβ1–42 fibril samples range from 200 to 400 Å, and the fibril diameter in projection is more or less constant, varying between 67 and 85 Å, whereas the average cross-over distance for the Aβ1–40 fibril samples is 1,425 Å and its diameter in projection varies widely along the fibril between 60 and 200 Å. Finally, the density corresponding to 1 protofilament in the cross-section of our Aβ1–42 map demonstrates a good fit with the prevailing hairpin atomic model for 1 Aβ peptide but not for the Aβ1–40 fibril structures. In the cross-sectional plane, each protofilament in our Aβ1–42 model contains only 1 Aβ peptide with a hairpin configuration, whereas in the Aβ1–40 model within each protofilament there are 2 nonequivalent Aβ peptides with fully-extended conformation side by side. These striking structural differences between Aβ1–40 and Aβ1–42 fibrils may be attributable to intrinsic differences in the biochemical properties of these 2 peptides or different growth conditions in various studies (i.e., different pHs of the buffers used in these 2 studies may alter the charge of certain Aβ residues so as to change the local interactions and β-sheets formations). Accumulating evidence suggests that fibril morphology exquisitely depends on the specific growth conditions (pH, agitation/quiescence, temperature, presence of metal ions, presence of seeds, etc.) of the fibril. In addition, it is possible that our model reveals but one of an ensemble of configurations for Aβ1–42 fibrils. Further studies will be required to determine how different fibril growth conditions influence ultimate fibril supramolecular structure.
Materials and Methods
Sample Preparation.
Dry synthetic human Aβ1–42 (American Peptide) was dissolved in neat trifluoroacetic acid, and then dried under an inert atmosphere of nitrogen. The peptide was washed in hexafluoroisopropanol twice, redissolved in DMSO to 5 mM, then diluted in 10 mM HCl to 100 μM, and vortexed for 30 s (pH ≈ 2.0) to form freshly soluble Aβ (sAβ). The solution was incubated at 37 °C for 1 month to form fibrillar Aβ (fAβ).
Agarose Gel Electrophoresis.
Agarose (1.5% wt/vol) was melted in running buffer (20 mM Tris, 200 mM glycine). During stirring of the melted agarose, 10% (wt/vol) SDS was added (drop by drop to avoid local solidification of the agarose) to a final concentration of 0.1%. sAb1-42 or fAb1-42 were incubated for 7 min in sample buffer [60 mM Tris·HCl (pH 6.8), 5% glycerol, 2% SDS, 0.05% bromophenol blue] at room temperature, resolved in a horizontal 1.5% agarose gel in a standard Tris/glycine/SDS buffer, transferred electrophoretically to a PVDF membrane, and probed with Aβ antibody 6E10 (Sigma).
Thioflavin-T Fluorescence Spectroscopy.
The fluorescence of Aβ1–42 fibril samples was determined in 50 mM glycine buffer (pH 9.0). Final fibril concentration was 0.05 mg/mL, and the thioflavin-T concentration was 10 μM. All fluorescence measurements were carried out at room temperature by using a Cytofluor (r) Fluorimeter (Perseptive Biosystems). Absorbance was measured at 440 excitation and 530 emission.
CD Spectroscopy.
The far-UV CD experiments were carried out on a JASCO J-715 spectropolarimeter at room temperature by using quartz cells of 1-mm path length. Spectra were recorded between 260 and 200 nm at 0.05-nm intervals, with a response time of 4 s. Data are expressed as mean residue ellipticity (deg·cm2·dmol−1) × 10−3.
Proteolytic Digestions.
Recombinant human thermolysin, trypsin, chymotrypsin, pepsin, and MMP-9 were used for digestion reactions using optimal conditions and buffers recommended by the manufacturer (R&D Systems). Pro-MMP-9 was activated with 1 mM p-aminophenylmercuric acetate (APMA) at 37 °C for 24 h before use. For sAβ digestions, 100 nM protease was incubated with 2.5 μM sAβ at 37 °C for varying periods of time (15 min to 4 h), then analyzed by MS. For fAβ digestions, 10 μL of fAβ was applied to a Microcon 50-kDa centrifugal ultrafiltration unit (YM-50; Millipore) and washed with 500 μL of 10 mM HCl 3 times, then equilibrated with 500 μL of MQ water. Thirty microliters of reaction buffer was added to the purified fAβ (retentate) and centrifuged; the filtrate was analyzed by MS (see below) to ensure that any contaminating Aβ monomer or fragments were washed away. Ten microliters of 300 nM protease and 20 μL of reaction buffer were added to the confirmed purified fAβ (retentate) and incubated at 37 °C for (1–24 h), then centrifuged and analyzed by MS to detect liberated fragments of fAβ.
