Abstract
Motivation: High-density tiling microarrays are increasingly used in combination with ChIP assays to study transcriptional regulation. To ease the analysis of the large amounts of data generated by this approach, we have developed ChIP-on-chip Analysis Suite (CoCAS), a standalone software suite which implements optimized ChIP-on-chip data normalization, improved peak detection, as well as quality control reports. Our software allows dye swap, replicate correlation and connects easily with genome browsers and other peak detection algorithms. CoCAS can readily be used on the latest generation of Agilent high-density arrays. Also, the implemented peak detection methods are suitable for other datasets, including ChIP-Seq output.
Availability: The software is available for download along with a sample dataset at http://www.ciml.univ-mrs.fr/software/ferrier.htm.
Contact: ferrier@ciml.univ-mrs.fr; andrau@ciml.univ-mrs.fr; spicuglia@ciml.univ-mrs.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In the last few years, coupling of chromatin immunoprecipitation with microarray technology (ChIP-on-chip; Ren et al., 2000) and computational analysis tools has resulted in major leaps in our understanding of transcriptional networks and of the dynamics of chromatin structure (Bock and Lengauer, 2008). Microarray analysis is a stepwise process which encompasses spot detection in scanned images, normalization of fluorescence intensities within and between arrays, as well as probeset to gene assignment. In the case of ChIP-on-chip (CoC), this process comprises the additional processing of binding events, also known as peak detection. Several CoC analysis software solutions already exist, often adapted for one specific microarray platform. To our knowledge, in the case of Agilent microarrays, only one application suite is currently available: DNA Analytics (http://chem.agilent.com), a licensed program. Here, we introduce a new standalone ChIP-on-chip Analysis Suite (CoCAS) that provides several additional functions, including new normalization options, flexible peak detection, quality control reports, as well as a compilation of replicate samples. CoCAS is free (GPL) software which runs independently on Windows XP/Vista, Mac OSX, Linux and builds upon existing packages in the Java and R programming languages (http://www.r-project.org), notably BioConductor (http://bioconductor.org). CoCAS uses Java as graphical user interface as well as peak detection, and R for the bulk of the calculations.
2 PROCEDURES
As input, CoCAS takes Feature Extraction files (Agilent Technologies) originating from scanner quantification. Microarray files are read in R using BioConductor. Since two-channel normalization methods tend to underestimate enrichment, we made variance stabilization normalization (Huber et al., 2002) available in our software, as opposed to other Agilent CoC analysis programs. We also adapted, implemented and validated a novel CoC optimized intra-normalization method (Peng et al., 2007) de novo in R (Supplementary Fig. S1). These methods can now be used along with other traditional intra- and inter-normalization methods: median, loess and quantile (Yang et al., 2002) (Supplementary Fig. S2). Background subtraction can be carried out using all options limma (Smyth, 2004) offers in this regard, or disabled. A per-spot P-value is systematically calculated according to the Rosetta error model (Weng et al., 2006), which can be used for peak detection. Multiple slide designs are handled as separate experiments until inter-array normalization, after which they are merged as one whole experiment. Experimental and/or biological replicates can be merged either using a mean of log ratios, or the Rosetta error model. Peak detection is automatically performed in Java following microarray processing. The peak detection tab can be called from within the main interface at any time for standalone peak detection. The algorithm is based on the neighbourhood effect (Zheng et al., 2007). Significantly enriched probes are first mapped above a given threshold based on background noise estimation as used by Ringo (Toedling et al., 2007) or MPeak (Zheng et al., 2007). Peaks are extended as long as the log ratio of contiguous probes is greater than the extension threshold. A score is given by calculation of the effective peak area.
3 RESULTS AND CONCLUSION
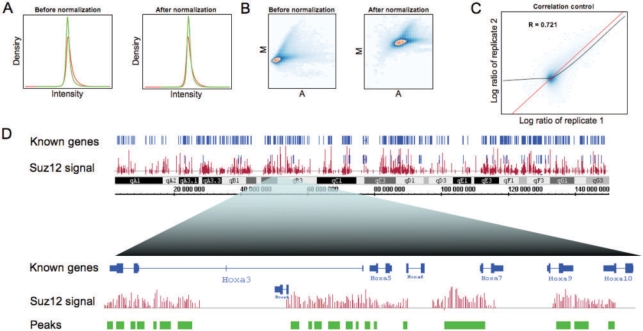
CoCAS features either a simple stepwise wizard with detailed help which facilitates analyses, or a user-parameterized interface allowing more flexibility (an example screenshot of the interface is shown in Supplementary Fig. S3). It can handle large files originating from new high-density microarrays (>1 000 000 probes). Dye swap can be carried out on a selection of slides and replicate correlation plots are displayed. As illustration, we provide genome-wide profiling of Suz12, a subunit of the Polycomb repressor complex, performed in mouse ES cells, and processed with CoCAS (Fig. 1 and Supplementary Material S1). Because Suz12 is located throughout the genome (Boyer et al., 2006), we applied median normalization in this case. A PDF Quality Control report is generated for global estimation of per-slide enrichment (Fig. 1A–C). Resulting output is written as several generic file formats that are readable on most genome browsers, such as Integrated Genome Browser (IGB), Ensembl (http://ensembl.org) or UCSC genome browser (http://genome.ucsc.edu) (Supplementary Fig. S4), a function supported by most CoC packages, except for the Agilent platform, as of yet (Supplementary Table S1). As expected, our software shows high Suz12 enrichment at the genome-wide scale, notably in the Hox cluster region (Fig. 1D and data not shown). Importantly, the peak detection methods implemented in CoCAS can be used for any set of data (in GFF format), including ChIP-Seq data (Supplementary Fig. S5), where signal processing is similar to that of CoC.
Fig. 1.
Stepwise data analysis of Suz12 ChIP-on-chip in CoCAS. Quality control reports include (A) density plots of immunoprecipitated (IP) DNA, in red, and Input DNA, in green, so as to detect any dye bias; (B) MA plots which allow assessment of normalization quality and probe enrichment; (C) replicate correlation plots, which also help estimate background noise (which shows no correlation at low intensities). (D) Chromosomal view (chromosome 6) of Suz12 IP over input log ratios (in red) via IGB (top), followed by peak detection (green track) on a close up in the Hox cluster region (bottom).
Funding: Inserm, CNRS, Association pour la Recherche sur le Cancer, Institut National du Cancer, Fondation de France, Association Laurette Fugain, Fondation Princesse Grace de Monaco and Commission of the European Communities (to Ferrier laboratory); Inserm, Université de la Méditerranée and Association pour la Recherche sur le Cancer (to Imbert laboratory); Agence Nationale de la Recherche (ANR-06-BYOS-0006 for collaboration between the two groups and to T.B.); fellowship from Institut National du Cancer (to P.C.); Marie Curie Research Training Network (RTN ‘Chromatin Plasticity’) from the Commission of the European Communities (to F.K.).
Conflict of Interest: none declared.
Supplementary Material
References
- Bock C, Lengauer T. Computational epigenetics. Bioinformatics. 2008;24:1–10. doi: 10.1093/bioinformatics/btm546. [DOI] [PubMed] [Google Scholar]
- Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Huber W, et al. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Peng S, et al. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 1. [DOI] [PubMed] [Google Scholar]
- Toedling J, et al. Ringo–an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L, et al. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- Yang YH, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, et al. ChIP-chip: data, model, and analysis. Biometrics. 2007;63:787–796. doi: 10.1111/j.1541-0420.2007.00768.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.