Abstract
Whole-cell quantitative proteomic analyses were conducted to investigate the change from an extracellular to intracellular lifestyle for Porphyromonas gingivalis, a Gram-negative intracellular pathogen associated with periodontal disease. Global protein abundance data for P. gingivalis strain ATCC 33277 internalized for 18 hours within human gingival epithelial cells and controls exposed to gingival cell culture medium were obtained at sufficient coverage to provide strong evidence that these changes are profound. A total of 385 proteins were over-expressed in internalized P. gingivalis relative to controls; 240 proteins were shown to be under-expressed. This represented in total about 28% of the protein encoding ORFs annotated for this organism, and slightly less than half of the proteins that were observed experimentally. Production of several proteases, including the classical virulence factors RgpA, RgpB, and Kgp, was decreased. A separate validation study was carried out in which a 16-fold dilution of the P. gingivalis proteome was compared to the undiluted sample in order to assess the quantitative false negative rate (all ratios truly alternative). Truly null (no change) abundance ratios from technical replicates were used to assess the rate of quantitative false positives over the entire proteome. A global comparison between the direction of abundance change observed and previously published bioinformatic gene pair predictions for P. gingivalis will assist with future studies of P. gingivalis gene regulation and operon prediction.
Keywords: Porphyromonas gingivalis, protein expression, invasion, internalization, proteomics
1 Introduction
Periodontal diseases are a group of inflammatory conditions that lead to the destruction of the supporting tissues of the teeth and are among the most common infections of humans [1]. Furthermore, an association is emerging between periodontal disease and serious systemic conditions including coronary artery disease and preterm delivery of low birth weight infants [2]. Foremost among a group of periodontal pathogens is the Gram-negative anaerobe Porphyromonas gingivalis. P. gingivalis is a highly invasive intracellular oral pathogen [3] that enters gingival epithelial cells through manipulation of host cell signal transduction and remains resident in the perinuclear area for extended periods without causing host cell death [4]. Indeed, intracellular P. gingivalis cells remain viable and capable of spreading between host cells [5], and epithelial cells can survive for prolonged periods after infection with P. gingivalis [6]. Moreover, epithelial cells recovered from the oral cavity show high levels of intracellular P. gingivalis [7–9], indicating that an intracellular location is an integral component of the lifestyle of this organism and contributes to persistence in the oral cavity.
The interaction of human gingival epithelial cells (GECs) with P. gingivalis is a valuable model system for studying host-pathogen interactions in general and bacterial invasion in particular. The completion of the P. gingivalis strain W83 genome sequence in 2001 [10] allowed the application of global expression measurements to the study of P. gingivalis invasion, including proteomic studies [11]. Here we report the observed proteome of P. gingivalis under two biological conditions: intracellular, consisting of organisms recovered from within gingival epithelial cells; and an extracellular reference state consisting of organisms in epithelial cell culture medium that approximates an extracellular milieu in which epithelial cells are physiologically active.
The experimental approach chosen for these studies was the combination of multiple dimension capillary HPLC and tandem mass spectrometry using a linear ion trap [12]. Since 2001, this separation approach has become widely known in the proteomics community as MudPIT (multidimensional protein identification technology) and has become increasingly popular because of its power to generate a vast amount of searchable data relative to other methods for the measurement of protein abundance [13]. Quantitation of abundance changes has been accomplished using stable isotopes [14] and, more recently, using various non-label approaches [12, 15–17]. The application of these techniques to the study of P. gingivalis invasion and pathogenicity was recently reviewed [11]. Thus far it has not been possible to measure the intracellular proteome of P. gingivalis using stable isotope methods for quantitation due to technical problems with growth media using a well-defined, exclusively 15N nitrogen source. Although generally regarded as the best approach for mass spectrometry-based quantitative proteomics, isotope labeling also has shortcomings, such as increased sample complexity and reduced qualitative proteome coverage [12, 18], especially in global studies where under-sampling is always a major concern. Therefore, an effective system was developed for measuring protein abundance change in P. gingivalis without the use of stable isotopes. A paper describing these quantitative methods based on spectral counting [19, 20] and signal intensity measurements in MS1 has been published [12]. We report our data here based on both types of calculation because it is not clear at present that either approach is better under all conditions. For proteins under approximately 20 kD and (or) proteins that are expressed at low abundance, intensity based measurements seem to be more useful. For proteins that are larger and (or) more abundant, spectral counting methods have shown a high degree of precision [16, 20] but may result in abundance ratios that are more compressed or invariant with relative concentration change compared to those based on ion intensity, even though the inherent dynamic range of spectral counting is believed to be wider than what can be achieved using MudPIT methods with ion intensity-based quantitation [16]. Spectral counting is based on the concept that the frequency with which peptides associated with a particular protein appear during the analysis is an indicator of protein abundance [20].
Previous protein- and mRNA- based reports have suggested that the intracellular phenotype of P. gingivalis differs greatly from the extracellular phenotype observed in the laboratory in terms of general metabolic pathways, adaptation to stress and expression of classical virulence factors [21, 22], see also supplemental Table S4. The data reported here confirm and extend these findings. The quantitative proteomics analysis indicates that there are hundreds of proteins undergoing significant abundance changes during internalization within the model host cells, a situation that can be quite challenging in terms of multiple hypothesis testing, as has long been recognized in the transcription microarray field [23]. Therefore, a separate control study was required to establish reasonable cut-off values for the q-value statistic that served as the final metric for determining significant abundance change [12].
Some of these abundance changes have been observed in previous studies of early stages of the invasion process, prior to internalization [21, 24, 25]. The importance of other proteins showing pronounced abundance changes has been validated by invasion studies using knock-out mutants of P. gingivalis in which the genes in question are not functional. Others have yet to be validated by orthogonal approaches such as functional studies, real time RT-PCR or Northern blot analysis. Because P. gingivalis operon and regulon structures are still largely unknown, we have also compared the directionality of protein abundance change observed experimentally with the bioinformatic gene pair predictions of Ermolaeva et al. [26], who examined spatially and genetically conserved gene pairs over 34 sequenced prokaryotic genomes, including P. gingivalis.
2 Materials and methods
More detailed descriptions of the HPLC fractionation, linear ion trap tandem mass spectrometry, post-acquisition data processing and especially the statistical analysis can be found in [12]. The description given here emphasizes certain refinements that have been made since the previous paper was published, and provides details regarding the biological controls and internalized P. gingivalis cell culturing that have not been published previously.
2.1 Bacterial and gingival epithelial cell culture
P. gingivalis strain ATCC 33277 was cultured anaerobically to mid log phase at 37°C in trypticase soy broth supplemented with yeast extract (1 mg ml −1), hemin (5 μg ml −1), and menadione (1 μg ml −1). HIGK cells (human HPV-immortalized gingival keratinocytes) [27] were cultured under 5% CO2 in keratinocyte serum-free medium (K-SFM, Gibco/Invitrogen, Carlsbad, CA) supplemented with 0.05 mM calcium chloride and 200 mM L-glutamine (Gibco/Invitrogen, Carlsbad, CA).
2.2 Sample preparation
P. gingivalis cells were added to HIGKs at a multiplicity of infection of 50 and incubated for 18h under normal HIGK cell culture conditions. HIGK cells were washed three times with PBS to remove free and surface bound P. gingivalis [3], then lysed in ice-cold sterile distilled H2O and internalized bacteria were recovered by centrifugation. The bacteria were washed in ice-cold sterile distilled H2O and pelleted. These samples will be referred to as biological replicates PG_PP1 and PG_PP2. For the control reference state, P. gingivalis cells were suspended in K-SFM, in which the organism remains metabolically active but does not replicate, and incubated for 18h under normal HIGK cell culture conditions. The bacteria were washed in ice-cold sterile distilled H2O and pelleted. These samples will be referred to as biological replicates PG_PPC1 and PG_PPC2. P. gingivalis pellets of ~ 109 cells were resuspended and lysed. DTT reduction, iodoacetamide alkylation, and trypsin digestion steps were then performed as described [12]. The supernatants from the digestion step were separated into five fractions with a 2.0 mm i.d. × 150 mm YMC polymer C18 S-6 HPLC column (Waters, Milford, MA, USA). Each fraction was concentrated and adjusted to 50 μl with 0.5% acetic acid and 5% acetonitrile (v/v). The insoluble portions were dissolved in urea and RapiGest (Waters), reduced with DTT, alkylated with iodoacetamide, and digested with trypsin (Promega, Madison, WI, USA) then desalted and fractionated as described [12]. Briefly, each of the five pre-fractions was separated further by 2D capillary HPLC, yielding a total of 35 fractions for each technical replicate. Two technical replicates were acquired for each biological replicate. This level of separation and data redundancy was adequate to minimize the impact of differences in sample complexity for the protein abundance ratios given in supplementary Table S1.
