Abstract
Quantitative proteome analysis of microbial systems generates large datasets that can be difficult and time consuming to interpret. Fortunately, many of the data display and gene clustering tools developed to analyze large transcriptome microarray datasets are also applicable to proteomes. Plots of abundance ratio versus total signal or spectral counts can highlight regions of random error and putative change. Displaying data in the physical order of the genes in the genome sequence can highlight potential operons. At a basic level of transcriptional organization, identifying operons can give insights into regulatory pathways as well as provide corroborating evidence for proteomic results. Classification and clustering algorithms can group proteins together by their abundance changes under different conditions, helping to identify interesting expression patterns, but often work poorly with noisy data like that typically generated in a large-scale proteome analysis. Biological interpretation can be aided more directly by overlaying differential protein abundance data onto metabolic pathways, indicating pathways with altered activities. More broadly, ontology tools detect altered levels of protein abundance for different metabolic pathways, molecular functions and cellular localizations. In practice, pathway analysis and ontology are limited by the level of database curation associated with the organism of interest.
Keywords: DAVID, Gene Ontology, GoMiner, bioinformatics, proteomics, Porphyromonas gingivalis, Methanococcus maripaludis, protein profiling, protein expression
INTRODUCTION
The advent of cDNA microarrays, allowing genome wide transcriptional analysis, has resulted in a large increase in the volume of data available to researchers. Large datasets, while providing tremendous amounts of new information, pose problems for efficient analysis. The result has been the development of programs and techniques for analyzing microarray data. Until recently the complexities of dealing with large datasets were not as much of a concern for proteomics, which was limited by the number of proteins that could be resolved by a 2D gel. Approaches based on the analysis of proteolytic digests by 2D capillary HPLC coupled with tandem mass spectrometry (Washburn et al., 2001, 2002) identify more proteins with more mass spectra per protein, yielding a greater level of proteome coverage and sampling depth than can be accomplished with gel electrophoresis, thus producing microarray size datasets. The LC/MS based approaches have been particularly useful for microbial systems, including those of interest to oral biologists (Lamont et al., 2006).
Manual in-depth inspection coupled with literature research is indispensable when interpreting whole genome data. However, data analysis tools can make the process easier and more efficient. The field of genome wide data analysis is large and a discussion of every available program and technique is beyond the scope of this review. Rather our goal is to provide a critical introduction to the types of data analysis tools available that would be of interest to researchers exploring proteomics data from microbial species, with an emphasis on those we have actually used. We describe the tools roughly in order of increasing complexity. In order to give real world examples, we have used data from studies of two organisms, Methanococcus maripaludis S2 and Porphyromonas gingivalis ATCC 33277. M. maripaludis is an anaerobic Archaeon that obtains energy through the methanogenesis pathway, which reduces CO2 to methane employing H2 as an electron donor (Hendrickson et al., 2004). Our ongoing studies of M. maripaludis have implications for global carbon cycles, alternative energy sources, and an overall understanding of Archaeal biology. P. gingivalis is a Gram-negative facultative intracellular bacterial pathogen associated with periodontal disease and potentially with serious systemic conditions (Lamont and Jenkinson, 1998). P. gingivalis research, in the context of oral biofilms and the many other organisms present therein, provides a window into the role of microbial communities in human disease. These two organisms were chosen because of the authors’ familiarity with them but also as examples of microbial species commonly used in research that are not well-established model systems like Escherichia coli. As will be discussed, some tools are less tractable for less well-documented species.
The tools discussed in this paper are primarily focused on extracting a biological interpretation from large datasets. Prior to looking for biological meaning, quantitative proteomics data needs to undergo several layers of processing. After initial acquisition with a mass spectrometer interfaced to some kind of high-resolution separations scheme (Washburn et al., 2001, 2002) the data need to be processed to assign peptide identifications to the fragments in a proteolytic digest (Eng et al., 1994), map the peptides back to their associated proteins (Tabb et al., 2002), and to determine the statistical validity of the results, both qualitatively and quantitatively (Keller et al., 2002; Nesvizhskii et al., 2004, Choi et al., 2008; Käll et al., 2008,). The earlier stages of data processing that create the input files for the tools reviewed here, with emphasis on the quantitative aspects, were recently reviewed with specific reference to microbial systems (Xia et al., 2007a).
As with transcriptome microarrays, whole genome proteomics is primarily comparative, looking for a change in protein abundance between two or more conditions. From the initial stages of data processing one generates a list of proteins, with an abundance ratio relative to a reference condition, and a measure of the data’s statistical strength. Statistical measurements of the likelihood of an abundance difference, such as a p-value or a q-value (Storey and Tibshirani, 2003), are needed to properly evaluate the results. The list of proteins with accompanying abundance ratios and statistical measurements is virtually identical to the final results tabulated from transcriptome microarrays, though representing protein rather than RNA levels, and most of the statistical and bio-informatic tools and concerns are the same, even though the physical detection method and the biological basis of each method are completely different. The protein abundance ratio lists serve as inputs into the various computational tools described in this review. The point of view expressed by the authors is that of experimentalists in that we are users of data mining tools driven by biological questions, not statisticians or algorithm developers. Regarding nomenclature, we have followed the lead of most of the papers cited herein and used the terms “expression” and “abundance” somewhat interchangeably, keeping in mind that what is actually being measured is the net result of various biosynthetic and destructive processes that can contribute to a protein relative abundance measurement.
PLOTS OF ABUNDANCE RATIO VERSUS SIGNAL OR SPECTRAL COUNTS
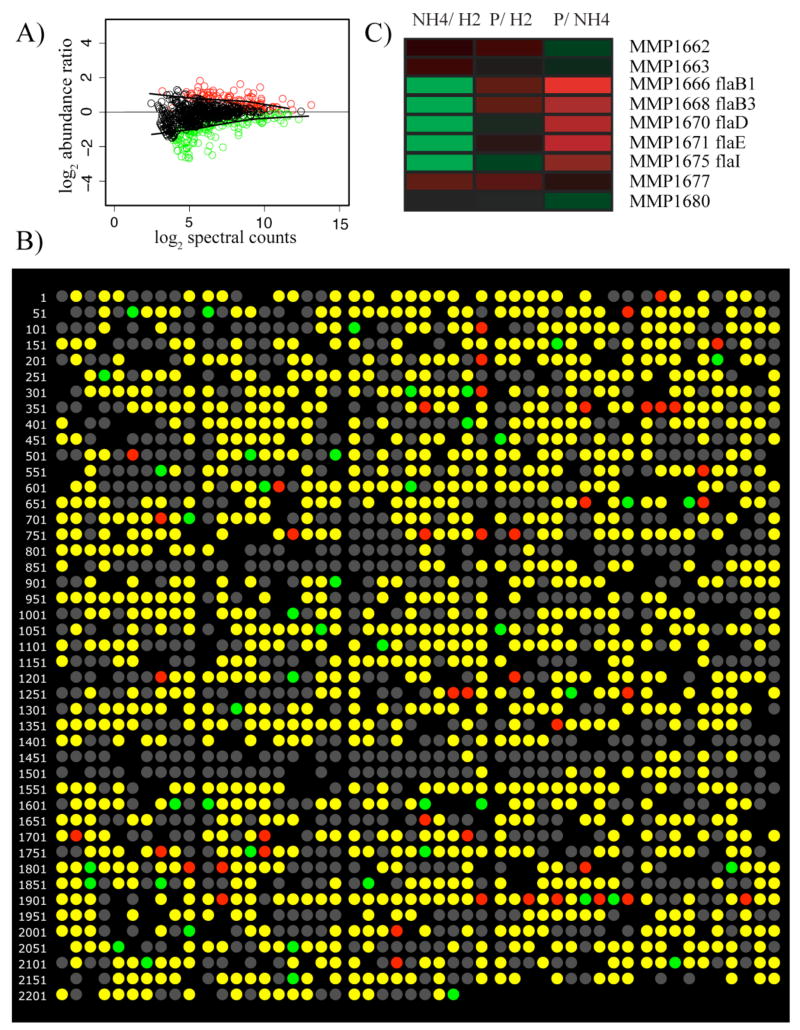
Transcriptome microarray analysis has long visualized the overall quality and uncertainty of microarray results using M versus A plots. These are plots of the abundance ratio for each measurement, the M, against the overall signal strength of the measurement, the A (Quackenbush, 2002). Similar plots can be used for proteomics results, see Fig. 1A. Such a graph of proteome data can serve several purposes, including visualizing statistically significant abundance changes as well as the data distribution where uncertainty is too great to identify statistically supported changes. It can also be used to evaluate the effectiveness of outlier detection algorithms. Perhaps most importantly in proteomics an M versus A plot serves to visualize the important relationships among sampling depth, the number of peptides per protein identified during the experiment, and the power to detect abundance change (Hackett, 2008). The data displayed in Fig. 1A shows P. gingivalis protein abundance when incubated alone and in the presence of Streptococcus gordonii and Fusobacterium nucleatum. The abundance ratios were obtained using spectral counting, a frequency measurement that correlates with protein abundance (Liu et al., 2004, Choi et al., 2008) and which is similar conceptually to the way SAGE (serial analysis of gene expression) data are analyzed. This approach is based on the number of peptides observed that map to a given ORF, rather than the intensity of the observed signals. The x-axis shows the log2 of the total spectral counts for each protein. The M versus A type plot gives an estimate of error, shown by the black lines in Fig. 1A, which were fitted to a comparison of identical samples using locally weighted scatterplot