Abstract
Integrin α5β1 is expressed on several types of cancer cells, including colon cancer, and plays an important role in tumor growth and metastasis. The ability to target the integrin α5β1 using an appropriate drug delivery nano-vector can significantly help in inhibiting tumor growth, reducing tumor metastasis, and decreasing deleterious side effects associated with different cancer therapies. Liposomes are nano-sized phospholipid bilayer vesicles that have been extensively studied as drug delivery carriers. The goal of this study is to design stealth liposomes (liposomes covered with polyethylene glycol (PEG)) that will target colon cancer cells that express the integrin α5β1. The PEG provides a steric barrier allowing the liposomes to circulate in the blood and the functionalizing moiety, PR_b peptide, will specifically recognize and bind to α5β1 expressing cells. PR_b is a novel peptide sequence that mimics the cell adhesion domain of fibronectin, and includes four building blocks, RGDSP (the primary recognition site for α5β1), PHSRN (the synergy site for α5β1), a (SG)5 linker, and a KSS spacer. In this study we have demonstrated that by varying the amount of PEG (PEG750 or PEG2000) and PR_b on the liposomal interface we can engineer nano-vectors that bind to CT26.WT, HCT116, and RKOcolon cancer cells in a specific manner and are internalized through most likely α5β1-mediated endocytosis. GRGDSP-targeted stealth liposomes bind to colon cancer cells and internalize, but they have much lesser efficiency than PR_b-targeted stealth liposomes, and more importantly they are not as specific since many integrins bind to RGD peptides. PR_b-targeted stealth liposomes are as cytotoxic as free 5-Fluorouracil (5-FU) and exert the highest cytotoxicity on CT26.WT cells compared to GRGDSP-targeted stealth liposomes and non-targeted stealth liposomes. Thus, the proposed targeted delivery system has the great potential to deliver a therapeutic load directly to colon cancer cells, in an efficient and specific manner.
1. Introduction
Frequently, the use of novel therapeutics in medicine is hindered by the lack of efficiency in delivering these therapeutic agents to the target organs. As a result, research has been focused towards the development of targeted drug delivery systems for the treatment of diseases. Liposomes are phospholipid bilayer vesicles that have great potential as drug delivery carriers and have been extensively investigated in the past. The regulatory FDA (Food and Drug Administration) approval of stealth liposomes (liposomes sterically stabilized with poly(ethylene glycol) (PEG)) for diseases like breast cancer and ovarian cancer (Torchilin, 2005) has fueled research in the development of targeted stealth liposomal systems. Upon administration, stealth liposomes accumulate in tumor regions through a phenomenon known as the enhanced permeation and retention (EPR) effect (Maeda et al., 2001), due to the presence of fenestrated endothelium in tumor blood vessels (passive targeting). By incorporating site-specific ligands on the surface of stealth liposomes, the vesicles upon accumulating in tumor regions, can specifically target the receptor of choice on the cell surface (active targeting).
Tumor growth and metastasis is critically dependent on the development of new blood vessels (angiogenesis) to supply nutrients, oxygen and growth factors (Folkman, 1995, Folkman, 1990). Angiogenesis itself is a highly complex process involving growth factors and interactions between integrin adhesion receptors and their ligand proteins from the extracellular matrix (ECM) (Aiyer and Varner, 2005, Ellis, 2003, Folkman, 1995, Goel and Languino, 2004, Risau, 1997, Stromblad and Cheresh, 1996, Varner and Cheresh, 1996). Proliferating endothelial cells express several integrin molecules, which are not expressed on quiescent endothelial cells in normal tissue or blood vessels, like α5β1, αvβ3, and α4β1 (Aiyer and Varner, 2005, Kim et al., 2000, Kim et al., 2000). In particular, integrin α5β1 is minimally expressed in normal vasculature but is significantly up-regulated in tumor vasculature (Kim et al., 2000) and on tumor cells such as prostate, breast, colon and rectal cancer (Chen et al., 2004, Ellis, 2003, Gong et al., 1997, Jayne et al., 2002, Jia et al., 2004, Kim et al., 2000, Livant et al., 2000). Previous studies suggest that peptide and antibody antagonists of the integrin α5β1 are potent inhibitors of tumor growth, tumor induced angiogenesis, and tumor metastasis (Jia et al., 2004, Kim et al., 2000, Livant et al., 2000, Meerovitch et al., 2003, Shannon et al., 2004, Stoeltzing et al., 2001, Stoeltzing et al., 2003, White et al., 2001, Yokoyama and Ramakrishnan, 2004). Additionally, ligands that bind to the integrin α5β1 with high affinity are capable of mediating cellular internalization (Hart et al., 1994, Kintarak et al., 2004, Uduehi et al., 2003, Xiong et al., 2005, Xiong et al., 2005, Xiong et al., 2005). Therefore, α5β1 is a strong candidate for targeting colon cancer, the third most common cancer type in this country.
Researchers have primarily employed RGD (the primary recognition site for α5β1) peptide-based techniques to target the integrin α5β1, but their success has been limited. RGD recognizes several integrins and therefore this makes specificity difficult to achieve. Also, the RGD design lacks the synergistic effect, which is provided by the PHSRN, the synergy site for α5β1 (enhances the cell adhesion activity of the RGD sequence) (Aota et al., 1994). Efforts in the past to construct a single peptide sequence incorporating both RGD and PHSRN domains have also met with limited success (Aucoin et al., 2002, Benoit and Anseth, 2005, Kao, 1999, Kim et al., 2002, Susuki et al., 2002). We have previously developed a novel peptide-amphiphile sequence referred as PR_b in the text that closely mimics the cell adhesion domain of fibronectin. The design features C16 dialkyl ester tail, a glutamic acid (Glu) tail connector, a –(CH2)2– tail spacer, and the peptide headgroup (Figure 1). The PR_b peptide headgroup consists of four building blocks: a spacer KSS, PHSRN (the synergy site for α5β1), a linker (SG)5, and RGDSP (the primary recognition site for α5β1) (Mardilovich et al., 2006). The distance between the PHSRN and RGD domains on native fibronectin is approximately 30–40 Å (Leahy et al., 1996) and the ratio of hydrophilic to hydrophobic residues between the two domains is close to one (Mardilovich and Kokkoli, 2004). The linker has been designed to mimic these two important criteria; the length of the (SG)5 linker is approximately 37 Å (10 amino acids × 3.7 Å per amino acid residue (Idiris et al., 2000, Kokkoli et al., 2004)), and the ratio of the smallest “hydrophilic” amino acid serine (S) (Fransson et al., 1982, Litowski et al., 1999, Trevino et al., 2007) to the smallest “hydrophobic” amino acid glycine (G) (Berezovsky et al., 2001, Han et al., 1999, Tzvetkov et al., 2005) is equal to one. The role of KSS spacer is to extend the bioactive region further away from the interface as the accessibility of the peptide can significantly affect the binding affinities to their target proteins (Dori et al., 2000, Kokkoli et al., 2005, Kokkoli et al., 2006, Mardilovich and Kokkoli, 2004). The performance of the PR_b peptide-amphiphile was evaluated with human umbilical vein endothelial cells in terms of cell adhesion, spreading, cytoskeletal organization, and extracellular fibronectin production. PR_b outperformed all other peptide sequences that were tested and performed comparably to fibronectin (Mardilovich et al., 2006). The specificity of the PR_b peptide for integrin α5β1 was established with blocking antibodies (Mardilovich et al., 2006).