2.3 Linear ion trap tandem mass spectrometry
Samples were analyzed as described [12] using a Thermo-Finnigan (San Jose, CA, USA) LTQ linear ion trap and a Michrom Magic 2002 HPLC modified for capillary operation with a 75 μm i.d. biphasic column consisting of an SCX resin (strong cation exchange) and reversed phase C18 material as originally described by Washburn et al. [13, 14] and modified as described [12, 28], except for the 16-fold dilution of PG_PPC2. A threshold of 5,000 counts for collision-induced-dissociation (CID, MS2) was chosen for this experiment rather than the 20,000 data system count threshold reported previously. The MS1 scan range was 400–2000 m/z units. After each main beam (MS1) scan, the 10 most intense signals above threshold were selected for CID scans with one CID scan collected for each of the 10 most abundant precursor ions. Default parameters under the Xcalibur 1.4 data acquisition software (Thermo-Finnigan, San Jose, CA, USA) were used, with the exception of an isolation width of 3.0 m/z units and a normalized collision energy of 40%. Dynamic exclusion was activated during all data acquisition.
2.4 SEQUEST and DTASelect
SEQUEST database matching [29] and DTASelect [30] filtering were performed as described [12]. Briefly, product ion (MS2) mass spectra were searched using TurboSEQUEST Cluster Version 3.2 (Thermo-Finnigan) on a 16-CPU computing cluster (Denali Advanced Integration, Seattle, WA, USA) against a combined fasta database that included bovine, human and P. gingivalis proteins, and the NIH-NCI MGC (National Cancer Institute Mammalian Gene Collection). The total database size was 119 Mbytes, the total number of entries was 38,695 proteins. The P. gingivalis ORF sequences consisted of Version 3.1 of the genome annotation by TIGR described by Nelson et al. [10] and dated June 8, 2001. See also http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=gpg and http://www.oralgen.lanl.gov/. Certain gene descriptors for P. gingivalis differ among the annotations by TIGR, LANL and common usage in the microbiological literature. The present paper uses the most commonly cited gene names in the literature in the event of a nomenclature conflict. In the supplemental Table S1, these ORF numbers have been cross-referenced to those assigned to the same W83 genome sequence by LANL (Los Alamos National Laboratory). The bovine database consisted of two portions, the bovine nrdb subset contained in Bioworks release 3.2 (Thermo-Finnigan) and the UCSC (University of California at Santa Cruz) bovine database, release date February 17, 2005. The human portion consisted of the entire NCBI nrdb human subset included with Bioworks 3.2. The MGC portion consisted of the entire collection of full-length clone sequences in the state of curation that existed on August 17, 2004. DTASelect Version 1.9 filtering criteria: peptides were fully tryptic; ΔCn/Xcorr values for different peptide charge states were 0.08/1.9 for +1, 0.08/2.0 for + 2, and 0.08/3.3 for +3; all spectra detected for each sequence were retained (t = 0 in DTASelect). Three peptides unique to a particular ORF were required for positive identification. This last requirement has its basis in the definition of uniqueness employed by the DTASelect software, in which the same sequence, in theory, could be covered by three different charge states and yield three different unique peptides, yet still fulfill certain definitions of a “one hit” identification. By setting the “distinct peptides” switch in DTASelect to a value of 3, in practice this “one hit” scenario did not happen, and it guaranteed that at least two unique peptide sequences, independent of charge state, were used to identify the protein. Peptides common to two or more ORFs were disregarded for purposes of both identification and quantitation. A positive qualitative identification using the above criteria was required prior to further filtering for purposes of generating abundance ratios. The DTASelect 1.9 filter data for all protein identifications are contained in supplementary Tables S5–S7. Prior to archiving in PRIDE (http://www.ebi.ac.uk/pride/) with the raw mass spectral data, these filter files will be available on the author’s server (http://depts.washington.edu/mhlab/) rather than on the Proteomics web site due to their large size. These files include details such as SEQUEST scores, peptide sequence, percentage of peptide coverage by observed ions in the CID spectrum, spectral counts, and other information described in the headers accompanying the filter files. More detail regarding the type of information contained in the filter files can be found in Tabb et al. [30].
2.5 Estimation of the qualitative random false positive rate
Random false positive assignments at the peptide level were assessed by searching P. gingivalis raw data against a concatenated database consisting of the elements described above and the reversed decoy database [31, 32] for each. The reversed protein sequences were generated using either a script in FileMaker 8 or the routine contained in Bioworks 3.3 (Thermo-Finnigan) that performs this function automatically. Search results that passed the criteria described above and matched an entry in the decoy database were counted as false positives. The qualitative random false positive rate for this study was thus estimated to be approximately 3%.
2.6 Data processing for protein abundance ratios
By referring to a Visual Basic 6 program “makems2,” provided by Michael J. MacCoss (Department of Genome Sciences, University of Washington), a Visual Basic 2005 program using the Xcalibur Raw File ActiveX Control was developed in-house under the Microsoft Visual Studio 2005 environment to convert the raw data files into plain text intensity files containing the file name header, full scan number, CID scan number, precursor m/z, and the intensities of that precursor ion in the surrounding ± 3 full scans (MS1). The source code (IntensityMaker Version 2.0, release date October 15, 2006) is available from the authors. A relational database was constructed in FileMaker Pro 8. The DTASelect-filter file and the intensity file described above were imported into FileMaker Pro 8 as separate tables. Identical multiple criteria relationships between each intensity table and the DTASelect-filter table were established by matching the file name header and the CID scan number. Then, a FileMaker script application, written in-house (QuantScripts, Version 1.0, March 2006), was used to update the intensity field in the DTASelect-filter table. Because the DTASelect-filter to intensity file relationships were defined to always sort the intensity file in descending order of signal intensity, the updated intensity field in the DTASelect-filter table was the value from the highest intensity mass peak for the corresponding precursor ion among the surrounding ± 3 full scans (MS1), or alternatively, 30 scans total (MS1 and MS2). It was these values that were used for the signal intensity based abundance ratio calculations described below, and constituted what in experiments done on a more conventional scale would be called a “peak list.”
2.7 Two relative quantitation methods for generating abundance ratios
The protein level spectral counts, defined here as the number of redundant unique peptides [12] associated with an ORF, were extracted from the DTASelect-filter table for each ORF (see supplemental Tables S5–S7). A redundant unique peptide is one that is specific for the ORF in question that may be measured many times in the course of the analysis, as required by a method that is based on the frequency of occurrence [20]. The ratio of each protein from two comparisons (PG_PP1/PG_PPC1 or PG_PP1/PG_PPC2) was then calculated from the two spectral count values after the normalization steps described below. The protein level intensity method used the intensity values from all unique peptides identified for a given protein, as described in the previous section, including redundant measurements. The sum of these intensity values in MS1 was used to represent the total signal intensity of each protein in the sample.
2.8 Data normalization
Following the normalization procedures described previously [12], PG_PP1 spectral counts were multiplied by 1.68 and 2.73 to normalize the dataset such that the total counts would be equal for the two samples in each of the two comparisons: PG_PP1/PG_PPC1 and PG_PP1/PG_PPC2, respectively. This normalization also had the effect of centering the ratio distributions at approximately zero on a log2 scale. The summed signal intensity over all peptides assigned to each ORF was multiplied by 1.75 for PG_PP1/PG_PPC1 and 2.96 for PG_PP1/PG_PPC2 for the same reasons, to insure that differences in absolute signal intensity due to differences in the amount of sample injected and slight changes in instrumental conditions did not unduly influence the abundance ratios.
2.9 Statistical significance testing and curve fitting
The G statistics, two sample t-test, p-value, and q-value calculations were performed using R (http://www.r-project.org/). The LOWESS (locally weighted scatter plot smoothing) curves [33] were also generated in R. The detailed procedures for use of each of the above statistics have been described [12]. The G-statistic is a likelihood ratio test for significance in discrete datasets [35] that has recently been applied to spectral counting in quantitative, mass spectrometry-based proteomics by several groups [12, 15, 18, 34]. We used a G-statistic as a test of whether or not a protein abundance ratio based on spectral counting was significantly different from zero on a log2 scale (one on a linear scale). The t-test was applied to the peptide signal intensity data for the same purpose.