smoothing, LOWESS (Cleveland, 1981), and superimposed on the experimental data. Because they fall within the data scatter for unchanged abundances the measurements lying within the lines are unlikely to show statistically significant change. Statistically significant abundance changes are shown as colored circles. Fig. 1A also shows the importance of sampling. As sampling depth increases the number of spectral counts observed for each protein generally increases, shifting the results further to the right on the x-axis. As shown by the black lines, the overall uncertainty generally decreases to the right of the graph. Since the abundance change needs to be larger than the uncertainty to identify a statistically significant change, the power to detect changes goes up as the uncertainty goes down. The greater the sampling depth, the further to the right on the x-axis the data will plot, and thus the smaller the change in abundance that can be reliably detected (Hackett, 2008). This type of plot can also serve as a general quality control tool for proteomics experiments. Every properly executed shotgun proteomics experiment we have evaluated to date shows the characteristic shape of Fig. 1A, a symmetry about y = 0 after normalization to take into account the absolute quantity of protein measured for each biological state being compared. Marked deviations from the pattern usually indicate a problem, e.g. with inadequate sampling or the normalization procedure. A common assumption is that most proteins will not show significant, non-random changes between conditions. In the majority of experiments, this assumption holds true. However, we have found that even in an experiment where a large fraction of the P. gingivalis proteome was changing dramatically (Xia et al., 2007b), the symmetry and general appearance of the normalized data were still similar to that shown in Fig. 1A.
Figure 1.
(A), Pseudo M versus A plot of 791 P. gingivalis proteins determined by spectral counting. Green indicates under-expression when P. gingivalis was co-incubated with S. gordonii and F. nucleatum; red, over-expression; black, no change in abundance. The solid black lines, LOWESS curves (Cleveland, 1981), indicate boundaries for the region of expected random error about an abundance change of zero, and can also be used to estimate the power to detect abundance change as a function of total spectral counts at any point on the x-axis. (B), An example of proteomics data displayed in genome order, P. gingivalis response to co-incubation with S. gordonii by spectral counting. Each circle represents an annotated protein-encoding ORF in the order that it is encoded in the genome. Colored circles show the results of the quantitative proteomic analysis. Green indicates under-expression in P. gingivalis cells co-incubated with S. gordonii compared to P. gingivalis alone; red, over-expression; yellow, proteins that were identified but showed no statistically significant abundance change; grey, no protein was detected. Blacked out regions indicate ORF numbers for which there is no TIGR annotation. (C), A section of the genomic representation of three M. maripaludis proteome experiments displayed in MEV (www.tm4.org/mev.html). Each row represents an ORF in the order that it is encoded in the genome (MMP1662-1680). Undetected proteins in this range are not shown. The annotation of each ORF is listed at the end of the row. The columns represent experimental conditions. NH4/H2, ammonia limiting compared to H2 limiting growth conditions; P/H2, phosphate limiting compared to H2 limiting growth conditions; P/NH4, phosphate limiting compared to ammonia limiting growth conditions. The colors represent a gradient of differential abundance between the conditions, from green for over-expression to red for under-expression, with black showing no change in abundance.
PLOTS OF PROTEIN ABUNDANCE TRENDS IN GENOME ORDER
The simplest analysis tool, sometimes referred to as “beads-on-a-string”, is shown in Fig. 1B. The output was generated using FilemakerPro, as in our previous work (Xia et al., 2007b). Here P. gingivalis proteomics data are displayed as a sequence of circles color-coded to show the difference in relative protein abundance between P. gingivalis co-incubated with another biofilm component and P. gingivalis by itself. Color codes are given in the caption and are completely arbitrary. The “beads-on-a-string” presented here uses a simple three-color display designed to emphasize statistically significant results. Proteins are only coded as over- or under- expressed if they meet a level of statistical significance set by the researcher. In this example statistical significance was calculated using an uncertainty estimate, q-values, specifically designed for testing large numbers of results (Storey and Tibshirani, 2003). By color-coding only results that are statistically likely to be truly changed, the three-color display provides a graphic handle on the underlying reliability of the observed changes. What is not displayed is the observed magnitude of the changes. However, as with transcription microarray data, the absolute value of the measured change is highly non-reproducible (Bammler et al., 2005). The trend, under- or over-expression, is more reproducible (Hendrickson et al., 2006; Xia et al., 2007a). By concentrating on statistically significant trends, rather than magnitude, the three-color display gives a more reliable picture of what is really different between samples. In Fig. 1B the circles are organized in the order they appear on the genome. By placing them in this order the format draws attention to potentially regulated operons, which can appear as a series of adjacent circles of the same color. The genome order display takes advantage of the operon structure of microbial transcription. Operons can be predicted computationally from annotated genome sequence data or identified experimentally using the tools of molecular biology. If an entire operon shows similar expression changes, then this provides increased confidence in the results. A potential limitation of this approach is that the sequenced strain is not always the choice for experimental investigation. In the case of P. gingivalis the genome sequence is from strain W83 and the experimental strain is ATCC 33277, which contains genes that are absent in W83. However, the genome sequence of P. gingivalis ATCC 33277 has recently been published and future analyses can employ that sequence (Naito et al., 2008). This should improve the quality of the genome order display significantly. While 82% of the ORF’s in W83 are also present in ATCC 33277, extensive genome rearrangements where observed between the strains.
HEAT MAPS
A slightly different display method is that of “heat maps”. Heat maps display data in small cells colored to represent relative abundance values (Fig. 1C). Here log2 ratios from M. maripaludis proteomics data are displayed using MEV (Multi-experiment Viewer) from TIGR (www.tm4.org/mev.html). The data came from three M. maripaludis proteomics experiments. MEV accepts a number of different file formats, including tab delimited text files, which were used for the M. maripaludis data. By using a color gradient to represent abundance differences, this style of display draws attention to the most differentially expressed proteins. However, the display contains no information about the reliability of the data. For any proteins with the same magnitude of observed change, statistically unsupported results are indistinguishable from statistically significant differences. This emphasis is in contrast to the three-color display (Fig. 1B). Three-color displays emphasize statistically significant changes but provide no information about the magnitude, while heat maps provide a graphical display of the magnitude of the changes, but no information on statistical significance. Three-color displays may describe the actual changes seen between samples more reliably, but heat maps can be useful for biological interpretations that depend on magnitude. A statistically well-supported change that is quite small, for example a 25% abundance increase, may be of little meaning from a biological perspective. Thus, looking at the magnitudes of the changes can provide an important perspective. As the sampling depth and quality of proteomics data improves, this issue becomes more significant due to the increased ability to detect small changes that may be “real” in an analytical or statistical sense, but that are not relevant biologically. The small section of a larger heat map shown in Fig. 1C is once again presented in genome order. In this case an operon encoding the flagellar genes in M. maripaludis is evident, MMP1666, 1668, 1670–71, and 1675. The other flagellar proteins were not consistently detected in this experiment.