Figure 1.

Structure of PR_b peptide-amphiphile.
In this study, we incorporated the PR_b peptide-amphiphile into stealth liposomes with the goal of targeting integrin α5β1 that is expressed on colon cancer cells. Integrin α5β1 is also expressed on the colon cancer endothelium but not on the endothelium of normal colonic tissue (Ellis, 2003, Kim et al., 2000), and is expressed only on proliferating endothelial cells and not on quiescent endothelial cells found in normal tissue or blood vessels (Aiyer and Varner, 2005, Kim et al., 2000, Kim et al., 2000). More importantly, highly invasive colon cancer cell lines express greater amount of integrin α5β1, and its level of expression is usually positively correlated with their invasiveness (Gong et al., 1997).
In the present study, we demonstrate that functionalizing stealth liposomes with our novel PR_b peptide-amphiphile (Mardilovich et al., 2006) is superior to conventional RGD targeting, allowing to achieve greater binding and larger uptake of liposomes on colon cancer cells. Also, we show that by varying the amount of PEG (PEG750 or PEG2000) and the amount of peptide-amphiphile incorporated in the liposomes we can achieve increased binding affinities. PR_b-targeted stealth liposomes are as affective as free 5-FU and more cytotoxic on CT26.WT cells compared to GRGDSP-targeted stealth liposomes and non-targeted stealth liposomes. Results are reported here mostly for the mouse colon carcinoma cell system (CT26.WT). We have repeated some experiments with other colon cancer cells such as the human colon cancer cell lines HCT116 and RKO, and have obtained similar results and trends.
2. Experimental Methods
2.1. Materials
Lipids, 1,2-Dipalmitoyl-sn-Glycero-3-Phosphocholine (DPPC), cholesterol (CHOL), 1,2-Dipalmitoyl-sn-Glycero-3-Phosphoethanolamine-N-(Methoxy(Polyethylene glycol)-750)-Ammonium Salt) (PEG750) and 1,2-Dipalmitoyl-sn-Glycero-3-Phosphoethanolamine-N-(Methoxy(Polyethylene glycol)-2000)-(Ammonium Salt) (PEG-2000) were purchased from Avanti Polar Lipids Inc. (Alabaster, AL). The extruder and the 100 nm polycarbonate membranes were obtained from Avestin Inc. (Ottawa, Canada). The peptide headgroups PR_b (KSSPHSRN(SG)5RGDSP) and GRGDSP (KAbuGRGDSPAbuK) were purchased in crude form from the Microchemical Facility at the University of Minnesota. The PR_b peptide-amphiphile ((C16)2-Glu-C2-KSSPHSRN(SG)5RGDSP), and GRGDSP peptide-amphiphile ((C16)2-Glu-C2-KAbuGRGDSPAbuK) were synthesized as described previously (Berndt et al., 1995, Mardilovich et al., 2006, Mardilovich and Kokkoli, 2004). CT26.WT (mouse colon cancer cell line) and human colon cancer cell lines HCT116 and RKO were obtained from ATCC (Manassas, VA). Hoechst 33342 nucleic stain, Alexa Fluor® 594 wheat germ aggutin (WGA) cell membrane stain, and ProLong Gold antifade reagent were purchased from Invitrogen Corporation (Carlsbad, CA). Primary polyclonal antibody anti-integrin α5β1 and secondary antibody donkey anti-Goat IgG FITC conjugated, were purchased from Chemicon International Inc. (Temecula, CA). Goat IgG isotype control was purchased from Sigma Aldrich Corporation (St. Louis, MO). Cell culture media was purchased from ATCC (Manassas, VA), fetal bovine serum (FBS) was purchased from Atlas Biologicals (Fort Collins, CO), and human fibronectin-coated round coverslips were purchased from BD Biosciences (San Jose, CA). The BCA (bicinchoninic acid) protein assay kit was purchased from Pierce (Rockford, IL). 5-Fluorouracil (5-FU) and all other reagents were purchased from Sigma Aldrich Corporation (St. Louis, MO) and were of biotechnology performance certified grade.
2.2. Cell culture
CT26.WT cell lines were grown in RGM (modified RPMI-1640 medium supplemented with 10% FBS, 2 mM L-Glutamine, 100 units/ml Penicillin, and 0.1 mg/ml Streptomycin). Cells were grown in T-75 flasks with a feeding cycle of 2 days. After cells became 80% confluent (usually after 5 days) they were trypsinized (0.25% Trypsin + 0.1% EDTA) and were suspended in RGM. Cells were washed twice and finally were frozen under liquid nitrogen in RGM containing 10% DMSO (dimethyl sulfoxide) for future use. For subsequent passages cells were seeded in fresh T-75 flasks at a density 10,000 cells/cm2 and were cultured in RGM with a feeding cycle of 2 days. HCT116 cell lines were grown in MGM (modified McCoy’s 5A medium, supplemented with 10% FBS, 2 mM L-Glutamine, 100 units/ml Penicillin, and 0.1 mg/ml Streptomycin) and RKO cells were grown in EGM (modified Eagle’s Minimum essential medium, supplemented with 10% FBS, 2 mM L-Glutamine, 100 units/ml Penicillin, and 0.1 mg/ml Streptomycin).
2.3. Liposome preparation and characterization
Liposomes were prepared as described elsewhere (Fenske et al., 2003). Briefly, lipids were dissolved in chloroform and peptide-amphiphiles were dissolved in methanol and water. Lipids and peptide-amphiphiles were combined at the ratios (65-x-y):35:x:y mol% of DPPC:CHOL:PEG:Peptide-Amphiphile, where x is the indicated mol% of PEG and y is the mol% of peptide-amphiphile. Solvents were removed by evaporating under a gentle stream of argon at 65°C, and lipids were dissolved again in chloroform to form a homogenous mixture. The lipid mixture was finally dried under a gentle stream of argon at 65°C until a uniform lipid film was formed, followed by drying under vacuum overnight. Fluorescently labeled liposomes were prepared by hydrating the lipid film with fluorescent HBSE buffer (10 mM Hepes, 150 mM NaCl, 0.1 mM EDTA, and 2 mM Calcein, pH 7.4) at 65°C and at a concentration of 10 mM total lipids. 5-FU encapsulated liposomes were prepared similarly by hydrating the lipid film with HBSE buffer containing 10 mg/ml 5-FU (pH 7.4). Hydrated lipids were freeze-thawed five times, then extruded for 21 cycles through two stacks of 100 nm polycarbonate membranes using the hand-held extruder. Liposomes were filtered over a Sepharose CL-4B gel filtration column to remove unencapsulated fluorescent dye or 5-FU, which did not get incorporated in the liposomes. Liposome diameter was determined by dynamic light scattering and ranged from 80–150 nm. Phospholipid concentration was determined using the phosphorus colorimetric assay described elsewhere (Chen et al., 1956, Fiske and Subbarow, 1925). Liposomes were stored at 4–8°C and were used within two weeks. Peptide concentration was determined using the BCA assay according to the manufacturer’s protocol. We were not able to accurately determine the PEG concentration due to experimental limitations (see supplementary data); therefore, in the text we address PEG concentration either as low when 2 mol% PEG was included in starting lipid concentration or as high when 5 mol% PEG was used in the starting lipid concentration. The amount of 5-FU encapsulated in liposomes was determined by dissolving liposomes in methanol (10% liposome formulation and 90% methanol) and measuring their absorbance at 260 nm. A calibration curve was generated by dissolving known amounts of free 5-FU in 90% methanol and 10% HBSE buffer.
