The genetic analysis of complex diseases has become a mainstream biomedical research goal. The methods for genetically dissecting human diseases have now evolved into an integrated science combining human genetics, functional genomics, high-throughput experimental technology and computational techniques. The inflammatory bowel diseases (IBDs) feature prominently in this field, and the realm of IBD genetics is continually and exponentially increasing. The present review will summarize and put into perspective the breadth of IBD genetics as it has evolved over the past 10 years, and will give the reader some sense of anticipation as to where it may be leading us.
EPIDEMIOLOGY – EVIDENCE FOR A GENETIC ETIOLOGY
A variety of epidemiological data suggest that genetic factors are intimately involved in the pathogenesis of IBD. For instance, IBD demonstrates a familial pattern of disease with a much higher disease frequency in first-degree relatives of affected individuals compared with the general population (1–4). The degree of ‘heritability’ or λs (the relative risk of the sibling of an affected individual) is estimated to range from 15 to 35 for Crohn’s disease (CD) and six to nine for ulcerative colitis (UC) (5–9). Twin studies strengthen the argument for a genetic basis for IBD, with a much higher rate of disease concordance demonstrated in monozygotic twins compared with dizygotic twins, particularly in CD (10–13). Marked ethnic and racial differences in disease prevalence have been noted, particularly in Ashkenazi Jews, and are also in keeping with genetic etiology (14–22).
IBD is said to be a complex genetic disorder with the disease appearing to occur in susceptible individuals carrying one or more risk alleles after exposure to one or more environmental triggers. Because there is an increased risk of UC in CD-affected families and vice versa, a genetic model is proposed in which CD and UC are related diseases sharing some susceptibility loci but perhaps differing at others, which results in diverging disease courses (23,24). Additional genetic ‘hits’ or environmental factors may determine the particular phenotype of the individual by influencing features such as disease site, disease behaviour or response to therapy, among others (Figure 1).
Figure 1).
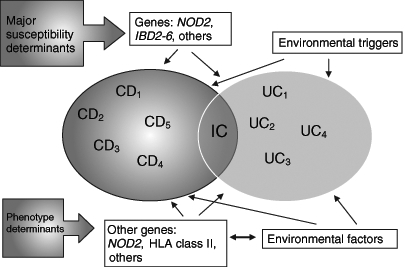
Gene-environment interactions in inflammatory bowel disease (IBD). CD Crohn’s disease; HLA Human leukocyte antigen; IC Indeterminate colitis; NOD2 Nucleotide oligomerization domain 2; UC Ulcerative colitis
GENE IDENTIFICATION TECHNIQUES
The quest for IBD susceptibility genes has used two broad approaches: linkage analysis using multiple-affected families and association using affected individuals in a case-control format. Genome-wide scans (GWSs) were traditionally used only with linkage analysis but recent progress has allowed a genome-wide approach by case-control association studies. Given the almost infinite number of possible genetic candidates, it is not surprising that the first major advances in this area were generated by linkage analysis: a ‘hypothesis-free’ technique in which the entire genome can be evaluated to look for areas of genetic susceptibility before trying to narrow down the search to specific genes. A genome-wide linkage analysis usually requires the ascertainment of numerous ‘sibling pair’ families, that is, two affected siblings and their parents (whether affected or not). Genotyping of all the members of each family is performed for polymorphic DNA microsatellite markers (approximately 300 microsatellite markers), which are located at intervals throughout the genome. Linkage analysis generates log of odds (LOD) scores which are measures of excess sharing of the same allele between affected siblings, suggesting a correlation between inheritance of disease and inheritance of that particular allele (Figure 1). This allows for the narrowing down of the search for susceptibility genes to a more specific part of the genome. Numerous genome-wide linkage studies in IBD have been published since 1996, identifying at least nine susceptibility loci (IBD1 – IBD9) thought to contain causal genes (Table 1).
TABLE 1.
Inflammatory bowel disease (IBD) susceptibility loci
| Chromosome | HUGO ID | Phenotype | Associations |
|---|---|---|---|
| 16q | IBD1 | CD | NOD2/CARD15 |
| 12q | IBD2 | IBD, especially UC | |
| 6p | IBD3 | IBD | HLA, TNF |
| 14q | IBD4 | CD | |
| 5q | IBD5 | IBD | IBD5 haplotype, OCTN1 and OCTN2 |
| 19p | IBD6 | IBD | |
| 1p | IBD7 | IBD | |
| 3p | IBD9 | IBD |
CD Crohn’s disease; HUGO ID Human genome organization identification; HLA Human leukocyte antigen; NOD2/CARD15 Nucleotide-binding oligomerization domain 2/Caspase activating recruitment domain 15; OCTN Organic cation transporter; TNF Tumour necrosis factor; UC Ulcerative colitis
On the other hand, candidate gene analysis to some extent is hypothesis driven. It is an ‘association test’ that examines differences in the allele frequencies of the nominated gene in affected patients compared with controls. A variety of different statistical techniques can be used and the controls can be ‘population-based’ controls who must be carefully matched, or family-based controls who are often the parents of the affected individual. The chosen ‘candidate’ is usually based on findings from an earlier linkage study putting that gene within the susceptibility region identified in a prior GWS (a positional candidate) or by the purported function of the gene such as tumour necrosis factor-alpha (TNF-α) (a functional candidate). These approaches are certainly complementary, and ideally, both approaches are required in tandem, as was illustrated with the identification of the first IBD susceptibility gene described below. More recently, single nucleotide polymorphisms (SNPs) have been used in the place of microsatellite markers for the purpose of performing genome-wide association studies. SNPs represent sites along the genome in which a single base pair variation occurs from person to person where the least frequent allele has a frequency of 1% or greater. Many SNPs occur within genes, with some representing variations that alter gene function and may even represent the genetic lesion of interest. Genome-wide association studies may represent the most powerful approach to identifying all of the IBD susceptibility genes that are relevant to disease pathophysiology because as many as 500,000 SNPs could be assayed for each patient in the study, making a massive amount of data available for analysis.
