Abstract
Uracil in DNA may result from incorporation of dUMP during replication and from spontaneous or enzymatic deamination of cytosine, resulting in U:A pairs or U:G mismatches, respectively. Uracil generated by activation-induced cytosine deaminase (AID) in B cells is a normal intermediate in adaptive immunity. Five mammalian uracil-DNA glycosylases have been identified; these are mitochondrial UNG1 and nuclear UNG2, both encoded by the UNG gene, and the nuclear proteins SMUG1, TDG and MBD4. Nuclear UNG2 is apparently the sole contributor to the post-replicative repair of U:A lesions and to the removal of uracil from U:G contexts in immunoglobulin genes as part of somatic hypermutation and class-switch recombination processes in adaptive immunity. All uracil-DNA glycosylases apparently contribute to U:G repair in other cells, but they are likely to have different relative significance in proliferating and non-proliferating cells, and in different phases of the cell cycle. There are also some indications that there may be species differences in the function of the uracil-DNA glycosylases.
Keywords: base excision repair, B-cell lymphoma, class switch recombination, cytosine deamination, somatic hypermutation, uracil-DNA glycosylase
1. Background
DNA is inherently unstable and decomposes spontaneously by hydrolysis (Lindahl 1993). In addition, it is damaged by environmental chemicals and radiation. Water and reactive oxygen species (ROS) are among the major contributors to spontaneous damage. An example of damage by ROS is 8-oxoguanine, which is a highly mutagenic lesion causing G:C to T:A transversions (Slupphaug et al. 2003). Hydrolytic depurination generates approximately 10 000 abasic sites per cell per day, while hydrolytic deamination of cytosine generates 70–200 uracil bases per day in DNA (Lindahl 1993). While the latter number is less impressive, uracils in U:G mismatches are 100 per cent mutagenic if not repaired, due to the lack of discrimination by DNA polymerases between U and T in the template. This gives rise to G:C to A:T transition mutations. This type of transition is a very common mutation in the mammalian genome, as well as in human tumours. G:C to A:T transitions are also generated by other mechanisms, e.g. replication across O6-methylguanine in the DNA template. The presence of uracil in RNA, but thymine (5-methyluracil) in DNA, makes sense since thymine is a much more stable component in the genome, where stability is more important than in the short-lived RNA molecules. Thus, deamination of cytosine to uracil in DNA signals damage, but, in RNA, the corresponding deamination would give rise to a normal RNA constituent. In the absence of uracil-DNA repair, the relatively rapid deamination of cytosine and the lack of toxicity of the resulting uracil would cause a genetic drift that would probably not be tolerable in an organism. In line with this hypothesis, all known organisms have evolved mechanisms to remove uracil from DNA, mostly employing DNA glycosylases to excise the base (Krokan et al. 2002).
Nature's ability to take into use most thinkable, and unthinkable, possibilities for creating advantage continues to amaze. Such is the case also for the generation of uracil in DNA. While it is a nuisance in most cells, it is taken into use as a tool to create diversity of genes where diversity is the business, namely the antigen-driven generation of antibodies in B cells. These processes are initiated by activation-induced cytosine deaminase (AID) discovered less than 10 years ago in Tasuku Honjo's group (Muramatsu et al. 1999). The discovery of AID and its functions have dramatically improved the possibility to obtain an in-depth understanding of the mechanism of adaptive immunity by offering a conceptual framework for the design of mechanistic studies. First thought to be an RNA-editing cytidine deaminase, the strongest evidence now indicates that AID is a DNA cytosine deaminase that introduces uracil in variable regions of immunoglobulin genes in B cells (Di Noia & Neuberger 2007). Replication over this uracil directly generates G:C to A:T transition mutations, whereas removal of uracil by uracil-DNA glycosylase creates abasic sites that generate a wider range of mutations with the help of translesion bypass polymerases, such as DNA polymerase η that has a particularly important role in this process (Di Noia & Neuberger 2007). Thus, uracil-DNA glycosylase that is normally an antimutator protein initiating error-free base excision repair (BER), instead becomes part of a mechanism that generates mutations in immunoglobulin gene variable regions (somatic hypermutation (SHM)) and DNA strand breaks to exchange effector regions (class switch recombination (CSR)). It should be pointed out that the action of uracil-DNA glycosylase at deaminated cytosines (at C:G pairs) does not explain the occurrence of mutations at T:A pairs. Such mutations are dependent on the mismatch repair factors MSH2/6 and DNA polymerase η. In the follow-up of the initial discoveries of the roles of AID and uracil-DNA glycosylase, it has become clear that deamination of cytosine to uracil has a wide biological significance; first, to create diversity of immunoglobulin genes and, second, to label viral DNA replication intermediates of retroviruses for degradation to free the infected organism of the intruder (Sousa et al. 2007).