2.4. Flow cytometry
Confluent cell monolayers were trypsinized (0.25% Trypsin + 0.1% EDTA) and resuspended in ice-cold FB (fluorescent buffer: phosphate buffered saline (PBS) supplemented with 0.02% sodium azide and 2.5% fetal bovine serum) containing liposomes at a lipid concentration of 250 μM and a cell concentration of 1 million/ml in 15 ml centrifuge tubes. Tubes were incubated at 4°C or 37°C over a rotary shaker for the specified duration of time. Cells were then pelleted and washed twice in FB. Flow cytometric analysis was carried out immediately. For peptide blocking experiments, the protocol specified above was used except cells were incubated with 200 μg/ml of free peptide-amphiphile in FB for one hour prior to incubating the cells with the liposomes. For integrin α5β1 expression studies confluent cell monolayers were trypsinized (0.25% Trypsin + 0.1% EDTA) and resuspended in ice-cold FB at a cell concentration of 1 million/ml in 15 ml centrifuge tubes. Tubes were incubated at 4°C with primary antibody (anti-integrin α5β1) or goat isotype control (goat IgG) over a rotary shaker for 35 min. Cells were then pelleted and washed twice in FB and then incubated again with the secondary antibody (anti-goat IgG FITC conjugated) for 35 min. Finally, cells were pelleted and washed again twice in FB. Flow cytometric analysis was carried out immediately. FACS Calibur located at the Flow Cytometry Core facility in the Cancer Research Center of the University of Minnesota was used. All flow cytometry experiments are representative of n=2, but results are presented from a single experiment.
2.5. Confocal microscopy
CT26.WT confluent cell monolayers grown on fibronectin coverslips were incubated with liposomes at a lipid concentration of 250 μM, in a 5% CO2 incubator at 37°C or at 4°C for the specified duration in RGM. Cell monolayers were then washed with ice-cold FB twice. Cells were later fixed with a fixation buffer (4% paraformaldhyde in PBS, pH ~ 7.4) for 15 min at 37°C. Nuclear staining was carried out using a cell membrane permeable blue-fluorescent Hoechst 33342 dye at a concentration of 2.0 μmole/ml, and the cell membrane was stained with a cell impermeable red-fluorescent Alexa Fluor® 594 wheat germ aggutin (WGA) at 5.0 μg/ml in FB for 10 min. Cells were washed three times with FB, and coverslips were mounted on glass slides over ProLong Gold antifade reagent. For every sample 40 z-scans (horizontal cross-section of a cell at a particular z height) were taken at 0.25 μm z-step height to cover the entire height of the cell. On the confocal images liposomes were labeled with green, cell membrane with red, and nucleus with a blue. Olympus Fluoview 1000 Confocal Laser Scanning Microscope at the Biomedical Image Processing Laboratory in the Department of Neuroscience at the University of Minnesota was used.
2.6. Cytotoxicity Studies
Cytotoxicity of liposomes encapsulating 5-FU was determined by the sulforhodamine B colorimetric assay (SRB assay) (Vichai and Kirtikara, 2006). CT26.WT confluent cell monolayers were trypsinized (0.25% Trypsin + 0.1% EDTA) and were suspended in RGM at a concentration of 100,000 cells/ml. 200 μl of cell suspension was transferred to each well on a 96-well tissue culture plate to give a final seeding density of 20,000 cells/well. Cells were allowed to attach to the plate overnight. The medium was then removed (t = 0 hours), and 200 μl of medium containing two-fold serial dilutions of 5-FU ranging from 50 μM to 0.5 μM (concentration either encapsulated in liposomes or free drug) was added to the cells. A negative control (no liposome or free drug added) was included. The plates were incubated in 5% CO2 incubator at 37°C for t = 6 hours. Plates were later washed 4 times with PBS and incubated with fresh RGM at 37°C in 5% CO2 incubator for a total of 72 hours (t = 72 hours). At t = 0 hours, one plate was set aside for a no-growth control, to account for initial seeding density. On the no-growth control plate, the medium was replaced with 200 μl of cold RGM and cells were fixed by adding 100 μl of cold 10% (w/v) TCA (trichloro acetic acid) and the plate was incubated for 1 hour at 4°C. Remaining plates were fixed similarly at the conclusion of the experiment (t = 72 hours). After fixation, plates were washed four times with de-ionized (DI) water and dried. Plates were stained with 100 μl 0.057% (w/v) SRB (sulforhodamine B solution in DI water) at room temperature for 1 hour. Plates were washed again four times with 1% (v/v) acetic acid solution to remove the unbound SRB dye and dried. 200 μl of 10 mM Tris base solution (pH 10.5) was later added to dissolve the bound dye and plates were incubated at room temperature for another hour. Finally the OD (optical density) was measured on a plate reader at 510 nm. For IC50 (concentration of a drug that is required for 50% inhibition of cell growth) determination, a dose-response curve was generated by plotting the 5-FU concentration of the formulations versus the percent growth inhibition. IC50 values were obtained by fitting a sigmoidal curve and obtaining the 5-FU concentration at 50% growth inhibition. Percent growth inhibition was calculated using the following formulas:
Results are shown from three independent experiments (n=3). Each experiment was performed in quadruplet.
3. Results and Discussion
3.1. Expression of integrin α5β1 on colon cancer cells
For a targeted drug delivery system to be effective the target should be upregulated on the cells of interest. Figure 2A shows the histogram for expression of α5β1 on CT26.WT cells. 93.5% of the cell population tested positive for integrin α5β1 expression. Isotype control binding was also characterized and was found to be minimal (shown on the same histogram). This result confirms that integrin α5β1 is upregulated on mouse colon carcinoma cells. Integrin α5β1 expression was also found to be upregulated on human colon cancer cells HCT116 (Figure 2B) and RKO (Figure 2C).
Figure 2.

Expression of integrin α5β1 on A) CT26.WT B) HCT116 and C) RKO. Cells were incubated with antibodies to integrin α5β1. Appropriate isotype control is included. The number on the marker represents the percentage of cells tested positive for integrin α5β1 expression. The results are representative for n=2 but are shown only from one single experiment.