CARD15/NOD2 AND CD SUSCEPTIBILITY
In 2001, two groups identified the first gene contributing to CD susceptibility within the IBD1 susceptibility locus on chromosome 16. This gene was known as caspase activating recruitment domain 15 (CARD15) or nucleotide-binding oligomerization domain 2 (NOD2) (25,26), and is the only confirmed susceptibility gene to date for IBD. Three major independent polymorphisms of this gene are associated with CD in Caucasians: Arg702Trp and Gly908Arg are both missense mutations resulting in an amino acid substitution, as well as Leu1007fsinsC, which is a frameshift mutation that results in shortening of the protein product (25,27–43). While the identified mutations are either rare or absent in Asian (32,33,44–46), Arab (47,48) and African (49) populations, it is estimated that 27% to 38% of Caucasian CD patients carry one of the major risk alleles (compared with approximately 20% of Caucasian controls) and an additional 8% to 17% carry two copies (compared with less than 1% of controls). Of note, allele frequency for the three common mutations in sporadic CD is comparable with that seen in familial CD. A recent meta-analysis (50) calculated that the overall relative risk of developing CD in Caucasian populations was 2.4 (95% CI 2.0 to 2.9) for carriers of one mutant allele and 17.1 (95% CI 10.7 to 27.2) for two or more mutant copies. The risk varied with each mutation, Leu1007fsinsC generally carrying the highest risk and Arg702Trp the lowest. The overall proportion of CD cases attributed to the presence of mutant alleles in Caucasian populations was approximately 22% (50).
Further evaluation of CARD15/NOD2 risk alleles have demonstrated a number of phenotypic associations. The major variants are associated with earlier age of onset (27,51,52) and ileal disease location (27–30,35,42,52–54). They are also suggested to be associated with fibrostenotic disease behaviour, (29,30,34,36,51,52,55) and negatively associated with pure colonic disease (27,30,35,42,43,51,52). In summary, the evidence to date suggests that the three risk alleles within the CARD15/NOD2 gene are correlated with CD which has an earlier onset, involves the ileum and is frequently fibrostenoic.
OTHER IMPORTANT IBD LOCI
To date, CARD15/NOD2 is the only confirmed IBD susceptibility gene identified. There are a number of other likely candidate genes and loci that have been described, although details remain less clear, and their roles less well characterized than CARD15/NOD2.
The IBD5 locus on chromosome 5q31
The IBD5 locus on chromosome 5q31 has received much attention recently. Aside from CARD15/NOD2, it is the next most relevant region for IBD susceptibility. The locus was initially identified as significant by two GWSs (56,57). Subsequent analyses refined the locus to a 250 kb risk haplotype (58). The association of this haplotype with IBD has been widely replicated in a number of independent populations (59–62). The IBD5 risk haplotype has been principally associated with CD, although there have been some suggestions of a weak association with UC as well. Polymorphisms within the organic cation transporter (OCTN1 and OCTN2) genes in the region have been recently put forward as the causative variant based on a combination of functional and genetic evidence (63). OCTN1 and OCTN2 are transporters that mediate transmembrane transport of carnitine and other organic cations (64,65). Although putative mechanisms have been suggested (65), no direct evidence is available to date to explain the role the variants of these genes could play in the pathobiology of IBD. Despite the efforts of several large centres, there has been an inability to replicate the findings that OCTN1 and OCTN2 polymorphisms are independently associated with CD, and these additional studies (66–69) support the possibility that the OCTN1 and OCTN2 SNPs are simply part of the extended haplotype in the region. This region is particularly difficult to fine map because it contains a significant degree of linkage disequilibrium, making it difficult to discern a causative allele from a marker allele coinherited with the disease-causing allele (58,70).
Phenotypically, the IBD5 locus has been associated with earlier onset disease (56) as well as perianal disease (61,67). While most commonly reported with CD, an association with UC has been reported (62) with the strength of both associations possibly increased in the presence of a known CARD15/NOD2 variant (60,71) suggesting a positive epistatic interaction (the influence of one gene on the expression of another). With the uncertainty over the role of OCTN1 and OCTN2, attention has focused on a number of additional genes located within the IBD5 locus. These have included genes within the cytokine cluster, as well as interferon regulatory factor 1, PDLIM and proline 4-hydroxylase alpha polypeptide II, all of which are equally likely to contain the IBD5 causal variant as the OCTN genes.
THE MAJOR HISTOCOMPATIBILITY COMPLEX REGION (IBD3) ON CHROMOSOME 6P
Linkage of IBD (both UC and CD) to the IBD3 region on chromosome 6 has been confirmed in a variety of GWSs (56,57,72–74). In a recent meta-analysis (75) of 10 such scans, it was the only locus that achieved genome-wide significance. The IBD3 region contains the major histocompatibility complex genes (also referred to as the human leukocyte antigen [HLA]). The HLA complex is divided into three regions (class I, class II and class III) and contains a total of 224 densely packed highly polymorphic gene loci (76). Not surprisingly, the study of this area has proved very challenging, with various conflicting results. Several linkage and association studies (56,73,74,77–80) have implicated the HLA region in both IBD susceptibility and phenotype, although it has not been consistently confirmed in replication studies (81). A recent meta-analysis of 20 studies (82) highlighted both positive and negative associations with a variety of class II DRB1 alleles. For example, DRB1*1502 is associated with UC across a variety of different populations. Despite extremely variable background prevalence (from less than 1% in Northern Europeans to approximately 25% in Japanese) the relative risk in each ethnic group is surprisingly similar (two to 4.5) (80,83–86). This pattern suggests the allele is a susceptibility variant.
However, genetic variation may not only contribute to susceptibility, but may also modify disease phenotype. Indeed, multiple-affected families show surprising concordance for disease phenotype, including age of onset, disease location, behaviour, need for surgery and extraintestinal manifestations (3,87–90). It is possible that HLA plays a greater role in determining final disease phenotype than initial disease susceptibility (76). For instance, DRB1*07 is the most consistently replicated association with CD. It is specifically associated with ileal disease in the absence of a major CARD15/NOD2 mutation (27,28,82,91). In comparison, DRB1*0103 is a rare allele that has been associated with both UC and CD. In UC, it is the most consistently replicated HLA association. In UC, the association has been reported across a variety of ethnic groups in combination with various haplotypes, and is particularly strong in patients with extensive or severe disease (80,83,92–95). In CD, the allele is strongly associated with isolated colonic disease (27,28,91,96). Interestingly, this allele (along with HLA-B*27) has also been associated with uveitis (97). In addition, DRB1*0103 (as well as B*27 and B*35) has been associated with type I peripheral arthropathy (acute, self-limiting, pauciarticular large-joint arthritis associated with IBD relapses) while HLA-B*44 has been associated with type II arthritis (symmetrical, seronegative, small-joint arthropathy unrelated to disease activity) (98).
Many of the genes in this region are also involved in the immune response and include the genes for TNF, lymphotoxin alpha and the heat shock proteins. Of these, TNF is certainly the most intensely studied. Associations have been found with CD and promoter polymorphisms at positions −308, −857, −863 and −1031; however, their functional significance remains unclear (27,99–106).