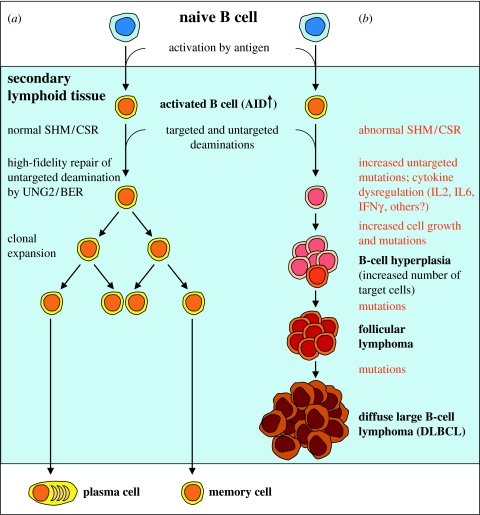
A role of uracil-DNA glycosylase encoded by the UNG gene family in adaptive immunity has been demonstrated using Ung knockout mice (Rada et al. 2002) and subsequently in human patients carrying inactivating mutations in the UNG gene (Imai et al. 2003). While these studies were principally in good agreement with respect to both skewed SHM and reduced CSR, there were also clear differences: CSR was reduced by 99 per cent in human B cells, but only approximately 50–70% in mouse cells. Therefore, knockout mice may not be an equally good model for all aspects of adaptive immunity, at least not as a quantitative model for various functions. Nevertheless, similarities may appear to be more obvious than differences. At the level of the organism, both UNG-defective mice (Nilsen et al. 2003; Andersen et al. 2005a) and humans (Imai et al. 2003) develop lymphoid hyperplasia. In addition, the gene-targeted mice also develop B-cell lymphomas of follicular type and diffuse large B-cell type in old age. Lymphoid hyperplasia may precede malignant B-cell lymphomas in both mouse and man. Possibly the mechanism could be that hyperproliferating B cells accumulate non-targeted deaminations by AID, resulting in an increase in mutations in an Ung-deficient background (figure 1). Interestingly, it has recently been reported that AID acts broadly on the genome during SHM, but widespread mutations are averted by gene-specific Ung2- and Msh2-dependent DNA repair. In the Myc oncogene, the mutation frequency increased more than 6-fold in Ung−/− mice and more than 18-fold in Ung−/−/Msh2−/− mice (Liu et al. 2008). There is no evidence indicating that mutations in the UNG gene are responsible for a significant fraction of B-cell lymphomas in man, but the general scheme (increased load of damage and reduced repair) might well be a contributing mechanism worth considering. The lack of functional uracil-DNA glycosylase due to inactivating mutations is apparently very rare in humans, and the few individuals identified are young, hence it is not known whether they are at risk of developing lymphomas. However, they do suffer from recurrent infections indicative of a significant immune deficiency (Imai et al. 2003).
Figure 1.
Model of B-cell lymphoma development in Ung-deficient mice. (a) The pathway illustrates the normal development of Ung+/+ B cells after translocation to secondary lymphoid tissues, leading to antibody-secreting plasma cells and memory cells. (b) The pathway models how cytokine dysregulation and increased accumulation of untargeted mutations in Ung−/− B cells result in lymphoid hyperplasia and increased number of cells having a mutator phenotype. Eventually, this may lead to the development of follicular lymphoma and progression to diffuse large B-cell lymphoma.
The focus of this short review is on the role of uracil-DNA glycosylases in processing of U:G mismatches in mammalian cells, but it should be remembered that the other route by which uracil may enter DNA, namely by incorporation of dUMP from the natural intermediate dUTP, is probably quantitatively dominant (Andersen et al. 2005b). This lesion has been assumed to be relatively harmless because it is present in a U:A match and therefore not thought to be mutagenic. However, this has not really been well documented. In addition, a physiological role of U:A matches in the immune system or in other cells remains a possibility that should not be discarded without further exploration.
2. Deamination of cytosine in DNA: quantitative aspects
The rate of deamination of cytosine to uracil has been measured by chemical and genetic methods and generally these are in relatively good agreement, as recently reviewed in Kavli et al. (2007). The main uncertainty stems from the 140–300-fold higher rate of deamination in single-stranded DNA when compared with double-stranded DNA. Single-stranded DNA exists near forks, in transcribed genes, breathing DNA and displacement loops (e.g. in mitochondrial DNA), but the fraction that is single-stranded at any one time is not known. It is probably higher in replicating cells than in non-replicating cells. If assuming that 0.1 per cent of DNA is single-stranded (probably a high estimate), the range of estimates is approximately 70–200 spontaneous cytosine deaminations per cell per day. Although this would appear to be a relatively low number compared with numbers for depurination and oxidative damage, it would probably increase mutation frequencies in proliferating cells substantially (overall approximately 100 mutations per cell per round of replication), unless repaired. Mammalian cells counteract this threat mainly by BER using different uracil-DNA glycosylases to initiate the process. The individual roles of these glycosylases in mouse and man are starting to be understood. We also know that we understand less than we thought a decade ago.