3.2. Effect of PR_b on liposome targeting
To test the effect of PR_b on the binding of liposomes to colon cancer cells, liposome formulations were initially prepared without PEG and with increasing amounts of PR_b peptide-amphiphile. The minimum PR_b concentration that was investigated was 0.7 mol% and the maximum peptide concentration was arbitrarily chosen at five times the minimum (3.5 mol%). Liposomes were incubated with cells for 3 hours at 4°C and 37°C. Cellular uptake of liposomes via endocytosis is inhibited at 4°C because the endocytotic pathways do not operate at lower temperatures (Kessner et al., 2001, Lee et al., 1993). Therefore, conducting experiments at 4°C allows studying the effect of peptide concentration on surface binding of liposomes to cells with no contributions from endocytosis. Figure 3 shows the effect of peptide concentration on liposome binding at 4°C to the following α5β1-expressing colon cancer cells: CT26.WT (Figure 3A), HCT116 (Figure 3B), and RKO (Figure 3C). Similar trends were observed at 37°C. For all three colon cancer cells, conventional liposomes (liposomes containing DPPC/Chol and no PR_b peptide-amphiphile) show no binding to cells since their fluorescent intensity overlaps with the auto-fluorescence of the cells. Even a small concentration of PR_b, 0.7 mol%, gives sufficient binding to the colon cancer cells (Figure 3). Increasing the peptide concentration improves further the binding of the liposomes to the cells with maximum binding observed at the highest concentrations of peptide studied, 2.2 and 3.5 mol%.
Figure 3.

Effect of concentration of PR_b peptide-amphiphile on binding of liposomes to A) CT26.WT, B) HCT116, and C) RKO cells at 4°C for 3 hours. Binding efficiency improves with increasing peptide concentration. Conventional liposomes show no binding to cells. The results are representative for n=2 but are shown only from one single experiment.
3.3. Effect of PR_b on stealth liposome targeting
Vatying both the PEG layer thickness and concentration and the peptide concentration is critical for a liposome delivery system to be effective. Longer PEG molecules (e.g. PEG2000) provide a better steric barrier but may mask the peptide molecule at certain concentrations. Shorter PEG molecules (e.g. PEG750) may provide sufficient access for the peptide molecule to bind to the target receptor but may reduce the steric barrier. Therefore, two different lengths of PEG were studied, PEG750 and PEG2000. The goal of this study was to determine the amount of both the peptide and PEG concentration on the liposome formulation that will give binding to cancer cells, with the idea that the total concentration of PEG and PR_b should not exceed 8–10 mol%, as work in our lab showed that when higher molecular weight molecules like PEG2000 and PR_b peptide-amphiphiles are incorporated at a concentration of 8–10 mol%, it may result in the destabilization of the liposome membrane. PEG was studied at two different concentrations, low (2 mol% initial lipid concentration) and high (5 mol% initial lipid concentration). Therefore, to avoid functionalizing liposomes at concentrations higher than 8–10 mol% in the presence of high PEG, PR_b was included at a concentration of 2.2–2.6 mol%. Half of that peptide amount was also used for comparison (1.1–1.2 mol% PR_b). The reported peptide concentrations were determined by the BCA assay on the purified liposome formulations, and were usually lower than the ones initially mixed in solution. Previous work in our lab has shown that Langmuir-Blodgett bilayers of peptide-amphiphiles with lipidated PEG are mixed at peptide concentrations less than 10 mol% or higher than 35 mol%, whereas for peptide-amphiphile concentrations between 10–35 mol% the molecules are phase separated (Mardilovich and Kokkoli, 2005). Therefore, we can speculate that for the peptide-amphiphile concentrations investigated here the peptides are probably well mixed, although we have not verified this in the present study.
Flow cytometry results for PR_b functionalized PEGylated liposomes targeted to colon cancer cells for 3 hours at 4°C are shown in Figure 4. Liposome binding to CT26.WT (Figure 4 A and B) and HCT116 (Figure 4 C and D) cells increased with increasing PR_b concentration and decreasing concentration of PEG750 (Figure 4 A and C) and PEG2000 (Figure 4 B and D). Overall, comparison between the PEG750 and PEG2000 formulations with similar peptide concentrations shows that the PR_b functionalized liposomes show similar trends, with the PEG750 formulations giving slightly higher fluorescent intensities than the PEG2000. Conventional stealth liposome formulations (with no peptide) showed minimal binding. From the concentrations that were investigated, the maximum binding efficiency was achieved for a system containing 2.2–2.6 mol% PR_b and low PEG concentration for both PEG750 and PEG2000. Liposome formulations with peptide concentrations of about 2.5 mol% PR_b and high PEG750 or PEG2000 concentration were the next best option in terms of binding. Stealth liposomes of PEG750 or PEG2000 with smaller peptide concentrations of about 1.2 mol% showed reduced binding by approximately an order of magnitude, with the exception of 1.2 mol% PR_b and low PEG2000. A high PEG2000 concentration on a liposome system with a 1.2 mol% peptide concentration, masked the peptide and reduced the binding to a minimum level.
Figure 4.

Binding of PR_b-targeted stealth liposomes to CT26.WT (A, B) and HCT116 (C, D) cells at 4°C for 3 hours. The effect of PR_b concentration and PEG concentration were investigated. Low (2 mol% in the initial lipid mixture) and high (5 mol% in the initial lipid mixture) concentrations of PEG750 (A, C) and PEG2000 (B, D) were considered. Significant binding affinities were achieved for liposomes functionalized with PR_b and PEG molecules compared to PEGylated liposomes with no peptide. For both high and low concentrations of PEG750 and PEG2000, a concentration of 2.2–2.6 mol% PR_b peptide-amphiphile gave highest binding affinity to the CT26.WT cells. The results are representative of n=2 but are shown only from one single experiment.
These results demonstrate the limitation of non-targeted stealth liposome systems currently being used in clinical practice. Functionalization of PEGylated liposomes with the PR_b peptide, designed to specifically target the integrin α5β1, can help achieve higher binding efficiencies. Both the PR_b peptide amphiphile and PEG molecules are incorporated in the liposome membrane and high binding efficiency can be achieved by varying the amounts of PEG and peptide in parallel.
3.4. PR_b versus GRGDSP targeting
PR_b functionalized stealth liposomes were compared to GRGDSP functionalized stealth liposomes in Figure 5. Figure 5A shows that when low concentrations of PEG750 are incorporated in the design, 3.3 mol% GRGDSP is less effective then 1.7 mol% PR_b, and 6.8 mol% GRGDSP shows similar or slightly better binding than 2.9 mol% PR_b. Addition of low concentration of PEG2000 to GRGDSP systems (Figure 5B) significantly decreases cell binding compared to PR_b formulations. A 2.5 mol% GRGDSP and low PEG2000 system shows minimal binding, while a 2.4 or 1.2 mol% concentration of PR_b and low PEG2000 shows significantly better performance with approximately two orders of magnitude increase of binding. A 4.6 mol% GRGDSP low PEG2000 system does show some increase in binding over a 2.5 mol% GRGDSP system but still is outperformed by both 1.2 and 2.4 mol% PR_b low PEG2000. These results demonstrate the superiority of our novel PR_b targeting over the GRGDSP-based targeting. Both the PR_b peptide amphiphile and PEG molecules are incorporated in the liposome membrane and high binding efficiency can be achieved by varying the amounts of PEG and peptide in parallel.