The q-value as employed here and in our previous methods paper [12] is a correction for multiple hypothesis testing applied to the better known p-value, which is based on the concept of false discovery rate [36]. It was originally developed for transcription microarray data but is also applicable to other types of large-scale gene expression studies. The q-value can be defined in the present context as the minimum false discovery rate when rejecting the null hypothesis of no significant change in protein abundance. In other words, the rate of false positive assignment (abundance is judged to be changing when in truth it is not) expected among all proteins judged to be significantly changed at a given p-value. For a more technical description of the q-value and false discovery rate, see Storey and Tibshirani [36]. The q-value correction was applied to the p-value calculated for each ORF using both the spectral counting and signal intensity calculations, as both the G-test and the t-test yield a p-value that in turn was used as an input into the QVALUE R package, see http://faculty.washington.edu/~jstorey/qvalue/. It is the q-value that is reported in the supplement (Table S1), along with the abundance ratios for each ORF. Methods for assessing the cut-off value of q for purposes of defining a subset of proteins showing significant abundance change are described in the next section. The LOWESS curve fitting procedure [33] has been used for many years to define boundaries in datasets and was used to define regions of random experimental error in abundance ratios calculated using spectral counting. The R source code for the implementation of the LOWESS procedure used here has been published [12].
2.10 Measurement of quantitative false positive and false negative rates for protein abundance ratios
To help assess the reliability of protein abundance ratios generated using the methods described we diluted a sample of the P. gingivalis proteome 16-fold (a convenient power of 2) and compared the observed and expected ratios for the diluted/undiluted sample of PG_PPC2. This was done using both spectral counting and intensity-based calculations without normalization. This comparison was particularly useful for examining false negatives (Type 2 error), given that the true ratio was known in each case to be non-zero on a log2 scale, that is truly alternative. Replicate analyses of the same sample (technical replicates) in which ratios were calculated for the same sample against itself were used to assess random errors using the LOWESS curve fitting procedure described above, which was particularly useful for the assessment of false positive abundance changes (Type 1 error), because the true ratio was zero on a log2 scale in each case, that is truly null. These approaches were used to set the q-value thresholds for significance testing in a way that directly reflects our experimental observations rather than error estimates based primarily on statistical theory, see Table 1. Cut-off values of q were selected to minimize both Type 1 and Type 2 error. In Table 1, the FPRs (false positive rates) were calculated by dividing the number of proteins called as alternative by all proteins tested at a given q-value cutoff, when all proteins were known to be truly null. Similarly, FNRs (false negative rates) were calculated by dividing the number of proteins called as null by all proteins tested when all proteins were known to be truly alternative.
Table 1.
Relation of different q-value cut-offs to the quantitative false positive rate (FPR) and false negative rate (FNR). The numbers in boldface were chosen to select the q-value cut-offs.
| q cut-off | Spectral Counting Ratio | Signal Intensity Ratio | ||
|---|---|---|---|---|
| PG_PPC2 run1/run2 | PG_PPC2/PG_PPC2_dilute 16 | PG_PPC2 run1/run2 | PG_PPC2/PG_PPC2_dilute 16 | |
| FPR (%)1 | FNR (%)2 | FPR (%)1 | FNR (%)2 | |
| 0.1 | 2.8 | 55 | 5 | 1.4 |
| 0.08 | 2.7 | 56 | 4.6 | 1.4 |
| 0.06 | 2.5 | 58 | 4.2 | 1.4 |
| 0.04 | 2.5 | 61 | 4.0 | 1.4 |
| 0.02 | 2.2 | 65 | 3.3 | 1.4 |
| 0.01 | 1.6 | 69 | 3.0 | 1.4 |
| 0.005 | 1.0 | 71 | 2.5 | 1.5 |
| 0.002 | 0.8 | 74 | 2.2 | 2.0 |
| 0.001 | 0.7 | 76 | 1.8 | 2.8 |
| 0.0005 | 0.7 | 77 | 1.6 | 4.1 |
| 0.0002 | 0.7 | 79 | 1.1 | 7.3 |
| 0.0001 | 0.7 | 81 | 1.0 | 9.3 |
| 0.00005 | 0.7 | 82 | 0.8 | 11.5 |
| 0.00002 | 0.6 | 84 | 0.7 | 14.5 |
| 0.00001 | 0.5 | 84 | 0.6 | 17.2 |
| 0.000005 | 0.5 | 85 | 0.6 | 19.7 |
| 0.000002 | 0.5 | 86 | 0.6 | 23.0 |
| 0.000001 | 0.5 | 87 | 0.6 | 25.4 |
Technical replicates of PG_PPC2, one of the two controls, in which all abundance ratios should represent random scatter about a true change of zero. The q-value cut-offs were not sensitive to sample or replicate identity, but they were highly sensitive to the quantitation approach employed, see text discussion.
PG_PPC2 and a 16-fold dilution of the same sample, in which all abundance ratios should be true non-zeros.
2.11 Calculation of codon bias
Codon adaptation index (CAI) and Karlin Index, E(g), were calculated for each P. gingivalis ORF as previously described. CAI [37] is a general predictor of gene expression levels, as is the Karlin index [38, 39]. This treatment of CAI and E(g) follows that of our previous work with the Archaeon Methanococcus maripaludis [28]. CAI and E(g) differ mainly in the way highly expressed genes are chosen for the reference set and the codon relative frequency calculation. The calculation of CAI involves a defined set of highly expressed genes (HEX), usually about 24 ORFs. Then, the relative frequency of each codon (rf) is calculated within this subset: the observed frequency of each codon in the HEX subset divided by the highest observed frequency within the set of codons for each amino acid. The CAI value for each ORF is then calculated as CAI = (Πrfi)1/L, where L is the total number of encoding codons for each ORF and the subscript i is used to designate each encoding codon. In other words, the geometric mean of rf values for each codon used to encode the entire protein. In Karlin’s E(g) calculation, the codon bias (B) of the target genes relative to the whole genome (C), and the target genes relative to three groups of highly expressed genes (usually ribosomal proteins (RPs), translational elongation factors (TFs) and chaperones (CHs)), are first calculated separately as B(g|C), B(g|RP), B(g|TF), and B(g|CH) [38, 39]. Then E(g) is calculated as E(g) = B(g|C) / (0.5 × B(g|RP) + 0.25 × B(g|TF) + 0.25 × B(g|CH)).
3 Results and discussion
3.1 Qualitative proteome coverage
A total of 1,223 P. gingivalis proteins were qualitatively identified in PG_PP1, PG_PPC1 and PG_PPC2: 987 proteins were identified in PG_PP1, 1068 proteins in PG_PPC1 and 1074 proteins in PG_PPC2. If we take into consideration the genome sequence differences between strains ATCC 33277 and W83 noted in the literature [40] and also the existence of several hundred annotated interposons and other mobile elements [10] that are typically not observed experimentally as proteins, our actual proteome qualitative coverage was on the order of 60% of the protein expressing ORFs. This was significantly higher coverage relative to other literature reports for intracellular pathogens. For example, in a recent study of Salmonella enterica serovar Typhimurium recovered from macrophages, 315 proteins were detected and 39 were described as differentially regulated [42].
About 7% of the genes in 33277 are reported to be highly divergent relative to the sequence strain, W83 [40]. As expected, these same highly divergent genes were under-represented in our dataset as expressed protein. Our ORF database used to search the mass spectral data was of necessity based on the annotation of the sequence strain, with the addition of the FimA sequence from 33277 [41]. Of 64 highly divergent genes noted for 33277 relative to the sequence strain, 16 were detected as expressed protein. Of the 16 highly divergent ORFs among four strains of P. gingivalis selected for PCR confirmation by Chen et al. [40], we detected two: PG0111 and PG0683.