METABOLIC MAPS
While displaying relative abundance results in genome order can highlight regulated operons, it does not provide any direct information about the functional differences between the tested conditions. Overlaying the abundance ratio data onto metabolic maps can make changes in metabolic processes readily apparent. As an example, we analyzed P. gingivalis proteomics data (Xia et al., 2007b) using BioCyc (available for online use or download from www.biocyc.org). For this example we used the web-based interface. Fig. 2 shows the metabolic map for P. gingivalis over which the data was displayed. Normally BioCyc displays the ratio data as expression gradients such as those seen with heat maps (Fig. 1C) and submitting protein abundance ratio data will produce a display with color gradients for the abundance levels. Users interested in a three-color display to emphasize statistically significant changes, as seen with the beads-on-a-string (Fig. 1B), can obtain this effect by submitting the data coded numerically with three integer values, by convention 1 for proteins with increased abundance, 0 for unchanged, and −1 for proteins with decreased abundance. BioCyc also has an internal function for three-color maps, but it employs fold-change only rather than statistical significance as the classification factor, a significant weakness in our view due to the weak theoretical underpinnings of calling either genes or proteins “changed” on the basis of magnitude alone (Kim and Lee, 2006), as noted in the previous section.
Figure 2.
The P. gingivalis metabolic pathway map from BioCyc (www.biocyc.org). This is the metabolic map prior to overlaying any data onto the pathways. The name Calvin cycle was added to the map to highlight the location of this metabolic pathway. Color examples of individual metabolic pathways with accompanying proteomics data can be found in the electronic supplement.
In addition to the metabolic map, BioCyc contains an extensive database and links to other databases to help analyze results. Clicking on any compound in a pathway will pull up a small map of the pathway results with the names of the compounds and proteins involved in the pathway. The small maps contain links to larger displays of the pathways, which in turn contain links to summary pages for all of the pathway’s substrates, products and enzymes. There are links to other summary pages and often other databases. Thus, while no pathway names appear on the metabolic map (Fig. 2) due to space concerns, a user can quickly identify any pathway or pathway component of interest. It should be noted that the metabolic map is available even without data to overlay and can provide a useful reference tool. BioCyc also provides genomic views of its database, as well as extensive search functions.
The major drawback to BioCyc, and the main difference with tools such as MEV, is the need for extensive curation prior to use. In order to map the data to a metabolic pathway in Biocyc two things need to occur. First, a metabolic map needs to exist. Only a limited number of organisms have pathway/genome databases in BioCyc and these are broken down into three levels of curation. The database for Escherichia coli K-12 is extensively curated and is the most accurate and complete database in BioCyc. Twenty other organisms (April, 2008) are what are termed Tier 2 and have limited manual curation. Most of the databases, including those for P. gingivalis W83 and M. maripaludis S2, are Tier 3 and solely computationally derived. Without manual curation, these pathway maps are incomplete, and often contain errors. In the case of M. maripaludis the metabolic map does not contain methanogenesis, its central metabolic pathway. As a result, BioCyc is presently of little use for examining M. maripaludis data. The second hurdle is that the data have to be correlated to the BioCyc database to be properly displayed. Fortunately, BioCyc accepts the genome sequence gene designations or ORF numbers, e.g. PG2181, and a simple tab delimited text file containing the gene designations and abundance ratios was used to upload the P. gingivalis data.
While the metabolic map for P. gingivalis is not complete and is based on the genome of strain W83 rather than the experimental strain ATCC 33277, displaying the proteomics data on the map was still informative. Using BioCyc, we detected increased expression of thiamine biosynthetic genes in internalized versus control cells. Thiamine, also known as vitamin B1, is important for the pentose phosphate pathway (Schenk et al., 1998). P. gingivalis encodes proteins for the non-oxidative portion of the pentose phosphate pathway, which feeds into glycolysis. Both the non-oxidative pentose phosphate pathway and glycolysis showed some induction in internalized cells. These results were not as consistent as those for the thiamine biosynthetic pathway. This is not uncommon given the noisiness of the proteome data and the complexity of metabolic interactions. However, these data are consistent with the overall model of internalized cells experiencing higher nutrient levels, accompanied by an increase in metabolic activity and cell component synthesis (Xia et al., 2007b).
A cautionary example of the kinds of artifacts often seen in this type of data analysis is illustrated by the BioCyc display of the Calvin cycle in P. gingivalis (Fig. 2). The Calvin cycle is found in photosynthesis and some aerobic metabolisms and is not likely to be present in P. gingivalis. As mentioned above, genomes such as that of P. gingivalis are computationally added to BioCyc without manual curation. Most of the proteins found in the Calvin cycle are also found in the pentose phosphate or glycolysis pathways. One protein, phosphoribulokinase, is unique to the Calvin cycle and considered diagnostic of its presence in an organism. In this case the computationally generated map identified gene PG1745 as phosphoribulokinase, and from this determined that the Calvin cycle must be present in P. gingivalis. However, a blast search using PG1745 shows that it merely has sequence homology to the phosphoribulokinase/uridine kinase family. It is usually difficult to determine function from homology to a broad family. This family includes uridine kinase, which has a role in pyrimidine salvage, in addition to phosphoribulokinase and is an extremely weak indicator of a pathway that is unlikely to exist in P. gingivalis. PG1745 also has homology to threonyl-tRNA synthetase. Given that P. gingivalis has tRNA charging and probably salvages pyrimidine, either of these functions is a more likely candidate for PG1745.
Another important consideration when dealing with web-based software resources like BioCyc is that they are constantly being updated. As this review goes to press, the P. gingivalis map has been changed and no longer shows the Calvin cycle. Such changes can appear without citation or other explanatory material, so the burden is often completely on the user to assess the accuracy of the pathway annotation.
For researchers interested in oral pathogens another source of metabolic maps, though not one that can overlay proteomics data, can be found at Los Alamos National Laboratory (http://www.oralgen.lanl.gov/). The LANL site contains a searchable database for oral pathogens including genome displays, sequence searches, and metabolic pathways drawn from the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (http://genome.jp/kegg). For P. gingivalis the Los Alamos database had fewer errors than BioCyc but did not seem to be as complete. The Calvin cycle, mistakenly presented in BioCyc, was not listed among the metabolic pathways in the Los Alamos P. gingivalis database. However, when comparing the thiamine biosynthesis pathway discussed above we found that only a subset of the proteins displayed as part of the pathway by BioCyc appeared on the same pathway at the Los Alamos site. The missing proteins were in the database, and their description included the fact that they are part of thiamine biosynthesis, but they simply did not appear on the thiamine biosynthesis pathway display. This shows an advantage of using multiple databases and display programs. They provide an opportunity to cross check databases against each other, helping to overcome the weaknesses in any one pathway tool.