Figure 5.

Comparison of binding affinities between PR_b-targeted liposomes and GRGDSP-targeted liposomes with A) PEG750 and B) PEG2000. CT26.WT colon cancer cells were incubated with different liposome formulations for 3 hours at 4°C. The results demonstrate that PR_b targeting is superior to GRGDSP targeting. The results are representative for n=2 but are shown only from one single experiment.
3.5. Blocking Experiments
We have previously reported that the PR_b peptide is a specific ligand for the integrin α5β1 (Mardilovich et al., 2006). In order to establish that the peptide is mediating the binding of the peptide functionalized stealth liposomes to colon cancer cells, cells were incubated with an excess of free PR_b peptide-amphiphile for 1 hour prior to incubating them with the liposomes for another 1 hour at 4°C. Flow cytometry studies without any blocking with free peptide were also performed for the same formulations for 1 hour at 4°C and results were similar to the ones shown in Figure 5B. Figure 6 shows flow cytometry results from blocking experiments for PR_b and GRGDSP functionalized stealth liposomes with low concentrations of PEG2000, using free PR_b peptide-amphiphile as the blocking agent at a concentration of 200 μg/ml. Comparison of Figures 5B and 6 shows that addition of the free peptide completely blocks the binding of the functionalized stealth liposomes to CT26.WT cells. Therefore the peptides are mediating the binding of the peptide functionalized stealth liposomes to the cells.
Figure 6.

Binding of PR_b and GRGDSP targeted liposomes (with low concentrations of PEG2000) to CT26.WT cells was blocked by incubating the cells with PR_b at a concentration of 200 μg/ml for 1 hour at 4°C before incubating the cells with liposomes for 1 hour at 4°C. Cell adhesion was completely blocked in the presence of the free peptide.
3.6. Endocytosis of PR_b and RGD targeted stealth liposomes by CT26.WT cells
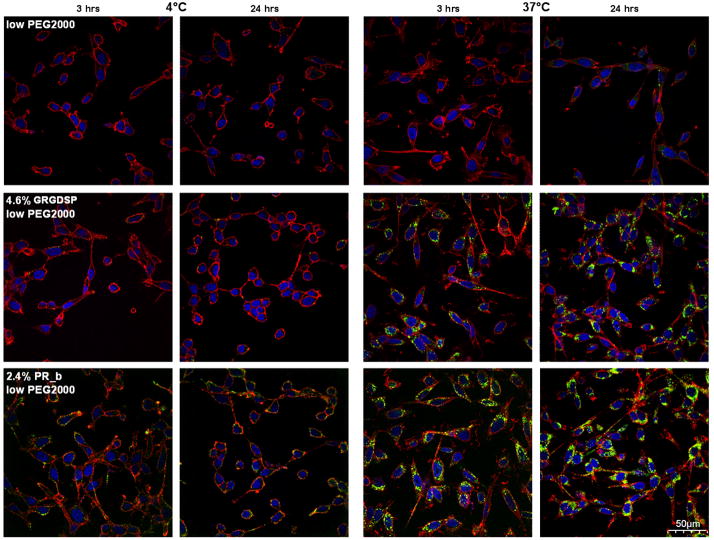
Binding of targeted liposomes is only one aspect for the development of an effective drug delivery system. These liposomes also need to be uptaken by the target cells. In order to characterize endocytosis as the most probable mechanism of internalization for the PR_b and GRGDSP targeted stealth liposomes by CT26.WT cells, a confocal laser scanning microscope was used. Figure 7 shows confocal images for a horizontal cell section nearly 2–3 μm above the coverslip and 7–8 μm below the cell surface. Binding to the cell surface on these images can be identified by co-localization of red (cell membrane stain) and green (liposome stain) signal. Internalization is marked by localization of green fluorescent dots (liposomes) between the blue nuclear region and the red cell membrane. Figure 7 demonstrates that at all the times and temperatures examined binding and internalization of stealth liposomes with low concentration of PEG750 (Figure 7A) and PEG2000 (Figure 7B) is minimal. GRGDSP stealth liposomes with low levels of PEG750 (Figure 7A) or PEG2000 (Figure 7B) show surface binding at 4°C, and evidence of internalization at 37°C. Internalization is seen only at 37°C, since endocytosis mechanisms do not operate at 4°C, and is greater at 24 hours compared to 3 hours. One possible explanation for this is that 24 hours of incubation allows more time for the recycling of integrins and thereby increasing the amount of endocytosed liposomes.
Figure 7.
Intracellular uptake of various stealth liposomes to mouse CT26.WT colon carcinoma cells. A) Liposomes functionalized with low levels of PEG750. B) Liposomes functionalized with low levels of PEG2000. First row shows PEGylated formulations, second row shows PEGylated formulations functionalized with GRGDSP and third row shows PEGylated formulations functionalized with PR_b. Internalization of different stealth liposomal formulations loaded with calcein was determined with confocal microscopy. Liposomes (shown with green) were incubated with CT26.WT at 4°C and 37°C, for 3 and 24 hour. 40 scans were taken (0.25μm step) across the body of the cells. Images shown are 2–3 μm above the coverslip, and 7–8 μm below the surface of the cells, and were merged with the nucleus (shown in blue) and cell membrane (shown in red). The scale bar is 50 μm for all images. Liposome binding to cell surface is represented by the orange colored cell membrane (co-localization of red cell membrane and green liposomes). Internalization can be identified by the green liposome signal in between the red cell membrane and blue nucleus. These images illustrate that PR_b-targeted stealth liposomes can be endocytosed by the colon cancer cells after binding to the integrin α5β1. GRGDSP-targeted stealth liposomes show smaller levels of binding and uptake.
Similar trends are observed for PR_b-targeted stealth liposomes with low levels of PEG750 (Figure 7A) or PEG2000 (Figure 7B); except there is significantly higher level of internalization at 37°C and surface binding at 4°C compared to the GRGDSP formulations. Internalization studies at 37°C show that even at 3 hours of incubation the amount of green fluorescence inside the red cell membrane is high, and at 24 hours this effect is even more pronounced compared to GRGDSP stealth liposomes. With the PR_b formulations almost the entire cytoplasmic region is stained with the green internalized vesicles.
These confocal images illustrate the internalization of PR_b-targeted stealth liposomes by the colon cancer cells, and show that when the PR_b peptide is used for targeting more stealth liposomes are internalized with only half the concentration of GRGDSP. These results support the surface binding studies from the flow cytometry experiments (Figure 5) and show that PR_b targeting can significantly improve the performance of stealth liposomes as compared to conventional RGD targeting techniques. Furthermore, qualitative comparisons between the lower right images on the third row, for PR_b-targeted stealth liposomes incubated with the colon cancer cells for 24 hours at 37°C, show that the PR_b PEG750 functionalized liposomes (Figure 7A) show more internalization compared to the PR_b PEG2000 functionalized liposomes (Figure 7B) consistent with the idea that shorter PEG molecules may show reduced steric barrier and thus allowing for more peptide functionalized stealth liposomes to bind and internalize.