DROSOPHILA DISCS LARGE HOMOLOGUE 5 (DLG5) AND IBD SUSCEPTIBILITY
The pericentromeric region of chromosome 10 was identified as a potential IBD susceptibility locus in a European GWS. More recently, the region was refined to a haplotype block that contained only one gene: drosophila discs large homologue 5 (DLG5) (107). The data suggested an overall association for DLG5 with IBD susceptibility rather than just CD, with a relative risk of approximately 1.5 (107). These findings have been, at least, partially replicated in some centres (108,109), but not in the majority of studies (67,110,111). DLG5 is an attractive susceptibility candidate because it is putatively involved in maintaining epithelial integrity, and thus, its dysfunction would be consistent with an etiological model of impaired barrier function. However, much work remains to be performed to ascertain the role of this gene in IBD susceptibility.
MULTIDRUG RESISTANCE-1 GENE AND IBD SUSCEPTIBILITY
The multidrug resistance-1 (MDR1) gene is a biologically plausible candidate gene for a number of reasons. First, MDR1-deficient mice are known to spontaneously develop enterocolitis when maintained in a specific pathogen-free environment (112). The gene’s product, P-glycoprotein 170, is highly expressed in various epithelial surfaces including the intestine. Langmann et al (113) recently demonstrated a marked downregulation of this product in the colonic tissue of UC patients but not CD patients. Finally, the MDR-1 gene maps to chromosome 7q22 in a region that was previously identified by a GWS as being suggestive for linkage to IBD (114). To date, the most consistently reported association is with UC (115–118). Ho et al (115) described two haplotypes, one associated with disease susceptibility and the other disease protective. Current data suggest a specific association with extensive UC (115). The exact physiological role of this protein within the gut remains controversial. A variety of other case-control studies (119,120) have failed to detect any association between this gene variant and UC.
CLINICAL IMPLICATIONS
The field of IBD genetics is in its infancy despite relatively rapid success in a number of areas. The integration of genetic testing into the clinic is still premature, however, it will not be long before genetic testing may become an important part of the initial workup of a patient with suspected or known IBD (Table 2).
TABLE 2.
Potential application of genetic testing in inflammatory bowel disease
| Prediction of disease susceptibility |
| Presymptomatic testing to detect ‘at risk’ individuals |
| Diagnosis |
| Prognosis |
| Choice of therapy and response to therapy |
| Disease prevention with specific interventions |
| Do not smoke? |
| Avoid unnecessary nonsteroidal anti-inflammatory drugs? |
| Probiotics administration? |
| Gene therapy? |
The use of genetic testing to predict disease in presymptomatic patients is still not possible due to the relative lack of sensitivity and specificity of CARD15/NOD2 testing. The same argument applies to genetic testing for diagnosis of IBD and the early presentation of IBD patients. It is more plausible that in the future, a panel of genetic and possibly serum markers will be tested to provide a measure of how likely a person is to develop IBD and what a patient’s diagnosis is after symptoms have begun. Prognostic testing in individuals diagnosed with IBD is an area where molecular testing of genetic and serum markers may have the most potential current value. For example, it is known that CARD15/NOD2-positive individuals are more likely to have ileal disease and fibrostenotic disease, and are potentially more likely to proceed to an early ileal resection (121). If this is confirmed, then testing may allow the identification of those at risk of this complication and potentially target earlier or more advanced therapies to these individuals to prevent such complications. Again, a panel of genetic markers will likely be more useful for this indication. This type of predictive testing is evolving in serological marker testing where combinations of markers are associated with more aggressive small-bowel CD with a predilection to advanced disease behaviour (such as fibrostenotic or internal penetrating disease). Indeed, it is possible that the presence of such serum markers is genetically mediated and that ultimately a combination of genetic and serum marker testing will predict the course of the disease.
Pharmacogenetics is the study of how genetic variation influences an individuals’ response to therapy (122,123). The hypothesis is that characterization of a specific genetic polymorphism will predict drug response and/or toxicity. It has been estimated that genetic variation can account for 20% to 90% of variability in drug disposition and effect (124). A classic example of the role of pharmacogenetics in IBD is the utility of thiopurine methyltransferase (TPMT) genotyping and the relationship to the metabolism of azathioprine/6-mercaptopurine. TPMT is one of the main degradation enzymes for these drugs, and mutations within this gene have been associated with toxic side effects. However, the appropriate clinical application of this knowledge is still debated. While one might hypothesize that knowledge of the genotype could avoid toxicity, it is important to note that TPMT mutations appear to only account for approximately 10% to 27% of observed toxic reactions (125–128). Recent work with MDR-1 introduced the potential ability to predict steroid resistance with the suggestion that there is an overexpression of MDR-1 in the peripheral blood lymphocytes of steroid-resistant IBD patients (129). Hoffmeyer et al (130) have demonstrated an association with a specific variant of this gene (SNP C3235T) and its in vivo expression levels. In the future, it may be possible to predict a patient’s steroid responsiveness from a genetic test (122,131). Conversely, as discussed earlier, an association was demonstrated with underexpression of MDR-1 and the development of colitis. Pharmacogenetic studies are being undertaken in virtually all current IBD therapies, however, to date, there are minimal data available that would yield an impact upon clinical care.
FUTURE DIRECTIONS
While each individual genetic risk factor identified thus far for IBD accounts for very little of the disease’s overall heritability, the interaction between these and other genes as well as between genes and environmental risk factors is likely to play a very important role in disease pathogenesis and outcome. With advances in genetic technology leading to genome-wide association testing about to be completed in IBD, it is much more likely that all of the important genetic variants that contribute to IBD susceptibility will be identified. This will initiate an era of tremendous promise in the field of IBD research with the unique opportunity to make real advances in understanding the cause of IBD and to enable us to reach the goal of disease prevention and possibly a cure. While this is surely to take many years of painstaking work, the discoveries in IBD genetics will provide many ways to improve the lives of those living with IBD and make more tools available to those treating IBD. With improvements in diagnostic and prognostic testing as well as the development of novel drugs or drug tools to enable us to use existing therapies more effectively, the field of IBD genetics places us at the dawn of a new horizon in our efforts to better understand and manage IBD.