3.2 Overview and functional classes
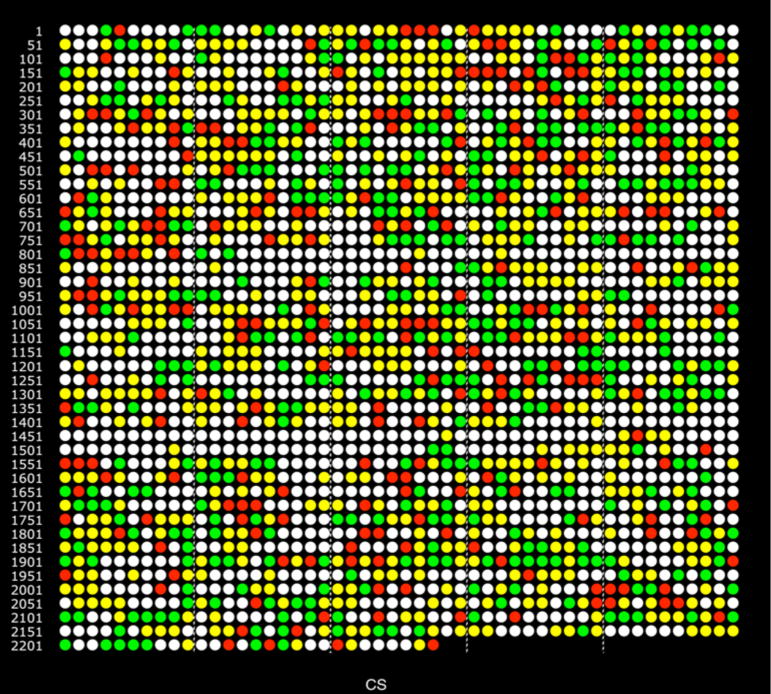
Overall, 385 over-expressed and 240 under-expressed proteins from internalized P. gingivalis were observed relative to controls, based on the consensus (CS) assignment for each ORF presented in the data summary given in Table S1 of the supplement. Two PDF versions of Table S1 are given, one in which the ORFs are sorted by direction of abundance change, and an alternative sorting by ORF order without regard to abundance status for all detected proteins. The color-coding of the CS variable was determined by examining the q-values from both comparisons and the two calculation methods, giving four inputs into the consensus calculation (SP1, SP2, IP1, IP2), also shown in Table S1: SP1 = PG_PP1/PG_PPC1 by spectral counting, SP2 = PG_PP1/PG_PPC2 by spectral counting, IP1 = PG_PP1/PG_PPC1 by summed signal intensity, IP2 = PG_PP1/PG_PPC2 by summed signal intensity. If at least two out of four ratios indicated the same direction of significant change, then the consensus was that direction, green for over-expression and red for under-expression. For the great majority of the results given in Table S1 the abundance trends were the same for all four ratios, but not necessarily significantly so according to the statistical analysis. Taking two reds or two greens to code the CS variable gave the best balance between throwing away too much data (quantitative false negative error) and retaining too many significant abundance changes (false positive error). More detail regarding the coding of the consensus assignments can be found in the supplement, see the notes for Table S1 and Fig. S1. In the five ORFs shown in Table S1 with a split decision, i.e. two reds and two greens, the consensus was coded as yellow: protein detected qualitatively but ambiguous with respect to abundance change. A whole proteome color ORF plot of the CS variable is given in Fig. 1. Similar color plots for SP1, SP2, IP1, and IP2 are given as supplementary Figures S2–S5.
Figure 1.
Genomic representation of the proteome, each circle represents an ORF in the order it is encoded in the genome. TIGR ORF PG0001 in the upper left, PG2227 second from the left, end of the bottom row. The last ORF is P13793, the FMA protein specific to strain ATCC 33277 [41]. Colored circles show the consensus result (CS) for the combined proteome analyses of two biological replicates using two calculation methods, see supplementary Table S1. Green indicates over-expression in PG_PP1, the internalized phenotype; red, under-expression; yellow, protein was identified qualitatively, but without statistically significant abundance level change; white, no protein was detected qualitatively from this locus.
Table 2 describes observed P. gingivalis protein abundance changes during internalization among TIGR functional role categories. A number of proteins in all functional classes showed evidence of differential abundance in internalized P. gingivalis, indicating that the phenotype of intracellular P. gingivalis differs significantly from that of bacteria in laboratory culture. This represents a paradigm shift in our understanding of the pathobiology of the organism, as discussed further below. An interesting observation was that while most functional classes contained approximately equal numbers of under- or over-expressed proteins, the Protein Synthesis class contained over 6-fold more over-expressed proteins compared to under-expressed, and the Transcription class contained 3-fold more over-expressed proteins. This is consistent with epithelial cell culture medium not being permissive to P. gingivalis replication, and further, shows that the intracellular environment of gingival epithelial cells is conducive to P. gingivalis growth and replication, in accord with results from intracellular survival assays [3].
Table 2.
ORF totals for each TIGR functional role category. Over-expressed (up), under-expressed (down), and unchanging protein abundance ratios (no change) in internalized P. gingivalis (PG_PP1) versus both controls according to the consensus trends (CS) in Table S1 and Fig. 1. The total numbers have some redundancy because TIGR has listed certain ORFs in multiple functional categories.
| Functional class | TIGR | Up in PP | Down in PP | No change | Changed (%) | Total |
|---|---|---|---|---|---|---|
| Energy metabolism | 126 | 39 | 33 | 44 | 57 | 116 |
| Fatty acid and phospholipid metabolism | 16 | 6 | 4 | 4 | 63 | 14 |
| Purines, pyrimidines, nucleosides and nucleotides | 44 | 14 | 10 | 19 | 55 | 43 |
| Amino acid biosynthesis | 18 | 5 | 2 | 8 | 39 | 15 |
| Protein synthesis | 114 | 56 | 9 | 32 | 57 | 97 |
| Transcription | 32 | 9 | 3 | 13 | 38 | 25 |
| Protein fate | 75 | 25 | 18 | 25 | 57 | 68 |
| DNA metabolism | 75 | 19 | 6 | 31 | 33 | 56 |
| Unknown function | 198 | 41 | 26 | 76 | 34 | 143 |
| Cellular processes | 50 | 16 | 7 | 20 | 46 | 43 |
| Central intermediary metabolism | 24 | 5 | 3 | 11 | 33 | 19 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 74 | 19 | 8 | 32 | 36 | 59 |
| Hypothetical proteins-conserved | 197 | 37 | 20 | 69 | 29 | 126 |
| Transport and binding proteins | 110 | 14 | 15 | 45 | 26 | 74 |
| Cell envelope | 119 | 32 | 13 | 48 | 38 | 93 |
| Hypothetical proteins | 808 | 50 | 62 | 112 | 14 | 224 |
| Regulatory functions | 44 | 8 | 5 | 11 | 30 | 24 |
| Other categories | 133 | 2 | 1 | 5 | 2 | 8 |
| Signal transduction | 12 | 4 | 0 | 3 | 33 | 7 |
| Disrupted reading frame | 41 | 2 | 0 | 2 | 5 | 4 |
| Total | 2310 | 403 | 245 | 610 | 28 | 1258 |
3.3 Setting the q-value threshold and estimating quantitative error
The cut-off values of q were determined using two technical replicate analyses of PG_PPC2 to control for the quantitative false positive rate (all values truly null) and by using an undiluted quantity of PG_PPC2 and 16-fold diluted PG_PPC2 to control the quantitative false negative rate (all values truly alternative). The null hypothesis in each case was an abundance ratio of zero on a log2 scale, indicating the measured spectral counts or signal intensities were approximately the same in the numerator and the denominator of the ratio calculation. The q-values were calculated using default parameters. Further details of the hypothesis testing are given in Materials and Methods and in Xia et al. [12]. The choice of a 16-fold dilution was largely arbitrary. Any value centered within the dynamic range of the method that allowed a broad range of proteins to be detected for both the diluted and undiluted proteome would have sufficed for our intended purpose of setting reasonable q-value cut-offs. For a broader discussion of the issues raised by the data shown in Table 1, e.g. statistical power calculations, sampling issues and their relationship to detection of abundance change, and results from similar experiments at other dilutions, the reader is referred to a recent review [43]. Table 1 summarizes the dilution results and illustrates why 0.01 was chosen for the spectral count ratios and 0.001 for the precursor ion signal intensity ratios. These values gave a reasonable balance between Type 1 and Type 2 error, although for the spectral counting data finding such a balance was harder and more subjective due to the high number of false negatives indicated in Table 1. This was consistent with previous observations suggesting that spectral counting has a lower statistical power (1 - FNR) for detecting abundance changes, relative to other approaches based on signal intensity differences, such as metabolic stable isotope labeling [18] or the non-label approach used in the present study.