3.7. Cytotoxicity of 5-FU encapsulated targeted stealth liposomes to CT26.WT cells
In this study we examined the cytotoxicity of different formulations that encapsulated 5-FU. 5-FU was chosen as an anti-cancer agent since it is commonly used for the treatment of colon cancer. CT26.WT cells were initially incubated with either 5-FU containing liposomes or free 5-FU for 6 hours, and allowed to grow for a total of 72 hours. Using a negative control (no liposomes or free drug added) and a no-growth control that determined the initial seeding density, the amount of growth inhibition was examined.
Table 1 summarizes the IC50 values for free drug and different liposome formulations. At 6 hours, PR_b-targeted stealth liposomes increased cytotoxicity from 11.14 ± 0.17 μM (IC50 for non targeted stealth liposomes) to 7.31 ± 0.14 μM, about 35% increase in 5-FU efficacy. When compared to GRGDSP-targeted stealth liposomes, PR_b-targeted stealth liposomes (with only half the concentration of peptide) show nearly 20% improvement in cytotoxicity. More importantly, PR_b-targeted stealth liposomes are as effective as free 5-FU.
Table 1.
Cytotoxicity of 5-FU encapsulated stealth liposomes targeted to CT26.WT cells for 6 hours. After washing cells were allowed to grow for a total of 72 hours. All the values are representatives of mean ± SD from three independent experiments (n=3). Each experiment was performed in quadruplet
| Formulation | IC50 (μM) (t = 6 hours) | |
|---|---|---|
| AVG | S.D. | |
| 5-Fluorouracil | 7.50 | 0.17 |
| low PEG2000 | 11.14 | 0.17 |
| 1.6–1.9% PR_b low PEG2000 | 7.31 | 0.14 |
| 2.9–3.8% GRGDSP low PEG2000 | 9.07 | 0.48 |
Recent studies reported that RGD-targeted stealth liposomes show increased cytotoxicity over non-targeted liposomes. For example, RGD-targeted liposomes containing the anti-cancer drug doxorubicin showed a 30–40% increase in efficacy over stealth liposomes delivered to murine B16 and human A375 melanoma cells after incubation for 8 hours (Xiong et al., 2005, Xiong et al., 2005).
The cytotoxicity studies in Table 1 support the binding and internalization data discussed in earlier sections, and indicate that using a PR_b peptide sequence, that has been shown to have higher affinity and specificity for the α5β1 than GRGDSP (Mardilovich et al., 2006), leads to significant improvement in both targeting capability and biological activity.
4. Conclusion
In this study we have engineered a targeted delivery system that can deliver a therapeutic load to colon cancer cells using a peptide sequence (PR_b) that has been shown previously to specifically target the integrin α5β1 with high affinity. We have optimized the delivery of the nano-vector by varying the amounts of both the peptide and PEG molecules on the liposome surface, and studying the effect of their concentration on binding to colon cancer cells. The PR_b-targeted stealth liposome system is capable of binding to the integrin α5β1 expressing colon carcinoma cells, and undergoes cellular internalization via most likely an endocytosis mechanism. Our results are well correlated and the trends are logical based on our understanding of the effect of PEG and peptide-amphiphile on the liposome interface. Increasing the amount of PR_b peptide enhances the binding affinity of liposomes and increasing the amount of PEG reduces it. When PEG2000 is incorporated in the peptide functionalized liposomes, binding and internalization of the liposomes slightly decreases compared to systems with similar concentrations of PEG750 and peptide. We have shown that by varying both the concentrations of peptide-amphiphile and PEG on the liposome interface, significant levels of binding can be achieved even when target functionality is implanted in parallel with PEG instead at the tip of the PEG chain. PR_b targeting is superior to GRGDSP targeting as shown by improved binding and internalization at lower concentrations of PR_b. For example, liposomes with 6.8 mol% GRGDSP and low levels of PEG750 are required to perform comparably to 2.9 mol% PR_b liposomes with low PEG750 concentrations, and similarly, liposomes with low concentrations of PEG2000 and 4.6 mol% GRGDSP are outperformed by low PEG 2000 formulations functionalized with 1.2 mol% PR_b. Furthermore, PR_b-targeted stealth liposomes can internalize in significantly higher amounts than the GRGDSP-targeted stealth liposomes. More importantly, PR_b has been shown in our previous work to be α5β1 specific whereas many integrins bind to RGD peptides. PR_b-targeted stealth liposomes show significantly higher cytotoxicity than the GRGDSP-targeted stealth liposomes, and the non-targeted stealth liposomes, and are as effective as free 5-FU. Based on the above findings, we conclude that PR_b-targeted stealth liposomes can be potentially used in-vivo, to deliver a therapeutic load directly to cancer cells, and may help overcome the deleterious side effects present in non-targeted treatments.
Supplementary Material
Acknowledgments
We thank Anastasia Mardilovich for synthesizing the GRGDSP peptide-amphiphile. This work was supported in part by the University of Minnesota Nanobiotechnology Initiative, The American Cancer Society Institutional Research Grant (IRG-58-001-46-IRG45), the National Science Foundation (CBET-0553682), the MRSEC Program of the National Science Foundation under Award Number DMR-0212302, the National Institute of Biomedical Imaging and Bioengineering (R03EB006125), and the National Cancer Institute (R01CA120383). We would like to acknowledge the assistance of the Flow Cytometry Core Facility at the University of Minnesota Cancer Center, a comprehensive cancer center designated by the National Cancer Institute, supported in part by P30 CA77598. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering, the National Cancer Institute, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- American Cancer Society. Cancer facts and figures. Atlanta: 2006. [Google Scholar]
- Aiyer A, Varner JA. Integrins in cancer progression and therapy. Science and Medicine. 2005;10:84–96. [Google Scholar]
- Aota S, Nomizu M, Yamada KM. The Short Amino-Acid-Sequence Pro-His-Ser-Arg-Asn in Human Fibronectin Enhances Cell-Adhesive Function. Journal of Biological Chemistry. 1994;269:24756–24761. [PubMed] [Google Scholar]