CLASSIFICATION AND CLUSTERING
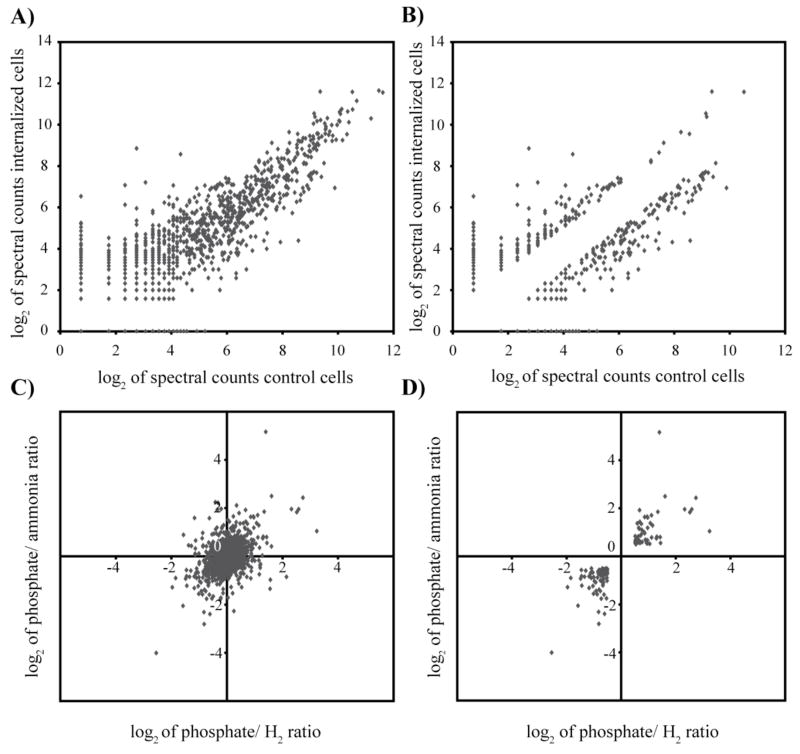
The graphical displays described above are primarily useful for examining individual experiments. Researchers dealing with experiments across multiple conditions are often interested in the patterns of abundance ratios across the experiments and grouping results with similar patterns. The expectation is that similarity with respect to protein levels is often indicative of similarity with respect to function (Eisen et al., 1998). Mathematical methods for grouping similar results have a long history in statistics and the transcriptome microarray community has borrowed and built on these methods to help identify patterns and groups of co-expressed genes from their large datasets. The result has been the development of a large number of algorithms for this purpose (Raychaudhuri et al., 2001; Chen et al., 2002; Boutros and Okey, 2005; Allison et al., 2006; Datta and Datta, 2006). These methods fall into two categories, classification and clustering, which can also be described as supervised and unsupervised methods, respectively (Allison et al., 2006). It should be noted that while classification and clustering can be of value, their use is based on an assumption that transcriptome or proteome data naturally fall into distinct groups. Evidence indicates that they do not (Bryan, 2004). As seen in Fig. 3A, C quantitative proteomics data from P. gingivalis and M. maripaludis show a continuous range of values. No obvious groups based on abundance ratio are evident. Data with natural groups would resemble something like Fig. 3B, D. Because real datasets usually show a continuum rather than obvious groups, classification and clustering can often be inappropriate tools for transcriptome and proteome data analysis. Despite this limitation, they can under some circumstances aid data interpretation given relatively high signal-to-noise datasets as inputs. Unfortunately, this requirement is often not met in practice.
Figure 3.
The absence of natural clusters in protein abundance ratio data. (A), P. gingivalis proteomic data displayed as the log2 of the spectral counts for internalized (y-axis) against control cells (x-axis). (B), As in (A), but displaying only points with at least a two-fold difference between internalized and control cells. (C), M. maripaludis proteomics data plotting the protein ratios of phosphate limited/ammonia limited samples (y-axis) against the ratios of phosphate limited/H2 limited samples (x-axis). In this display proteins regulated by phosphate limitation fall along the diagonal. (D), As in (C), but displaying only points with at least a 1.4-fold difference between conditions for both phosphate/ammonia limitation and phosphate/H2 limitation.
Classification requires the user to define training sets to be used as the basis of the groupings. Training sets consist of subsets of the data, either proteins or entire datasets, which have been defined as belonging to a specific group. This requires prior knowledge in order to properly assign members of the training set. Because the training sets are used to drive the analysis they are considered to be “supervising” the analysis (Raychaudhuri et al., 2001; Allison et al., 2006). Classification algorithms are generally used in one of two ways in proteomics and functional genomics. One use is to classify entire datasets, for example whether a protein dataset was derived from an internalized intracellular pathogen or an externally grown control. The more common use is to classify subsets of proteins according to their abundance patterns across different datasets. If a particular set of proteins was of interest and known to be co-expressed, classification could be used to find other proteins in the dataset that matched their abundance pattern.
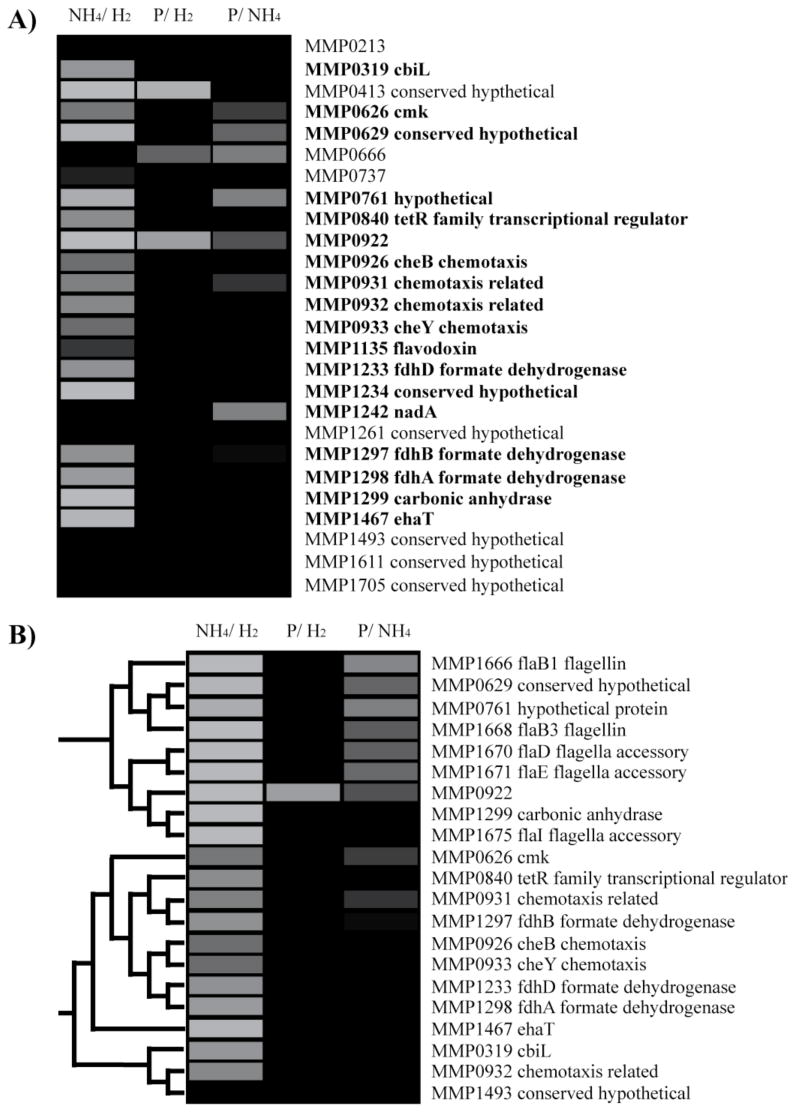
There are numerous classification and clustering algorithms incorporated into MEV. A comprehensive review of different classification techniques is beyond the scope of this paper. Other analysis packages can be found at http://ihome.cuhk.edu.hk/~b400559/arraysoft_mining_specific.html; classification and clustering algorithms are also available as part of commercial systems such as Elucidator from Rosetta Inpharmatics (http://www.rosettabio.com/products/elucidator/default.htm). As an example of classifying proteins according to their abundance patterns, we conducted a classification to find proteins with patterns like those of the M. maripaludis flagella proteins (Fig. 4A). For this example, we chose to use the K nearest neighbor algorithm (Soukas et al., 2000) implemented in MEV. K nearest neighbor compares the abundance of each protein under different conditions with members of the training sets and calculates a mathematical “distance” between the protein’s abundance profiles. There are a number of different methods used for calculating distance. The default settings for the K nearest neighbor classification in MEV use Euclidean distance (http://www.tmr.org/documentation/MeV_Manual_4_1.pdf). The K nearest neighbor algorithm then uses the class of the K nearest proteins as determined by the Euclidean or other distance metric, with K being set by the user, and assigns the protein to the class that has the highest representation amongst the K nearest proteins. In this example we used the same M. maripaludis proteomics dataset employed previously (Fig. 1C). We set up the example to identify proteins showing the same regulatory pattern as the flagella operon, which showed decreased abundance under ammonia-limited growth. There were two classes, those with patterns matching the flagella proteins, and those with other abundance patterns. Constructing a class required designating certain proteins as known members of the class. The training set for the flagella protein class included all the flagellar proteins. The training set for the second class included proteins from the methanogenesis and related pathways expected to respond primarily to H2, MMP0127, MMP1382-5, and MMP1692-1696; phosphate binding proteins known to respond to phosphate limitation, MMP1095, MMP1098-1099; and nitrogen metabolism proteins known to show increased abundance under ammonia limitation, the opposite regulation trend of the flagella genes, MMP0064-66 (Cohen-Kupiec et al., 1997; Hendrickson et al., 2007). The resulting flagella protein class (Fig. 4A) included 26 proteins. Many of these are chemotaxis or putative chemotaxis genes, consistent with a class based on flagella proteins. Interestingly, the list also includes a putative transcriptional regulator that may play a role in reduced abundance under ammonia limitation. Surprisingly, a number of proteins for formate dehydrogenase, which feeds into the methanogenesis pathway, also cluster with the flagella proteins. Previous studies suggested that formate dehydrogenase would respond to H2 rather then ammonia-limiting conditions (Hendrickson et al., 2007).