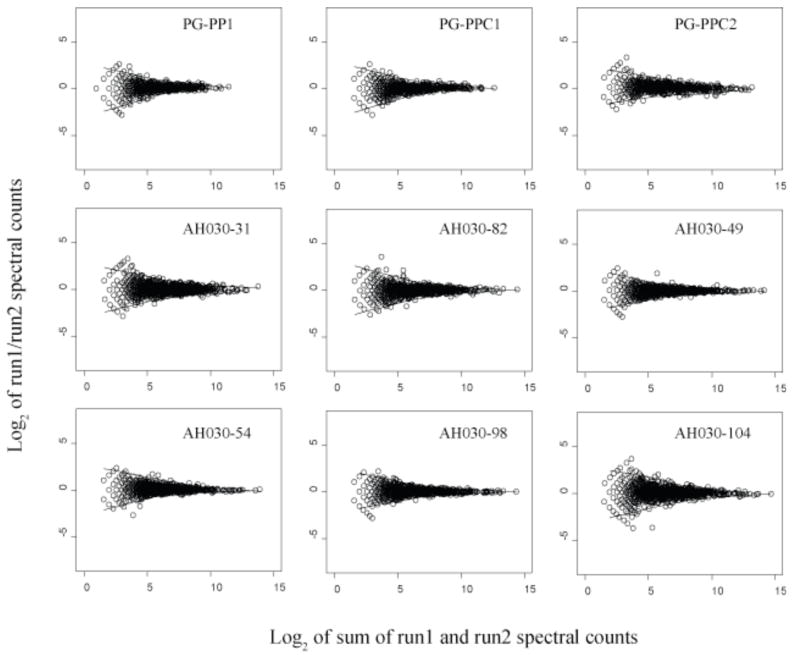
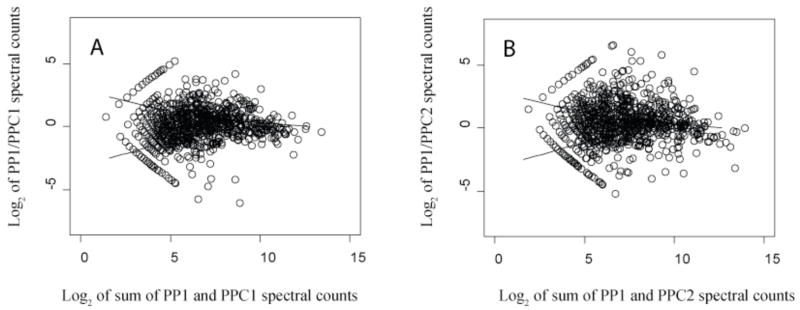
Fig. 2 shows scatter plots of technical replicates from two organisms, P. gingivalis and M. maripaludis, distributed among nine samples, to further support our previous observation [12] that this type of scatter plot can be used to define regions of random quantitative error, as it has been used in the microarray field for similar purposes [43]. The LOWESS curves illustrated in Fig. 2 for the replicate data can be superimposed on similar plots comparing two biologically different states for purposes of assessing which abundance ratios fall outside the boundaries established for random scatter about an abundance change of zero (log2 scale). Fig. 3 shows such LOWESS curves superimposed over the abundance ratio data for PG_PP1/PG_PPC1 and PG_PP1/PG_PPC2. The LOWESS curves shown in Fig. 3 served as a check on the q-value calculations [12].
Figure 2.
Scatter plots of protein abundance ratio versus total counts for technical replicates, log2 of spectral count ratios versus log2 of the total spectral counts from both technical replicates for nine biological samples. The two solid curves shown above and below the zero axis in each plot are the LOWESS smoothing curves [33] fit to the upper and lower boundaries of the random scatter about an abundance change of zero. The LOWESS curves shown for PG_PPC1 and PG_PPC2 were used to indicate the regions of random error in Fig. 3. PG_PP1 is the internalized biological replicate used for the abundance ratio calculations along with the two external controls, PG_PPC1 and PG_PPC2 (see text). The remaining plots are from nutrient limitation studies of the methanogenic Archaeon Methanococcus maripaludis. These plots serve as a useful graphical overview of the quantitative reproducibility of protein abundance ratios determined by spectral counting under the experimental conditions described. The similarity of these curves for different organisms under different conditions suggests that the region of random error within the upper and lower boundaries is determined largely by instrumental conditions.
Figure 3.
Scatter plots of protein abundance ratio versus total counts, biological replicates; (A), Log2 of PG_PP1/PG_PPC1 spectral count ratios vs. log2 summed protein level spectral counts; (B), the second biological replicate, PG_PP1/PG_PPC2. The solid curves shown are the LOWESS smoothing curves of the upper and lower boundary of the log2 ratios of PG_PPC1 run1/run2 and and PG_PPC2 run1/run2 technical replicates shown in Fig. 2.
For the intensity calculations, the level of random scatter in the normalized abundance ratios was generally higher, requiring the stricter cut-off value of 0.001. Table 1 shows the effect of different q-value cut-offs on the quantitative FPR and FNR. Literature q-value cut-offs for transcription microarray data have been determined using, among other methods, a plot of q-values versus the number of significantly changed ORFs associated with each q-value [36]. However, this approach did not work well for the present study (data not shown), suggesting the data were either too noisy, or too many proteins were changing, for this graphical method to be useful. Therefore, we chose an alternative approach: measuring the actual false positives and false negatives in data from experiments where these values were truly known over the entire proteome. The data from these analytical experiments (Table 1, qFig. 2, Fig. 3) were then used to generate cut-off values given above that were applied to the biological data summarized in Fig. 1, Table 2 and Table S1 in the supplement. The choice of -value cut-off was not sensitive to sample identity or human background contamination differences between the extracellular and intracellular samples. Indirect evidence that this should be the case is given by the overall similarity in the technical replicate plots for PG_PP1, PG_PPC1 and PG_PPC2 shown in Fig. 2, and the biological replicate plots shown in Figs. 4 and 5. However, as is clear from Table 1, the cut-off value was highly sensitive to the choice of quantitation method, spectral counting or signal intensity.
Figure 4.
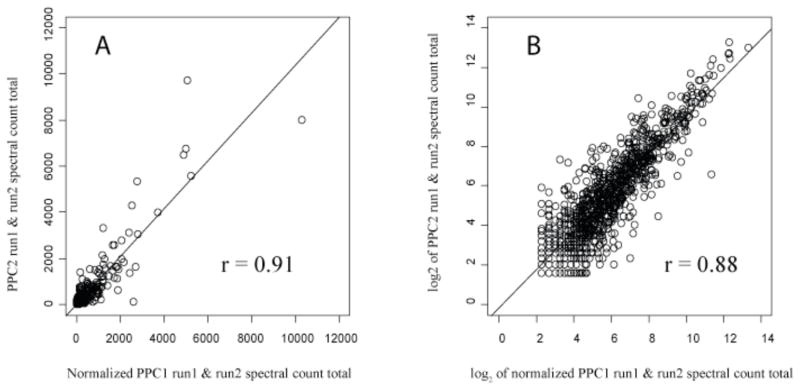
Reproducibility of biological replicates, extracellular P. gingivalis; proteins found in two extracellular controls were used to generate the plots of PG_PPC1 total spectral counts versus PG_PPC2 total spectral counts for each ORF. (A), The Pearson correlation coefficient was 0.91 on a linear scale. (B), The same data as a log2 transformation. A total of 963 proteins are plotted.
Figure 5.
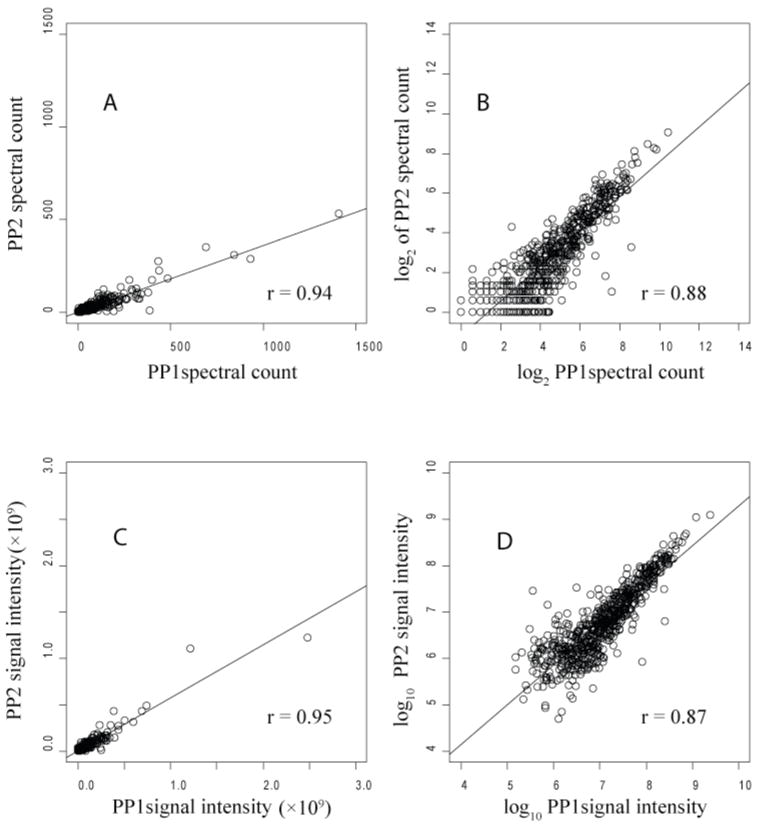
Spectral count (A, B) and signal intensity (C, D) correlations for two biological replicates, PG_PP1 and PG_PP2, of intracellular P. gingivalis. The Pearson correlation coefficient was 0.94 (A) for spectral counts plotted on a linear scale; on a log2 scale (B) it was 0.88. For the signal intensity measurements the results were about the same, 0.95 on a linear scale (C), and 0.87 on a log10 scale (D). A total of 649 proteins are plotted, representing the most abundant proteins detected and quantified in both replicates.