REFERENCES
- 1.Binder V. Genetic epidemiology in inflammatory bowel disease. Dig Dis. 1998;16:351–5. doi: 10.1159/000016891. [DOI] [PubMed] [Google Scholar]
- 2.Russel MG, Pastoor CJ, Janssen KM, et al. Familial aggregation of inflammatory bowel disease: A population-based study in South Limburg, The Netherlands. The South Limburg IBD Study Group. Scand J Gastroenterol Suppl. 1997;223:88–91. [PubMed] [Google Scholar]
- 3.Peeters M, Nevens H, Baert F, et al. Familial aggregation in Crohn’s disease: Increased age-adjusted risk and concordance in clinical characteristics. Gastroenterology. 1996;111:597–603. doi: 10.1053/gast.1996.v111.pm8780562. [DOI] [PubMed] [Google Scholar]
- 4.Farmer RG, Michener WM, Mortimer EA. Studies of family history among patients with inflammatory bowel disease. Clin Gastroenterol. 1980;9:271–7. [PubMed] [Google Scholar]
- 5.Satsangi J, Jewell DP, Rosenberg WM, Bell JI. Genetics of inflammatory bowel disease. Gut. 1994;35:696–700. doi: 10.1136/gut.35.5.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Probert CS, Jayanthi V, Hughes AO, Thompson JR, Wicks AC, Mayberry JF. Prevalence and family risk of ulcerative colitis and Crohn’s disease: An epidemiological study among Europeans and south Asians in Leicestershire. Gut. 1993;34:1547–51. doi: 10.1136/gut.34.11.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuster W, Pascoe L, Purrmann J, Funk S, Majewski F. The genetics of Crohn disease: Complex segregation analysis of a family study with 265 patients with Crohn disease and 5387 relatives. Am J Med Genet. 1989;32:105–8. doi: 10.1002/ajmg.1320320122. [DOI] [PubMed] [Google Scholar]
- 8.Meucci G, Vecchi M, Torgano G, et al. Familial aggregation of inflammatory bowel disease in northern Italy: A multicenter study. The Gruppo di Studio per le Malattie Infiammatorie Intestinali (IBD Study Group) Gastroenterology. 1992;103:514–9. doi: 10.1016/0016-5085(92)90841-l. [DOI] [PubMed] [Google Scholar]
- 9.Orholm M, Munkholm P, Langholz E, Nielsen OH, Sorensen TI, Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84–8. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 10.Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: A long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–73. doi: 10.1016/s0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- 11.Orholm M, Binder V, Sorensen TI, Rasmussen LP, Kyvik KO. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol. 2000;35:1075–81. doi: 10.1080/003655200451207. [DOI] [PubMed] [Google Scholar]
- 12.Thompson NP, Driscoll R, Pounder RE, Wakefield AJ. Genetics versus environment in inflammatory bowel disease: Results of a British twin study. BMJ. 1996;312:95–6. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tysk C, Lindberg E, Jarnerot G, Floderus-Myrhed B. Ulcerative colitis and Crohn’s disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–6. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas GA, Millar-Jones D, Rhodes J, Roberts GM, Williams GT, Mayberry JF. Incidence of Crohn’s disease in Cardiff over 60 years: 1986–1990 an update. Eur J Gastroenterol Hepatol. 1995;7:401–5. [PubMed] [Google Scholar]
- 15.Yapp TR, Stenson R, Thomas GA, Lawrie BW, Williams GT, Hawthorne AB. Crohn’s disease incidence in Cardiff from 1930: An update for 1991–1995. Eur J Gastroenterol Hepatol. 2000;12:907–11. doi: 10.1097/00042737-200012080-00010. [DOI] [PubMed] [Google Scholar]
- 16.Trallori G, Palli D, Saieva C, et al. A population-based study of inflammatory bowel disease in Florence over 15 years (1978–92) Scand J Gastroenterol. 1996;31:892–9. doi: 10.3109/00365529609051998. [DOI] [PubMed] [Google Scholar]
- 17.Ogunbi SO, Ransom JA, Sullivan K, Schoen BT, Gold BD. Inflammatory bowel disease in African-American children living in Georgia. J Pediatr. 1998;133:103–7. doi: 10.1016/s0022-3476(98)70187-8. [DOI] [PubMed] [Google Scholar]
- 18.Loftus EV, Jr, Silverstein MD, Sandborn WJ, Tremaine WJ, Harmsen WS, Zinsmeister AR.Crohn’s disease in Olmsted County, Minnesota, 1940–1993: Incidence, prevalence, and survival Gastroenterology 19981141161–8.(Erratum in 1999;116:1507). [DOI] [PubMed] [Google Scholar]
- 19.Andres PG, Friedman LS.Epidemiology and the natural course of inflammatory bowel disease Gastroenterol Clin North Am 199928255–81.vii. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein CN, Blanchard JF, Rawsthorne P, Wajda A. Epidemiology of Crohn’s disease and ulcerative colitis in a central Canadian province: A population-based study. Am J Epidemiol. 1999;149:916–24. doi: 10.1093/oxfordjournals.aje.a009735. [DOI] [PubMed] [Google Scholar]
- 21.Yang SK, Loftus EV, Jr, Sandborn WJ. Epidemiology of inflammatory bowel disease in Asia. Inflamm Bowel Dis. 2001;7:260–70. doi: 10.1097/00054725-200108000-00013. [DOI] [PubMed] [Google Scholar]
- 22.Carr I, Mayberry JF. The effects of migration on ulcerative colitis: A three-year prospective study among Europeans and first- and second-generation South Asians in Leicester (1991–1994) Am J Gastroenterol. 1999;94:2918–22. doi: 10.1111/j.1572-0241.1999.01438.x. [DOI] [PubMed] [Google Scholar]
- 23.Satsangi J, Jewell DP, Bell JI. The genetics of inflammatory bowel disease. Gut. 1997;40:572–4. doi: 10.1136/gut.40.5.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad T, Satsangi J, McGovern D, Bunce M, Jewell DP. Review article: The genetics of inflammatory bowel disease. Aliment Pharmacol Ther. 2001;15:731–48. doi: 10.1046/j.1365-2036.2001.00981.x. [DOI] [PubMed] [Google Scholar]
- 25.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 26.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad T, Armuzzi A, Bunce M, et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology. 2002;122:854–66. doi: 10.1053/gast.2002.32413. (Erratum in 2003;125:281). [DOI] [PubMed] [Google Scholar]
- 28.Newman B, Silverberg MS, Gu X, et al. CARD15 and HLA DRB1 alleles influence susceptibility and disease localization in Crohn’s disease. Am J Gastroenterol. 2004;99:306–15. doi: 10.1111/j.1572-0241.2004.04038.x. [DOI] [PubMed] [Google Scholar]