MS.
Samples were first passed through a reverse-phase C18 Ziptip (Millipore), according to the manufacturer's instructions, then diluted 1:1 with matrix solution (a saturated solution of 3, 5-dimethoxy-4-hydroxycinnamic acid in 50% acetonitrile with 0.1% trifluoroacetic acid in water), loaded onto a plate, and allowed to dry. The sample was then analyzed on an Applied Biosystems Voyager DE-STR MALDI-TOF mass spectrometer operated in linear mode.
AFM.
fAβ1–42 was diluted 1:40 in 200 mM HCl, and 20 μL of the diluted solution was placed on freshly-cleaved mica and allowed to adhere for 10 min. The mica was gently rinsed with the 200 mM HCl solution before placing it on the AFM and imaging it in the same solution. All images were acquired by using magnetic actuation of the cantilever (iDrive) in AC mode using silicon nitride cantilevers, model TR400PB. All final images were first-order flattened before being rendered into 3D with Asylum Research's Argyle software. Images were acquired in fluid with AC mode by using the Asylum Research MFP-3D-SA AFM.
CryoEM.
A Vitrobot (FEI) was used to flash-freeze a 2.5-μL aliquot of sample onto a copper Quantifoil R2/1 grid. The sample was loaded onto a Gatan 626 cryoholder and imaged in a JEM 2010F electron microscope operated at 200 kV and at a specimen temperature near liquid N2. A 70-μm condenser aperture, spot size 2, and a 60-μm objective aperture were used. Approximately 200 images were collected at a sampling value of 1.81 Å per pixel on a Gatan 4k CCD camera with a dose of 18–20 e/Å2 and a defocus range of 1.5–3 μm at a final detector magnification ≈82,800.
Image Processing.
The pipeline of the whole process is shown in Fig. S1. Most of the processing steps were done with EMAN software (31) based on the principle of IHRSR algorithm (16). Long, straight nonoverlapping fibril segments were first boxed out from the CCD frames by using the helixboxer program and then cut into short square boxes (288 × 288 pixels) with 90% overlap between successive boxes and masked by a cosine-edge rectangle. Each isolated box was treated as a single particle. The optimal box size was empirically determined to be 288 pixels so that it was neither too long to include substantial bending nor too short to cause ambiguity in image alignment. Contrast transfer function correction was done for each raw particle by simple binary phase flipping. The whole heterogeneous dataset was preseparated on the basis of their pitch lengths (Fig. 3). The 3D reconstruction process was then carried out on one of the homogenous subgroups with a constant pitch length by using IHRSR (16). First, reference projections with 2° angular step size around the fibril axis including out-of-plane tilts up to ± 9° were generated from a 3D model, which was a featureless solid cylinder at the beginning and resolved into 2 twisted protofilaments after a few iterations. Orientation for each particle image was assigned by projection matching with these reference projections. A 3D reconstruction was done with C2 symmetry around the fibril axis imposed. The C2 symmetry was validated by a previous asymmetric reconstruction that suggested a 2-fold axis (Fig. S4). Programs in the IHSHR package were used to search for helical parameters (azimuthal angle Δθ and axial rise Δz) in the 3D map with which a helical symmetry was then imposed on the model. This helical model was used as the reference in the subsequent iteration. The resolution of the final map was assessed by Fourier shell correlation (0.5 threshold) between 2 independently reconstructed volumes by dividing the whole dataset into halves (32).
Supplementary Material
Acknowledgments.
We thank Dr. Edward Egelman at the University of Virginia (Charlottesville) for providing programs and valuable advice. This work was supported by National Institutes of Health Grant P41RR02250 through the National Center for Research Resources and National Institutes of Health Grant PN1EY016525 through the Roadmap Initiative for Medical Research (to W.C.), National Institutes of Health Grants R01 NS048283 and P01 NS032636 (to J.-M.L.), National Institutes of Health Grant DK13332 (to C.F.), the Robert Welch Foundation (W.C.), and the Hope Center for Neurological Disorders (J.-M.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryoEM map has been deposited with the European Bioinformatics Institute, http://www.ebi.ac.uk/ (accession code EMD-5052).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901085106/DCSupplemental.