3.4 Reproducibility of biological replicates
Fig. 4 shows the correlation of spectral counts for each common protein in PG_PPC1 and PG_PPC2 on both linear and log2 scales. The Pearson correlations of 0.91 and 0.88 demonstrate that PG_PPC2 is a reasonably consistent biological replicate of PG_PPC1. There were 963 common proteins out of 1068 identified in PG_PPC1 and 1074 in PG_PPC2, indicating good qualitative agreement as well. Fig. 5 shows similar results for two biological replicates of internalized P. gingivalis, PG_PP1 and PG_PP2, with details given in the caption. As shown in Fig. 2, the random scatter observed with technical replicates of internalized P. gingivalis was the same as that observed for the external controls and replicates from another organism, suggesting that our random errors were constant, well characterized and predictably within bounds of the LOWESS curves shown. The extensive pre-fractionation employed in these studies, as described in Materials and Methods, the lack of sequence similarity between the P. gingivalis and human proteome and the highly reproducible response of P. gingivalis to the intracellular environment were probably the most important factors that allowed a straightforward abundance comparison between the intracellular and extracellular state. This was despite differences in sample complexity due to the presence of human proteins in the intracellular P. gingivalis proteome extracts that were not present in the extracellular controls. In a separate study performed to evaluate the pre-fractionation procedure (data not shown) and sampling issues with the internalized P. gingivalis proteins, we observed that the mass spectral data sampling was relatively insensitive to human and bovine background composition for all but the least abundant bacterial proteins so long as the percentage of spectral counts in the entire sample assigned to P. gingivalis proteins was at least approximately 8%. By way of comparison, the percentage of P. gingivalis spectral counts in PG_PP1 was about 26%. PG_PP1 was selected from among several biological replicates of internalized P. gingivalis for the abundance ratio calculations because it was more highly enriched in P. gingivalis proteins than other preps, leading to slightly more favorable sampling properties. As shown in Fig. 5 for the 649 most abundant proteins common to PG_PP1 and PG_PP2, the two biological replicates were essentially the same. They differed only in terms of the greater relative probabilities with which less abundant proteins could be sampled in PG_PP1, thus our choice to base the reported ratios in supplemental Table S1 on this internalization replicate. This decision was especially important in light of the fact that all proteomics approaches with which the authors are familiar tend to be biased in varying degrees towards the detection of proteins that are large and (or) abundant. For P. gingivalis in particular under these experimental conditions, this bias has been shown to be minimal at the level of qualitative identification [25]. However, such a bias clearly applies at the level of quantitation, where many less abundant proteins show too much scatter in the data and too few proteolytic fragments to be reliably quantified in terms of abundance ratios, using the non-label methods employed. These same bias relationships seem to also hold true of the other microbial systems under investigation in the senior author’s laboratory [43].
3.5 Abundance of known P. gingivalis invasins
Entry into and survival within gingival epithelial cells by P. gingivalis is a complex multistep process [22, 44–46]. Surface molecules such as the long fimbriae, FimA, mediate attachment and initiate a host cell signaling cascade necessary for internalization. A phosphatase, SerB, also provides necessary input into host cell signaling. Tight control of intracellular production of bacterial proteolytic enzymes (RgpA, Kgp and PepO) is necessary for intracellular survival. Clp stress-related proteins and ATPases (YjjK and ZntA) are also required for the process of entry and survival. The functionality of these proteins has been established through mutation analyses of the corresponding genes (Table 3). Seven out of eight of these invasins were found to be differentially expressed, the lone exception being ZntA, a cation transporting ATPase. This increases confidence that the proteins determined herein to be differentially expressed can be predicted to play a functional role in the interactions between P. gingivalis and host epithelial cells. Conversely, the role of proteins where we did not find an abundance change cannot be predicted. This ambiguity may result in part from sensitivity issues with protein detection and (or) post-translational control of protein activity in P. gingivalis. For example, the data in supplemental Table S1 do not distinguish between phosphorylated and de-phosphorylated forms of the same protein.
Table 3.
Comparison of protein level abundance and invasion assay results for selected mutant strains of P. gingivalis lacking in functional genes at the locus shown.
| ORF (TIGR) | Gene Name | Abundance Change1 | Mutant Invasion Phenotype2 | Reference |
|---|---|---|---|---|
| PG0159 | pepO | ↓ | ↓ | Park et al., 2004 [22] |
| PG0653 | serB | ↑ | ↓ | Tribble et al. 2006 [44] |
| PG1118 | clpB | ↑ | ↓ | Unpublished (Lamont et al.) |
| PG1642 | zntA | nd | ↓ | Park et al., 2004 [22] |
| PG1844 | kgp | ↓ | ↓ | Park and Lamont, 1998 [45] |
| FMA3 | fimA | ↓ | ↓ | Weinberg et al., 1997 [46] |
| PG2024 | rgpA | ↓ | ↓ | Park and Lamont, 1998 [45] |
| PG2206 | yjjK | ↑ | ↓ | Park et al., 2004 [22] |
Protein abundance in intracellular P. gingivalis (see supplementary Table S1): ↑ represents over-expression (green dot in the supplement Table S1 and Fig. 1), ↓ represents under-expression (red dot in Table S1), nd represents a qualitative non-detect (white dot in Table S1).
Ability of a mutant in the regulated gene to invade and survive in gingival epithelial cells as determined by antibiotic protection assays. ↓ represents a reduction in intracellular invasion/survival (p < 0.01). For methods used in the antibiotic protection assays, see [3].
The ATCC 33277 strain FimA sequence was used in the present studies [41]. The equivalent TIGR ORF for W83 is PG2132.
3.6 Insights into the intracellular lifestyle of P. gingivalis
i) Adhesins. The major adhesin for gingival epithelial cells is the long fimbriae comprised of the FimA subunit protein [41, 47]. FimA was under-expressed intracellularly, indicating that the organism dispenses with production of this molecule after entry has been accomplished. Loss of a major surface protein such as FimA may also have implications for recognition of the organism by intracellular microbial detection systems such as the Nod-like receptors [48]. ii) Proteases. P. gingivalis as an asaccharolytic organism produces a variety of proteolytic enzymes to obtain peptides for growth. P. gingivalis cells in the nutritionally rich environment of the cytoplasm showed lowered abundance of a number of proteinases including RgpA, RgpB and Kgp, that are considered major virulence factors [4]. Expression of bacterial proteases inside host cells will damage host proteins and, in addition, P. gingivalis proteases can induce apoptotic cell death [49]. Hence, reduced protease expression will contribute to the long-term survival of the host cells. In support of this concept, epithelial cells infected with P. gingivalis do not undergo apoptosis [50] and can survive for up to eight days after infection [6]. Thus, intracellular P. gingivalis appear to be adapted for long-term survival and co-existence with the host, and may not be overtly pathogenic. iii) Iron acquisition systems. Lacking siderophores, P. gingivalis employs specific outer membrane receptors, lipoproteins and proteases to acquire heme and iron [4, 55]. Many of these systems were under-expressed, including: HmuR, a TonB-linked outer membrane receptor required for both hemoglobin and hemin utilization, and HmuY, a hemin binding protein in the same operon [51, 52]; FetB (IhtB), a hemin binding lipoprotein that is located in an iron transport operon along with a TonB-linked outer membrane receptor, IhtA, [53] that was also under-expressed; and Kgp, that can degrade host iron- and heme-containing proteins, and possibly act as a hemophore, shuttling heme back to outer membrane receptors [55]. FeoB proteins [54] were not detected in these studies. These results indicate that the HIGK intracellular iron and hemin concentrations are in excess for P. gingivalis, as is also reflected in the finding that abundance of ferritin, an intracellular iron storage protein, was increased. iv) Stress. A reduction in free iron within P. gingivalis, as would result from under-expression of iron acquisition systems and over-expression of ferritin, could also be indicative of oxidative stress. Lower levels of iron in bacteria will limit the production of OH− from H2O2 via Fenton chemistry [56]. Consistent with the concept that intracellular P. gingivalis are under a degree of oxidative stress is the over-expression of two alkyl hydroperoxide reductases, a thiol peroxidase and rubrerythrin, all of which can be involved in protection against oxidative stress [57, 58]. In addition, there was over-expression of ClpB and ClpC, two components of the Clp family, a general protection system against stress, along with heat shock proteins GroEL, GroES, HtpG, HtrA, and DnaK. In any event, regardless of the nature or extent of the stress, which may be a more natural environment for P. gingivalis, intracellular P. gingivalis are clearly metabolically active and capable of protein synthesis. v) Transcription factors. Intracellular P. gingivalis over-expressed two predicted activators (PG0016 and PG0747) of the alternative sigma factor, sigma-54. A physiological theme for sigma-54-dependent genes has yet to emerge, as the target genes participate in a wide range of processes. However, in plant pathogens, sigma-54 controls type III secretion [59], a major pathway for delivery of bioactive bacterial proteins into host cells that contributes to bacterial invasion and survival. Over-expression of a TetR family transcriptional regulator was also observed. In other organisms TetR is involved in the transcriptional control of efflux pumps and responses to osmotic stress [60], thus TetR may play a role in regulating physiological homeostasis of intracellular P. gingivalis.