- Aucoin L, Griffith CM, Pleizier G, Deslandes Y, Sheardown H. Interactions of corneal epithelial cells and surfaces modified with cell adhesion peptide combinations. Journal of Biomaterials Science-Polymer Edition. 2002;13:447–462. doi: 10.1163/156856202320253956. [DOI] [PubMed] [Google Scholar]
- Benoit DSW, Anseth KS. The effect on osteoblast function of colocalized RGD and PHSRN epitopes on PEG surfaces. Biomaterials. 2005;26:5209–5220. doi: 10.1016/j.biomaterials.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Kirzhner VM, Kirzhner A, Trifonov EN. Protein folding: Looping from hydrophobic nuclei. Proteins-Structure Function and Genetics. 2001;45:346–350. doi: 10.1002/prot.1155. [DOI] [PubMed] [Google Scholar]
- Berndt P, Fields GB, Tirrell M. Synthetic Lipidation of Peptides and Amino-Acids: Monolayer Structure and Properties. Journal of the American Chemical Society. 1995;117:9515–9522. [Google Scholar]
- Chen J, De S, Brainard J, Byzova TV. Metastatic properties of prostate cancer cells are controlled by VEGF. Cell Communication and Adhesion. 2004;11:1–11. doi: 10.1080/15419060490471739. [DOI] [PubMed] [Google Scholar]
- Chen PS, Toribara TY, Warner H. Microdetermination of phosphorus. Analytical Chemistry. 1956;28:1756–1758. [Google Scholar]
- Dori Y, Bianco-Peled H, Satija SK, Fields GB, McCarthy JB, Tirrell M. Ligand accessibility as means to control cell response to bioactive bilayer membranes. Journal of Biomedical Materials Research. 2000;50:75–81. doi: 10.1002/(sici)1097-4636(200004)50:1<75::aid-jbm11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Ellis LM. A targeted approach for antiangiogenic therapy of metastatic human colon cancer. American Surgeon. 2003;69:3–10. [PubMed] [Google Scholar]
- Fenske DB, Maurer N, Cullis PR. Encapsulation of weakly-basic drugs, antisense oligonucleotides, and plasmid DNA within large unilamellar vesicles for drug delivery applications. Oxford university press; New York: 2003. [Google Scholar]
- Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. Journal of Biological Chemistry. 1925;66:375–400. [Google Scholar]
- Folkman J. Angiogenesis in Cancer, Vascular, Rheumatoid and Other Disease. Nature Medicine. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- Folkman J. What Is the evidence that tumors are angiogenesis dependent. Journal of the National Cancer Institute. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- Fransson B, Ragnarsson U, Zetterqvist O. Separation of Basic, Hydrophilic Peptides by Reversed-Phase Ion-Pair Chromatography. Analytical Biochemistry. 1982;126:174–178. doi: 10.1016/0003-2697(82)90125-7. [DOI] [PubMed] [Google Scholar]
- Goel HL, Languino LR. Integrin signaling in cancer. Cancer Treatment and Research. 2004;119:15–31. doi: 10.1007/1-4020-7847-1_2. [DOI] [PubMed] [Google Scholar]
- Gong J, Wang D, Sun L, Zborowska E, Willson JKV, Brattain MG. Role of alpha(5)beta(1) integrin in determining malignant properties of colon carcinoma cells. Cell Growth and Differentiation. 1997;8:83–90. [PubMed] [Google Scholar]
- Han X, Steinhauer DA, Wharton SA, Tamm LK. Interaction of mutant influenza virus hemagglutinin fusion peptides with lipid bilayers: Probing the role of hydrophobic residue size in the central region of the fusion peptide. Biochemistry. 1999;38:15052–15059. doi: 10.1021/bi991232h. [DOI] [PubMed] [Google Scholar]
- Hart SL, Knight AM, Harbottle RP, Mistry A, Hunger H, Cutler DF, Williamson R, Coutelle C. Cell-binding and internalization by filamentous phage displaying a cyclic Arg-Gly-Asp-containing peptide. Journal of Biological Chemistry. 1994;269:12468–12474. [PubMed] [Google Scholar]
- Idiris A, Alam MT, Ikai A. Spring mechanics of alpha-helical polypeptide. Protein Engineering. 2000;13:763–770. doi: 10.1093/protein/13.11.763. [DOI] [PubMed] [Google Scholar]
- Jayne DG, Heath RM, Dewhurst O, Scott N, Guillou PJ. Extracellular matrix proteins and chemoradiotherapy: alpha(5)beta(1) integrin as a predictive marker in rectal cancer. European Journal of Surgical Oncology. 2002;28:30–36. doi: 10.1053/ejso.2001.1182. [DOI] [PubMed] [Google Scholar]
- Jia Y, Zeng Z, Markwart SM, Rockwood KF, Ignatoski KMW, Ethier SP, Livant DL. Integrin fibronectin receptors in matrix metalloproteinase-1-dependent invasion by breast cancer and mammary epithelial cells. Cancer Research. 2004;64:8674–8681. doi: 10.1158/0008-5472.CAN-04-0069. [DOI] [PubMed] [Google Scholar]
- Kao WJ. Evaluation of protein-modulated macrophage behavior on biomaterials: designing biomimetic materials for cellular engineering. Biomaterials. 1999;20:2213–2221. doi: 10.1016/s0142-9612(99)00152-0. [DOI] [PubMed] [Google Scholar]
- Kessner S, Krause A, Rothe U, Bendas G. Investigation of the cellular uptake of E-Selectin-targeted immunoliposomes by activated human endothelial cells. Biochimica et Biophysica Acta. 2001;1514:177–190. doi: 10.1016/s0005-2736(01)00368-6. [DOI] [PubMed] [Google Scholar]
- Kim S, Bell K, Mousa SA, Varner JA. Regulation of angiogenesis in vivo by ligation of integrin alpha(5)beta(1) with the central cell-binding domain of fibronectin. American Journal of Pathology. 2000;156:1345–1362. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Harris M, Varner JA. Regulation of integrin alpha(v)beta(3)-mediated endothelial cell migration and angiogenesis by integrin alpha(5)beta(1) and protein kinase A. Journal of Biological Chemistry. 2000;275:33920–33928. doi: 10.1074/jbc.M003668200. [DOI] [PubMed] [Google Scholar]
- Kim TI, Jang JH, Lee YM, Ryu IC, Chung CP, Han SB, Choi SM, Ku Y. Design and biological activity of synthetic oligopeptides with Pro-His-Ser-Arg-Asn (PHSRN) and Arg-Gly-Asp (RGD) motifs for human osteoblast-like cell (MG-63) adhesion. Biotechnology Letters. 2002;24:2029–2033. [Google Scholar]
- Kintarak S, Whawell SA, Speight PM, Packer S, Nair SP. Internalization of Staphylococcus aureus by human keratinocytes. Infection and Immunity. 2004;72:5668–5675. doi: 10.1128/IAI.72.10.5668-5675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkoli E, Kasinskas RW, Mardilovich A, Garg A. Fractalkine targeting with a receptor-mimicking peptide-amphiphile. Biomacromolecules. 2005;6:1272–1279. doi: 10.1021/bm0493537. [DOI] [PubMed] [Google Scholar]
- Kokkoli E, Mardilovich A, Wedekind A, Rexeisen EL, Garg A, Craig JA. Self-assembly and applications of biomimetic and bioactive peptide-amphiphiles. Soft Matter. 2006;2:1015–1024. doi: 10.1039/b608929a. [DOI] [PubMed] [Google Scholar]