- 29.Linskens RK, Mallant-Hent RC, Murillo LS, von Blomberg BM, Alizadeh BZ, Pena AS. Genetic and serological markers to identify phenotypic subgroups in a Dutch Crohn’s disease population. Dig Liver Dis. 2004;36:29–34. doi: 10.1016/j.dld.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Vavassori P, Borgiani P, Biancone L, et al. CARD15 mutation analysis in an Italian population: Leu1007fsinsC but neither Arg702Trp nor Gly908Arg mutations are associated with Crohn’s disease. Inflamm Bowel Dis. 2004;10:116–21. doi: 10.1097/00054725-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 31.Buning C, Genschel J, Buhner S, et al. Mutations in the NOD2/CARD15 gene in Crohn’s disease are associated with ileocecal resection and are a risk factor for reoperation. Aliment Pharmacol Ther. 2004;19:1073–8. doi: 10.1111/j.1365-2036.2004.01967.x. [DOI] [PubMed] [Google Scholar]
- 32.Yamazaki K, Takazoe M, Tanaka T, Kazumori T, Nakamura Y. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn’s disease. J Hum Genet. 2002;47:469–72. doi: 10.1007/s100380200067. (Erratum in 2003;48:397). [DOI] [PubMed] [Google Scholar]
- 33.Inoue N, Tamura K, Kinouchi Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology. 2002;123:86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 34.Heresbach D, Gicquel-Douabin V, Birebent B, et al. NOD2/CARD15 gene polymorphisms in Crohn’s disease: A genotype-phenotype analysis. Eur J Gastroenterol Hepatol. 2004;16:55–62. doi: 10.1097/00042737-200401000-00009. [DOI] [PubMed] [Google Scholar]
- 35.Cuthbert AP, Fisher SA, Mirza MM, et al. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology. 2002;122:867–74. doi: 10.1053/gast.2002.32415. [DOI] [PubMed] [Google Scholar]
- 36.Abreu MT, Taylor KD, Lin YC, et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn’s disease. Gastroenterology. 2002;123:679–88. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Z, Lin XY, Akolkar PN, et al. Variation at NOD2/CARD15 in familial and sporadic cases of Crohn’s disease in the Ashkenazi Jewish population. Am J Gastroenterol. 2002;97:3095–101. doi: 10.1111/j.1572-0241.2002.07105.x. [DOI] [PubMed] [Google Scholar]
- 38.Cavanaugh JA, Adams KE, Quak EJ, et al. CARD15/NOD2 risk alleles in the development of Crohn’s disease in the Australian population Ann Hum Genet 200367Pt 135–41. [DOI] [PubMed] [Google Scholar]
- 39.Bonen DK, Ogura Y, Nicolae DL, et al. Crohn’s disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology. 2003;124:140–7. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 40.Hampe J, Cuthbert A, Croucher PJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–8. doi: 10.1016/S0140-6736(00)05063-7. (Erratum in 2002;360:806). [DOI] [PubMed] [Google Scholar]
- 41.Vavassori P, Borgiani P, D’Apice MR, et al. 3020insC mutation within the NOD2 gene in Crohn’s disease: Frequency and association with clinical pattern in an Italian population Dig Liver Dis 200234153(Lett) [DOI] [PubMed] [Google Scholar]
- 42.Vermeire S, Wild G, Kocher K, et al. CARD15 genetic variation in a Quebec population: Prevalence, genotype-phenotype relationship, and haplotype structure. Am J Hum Genet. 2002;71:74–83. doi: 10.1086/341124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esters N, Pierik M, van Steen K, et al. Transmission of CARD15 (NOD2) variants within families of patients with inflammatory bowel disease. Am J Gastroenterol. 2004;99:299–305. doi: 10.1111/j.1572-0241.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- 44.Guo QS, Xia B, Jiang Y, Qu Y, Li J. NOD2 3020insC frameshift mutation is not associated with inflammatory bowel disease in Chinese patients of Han nationality. World J Gastroenterol. 2004;10:1069–71. doi: 10.3748/wjg.v10.i7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Croucher PJ, Mascheretti S, Hampe J, et al. Haplotype structure and association to Crohn’s disease of CARD15 mutations in two ethnically divergent populations. Eur J Hum Genet. 2003;11:6–16. doi: 10.1038/sj.ejhg.5200897. [DOI] [PubMed] [Google Scholar]
- 46.Leong RW, Armuzzi A, Ahmad T, et al. NOD2/CARD15 gene polymorphisms and Crohn’s disease in the Chinese population. Aliment Pharmacol Ther. 2003;17:1465–70. doi: 10.1046/j.1365-2036.2003.01607.x. [DOI] [PubMed] [Google Scholar]
- 47.Karban A, Dagan E, Eliakim R, et al. Prevalence and significance of mutations in the familial Mediterranean fever gene in patients with Crohn’s disease. Genes Immun. 2005;6:134–9. doi: 10.1038/sj.gene.6364156. [DOI] [PubMed] [Google Scholar]
- 48.Zouiten-Mekki L, Zaouali H, Boubaker J, et al. CARD15/NOD2 in a Tunisian population with Crohn’s disease. Dig Dis Sci. 2005;50:130–5. doi: 10.1007/s10620-005-1290-0. [DOI] [PubMed] [Google Scholar]
- 49.Zaahl MG, Winter T, Warnich L, Kotze MJ. Analysis of the three common mutations in the CARD15 gene (R702W, G908R and 1007fs) in South African colored patients with inflammatory bowel disease. Mol Cell Probes. 2005;19:278–81. doi: 10.1016/j.mcp.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn’s disease risk and phenotype in diverse populations: A metaanalysis. Am J Gastroenterol. 2004;99:2393–404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 51.Lesage S, Zouali H, Cezard JP, et al. EPWG-IBD Group. EPIMAD Group. GETAID Group CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–57. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brant SR, Picco MF, Achkar JP, et al. Defining complex contributions of NOD2/CARD15 gene mutations, age at onset, and tobacco use on Crohn’s disease phenotypes. Inflamm Bowel Dis. 2003;9:281–9. doi: 10.1097/00054725-200309000-00001. [DOI] [PubMed] [Google Scholar]
- 53.Helio T, Halme L, Lappalainen M, et al. CARD15/NOD2 gene variants are associated with familially occurring and complicated forms of Crohn’s disease. Gut. 2003;52:558–62. doi: 10.1136/gut.52.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hampe J, Grebe J, Nikolaus S, et al. Association of NOD2 (CARD 15) genotype with clinical course of Crohn’s disease: A cohort study. Lancet. 2002;359:1661–5. doi: 10.1016/S0140-6736(02)08590-2. [DOI] [PubMed] [Google Scholar]