3.7 Transition of P. gingivalis from extracellular to intracellular
Our previous global analysis [21] compared the protein expression of P. gingivalis in keratinocyte growth medium (KGM) with conditioned keratinocyte growth medium (cKGM) that had been exposed to GECs. The intention was to examine changes in protein abundance that occur under conditions simulating the early stages of invasion, prior to internalization of the bacteria. The results of a comparison between the previous study and the data given in Table S1 are listed in supplementary Tables S2 and S3. A large number of the proteins were differentially expressed under both sets of experimental conditions, indicating that P. gingivalis rapidly senses the host cells and adapts to become “intracellular ready”. There were also proteins that responded differently to the two environments, which supports the hypothesis that P. gingivalis can also sense and respond to epithelial cell intracellular cues.
3.8 Codon Adaptation Index (CAI) and Karlin index E(g)
To explore the relationships among our quantitative results in terms of the relative abundance of proteins within a given sample, as opposed to abundance ratios comparing two samples, we examined the spectral counts for each ORF in light of two theoretical predictors of gene expression. Linear regressions of spectral counts versus CAI, [37] and Karlin index, E(g), [38, 39] were generated to demonstrate correlations between our experimental observations and these bioinformatic predictors of protein abundance (data not shown). The moderate correlations (r = 0.39 to 0.58) observed with spectral counting were not improved by corrections for molecular weight, a correction that has improved such correlations in studies of another prokaryote, Methanococcus maripaludis [28]. Although the use of spectral counting methods for the determination of abundance ratios rests on a somewhat firmer basis, there is less evidence to support the validity of using such measurements to make statements about the relative composition of proteomes within a given sample. This is analogous to the absence of any absolute interpretation of the laser induced fluorescence signal in a transcription microarray experiment, in which the normalized signal intensity ratio between two states is the usual endpoint [23]. We know, for example, that any such “within sample” interpretation will be compromised by major differences in quantitative detectability among proteins [17], depending on such factors as their true abundance, molecular weight, stability during the extraction and analytical procedures, hydrophobicity, isoelectric point and the degree to which the protein is intrinsic within the cell membrane. While purely qualitative detectability is remarkably consistent across the proteome for P. gingivalis [25], it has not been possible in the present studies to demonstrate an unequivocal and straightforward relationship between a protein’s spectral count and its absolute quantity.
3.9 Comparison of protein abundance ratios with theoretical gene pair predictions
Fig. S1 in the supplement shows the alignment of the consensus protein abundance ratio results with the directons predicted computationally by TIGR and the University of Maryland [26], see also www.cbcb.umd.edu/cgi-bin/operons/paris.cgi?taxon_id=20119. The authors of this study defined a directon as the maximal number of conserved adjacent genes located on the same DNA strand. A directon may contain one or more operons. From Fig. S1 it can be seen that many of the ORFs within a directon show the same trend in abundance change. For example, the ORFs in the directon from PG0775 to PG0785 are either over-expressed (green) or ambiguous (yellow); the ORFs in the directon from PG1063 to PG1080 are either under-expressed (red) or ambiguous. We found that the proposed LuxS operon (PG0497 to PG0504) [61] was consistent with both the directon and our experimental observations at the protein level. PG1910–1942 was exceptional in that it is the longest directon prediction. From the experimental results, it is possible that this directon consists of several operons that show different expression trends. However, it would be difficult to interpret the data presented here in terms of a common regulatory structure for the entire directon.
4 Concluding remarks
Organisms that colonize human mucosal surface have engaged in a long standing evolutionary association with the host and many, including opportunistic pathogens such as P. gingivalis, have become host adapted [62]. Indeed, the ability of P. gingivalis to survive within gingival epithelial cells, without causing excessive harm to the host, indicates an extensive degree of adaptation. Global protein profiling provides insights into the nature of host adaptation by P. gingivalis. The proteomic methods employed for these studies have reached a level of maturity such that it is now possible to make reliable quantitative measurements that are truly global in scope at the cellular level, although the quantitative results were biased towards false negatives (see Table 1). More genes are probably changing their protein abundance patterns in response to the host environment than what we have described here. Global transcription studies of intracellular pathogens isolated from within human target cells have also been problematic in terms of high noise and low signal due to the high rates of cross-hybridization and non-specific hybridization that occur in such experiments, most of which have focused on the host response rather than the internalized pathogen [63]. Making direct comparisons of efficacy is difficult due to, among other reasons, problems associated with measuring false negative rates in the absence of easily accessible absolute detection information, a situation common to most microarray studies. In proteomics, as applied here, absolute and highly specific detection is a precondition that must be met prior to generation of relative abundance ratios [43].
Our most recent studies of extracellular P. gingivalis interactions with Fusobacterium nucleatum and Streptococcus gordonii, in which changes to the P. gingivalis proteome are confined to a few specific ORFs, have provided additional negative controls confirming that the widespread changes observed during internalization are not sampling artifact (T. Wang, Q. Xia, et al., manuscript in preparation) due to high background levels of non-P. gingivalis proteins. As more work is carried out, our best judgment is that the results reported here are conservative. Nonetheless, despite the quantitative false negative error issue, what emerges from these measurements is a biologically consistent picture of an organism that down-regulates production of destructive proteases, coordinates responses to elevated oxygen tension and iron availability, dispenses with production of energetically costly surface molecules that have outlived their use, and ramps up protein production to thrive in a nutritionally rich intracellular environment. Unless P. gingivalis possesses a mechanism to restrain growth and division, in the long-term the increase in bacterial numbers may tip the ecological balance away from health and toward cell and tissue destruction.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Kevin Wheeler and Jim Shofstahl for their assistance, Michrom Bioresources for equipment support, Michael J. MacCoss for comments and makems2, David L. Tabb, John R. Yates 3rd for DTASelect and Contrast software, Erik L. Hendrickson and Martin Handfield for their insights into global expression analysis. This work was supported by the NIH NIDCR under DE014372 (M. H.) and DE011111 (R. J. L.).