- Kokkoli E, Ochsenhirt SE, Tirrell M. Collective and single-molecule interactions of alpha(5)beta(1) integrins. Langmuir. 2004;20:2397–2404. doi: 10.1021/la035597l. [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP. 2.0 angstrom crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Lee KD, Nir S, Papahadjopoulos D. Quantitative-analysis of liposome-cell interactions in vitro - rate constants of binding and endocytosis with suspension and adherent J774-cells and human monocytes. Biochemistry. 1993;32:889–899. doi: 10.1021/bi00054a021. [DOI] [PubMed] [Google Scholar]
- Litowski JR, Semchuk PD, Mant CT, Hodges RS. Hydrophilic interaction/cation-exchange chromatography for the purification of synthetic peptides from closely related impurities: serine side-chain acetylated peptides. Journal of Peptide Research. 1999;54:1–11. doi: 10.1034/j.1399-3011.1999.00066.x. [DOI] [PubMed] [Google Scholar]
- Livant DL, Brabec RK, Pienta KJ, Allen DL, Kurachi K, Markwart S, Upadhyaya A. Anti-invasive, antitumorigenic, and antimetastatic activities of the PHSCN sequence in prostate carcinoma. Cancer Research. 2000;60:309–320. [PubMed] [Google Scholar]
- Maeda H, Sawa T, Konno T. Mechanism of tumor-targeted delivery of macromolecular drugs, including the EPR effect in solid tumor and clinical overview of the prototype polymeric drug SMANCS. Journal of Controlled Release. 2001;74:47–61. doi: 10.1016/s0168-3659(01)00309-1. [DOI] [PubMed] [Google Scholar]
- Mardilovich A, Craig JA, McCammon MQ, Garg A, Kokkoli E. Design of a novel fibronectin-mimetic peptide-amphiphile for functionalized biomaterials. Langmuir. 2006;22:3259–3264. doi: 10.1021/la052756n. [DOI] [PubMed] [Google Scholar]
- Mardilovich A, Kokkoli E. Biomimetic peptide-amphiphiles for functional biomaterials: The role of GRGDSP and PHSRN. Biomacromolecules. 2004;5:950–957. doi: 10.1021/bm0344351. [DOI] [PubMed] [Google Scholar]
- Mardilovich A, Kokkoli E. Patterned biomimetic membranes: Effect of concentration and pH. Langmuir. 2005;21:7468–7475. doi: 10.1021/la0468085. [DOI] [PubMed] [Google Scholar]
- Meerovitch K, Bergeron F, Leblond L, Grouix B, Poirier C, Bubenik M, Chan L, Gourdeau H, Bowlin T, Attardo G. A novel RGD antagonist that targets both alpha(v)beta(3) and alpha(5)beta(1) induces apoptosis of angiogenic endothelial cells on type I collagen. Vascular Pharmacology. 2003;40:77–89. doi: 10.1016/s1537-1891(02)00339-7. [DOI] [PubMed] [Google Scholar]
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- Shannon KE, Keene JL, Settle SL, Duffin TD, Nickols MA, Westlin M, Schroeter S, Ruminski PG, Griggs DW. Anti-metastatic properties of RGD-peptidomimetic agents S137 and S247. Clinical and Experimental Metastasis. 2004;21:129–138. doi: 10.1023/b:clin.0000024764.93092.5f. [DOI] [PubMed] [Google Scholar]
- Stoeltzing O, Liu W, Reinmuth N, Fan F, Livant DL, Mazar AP, Ellis LM. Reduction of colon cancer growth by a novel antiangiogenic agent that targets the integrin alpha(5)beta(1) Clinical Cancer Research. 2001;7:3656s–3656s. [Google Scholar]
- Stoeltzing O, Liu W, Reinmuth N, Fan F, Parry GC, Parikh AA, McCarty MF, Bucana CD, Mazar AP, Ellis LM. Inhibition of integrin alpha(5)beta(1) function with a small peptide (ATN-161) plus continuous 5-FU infusion reduces colorectal liver metastases and improves survival in mice. International Journal of Cancer. 2003;104:496–503. doi: 10.1002/ijc.10958. [DOI] [PubMed] [Google Scholar]
- Stromblad S, Cheresh DA. Integrins, angiogenesis and vascular cell survival. Chemistry and Biology. 1996;3:881–885. doi: 10.1016/s1074-5521(96)90176-3. [DOI] [PubMed] [Google Scholar]
- Susuki Y, Hojo K, Okazaki I, Kamata H, Sasaki M, Maeda M, Nomizu M, Yamamoto Y, Nakagawa S, Mayumi T, Kawasaki K. Preparation and biological activities of a bivalent poly (ethylene glycol) hybrid containing an active site and its synergistic site of fibronectin. Chemical and Pharmaceutical Bulletin. 2002;50:1229–1232. doi: 10.1248/cpb.50.1229. [DOI] [PubMed] [Google Scholar]
- Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nature Reviews Drug Discovery. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- Trevino SR, Scholtz JM, Pace CN. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. Journal of Molecular Biology. 2007;366:449–460. doi: 10.1016/j.jmb.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzvetkov G, Ramsey MG, Netzer FP. Glycine-ice nanolayers: Morphology and surface energetics. Journal of Chemical Physics. 2005;122:114712. doi: 10.1063/1.1878592. [DOI] [PubMed] [Google Scholar]
- Uduehi A, Mailhos C, Truman H, Thrasher AJ, Kinnon C, Hart SL. Enhancement of integrin-mediated transfection of haematopoietic cells with a synthetic vector system. Biotechnology and Applied Biochemistry. 2003;38:201–209. doi: 10.1042/BA20030062. [DOI] [PubMed] [Google Scholar]
- Varner JA, Cheresh DA. Integrins and cancer. Current Opinion in Cell Biology. 1996;8:724–730. doi: 10.1016/s0955-0674(96)80115-3. [DOI] [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nature Protocols. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- White ES, Livant DL, Markwart S, Arenberg DA. Monocyte-fibronectin interactions, via alpha(5)beta(1) integrin, induce expression of CXC chemokine-dependent angiogenic activity. Journal of Immunology. 2001;167:5362–5366. doi: 10.4049/jimmunol.167.9.5362. [DOI] [PubMed] [Google Scholar]
- Xiong X, Huang Y, Lu W, Zhang H, Zhang X, Zhang Q. Enhanced intracellular uptake of sterically stabilized liposomal doxorubicin in vitro resulting in improved antitumor activity in vivo. Pharmaceutical Research. 2005;22:933–939. doi: 10.1007/s11095-005-4588-x. [DOI] [PubMed] [Google Scholar]
- Xiong X, Huang Y, Lu W, Zhang X, Zhang H, Nagai T, Zhang Q. Enhanced intracellular delivery and improved antitumor efficacy of doxorubicin by sterically stabilized liposomes modified with a synthetic RGD mimetic. Journal of Controlled Release. 2005;107:262–275. doi: 10.1016/j.jconrel.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Xiong X, Huang Y, Lu W, Zhang X, Zhang H, Nagai T, Zhang Q. Intracellular delivery of doxorubicin with RGD-modified sterically stabilized liposomes for an improved antitumor efficacy: In vitro and in vivo. Journal of Pharmaceutical Sciences. 2005;94:1782–1793. doi: 10.1002/jps.20397. [DOI] [PubMed] [Google Scholar]
- Yokoyama Y, Ramakrishnan S. Addition of integrin binding sequence to a mutant human endostatin improves inhibition of tumor growth. International Journal of Cancer. 2004;111:839–848. doi: 10.1002/ijc.20336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.