- 55.Radlmayr M, Torok HP, Martin K, Folwaczny C.The c-insertion mutation of the NOD2 gene is associated with fistulizing and fibrostenotic phenotypes in Crohn’s disease Gastroenterology 20021222091–2.(Lett) [DOI] [PubMed] [Google Scholar]
- 56.Rioux JD, Silverberg MS, Daly MJ, et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet. 2000;66:1863–70. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma Y, Ohmen JD, Li Z, et al. A genome-wide search identifies potential new susceptibility loci for Crohn’s disease. Inflamm Bowel Dis. 1999;5:271–8. doi: 10.1097/00054725-199911000-00005. [DOI] [PubMed] [Google Scholar]
- 58.Rioux JD, Daly MJ, Silverberg MS, et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–8. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 59.Negoro K, McGovern DP, Kinouchi Y, et al. Analysis of the IBD5 locus and potential gene-gene interactions in Crohn’s disease. Gut. 2003;52:541–6. doi: 10.1136/gut.52.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirza MM, Fisher SA, King K, et al. Genetic evidence for interaction of the 5q31 cytokine locus and the CARD15 gene in Crohn disease. Am J Hum Genet. 2003;72:1018–22. doi: 10.1086/373880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armuzzi A, Ahmad T, Ling KL, et al. Genotype-phenotype analysis of the Crohn’s disease susceptibility haplotype on chromosome 5q31. Gut. 2003;52:1133–9. doi: 10.1136/gut.52.8.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giallourakis C, Stoll M, Miller K, et al. IBD5 is a general risk factor for inflammatory bowel disease: Replication of association with Crohn disease and identification of a novel association with ulcerative colitis. Am J Hum Genet. 2003;73:205–11. doi: 10.1086/376417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peltekova VD, Wintle RF, Rubin LA, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–5. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 64.Yabuuchi H, Tamai I, Nezu J, et al. Novel membrane transporter OCTN1 mediates multispecific, bidirectional, and pH-dependent transport of organic cations. J Pharmacol Exp Ther. 1999;289:768–73. [PubMed] [Google Scholar]
- 65.Newman B, Siminovitch KA. Recent advances in the genetics of inflammatory bowel disease. Curr Opin Gastroenterol. 2005;21:401–7. [PubMed] [Google Scholar]
- 66.Noble CL, Nimmo ER, Drummond H, et al. The contribution of OCTN1/2 variants within the IBD5 locus to disease susceptibility and severity in Crohn’s disease. Gastroenterology. 2005;129:1854–64. doi: 10.1053/j.gastro.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 67.Vermeire S, Pierik M, Hlavaty T, et al. Association of organic cation transporter risk haplotype with perianal penetrating Crohn’s disease but not with susceptibility to IBD. Gastroenterology. 2005;129:1845–53. doi: 10.1053/j.gastro.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 68.Silverberg MS, Lewinger JP, Walters TD, Sherman PS, Griffiths AM. The IBD5 (5q31) locus is strongly associated with pediatric onset inflammatory bowel disease. Gastroenterology. 2005;128 [Google Scholar]
- 69.Silverberg MS, Cho JH, Duerr RH, et al. Association of the IBD5 locus With inflammatory bowel disease in a very large north American population Gastroenterology 20051284 Supp 2A-137(Abst) [Google Scholar]
- 70.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–32. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 71.McGovern DP, Van Heel DA, Negoro K, Ahmad T, Jewell DP.Further evidence of IBD5/CARD15 (NOD2) epistasis in the susceptibility to ulcerative colitis Am J Hum Genet 2003731465–6.(Lett) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dechairo B, Dimon C, van Heel D, et al. Replication and extension studies of inflammatory bowel disease susceptibility regions confirm linkage to chromosome 6p (IBD3) Eur J Hum Genet. 2001;9:627–33. doi: 10.1038/sj.ejhg.5200687. [DOI] [PubMed] [Google Scholar]
- 73.Hampe J, Schreiber S, Shaw SH, et al. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet. 1999;64:808–16. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hampe J, Shaw SH, Saiz R, et al. Linkage of inflammatory bowel disease to human chromosome 6p. Am J Hum Genet. 1999;65:1647–55. doi: 10.1086/302677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Heel DA, Fisher SA, Kirby A, Daly MJ, Rioux JD, Lewis CM, Genome Scan Meta-Analysis Group of the IBD International Genetics Consortium Inflammatory bowel disease susceptibility loci defined by genome scan meta-analysis of 1952 affected relative pairs. Hum Mol Genet. 2004;13:763–70. doi: 10.1093/hmg/ddh090. [DOI] [PubMed] [Google Scholar]
- 76.Yap LM, Ahmad T, Jewell DP. The contribution of HLA genes to IBD susceptibility and phenotype. Best Pract Res Clin Gastroenterol. 2004;18:577–96. doi: 10.1016/j.bpg.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 77.Yang H, Plevy SE, Taylor K, et al. Linkage of Crohn’s disease to the major histocompatibility complex region is detected by multiple non-parametric analyses. Gut. 1999;44:519–26. doi: 10.1136/gut.44.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reinshagen M, Loeliger C, Kuehnl P, et al. HLA class II gene frequencies in Crohn’s disease: A population based analysis in Germany. Gut. 1996;38:538–42. doi: 10.1136/gut.38.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danze PM, Colombel JF, Jacquot S, et al. Association of HLA class II genes with susceptibility to Crohn’s disease. Gut. 1996;39:69–72. doi: 10.1136/gut.39.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trachtenberg EA, Yang H, Hayes E, et al. HLA class II haplotype associations with inflammatory bowel disease in Jewish (Ashkenazi) and non-Jewish caucasian populations. Hum Immunol. 2000;61:326–33. doi: 10.1016/s0198-8859(99)00134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Annese V, Piepoli A, Latiano A, et al. HLA-DRB1 alleles may influence disease phenotype in patients with inflammatory bowel disease: A critical reappraisal with review of the literature Dis Colon Rectum 20054857–64.discussion 64–5. [DOI] [PubMed] [Google Scholar]