Figure 4.
Classification and clustering of the M. mairpaludis proteome. Each row represents an ORF from M. maripaludis. The annotation of each ORF is listed at the end of the row. The columns represent experimental conditions. NH4/H2, ammonia limiting compared to H2 limiting growth conditions; P/H2, phosphate limiting compared to H2 limiting growth conditions; P/NH4, phosphate limiting compared to ammonia limiting growth conditions. The abundance changes are shown in gray scale with black showing no change in abundance. A color version of the figure can be found in the electronic supplement. (A), Flagella protein expression class. Proteins were classified as matching the abundance pattern of flagella proteins using the K nearest neighbor algorithm in MEV (www.tm4.org/mev.html). A leave-one-out validation was conducted for the classification (Theilhaber et al., 2002). The procedure involves rerunning the classification leaving out one of the original training group proteins, and doing this for all of the members of the original training group. Proteins that were present in the class across the entire leave-one-out validation are indicated in bold. (B), A subsection of the agglomerative hierarchical clustering of the M. maripaludis proteome in MEV. The dendrogram shows the protein relationships in the clusters.
Classification techniques are subject to several sources of error. In brief, the major concerns are inappropriate choices for the training sets, over-fitting, and selection bias (Raychaudhuri et al., 2001; Ambroise and McLachlan, 2002; Allison et al., 2006). If the samples chosen for the training set are assigned to the set in error or cannot explain the phenotype of interest, then the classification is likely to fail. For the example (Fig. 4A) the training set for proteins with non-flagella abundance patterns included several proteins expected to respond to H2 limitation. Examining the abundance patterns (data not shown) of the proteins representing H2 limitation demonstrated that they do not in fact have patterns like the flagella proteins. However, the classification results found that formate dehydrogenase proteins, also expected to respond to H2 limitation, are grouped with the flagella proteins. Because they were expected to respond to H2 limitation these proteins could easily have been mistakenly placed in the non-flagella training set. To demonstrate such a situation, the classification was rerun adding formate dehydrogenase and carbonic anhydrase to the non-flagella class. The flagella class was reduced to only three proteins (data not shown). Over-fitting occurs when the algorithm becomes so fitted to the training set that its predictive functions perform well only within the training set and poorly with new data. This will exclude many real new members of the class. Over-fitting is usually only a problem when the number of parameters used for a comparison is high. Given that we are comparing proteins using only three datasets, over-fitting is of little concern in our example and for most classifications of proteins by abundance patterns. When presented with a large number of parameters, as is generally the case when classifying datasets rather than proteins, one can guard against over-fitting by using only a subset of the data. However, selection bias can occur if there is bias in the subset of data chosen. Selection bias can significantly impact the reliability of the results (Ambroise and McLachlan, 2002; Fu et al., 2005). For example, if a study compared proteins from a number of culture grown samples and a number of different disease isolates, using only the culture grown samples for the classification rule would eliminate any information about disease from the results. Our example employs only three conditions and there is no reason to leave any of the conditions out of the model, thus selection bias cannot be a problem for this classification. Given the possible pitfalls of classification, the results are often checked by cross-validation methods (Ambroise and McLachlan, 2002; Fu et al., 2005; Allison et al., 2006). Briefly, cross-validation in this context refers to a broad class of statistical methods that involve partitioning the dataset into training sets and partitions that are reserved for validation of the results contained in the training set (Theilhaber et al., 2002).
Because it draws on pre-existing knowledge, classification tends to be used to ask specific questions. The discovery of patterns in the data, especially unexpected patterns, is the strength of clustering, or unsupervised algorithms. Unlike supervised methods, clustering does not require any prior knowledge of the proteins or conditions in question. Instead, the clusters are organized according to their similarity to all other proteins or conditions under consideration. Many different clustering algorithms have been developed (Sherlock, 2000; Raychaudhuri et al., 2001; Allison et al., 2006). As with the K nearest neighbor classification discussed above, they all use some measure of mathematical distance between the proteins or conditions to be compared. Hierarchical algorithms construct trees of the entire dataset grouping proteins with other proteins based on the calculated distance. Some algorithms require the user to define the number of clusters into which the data will be grouped (Sherlock, 2000). The algorithm then uses the calculated distance to split the proteins into that many groups. Finally, some methods employ a user defined minimum correlation for a cluster (Sherlock, 2000). Such algorithms use the calculated distances to construct groups where the proteins in each group possess the user defined minimum correlation or higher. Unlike the other clustering techniques, minimum correlation algorithms will not necessarily cluster all the proteins in the dataset.
For an example of a commonly employed clustering technique, we chose agglomerative hierarchical clustering. The agglomerative hierarchical clustering algorithm from the MEV program was applied to the M. maripaludis dataset (Fig. 4B). Agglomerative hierarchical clustering compares all the proteins with each other and then pairs the two proteins with the closest abundance patterns. The proteins are merged into a ‘node’ treating them as a unit with an averaged abundance profile. This process continues until all the elements have been joined, resulting in a hierarchical tree, clustering elements with similar patterns together (Eisen et al., 1998). For this example the default parameters were used, Euclidean distances and average linkage clusters. As with all hierarchical clustering, the result was a tree including all proteins detected in the study. A subsection of the tree is presented in Fig. 4B. The subsection contains the same flagella proteins that were used to construct the classification (Fig. 4A). As expected, the flagella proteins cluster together. However, the carbonic anhydrase protein, which might be expected to cluster with the other proteins from the formate dehydrogenase operon, is also part of the cluster. The neighboring cluster with a similar overall abundance pattern is shown below (Fig. 4B). This cluster contains predominantly the chemotaxis and formate dehydrogenase proteins as seen previously (Fig. 4A). All of the proteins in this cluster were classified with the flagella proteins in the classification example discussed above. The advantage of clustering is that instead of specifically looking for proteins with regulation similar to the flagella proteins, one can run the clustering algorithm and then look at any interesting patterns that are found. In this example the overall results were similar to that produced in the classification example, but without the requirement of deciding what to look for before hand or the risk of designing inappropriate training sets. Clustering also has limitations, however. Clustering algorithms will force the creation of clusters, whether they are biologically meaningful or not. Clusters will be produced even if the algorithms are given random noise as an input. Because of this, and because clustering normally has no default result to test against, clustering has no correct answer (Garge et al., 2005). This also means that different clustering algorithms produce different results and there is no real consensus as to the best choice of method for specific situations (Sherlock, 2000; Yeung et al., 2001; Datta and Datta, 2003). Clustering methods are also poorly reproducible. All common clustering methods generally show poor reproducibility when using less then 50 samples (Yeung et al., 2004; Garge et al., 2005). To put this into context, the clustering example (Fig. 4B) uses only 3 samples. To deal with the problem of poor reproducibility, resampling techniques have emerged as a method for testing the reproducibility of clustering within a dataset (Datta and Datta, 2003; Bryan, 2004; Garge et al., 2005). In these procedures, subsets of the original sample are resampled and the clustering algorithm applied. The consistency of the results provides a measure of reproducibility. However, even these techniques are of limited use when dealing with small datasets (Bryan, 2004). The M. maripaludis proteomics set covers only three conditions with two replicates each for a total of six measurements for each ORF, hardly appropriate for resampling techniques, but typical of the limited number of samples and replicates found in such studies.