References
- 1.Burdick D, et al. Assembly and aggregation properties of synthetic Alzheimer's A4/β amyloid peptide analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 2.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxyl terminus of the β amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer's disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 3.Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB. Amyloid β-protein fibrillogenesis. Detection of a protofibrillar intermediate. J Biol Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 4.McGowan E, et al. Aβ42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005;47:191–199. doi: 10.1016/j.neuron.2005.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 6.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 7.Petkova AT, et al. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer's β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 9.Balbach JJ, et al. Supramolecular structure in full-length Alzheimer's β-amyloid fibrils: Evidence for a parallel β-sheet organization from solid-state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda Y, et al. Verification of the turn at positions 22 and 23 of the β-amyloid fibrils with Italian mutation using solid-state NMR. Bioorg Med Chem. 2005;13:6803–6809. doi: 10.1016/j.bmc.2005.07.071. [DOI] [PubMed] [Google Scholar]
- 11.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ippel JH, Olofsson A, Schleucher J, Lundgren E, Wijmenga SS. Probing solvent accessibility of amyloid fibrils by solution NMR spectroscopy. Proc Natl Acad Sci USA. 2002;99:8648–8653. doi: 10.1073/pnas.132098999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whittemore NA, et al. Hydrogen-deuterium (H/D) exchange mapping of Aβ1–40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 14.Olofsson A, Sauer-Eriksson AE, Ohman A. The solvent protection of Alzheimer amyloid-β-(1–42) fibrils as determined by solution NMR spectroscopy. J Biol Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 15.Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Ohman A. Amide solvent protection analysis demonstrates that amyloid-β(1–40) and amyloid-β(1–42) form different fibrillar structures under identical conditions. Biochem J. 2007;404:63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 17.Goldsbury C, Frey P, Olivieri V, Aebi U, Muller SA. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J Mol Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 18.Goldsbury CS, et al. Studies on the in vitro assembly of a β1–40: Implications for the search for a β fibril formation inhibitors. J Struct Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 19.Jimenez JL, et al. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachse C, et al. Quaternary structure of a mature amyloid fibril from Alzheimer's Aβ(1–40) peptide. J Mol Biol. 2006;362:347–354. doi: 10.1016/j.jmb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Luhrs T, et al. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalewski T, Holtzman DM. In situ atomic force microscopy study of Alzheimer's β-amyloid peptide on different substrates: New insights into mechanism of β-sheet formation. Proc Natl Acad Sci USA. 1999;96:3688–3693. doi: 10.1073/pnas.96.7.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, et al. AFM and STM study of β-amyloid aggregation on graphite. Ultramicroscopy. 2003;97:73–79. doi: 10.1016/S0304-3991(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 24.Arimon M, et al. Fine structure study of Aβ1–42 fibrillogenesis with atomic force microscopy. FASEB J. 2005;19:1344–1346. doi: 10.1096/fj.04-3137fje. [DOI] [PubMed] [Google Scholar]
- 25.Sato T, et al. Inhibitors of amyloid toxicity based on β-sheet packing of Aβ40 and Aβ42. Biochemistry. 2006;45:5503–5516. doi: 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serpell LC, Smith JM. Direct visualization of the β-sheet structure of synthetic Alzheimer's amyloid. J Mol Biol. 2000;299:225–231. doi: 10.1006/jmbi.2000.3650. [DOI] [PubMed] [Google Scholar]
- 27.Serpell LC, et al. The protofilament substructure of amyloid fibrils. J Mol Biol. 2000;300:1033–1039. doi: 10.1006/jmbi.2000.3908. [DOI] [PubMed] [Google Scholar]
- 28.Serpell LC, et al. Examination of the structure of the transthyretin amyloid fibril by image reconstruction from electron micrographs. J Mol Biol. 1995;254:113–118. doi: 10.1006/jmbi.1995.0604. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez JL, et al. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachse C, Fandrich M, Grigorieff N. Paired β-sheet structure of an Aβ(1–40) amyloid fibril revealed by electron microscopy. Proc Natl Acad Sci USA. 2008;105:7462–7466. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 32.Van Heel M. Angular reconstitution: A posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.