- 82.Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ. HLA-DR and -DQ phenotypes in inflammatory bowel disease: A meta-analysis. Gut. 1999;45:395–401. doi: 10.1136/gut.45.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahmad T, Armuzzi A, Neville M, et al. The contribution of human leucocyte antigen complex genes to disease phenotype in ulcerative colitis. Tissue Antigens. 2003;62:527–35. doi: 10.1046/j.1399-0039.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 84.Futami S, Aoyama N, Honsako Y, et al. HLA-DRB1*1502 allele, subtype of DR15, is associated with susceptibility to ulcerative colitis and its progression. Dig Dis Sci. 1995;40:814–8. doi: 10.1007/BF02064985. [DOI] [PubMed] [Google Scholar]
- 85.Yoshitake S, Kimura A, Okada M, Yao T, Sasazuki T. HLA class II alleles in Japanese patients with inflammatory bowel disease. Tissue Antigens. 1999;53(4 Pt 1):350–8. doi: 10.1034/j.1399-0039.1999.530405.x. [DOI] [PubMed] [Google Scholar]
- 86.Myung SJ, Yang SK, Jung HY, et al. HLA-DRB1*1502 confers susceptibility to ulcerative colitis, but is negatively associated with its intractability: A Korean study. Int J Colorectal Dis. 2002;17:233–7. doi: 10.1007/s00384-001-0381-4. [DOI] [PubMed] [Google Scholar]
- 87.Parkes M, Satsangi J, Lathrop GM, Bell JI, Jewell DP.Susceptibility loci in inflammatory bowel disease Lancet 19963481588(Lett) [DOI] [PubMed] [Google Scholar]
- 88.Bayless TM, Tokayer AZ, Polito JM, II, Quaskey SA, Mellits ED, Harris ML. Crohn’s disease: Concordance for site and clinical type in affected family members – Potential hereditary influences. Gastroenterology. 1996;111:573–9. doi: 10.1053/gast.1996.v111.pm8780559. [DOI] [PubMed] [Google Scholar]
- 89.Colombel JF, Grandbastien B, Gower-Rousseau C, et al. Clinical characteristics of Crohn’s disease in 72 families. Gastroenterology. 1996;111:604–7. doi: 10.1053/gast.1996.v111.pm8780563. [DOI] [PubMed] [Google Scholar]
- 90.Annese V, Andreoli A, Astegiano M, et al. Clinical features in familial cases of Crohn’s disease and ulcerative colitis in Italy: A GISC study. Italian Study Group for the Disease of Colon and Rectum. Am J Gastroenterol. 2001;96:2939–45. doi: 10.1111/j.1572-0241.2001.04685.x. [DOI] [PubMed] [Google Scholar]
- 91.Fernandez L, Mendoza JL, Martinez A, et al. IBD1 and IBD3 determine location of Crohn’s disease in the Spanish population. Inflamm Bowel Dis. 2004;10:715–22. doi: 10.1097/00054725-200411000-00004. [DOI] [PubMed] [Google Scholar]
- 92.Yamamoto-Furusho JK, Uscanga LF, Vargas-Alarcon G, et al. Clinical and genetic heterogeneity in Mexican patients with ulcerative colitis. Hum Immunol. 2003;64:119–23. doi: 10.1016/s0198-8859(02)00772-3. [DOI] [PubMed] [Google Scholar]
- 93.de la Concha EG, Fernandez-Arquero M, Martinez A, et al. Amino acid polymorphism at residue 71 in HLA-DR beta chain plays a critical role in susceptibility to ulcerative colitis. Dig Dis Sci. 1999;44:2324–9. doi: 10.1023/a:1026629409481. [DOI] [PubMed] [Google Scholar]
- 94.Roussomoustakaki M, Satsangi J, Welsh K, et al. Genetic markers may predict disease behavior in patients with ulcerative colitis. Gastroenterology. 1997;112:1845–53. doi: 10.1053/gast.1997.v112.pm9178675. [DOI] [PubMed] [Google Scholar]
- 95.Bouma G, Crusius JB, Garcia-Gonzalez MA, et al. Genetic markers in clinically well defined patients with ulcerative colitis (UC) Clin Exp Immunol. 1999;115:294–300. doi: 10.1046/j.1365-2249.1999.00797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Silverberg MS, Mirea L, Bull SB, et al. A population- and family-based study of Canadian families reveals association of HLA DRB1*0103 with colonic involvement in inflammatory bowel disease. Inflamm Bowel Dis. 2003;9:1–9. doi: 10.1097/00054725-200301000-00001. [DOI] [PubMed] [Google Scholar]
- 97.Orchard TR, Chua CN, Ahmad T, Cheng H, Welsh KI, Jewell DP. Uveitis and erythema nodosum in inflammatory bowel disease: Clinical features and the role of HLA genes. Gastroenterology. 2002;123:714–8. doi: 10.1053/gast.2002.35396. [DOI] [PubMed] [Google Scholar]
- 98.Orchard TR, Thiyagaraja S, Welsh KI, Wordsworth BP, Hill Gaston JS, Jewell DP. Clinical phenotype is related to HLA genotype in the peripheral arthropathies of inflammatory bowel disease. Gastroenterology. 2000;118:274–8. doi: 10.1016/s0016-5085(00)70209-5. [DOI] [PubMed] [Google Scholar]
- 99.Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376–88. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 100.van Heel DA, Udalova IA, De Silva AP, et al. Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCTN1 and NF(-kappa)B transcription factors. Hum Mol Genet. 2002;11:1281–9. doi: 10.1093/hmg/11.11.1281. [DOI] [PubMed] [Google Scholar]
- 101.O’Callaghan NJ, Adams KE, van Heel DA, Cavanaugh JA. Association of TNF-alpha-857C with inflammatory bowel disease in the Australian population. Scand J Gastroenterol. 2003;38:533–4. [PubMed] [Google Scholar]
- 102.Negoro K, Kinouchi Y, Hiwatashi N, et al. Crohn’s disease is associated with novel polymorphisms in the 5′-flanking region of the tumor necrosis factor gene. Gastroenterology. 1999;117:1062–8. doi: 10.1016/s0016-5085(99)70390-2. [DOI] [PubMed] [Google Scholar]
- 103.Mascheretti S, Hampe J, Kuhbacher T, et al. Pharmacogenetic investigation of the TNF/TNF-receptor system in patients with chronic active Crohn’s disease treated with infliximab. Pharmacogenomics J. 2002;2:127–36. doi: 10.1038/sj.tpj.6500091. [DOI] [PubMed] [Google Scholar]
- 104.Louis E, Peeters M, Franchimont D, et al. Tumour necrosis factor (TNF) gene polymorphism in Crohn’s disease (CD): Influence on disease behaviour? Clin Exp Immunol. 2000;119:64–8. doi: 10.1046/j.1365-2249.2000.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ferreira AC, Almeida S, Tavares M, et al. NOD2/CARD15 and TNFA, but not IL1B and IL1RN, are associated with Crohn’s disease. Inflamm Bowel Dis. 2005;11:331–9. doi: 10.1097/01.mib.0000158153.71579.b4. [DOI] [PubMed] [Google Scholar]