ONTOLOGY TOOLS
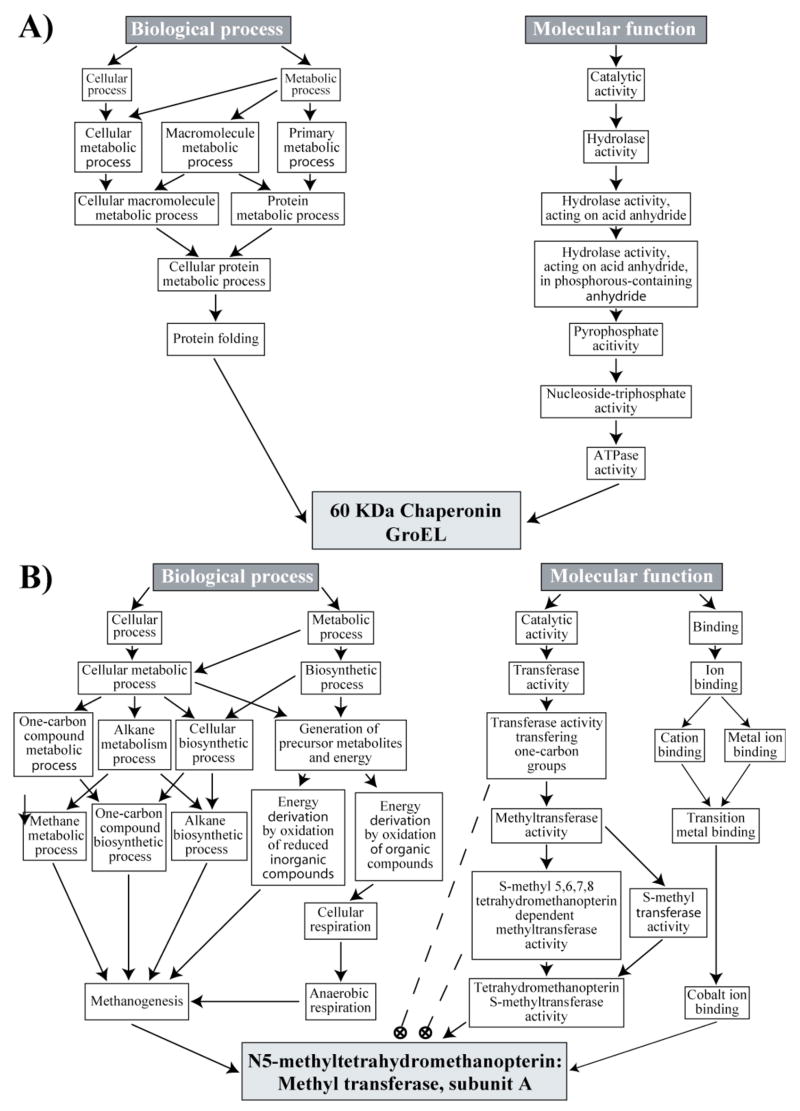
The newest set of tools being developed for transcriptome and proteome analysis employ the concept of ontology. There are several definitions that apply in different fields, but here we are concerned with the concept of ontology as it has evolved in computer science and computational biology. For our purposes ontology can be thought of as an attempt to describe a part, or possibly all, of the universe of interest to a scientific discipline using a system of highly specific, hierarchical categories and their relationships (Mizoguchi, 2001). The categories and relationships must be so well defined as to be machine-readable and easily manipulated by computer. While a few other specialized biological ontologies have been developed, for transcriptome and proteome analysis the primary ontology is the Gene Ontology (GO) (http://www.geneontology.org/(Ashburner et al., 2000)), which evolved from the human gene ontology (HUGO). Examples of the Gene Ontology are shown for both a P. gingivalis and an M. maripaludis protein (Fig. 5). The ontology for the 60 kDa chaperonin GroEL protein of P. gingivalis (PG0520) (Fig. 5A) is taken from the GO website given above. The hierarchical structure is clearly evident. “Protein folding” is a “Cellular protein metabolic process” is a “Protein metabolic process”, etc. It also shows that ontologies are not classification systems like phylogenies. Categories in a specific ontology can have multiple relationships, such that “Cellular protein metabolic process” is also a “Cellular macromolecule metabolic process”. It should be noted that despite the ability to assign multiple relationships, GroEL was only directly assigned the categories leading through the “Protein folding” category of “Biological process”. GroEL is commonly annotated as a heat shock protein (Maeda and Miyamoto et al., 1994). GO has a Biological process category “Response to heat”, but GroEL has not been assigned to this category. GroEL’s placement in the ontology is therefore either incomplete or the curator felt that GroEL really didn’t belong to the “Response to heat” category, despite the wide spread view of GroEL as a heat shock protein. Thus, even for proteins fortunate enough to be placed in the ontology, the categories might not be what a user expects.
Figure 5.
Ontology diagrams of proteins from P. gingivalis and M. maripaludis. (A), Ontology of the 60 kDa chaperonin GroEL (PG0520) from P. gingivalis. The position of GroEL is shown for two of the three ontologies in the Gene Ontology (http://www.geneontology.org/), Biological process and Molecular function. The light gray box represents the protein. All other boxes represent ontology terms. The arrows indicate category relationships. (B), Ontology of the protein N5-methyltetrahydromethanopterin: Methyl transferase A (MtrA) (MMP1564) from M. maripaludis. The position of the methanogenesis protein MtrA as given in GoMiner (http://discover.nci.nih.gov/gominer/) is shown for two of the three ontologies in the Gene Ontology, Biological process and Molecular function. The light gray box represents the protein. All other boxes represent ontology terms. The arrows indicate category relationships. Dashed lines and circled crosses indicate categories to which the protein was not assigned (see text discussion).
Most users will not interact directly with GO, but will use ontology tools that employ GO. An example of GO executed through an ontology tool, GoMiner, discussed in more detail below, is given for the M. maripaludis methanogenesis protein N5- methyltetrahydromethanopterin: Methyl transferase A (MtrA, MMP1564) (Fig. 5B). While in theory an entry inherits all of the higher categories in the ontology this is not the case for this example. MtrA is not assigned to several categories under “Molecular function” shown by the dashed lines ending in circled crosses (Fig. 5B). This is not an inherent property of the ontology. Presumably the inconsistency arose from either poor placement of the protein into the ontology or GoMiner’s method for extracting information from GO. Such inconsistency could effect the results generated by using an ontology tool and should be kept in mind by users. Those interested in a critique of the actual structure of GO should consult Schulze-Kremer, 2002. It should also be noted that GO is not a single ontology, but three separate ontologies. “Biological process” covers events accomplished by one or more ordered assemblies of molecular functions, for example signal transduction. “Molecular function” describes activities at the molecular level such as catalysis or binding. “Cellular component” (not shown in Fig. 5) covers the components of the cell, for example a specific organelle or location.
There are several uses for ontologies, such as integrating the large number of different, autonomous databases of biological information or clearing up the semantic ambiguity in biological nomenclature (Schulze-Kremer, 2002). However, probably the most important aspect of ontologies for proteomics and functional genomics is that they provide a consistent structure for categorizing genes and proteins that yields readily to machine searchable annotation. A protein assigned to the “One-carbon compound biosynthetic process” will never be called “Biosynthetic process one-carbon compound” or “One-carbon compound process”. Thus all the proteins assigned to this category will have the exact same label for purposes of computation. The ontology tools take advantage of this machine searchable aspect to look for categories that show abundance change within a dataset (Zeeberg et al., 2003; Pavlidis et al., 2004; Subramanian et al., 2005; Huang et al., 2007). If a large number of proteins in the category “One-carbon compound biosynthetic process” displayed altered abundance in an experiment the ontology tool would list “One-carbon compound biosynthetic process” as altered. The primary goal is to provide a more efficient way to mine large quantities of data.