Abbreviations
- ATCC
American type culture collection
- CAI
codon adaptation index
- cKGM
conditioned keratinocyte growth medium
- CS
consensus ratio of PG_PP1/PG_PPC1 and PG_PP1/PG_PPC2
- E(g)
Karlin index or general expression measure
- FDR
false discovery rate
- FPR
false positive rate
- FNR
false negative rate
- GECs
human gingival epithelial cells
- HIGK
human immortalized gingival keratinocyte
- hmu
haemin-uptake locus
- IP1
intensity ratio of PG_PP1/PG_PPC1
- IP2
intensity ratio of PG_PP1/PG_PPC2
- KGM
keratinocyte growth medium
- LOWESS
locally weighted scatter plot smoothing
- LTQ
Thermo-Finnigan linear ion trap mass spectrometer
- MS1
first dimension of mass spectrometry
- MS2
second dimension of mass spectrometry
- MudPIT
multidimensional protein identification technology
- nrdb
non-redundant database
- PG
Porphyromonas gingivalis
- PP cells
synonym for HIGK, see definition above
- PG_PP1
PG_PP2, P. gingivalis internalized within PP cells
- PG_PPC1
PG_PPC2, P. gingivalis incubated in media optimized for HIGK
- SFM
serum free medium
- SP1
spectral count ratio of PG_PP1/PG_PPC1
- SP2
spectral count ratio of PG_PP1/PG_PPC2
- TIGR
The Institute for Genomic Research
References
- 1.Albandar JM. Epidemiology and risk factors of periodontal diseases. Dent Clin North Am. 2005;49:517–532. v–vi. doi: 10.1016/j.cden.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Garcia RI, Henshaw MM, Krall EA. Relationship between periodontal disease and systemic health. Periodontol 2000. 2001;25:21–36. doi: 10.1034/j.1600-0757.2001.22250103.x. [DOI] [PubMed] [Google Scholar]
- 3.Lamont RJ, Chan A, Belton CM, Izutsu KT, et al. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun. 2006;74:703–710. doi: 10.1128/IAI.74.1.703-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madianos PN, Papapanou PN, Nannmark U, Dahlen G, Sandros J. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect Immun. 1996;64:660–664. doi: 10.1128/iai.64.2.660-664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitkov L, Krautgartner WD, Hannig M. Bacterial internalization in periodontitis. Oral Microbiol Immunol. 2005;20:317–321. doi: 10.1111/j.1399-302X.2005.00233.x. [DOI] [PubMed] [Google Scholar]
- 8.Rudney JD, Chen R, Sedgewick GJ. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect Immun. 2001;69:2700–2707. doi: 10.1128/IAI.69.4.2700-2707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noiri Y, Ozaki K, Nakae H, Matsuo T, Ebisu S. An immunohistochemical study on the localization of Porphyromonas gingivalis, Campylobacter rectus and Actinomyces viscosus in human periodontal pockets. J Periodontal Res. 1997;32:598–607. doi: 10.1111/j.1600-0765.1997.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 10.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamont RJ, Meila M, Xia Q, Hackett M. Mass spectrometry-based proteomics and its application to studies of Porphyromonas gingivalis invasion and pathogenicity. Infect Disord Drug Targets. 2006;6:311–325. doi: 10.2174/187152606778249935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia Q, Wang T, Park Y, Lamont RJ, Hackett M. Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: Non-label methods comparison, q-values and LOWESS curve fitting. Int J Mass Spec. 2007;259:105–116. doi: 10.1016/j.ijms.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 14.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., 3rd Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 15.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Arnold RJ, Alves P, Xun Z, et al. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickson EL, Xia Q, Wang T, Leigh JA, Hackett M. Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Analyst. 2006;131:1335–1341. doi: 10.1039/b610957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Opiteck GJ, Friedrichs MS, Dongre AR, Hefta SA. Changes in the protein expression of yeast as a function of carbon source. J Proteome Res. 2003;2:643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Wang T, Chen W, Yilmaz O, et al. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics. 2005;5:198–211. doi: 10.1002/pmic.200400922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park Y, Yilmaz O, Jung IY, Lamont RJ. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infect Immun. 2004;72:3752–3758. doi: 10.1128/IAI.72.7.3752-3758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Laidig KE, Park Y, Park K, et al. Searching the Porphyromonas gingivalis genome with peptide fragmentation mass spectra. Analyst. 2001;126:52–57. doi: 10.1039/b008012h. [DOI] [PubMed] [Google Scholar]
- 25.Wang T, Zhang Y, Chen W, Park Y, et al. Reconstructed protein arrays from 3D HPLC/tandem mass spectrometry and 2D gels: complementary approaches to Porphyromonas gingivalis protein expression. Analyst. 2002;127:1450–1456. doi: 10.1039/b206157k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ermolaeva MD, White O, Salzberg SL. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oda D, Bigler L, Lee P, Blanton R. HPV immortalization of human oral epithelial cells: a model for carcinogenesis. Exp Cell Res. 1996;226:164–169. doi: 10.1006/excr.1996.0215. [DOI] [PubMed] [Google Scholar]
- 28.Xia Q, Hendrickson EL, Zhang Y, Wang T, et al. Quantitative proteomics of the archaeon Methanococcus maripaludis validated by microarray analysis and real time PCR. Mol Cell Proteomics. 2006;5:868–881. doi: 10.1074/mcp.M500369-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J Am Soc Mass Spectrum. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 30.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 32.Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat Biotechnol. 2004;22:214–219. doi: 10.1038/nbt930. [DOI] [PubMed] [Google Scholar]
- 33.Cleveland WS. LOWESS-a program for smoothing scatterplots by robust locally weighted regression. The American Statistician. 1981;35:54–54. [Google Scholar]
- 34.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 35.Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 3. W. H. Freeman; New York: 1995. [Google Scholar]
- 36.Storey JD, Tibshirani R. Statistical significance for genome wide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlin S, Mrazek J. Predicted highly expressed genes of diverse prokaryotic genomes. J Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlin S, Mrazek J, Ma J, Brocchieri L. Predicted highly expressed genes in archaeal genomes. Proc Natl Acad Sci U S A. 2005;102:7303–7308. doi: 10.1073/pnas.0502313102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T, Hosogi Y, Nishikawa K, Abbey K, et al. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004;186:5473–5479. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiwara T, Morishima S, Takahashi I, Hamada S. Molecular cloning and sequencing of the fimbrilin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem Biophys Res Commun. 1993;197:241–247. doi: 10.1006/bbrc.1993.2467. [DOI] [PubMed] [Google Scholar]
- 42.Shi L, Adkins JN, Coleman JR, Schepmoes AA, et al. Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages. J Biol Chem. 2006;281:29131–29140. doi: 10.1074/jbc.M604640200. [DOI] [PubMed] [Google Scholar]
- 43.Xia Q, Hendrickson EL, Wang T, Lamont RJ, Leigh JA, Hackett M. Protein abundance ratios for global studies of prokaryotes. Proteomics. 2007;7:2904–2919. doi: 10.1002/pmic.200700267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci U S A. 2006;103:11027–11032. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park Y, Lamont RJ. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect Immun. 1998;66:4777–4782. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinberg A, Belton CM, Park Y, Lamont RJ. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae- mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 48.Kufer TA, Banks DJ, Philpott DJ. Innate immune sensing of microbes by Nod proteins. Ann N Y Acad Sci. 2006;1072:19–27. doi: 10.1196/annals.1326.020. [DOI] [PubMed] [Google Scholar]
- 49.Sheets SM, Potempa J, Travis J, Fletcher HM, Casiano CA. Gingipains from Porphyromonas gingivalis W83 synergistically disrupt endothelial cell adhesion and can induce caspase-independent apoptosis. Infect Immun. 2006;74:5667–5678. doi: 10.1128/IAI.01140-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakhjiri SF, Park Y, Yilmaz O, Chung WO, et al. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol Lett. 2001;200:145–149. doi: 10.1111/j.1574-6968.2001.tb10706.x. [DOI] [PubMed] [Google Scholar]
- 51.Lewis JP, Plata K, Yu F, Rosato A, Anaya C. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology. 2006;152:3367–3382. doi: 10.1099/mic.0.29011-0. [DOI] [PubMed] [Google Scholar]
- 52.Olczak T, Dixon DW, Genco CA. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J Bacteriol. 2001;183:5599–5608. doi: 10.1128/JB.183.19.5599-5608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dashper SG, Hendtlass A, Slakeski N, Jackson C, et al. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol. 2000;182:6456–6462. doi: 10.1128/jb.182.22.6456-6462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dashper SG, Butler CA, Lissel JP, Paolini RA, et al. A novel Porphyromonas gingivalis FeoB plays a role in manganese accumulation. J Biol Chem. 2005;280:28095–28102. doi: 10.1074/jbc.M503896200. [DOI] [PubMed] [Google Scholar]
- 55.Olczak T, Simpson W, Liu X, Genco CA. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev. 2005;29:119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx(-) mutants of Escherichia coli. Proc Natl Acad Sci U S A. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okano S, Shibata Y, Shiroza T, Abiko Y. Proteomics-based analysis of a counter-oxidative stress system in Porphyromonas gingivalis. Proteomics. 2006;6:251–258. doi: 10.1002/pmic.200401338. [DOI] [PubMed] [Google Scholar]
- 58.Mydel P, Takahashi Y, Yumoto H, Sztukowska M, et al. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS Pathog. 2006;2:712–725. doi: 10.1371/journal.ppat.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev. 2005;69:527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.James CE, Hasegawa Y, Park Y, Yeung V, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74:3834–3844. doi: 10.1128/IAI.01768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galan JE, Zhou D. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc Natl Acad Sci U S A. 2000;97:8754–8761. doi: 10.1073/pnas.97.16.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mans JJ, Lamont RJ, Handfield M. Microarray analysis of human epithelial responses to bacterial interaction. Infect Disord Drug Targets. 2006;6:299–309. doi: 10.2174/187152606778249926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.