- 106.Levine A, Karban A, Eliakim R, et al. A polymorphism in the TNF-alpha promoter gene is associated with pediatric onset and colonic location of Crohn’s disease. Am J Gastroenterol. 2005;100:407–13. doi: 10.1111/j.1572-0241.2005.41126.x. [DOI] [PubMed] [Google Scholar]
- 107.Stoll M, Corneliussen B, Costello CM, et al. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476–80. doi: 10.1038/ng1345. [DOI] [PubMed] [Google Scholar]
- 108.Yamazaki K, Takazoe M, Tanaka T, et al. Association analysis of SLC22A4, SLC22A5 and DLG5 in Japanese patients with Crohn disease. J Hum Genet. 2004;49:664–8. doi: 10.1007/s10038-004-0204-x. [DOI] [PubMed] [Google Scholar]
- 109.Daly MJ, Pearce AV, Farwell L, et al. Association of DLG5 R30Q variant with inflammatory bowel disease. Eur J Hum Genet. 2005;13:835–9. doi: 10.1038/sj.ejhg.5201403. [DOI] [PubMed] [Google Scholar]
- 110.Torok HP, Glas J, Tonenchi L, et al. Polymorphisms in the DLG5 and OCTN cation transporter genes in Crohn’s disease. Gut. 2005;54:1421–7. doi: 10.1136/gut.2005.066340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Noble CL, Nimmo ER, Drummond H, Smith L, Arnott ID, Satsangi J. DLG5 variants do not influence susceptibility to inflammatory bowel disease in the Scottish population. Gut. 2005;54:1416–1420. doi: 10.1136/gut.2005.066621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Panwala CM, Jones JC, Viney JL. A novel model of inflammatory bowel disease: Mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J Immunol. 1998;161:5733–44. [PubMed] [Google Scholar]
- 113.Langmann T, Moehle C, Mauerer R, et al. Loss of detoxification in inflammatory bowel disease: Dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26–40. doi: 10.1053/j.gastro.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 114.Satsangi J, Parkes M, Louis E, et al. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 115.Ho GT, Nimmo ER, Tenesa A, et al. Allelic variations of the multidrug resistance gene determine susceptibility and disease behavior in ulcerative colitis. Gastroenterology. 2005;128:288–96. doi: 10.1053/j.gastro.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 116.Glas J, Torok HP, Schiemann U, Folwaczny C.MDR1 gene polymorphism in ulcerative colitis Gastroenterology 2004126367(Lett) [DOI] [PubMed] [Google Scholar]
- 117.Schwab M, Schaeffeler E, Marx C, et al. Association between the C3435T MDR1 gene polymorphism and susceptibility for ulcerative colitis. Gastroenterology. 2003;124:26–33. doi: 10.1053/gast.2003.50010. [DOI] [PubMed] [Google Scholar]
- 118.Brant SR, Panhuysen CI, Nicolae D, et al. MDR1 Ala893 polymorphism is associated with inflammatory bowel disease Am J Hum Genet 2003731282–92.(Erratum in 2004;74:1080). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Croucher PJ, Mascheretti S, Foelsch UR, Hampe J, Schreiber S.Lack of association between the C3435T MDR1 gene polymorphism and inflammatory bowel disease in two independent Northern European populations Gastroenterology 20031251919–20.(Lett) [DOI] [PubMed] [Google Scholar]
- 120.Gazouli M, Zacharatos P, Gorgoulis V, Mantzaris G, Papalambros E, Ikonomopoulos J.The C3435T MDR1 gene polymorphism is not associated with susceptibility for ulcerative colitis in Greek population Gastroenterology 2004126367–9.(Lett) [DOI] [PubMed] [Google Scholar]
- 121.Kugathasan S, Collins N, Maresso K, et al. CARD15 gene mutations and risk for early surgery in pediatric-onset Crohn’s disease. Clin Gastroenterol Hepatol. 2004;2:1003–9. doi: 10.1016/s1542-3565(04)00452-5. [DOI] [PubMed] [Google Scholar]
- 122.Ho GT, Lees C, Satsangi J. Pharmacogenetics and inflammatory bowel disease: Progress and prospects. Inflamm Bowel Dis. 2004;10:148–58. doi: 10.1097/00054725-200403000-00014. [DOI] [PubMed] [Google Scholar]
- 123.Vogel F. Moderne probleme der Humangenetik. Ergeb Inn Med Kinderheilkd. 1959;12:52–125. [Google Scholar]
- 124.Kalow W, Tang BK, Endrenyi L. Hypothesis: Comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8:283–9. doi: 10.1097/00008571-199808000-00001. [DOI] [PubMed] [Google Scholar]
- 125.Colombel JF, Ferrari N, Debuysere H, et al. Genotypic analysis of thiopurine S-methyltransferase in patients with Crohn’s disease and severe myelosuppression during azathioprine therapy. Gastroenterology. 2000;118:1025–30. doi: 10.1016/s0016-5085(00)70354-4. [DOI] [PubMed] [Google Scholar]
- 126.Kader HA, Wenner WJ, Jr, Telega GW, Maller ES, Baldassano RN. Normal thiopurine methyltransferase levels do not eliminate 6-mercaptopurine or azathioprine toxicity in children with inflammatory bowel disease. J Clin Gastroenterol. 2000;30:409–13. doi: 10.1097/00004836-200006000-00011. [DOI] [PubMed] [Google Scholar]
- 127.Black AJ, McLeod HL, Capell HA, et al. Thiopurine methyltransferase genotype predicts therapy-limiting severe toxicity from azathioprine. Ann Intern Med. 1998;129:716–8. doi: 10.7326/0003-4819-129-9-199811010-00007. [DOI] [PubMed] [Google Scholar]
- 128.Spire-Vayron de la Moureyre C, Debuysere H, Mastain B, et al. Genotypic and phenotypic analysis of the polymorphic thiopurine S-methyltransferase gene (TPMT) in a European population. Br J Pharmacol. 1998;125:879–87. doi: 10.1038/sj.bjp.0702152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Farrell RJ, Murphy A, Long A, et al. High multidrug resistance (P-glycoprotein 170) expression in inflammatory bowel disease patients who fail medical therapy. Gastroenterology. 2000;118:279–88. doi: 10.1016/s0016-5085(00)70210-1. [DOI] [PubMed] [Google Scholar]
- 130.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;97:3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sakaeda T, Nakamura T, Okumura K. Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics. 2003;4:397–410. doi: 10.1517/phgs.4.4.397.22747. [DOI] [PubMed] [Google Scholar]