Ontology tools fall into of one of two general groups (Pavlidis et al., 2004). One type of tool looks for over-representation of changed proteins in a category. The user supplies the tool with two lists, one of all proteins in the experiment and the other of those proteins that are changed in the condition of interest. The ontology is used to determine the categories for each protein. Then the tool calculates how likely the number of changed proteins in each category is to arise by chance from the overall list. Those categories that are over-represented in changed proteins are then reported. The second type of tool uses what has been called “functional class scoring” (Pavlidis et al., 2002; Pavlidis et al., 2004). These tools take into consideration the statistical likelihood that any protein is changed in the condition of interest, most commonly given as a p-value. The tools then calculate the likelihood that any category is changed using the statistics for every protein in the category. The advantage of this method is there is no need to group the proteins into changed and unchanged, instead the program uses the statistical power of the results for all the proteins in a category to determine the likelihood that a category is altered. To our knowledge only two ontology tools use “functional class scoring”, erminej (http://www-bioinformatics-ubc-ca/ermineJ/ (Pavlidis et al., 2004)) and GOdist (http://basalganglia.huji.ac.il/links.htm (Ben-Shaul et al., 2005)), and at this time neither program can easily be made to accept microbial proteomic datasets as inputs.
We tested two ontology tools that employ over-representation algorithms, GoMiner (http://discover.nci.nih.gov/gominer/(Zeeberg et al., 2003; Zeeberg et al., 2005)) and DAVID (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/(Huang et al., 2007)). More ontology tools can be found at http://www.geneontology.org/GO.tools.shtml. Like BioCyc, both of these tools require files identifying the proteins to be associated with the databases underlying the programs. While both tools employed a number of different protein identifiers, neither took the gene designations from the sequencing projects, e.g. PG2181. For GoMiner we obtained the Universal Protein Resource (UniProt) identity numbers for the proteins from http://www.ebi.ac.uk/trembl/. For DAVID, entrez gene numbers (http://www.ncbi.nlm.nih.gov/sites/entrez) were used. Even when using protein identifiers recognized by the tool, only a subset of P. gingivalis and M. maripaludis proteins could be mapped onto the databases. The Gene Ontology came out of the Human Gene Ontology and the consortium working on GO is primarily focused on eukaryotic organisms, although the J. Craig Venter Institute and the Plant-Associated Microbe Gene Ontology consortium are also associated with the project (http://www.geneontology.org/GO.consortium.shtml#fulllist). The consequence is that GO is not well curated for many microbial species. Thus, not every protein from these organisms has been entered into GO and not all of the entries are complete.
We analyzed the same P. gingivalis dataset used with BioCyc (Fig. 4) (Xia et al., 2007b) with GoMiner. GoMiner takes two files. The first is the overall list of proteins, a simple text file listing the protein identifiers. In this case we used the list of detected proteins from the proteomics measurements rather than the list of all the proteins from the genome sequence. This is because GoMiner will look for over-representation of the changed proteins in a category compared to the number of proteins in that category in the first file, so changing the number of proteins in a category changes the statistics. The second file contains the proteins that statistics indicate have altered abundance. Of the 1195 protein identifiers in the overall list, only 868 had matches in GO, demonstrating the incomplete nature of the GO entries for an organism like P. gingivalis. The list of changed proteins included the direction of change. With this information GoMiner generated three sets of results. One looks at overall change in a category, regardless of direction. The others look only at over-representation of proteins with increased abundance or decreased abundance, respectively. The results provided by GoMiner are lists containing each GO category, the total number of proteins in that category in the overall protein list, the number of changed proteins in that category, and measures of the statistical likelihood that the category shows altered abundance between the samples. The top 13 categories for under expression in internalized P. gingivalis cells are shown in Table 1. Several of the categories in Table 1 are consistent with the previous proteomics analysis (Xia et al., 2007b). Internalized P. gingivalis is noted for the reduction of major surface proteins. This is consistent with the top category for reduced abundance, outer membrane. Previous studies have also shown a significant decrease in iron acquisition systems, especially outer membrane receptors and lipoproteins, and this was reflected in several categories, including metal cluster binding, iron binding and iron-sulfur cluster binding.
Table 1.
Example of Tabulated Output From GoMiner, Internalized P. gingivalis
| GO category | Total Proteins in category | Under-expressed proteins | p-valuea | FDRb |
|---|---|---|---|---|
| Outer membrane | 21 | 13 | 0.0000 | 0.0000 |
| Receptor activity | 13 | 9 | 0.0001 | 0.0000 |
| 4 iron, 4 sulfur cluster binding | 18 | 9 | 0.0018 | 0.0000 |
| Metal cluster binding | 25 | 11 | 0.0021 | 0.0000 |
| Iron-sulfur cluster binding | 25 | 11 | 0.0021 | 0.0000 |
| Molecular transducer activity | 20 | 9 | 0.0045 | 0.0250 |
| Signal transducer activity | 20 | 9 | 0.0045 | 0.0250 |
| Electron carrier activity | 20 | 9 | 0.0045 | 0.0250 |
| Iron ion binding | 28 | 11 | 0.0061 | 0.0667 |
| Transporter activity | 78 | 22 | 0.0142 | 0.2600 |
| Carbohydrate metabolic process | 71 | 20 | 0.0196 | 0.3333 |
| Cofactor binding | 71 | 20 | 0.0196 | 0.3333 |
| Cellular catabolic process | 29 | 10 | 0.0243 | 0.5067 |
probability that category is actually unchanged between conditions
false discovery rate (Benjamini and Hochberg, 1995)
The P. gingivalis example also shows one of the difficulties in interpreting the output from an ontology program. Several of the under-expressed categories are subsets of each other. GoMiner may be reporting the same result several times. In addition to the lists of GO categories, GoMiner also has a hierarchical display of the results, making it easier to visualize the relationships among the different categories. The hierarchical display (not shown) indicated that the 4 iron, 4 sulfur cluster binding category is a member of the iron-sulfur cluster binding category that is in turn part of the metal cluster binding category. The output also gave the number of proteins in each category. The numbers of proteins in the three categories were very similar, in fact both metal cluster binding and iron-sulfur cluster binding contained the same 25 proteins. Thus, the three iron categories are effectively the same result displayed three times. Users can try to cut down on this confusion by restricting GoMiner to use only certain levels of the ontology, but this requires knowledge of the overall ontology structure and the scope of the results of interest and we did not use this function.
The Database for Annotation, Visualization and Integrated Discovery (DAVID) was also tested using the P. gingivalis and M. maripaludis datasets. The results were roughly similar to those obtained with GoMiner, but there were differences worth noting. DAVID only takes lists of proteins and lists of changed proteins, unlike GoMiner, that can also accept information about the direction of the abundance change for a protein. This means that to generate the same three types of analyses seen in GoMiner, overall change, increased abundance, and decreased abundance, three separate analyses have to be run in DAVID. While GoMiner uses only the Gene Ontology, DAVID employs the GO and other databases to provide categories. The databases can be selected by the user and include Uniprot and KEGG as options. Thus DAVID is more likely to find a category for any submitted protein. The same dataset that yielded 868 categorized proteins using GO yielded a category for every protein, 1199, in DAVID. The protein annotations may also be more complete in databases other than GO. For example, GO only recognized 18 of the 36 methanogenesis proteins in a simulated M. maripaludis dataset. When this same dataset was run with DAVID, one of the over-represented categories was the KEGG folate pathway, which contains the methanogenesis pathway. This included a link to KEGG with the changed proteins highlighted, clearly showing that all of the methanogenesis proteins were identified and had altered abundance. However, there are negatives associated with using multiple databases. Just like BioCyc’s computational pathway assignments, trying to match every protein to a broad array of databases can yield some categories inconsistent with the known biology of the organism. Indeed, one category that DAVID found with increased abundance for internalized P. gingivalis was photosynthesis, as was seen with BioCyc. Until organisms like P. gingivalis are better represented in GO, however, the benefits of using DAVID are likely to outweigh the drawbacks.
CONCLUDING REMARKS
The wealth of data available from whole genome transcriptome analysis and whole cell proteomics presents opportunities to greatly increase our understanding of biological systems. However, interpreting large datasets can be a substantial undertaking. Combing through thousands of data points by eye is time consuming and prone to personal biases and missed results. To make analyzing large datasets more efficient, a number of interpretive aids have evolved. The simplest of the analysis tools help identify obvious patterns in the data. Beads-on-a-string, clustering, and classification can help identify groups of co-expressed proteins. Co-expression can give clues to the biological changes being examined and the role of unknown proteins. Visualization tools such as MEV are insensitive to the type of input and thus are readily adaptable to proteomics data, despite their origins in transcription analysis. With these tools, drawing biological conclusions from patterns is entirely in the hands of the user. Other tools seek to directly provide biological interpretation. Mapping relative abundance data onto biochemical pathways can highlight changes in metabolism and biological processes. Ontology tools, the latest set of interpretive aids, try to identify biologically relevant categories that undergo change in the experiment. The more direct biological interpretation tools are heavily dependent on the accuracy and completeness of their underlying databases. They are generally more accurate and complete for a few well-studied organisms, such as humans or Escherichia coli. For less studied microorganisms the tools can still be useful, provided the user is careful about validating the results and is knowledgeable regarding the organism in question. None of these tools can be used naively with the expectation of generating valid information. Biocyc, GoMiner, DAVID, etc. are best viewed as tools to speed the process of discovery, not as replacements for skilled human judgment.
Acknowledgments
This work was supported by the NIDCR under RO1 DE014372 and RO1 DE011111, with additional support provided under RO1 GM074783. We also thank Todd Kitten, Qiangwei Xia, Tiansong Wang, Fred Taub, Gundula Bosch, and John A. Leigh for their assistance.
References
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci U S A. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg YJ. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Royal Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- Ben-Shaul Y, Bergman H, Soreq H. Identifying subtle interrelated changes in functional gene categories using continuous measures of gene expression. Bioinformatics. 2005;21:1129–1137. doi: 10.1093/bioinformatics/bti149. [DOI] [PubMed] [Google Scholar]
- Boutros PC, Okey AB. Unsupervised pattern recognition: an introduction to the whys and wherefores of clustering microarray data. Brief Bioinform. 2005;6:332–343. doi: 10.1093/bib/6.4.331. [DOI] [PubMed] [Google Scholar]
- Bryan J. Problems in gene clustering based on gene expression data. J Multivariate Anal. 2004;90:44–66. [Google Scholar]
- Chen G, Jaradat SA, Banerjee N, Tanaka TS, Ko MSH, Zhang MQ. Evaluation and comparison of clustering algorithms in analyzing ES cell gene expression data. Stat Sinica. 2002;12:241–262. [Google Scholar]
- Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M800203-MCP200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland WS. LOWESS-a program for smoothing scatterplots by robust locally weighted regression. The American Statistician. 1981;35:54–54. [Google Scholar]
- Cohen-Kupiec R, Blank C, Leigh JA. Transcriptional regulation in Archaea: In vivo demonstration of a repressor binding site in a methanogen. Proc Natl Acad Sci U S A. 1997;94:1316–1320. doi: 10.1073/pnas.94.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Datta S. Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics. 2003;19:459–466. doi: 10.1093/bioinformatics/btg025. [DOI] [PubMed] [Google Scholar]
- Datta S, Datta S. Methods for evaluating clustering algorithms for gene expression data using a reference set of functional classes. BMC Bioinformatics. 2006;7:397. doi: 10.1186/1471-2105-7-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M, Spellman P, Brown P, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectra of peptides with amino acid sequences in a protein database. J Amer Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fu WJ, Carroll RJ, Wang S. Estimating misclassification error with small samples via bootstrap cross-validation. Bioinformatics. 2005;21:1979–1986. doi: 10.1093/bioinformatics/bti294. [DOI] [PubMed] [Google Scholar]
- Garge NR, Page GP, Spraque AP, Gorman BS, Allison DB. Reproducible clusters from microarray research: Whither? BMC Bioinformatics. 2005;6(Suppl 2):S10. doi: 10.1186/1471-2105-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett M. Science, marketing and wishful thinking in quantitative proteomics. Proteomics. 2008 doi: 10.1002/pmic.200800358. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanoccoccus maripaludis. J Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson EL, Xia Q, Wang T, Leigh JA, Hackett M. Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Analyst. 2006;131:1335–1341. doi: 10.1039/b610957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson EL, Haydock AK, Moore BC, Whitman WB, Leigh JA. Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc Natl Acad Sci U S A. 2007;104:8930–8934. doi: 10.1073/pnas.0701157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kim YS, Lee JW. Comparison of various statistical methods for identifying differential gene expression in replicated microarray data. Stat Methods Med Res. 2006;15:3–20. doi: 10.1191/0962280206sm423oa. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line, Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Meila M, Xia Q, Hackett M. Mass spectrometry-based proteomics and its application to studies of Porphyromonas gingivalis invasion and pathogenicity. Infect Disord Drug Targets. 2006;6:311–325. doi: 10.2174/187152606778249935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Maeda H, Miyamoto M, Hongyo H, Nagai A, Kurihara H, Muryama Y. Heat shock protein 60 (GroEL) from Porphyromonas gingivalis: Molecular cloning and sequence analysis of its gene and purification of the recombinant protein. FEMS Microbiol Lett. 1994;119:129–136. doi: 10.1111/j.1574-6968.1994.tb06879.x. [DOI] [PubMed] [Google Scholar]
- Mizoguchi R. Ontological Engineering: Foundation of the next generation knowledge processing. Lect Notes Comput Sci. 2001;2198:44–57. [Google Scholar]
- Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, et al. Determination of the Genome Sequence of Porphyromonas gingivalis Strain ATCC 33277 and Genomic Comparison with Strain W83 Revealed Extensive Genome Rearrangements in P. gingivalis. DNA Res. 2008 doi: 10.1093/dnares/dsn013. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Aebersold R. Analysis, statistical validation and dissemination of large-scale proteomics datasets generated by tandem MS. Drug Discov Today. 2004;9:173–181. doi: 10.1016/S1359-6446(03)02978-7. [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Lewis DP, Noble WS. Exploring gene expression data with class scores. Proceedings of the Pacific Symposium on Biocomputing; Jan 3–7, 2002; Lihue, HI. 2002. pp. 474–485. [PubMed] [Google Scholar]
- Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Sutphin PD, Chang JT, Altman RB. Basic microarray analysis: grouping and feature reduction. Trends Biotechnol. 2001;19:189–193. doi: 10.1016/s0167-7799(01)01599-2. [DOI] [PubMed] [Google Scholar]
- Schenk G, Duggleby RG, Nixon PF. Properties and functions of the thiamine diphosphate dependent enzyme transketolase. Int J Biochem Cell Biol. 1998;30:1297–1318. doi: 10.1016/s1357-2725(98)00095-8. [DOI] [PubMed] [Google Scholar]
- Schulze-Kremer S. Ontologies for molecular biology and bioinformatics. In Silico Biol. 2002;2:179–193. [PubMed] [Google Scholar]
- Sherlock G. Analysis of large-scale gene expression data. Curr Opin Immunol. 2000;12:201–205. doi: 10.1016/s0952-7915(99)00074-6. [DOI] [PubMed] [Google Scholar]
- Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;14:963–980. [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilhaber J, Connolly T, Roman-Roman S, Buxhnell S, Jackson A, Call K, et al. Finding genes in the C2C12 osteogenic pathway by k-nearest neighbor classification of expression data. Genome Res. 2002;12:165–176. doi: 10.1101/gr.182601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., III Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- Xia Q, Hendrickson EL, Wang T, Lamont RJ, Leigh JA, Hackett M. Protein abundance ratios for global studies of prokaryotes. Proteomics. 2007a;7:2904–2919. doi: 10.1002/pmic.200700267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Wang T, Taub F, Park Y, Capestany CA, Lamont RJ, et al. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics. 2007b;7:4323–4337. doi: 10.1002/pmic.200700543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
- Yeung KY, Medvedovic M, Bumgarner RE. From co-expression to co-regulation: how many microarray experiments do we need? Genome Biol. 2004;5:R48. doi: 10.1186/gb-2004-5-7-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, et al. High- throughput GoMiner, an ‘industrial- strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of common variable immune deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]