3. Mammalian uracil-DNA glycosylases and their assumed functions
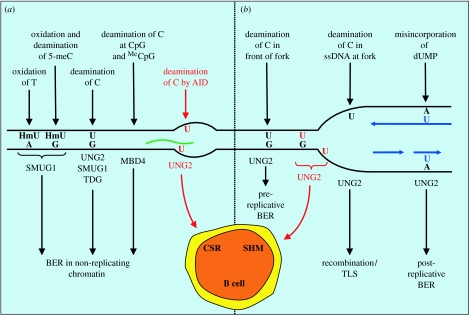
Mammalian cells have at least five distinct uracil-DNA glycosylases. Briefly, nuclear UNG2 is central to the repair of incorporated uracil (U:A context), whereas UNG2, SMUG1, TDG and probably MBD4 all contribute to U:G repair. UNG1 is apparently the only mitochondrial uracil-DNA glycosylase. The sources of uracil in mammalian nuclear genomes, and the uracil-DNA glycosylases processing these uracils, are modelled in figure 2.
Figure 2.
Model showing sources for the generation of uracil in DNA and the repair pathways of the lesions (a) in non-replicating chromatin and (b) at the replication fork. Uracil excision by UNG2 in single-stranded DNA (ssDNA) at the replication fork will probably result in stalled replication that will initiate recombination or translesion synthesis (TLS). The uracil-initiated diversification pathways in activated B cells are highlighted in red, newly replicated DNA is in blue and RNA is in green. U, uracil; HmU, hydroxymethyluracil.
(a) Members of the UNG family
Nuclear UNG2 and mitochondrial UNG1 are both encoded by the UNG gene (Ung in the mouse), which is representative of the classical large family of conserved uracil-DNA glycosylases found in vertebrates, yeast, most bacteria and some viruses (herpes and pox families), but not in insect cells (Krokan et al. 1997, 2002). Alternative N-terminal sequences in UNG2 and UNG1 determine nuclear and mitochondrial targeting, respectively. In addition, the N-terminal sequence of UNG2 interacts with proliferating cell nuclear antigen and replication protein A (RPA; Otterlei et al. 1999). In most bacteria and yeast, this is the sole uracil-DNA glycosylase. UNG1 is also apparently the only uracil-DNA glycosylase in mitochondria, which was recently found to have capacity for both short-patch BER (insertion of one nucleotide) and long-patch BER (insertion of two to eight nucleotides) (Akbari et al. 2008). All representatives of this family examined so far are active on both single-stranded and double-stranded DNA, but with a preference for uracil in single-stranded DNA. They also have a weak preference for U:G contexts over U:A contexts, but this preference is sequence dependent and not absolute (Eftedal et al. 1993; Slupphaug et al. 1995). Compared with other uracil-DNA glycosylases, UNG2 (and UNG1) has a very high turnover number, compatible with the dominant role of UNG2 in post-replicative repair of incorporated uracil (U:A context), where repair has to keep up with the rapid movement of the replication fork (Otterlei et al. 1999; Nilsen et al. 2000; Akbari et al. 2004). UNG2 also appears to have an important role in the repair of U:G mismatches, at least in human cells in vitro (Kavli et al. 2002; Akbari et al. 2004). In mouse cells, Smug1 was reported to have a central role in the repair of deaminated cytosines (U:G context) in vitro, while Ung2 apparently had only a minor role (Nilsen et al. 2001). This is not fully in agreement with results from human cells, and it is possible that UNG2- and SMUG1-type enzymes may have taken on quantitatively, and possibly qualitatively, different roles in mouse and man. This is an important aspect to clarify, since mouse is the favourite model for studies of gene functions at the level of the organism. Importantly, UNG2 is apparently the only uracil-DNA glycosylase involved in uracil removal in B cells as part of SHM and CSR (Imai et al. 2003). SMUG1 cannot compensate for UNG2 in these processes. This is probably caused by the much lower catalytic efficiency of SMUG1 than UNG2 against uracil in single-stranded DNA, as demonstrated in extracts from lymphoblastoid cell lines derived from patients with hyper IgM syndrome due to inactivating mutations in the UNG gene (Kavli et al. 2005). Furthermore, these UNG-deficient cells accumulate uracil in their genome (Kavli et al. 2005).
UNG2 is cell-cycle regulated with the highest protein level in early to mid-S-phase, in agreement with its role in the repair of incorporated uracils. UNG2 also undergoes sequential phosphorylations at Ser23, Thr60 and Ser64 during the cell cycle. Monophosphorylation at Ser23 in the G1/early S-phase apparently increases association with RPA and replicating chromatin and markedly increases the catalytic turnover number. Furthermore, the triple phosphorylated form is mono-ubiquitinylated late in the S-phase and then rapidly degraded in the G2-phase. Interestingly, the sequence encompassing the phosphorylated Thr60–Ser64 forms a motif that is very similar to a phosphodegron, probably explaining the rapid degradation in G2 (Hagen et al. 2008). Why UNG2 must be degraded remains obscure, but some observations indicate a toxicity of unregulated expression of the enzyme (Henrik Sahlin Pettersen 2007, unpublished data).
(b) Uracil-DNA glycosylase SMUG1
This enzyme was described as a single-strand-selective monofunctional uracil-DNA glycosylase, but, at least, the mammalian SMUG1 works well with double-stranded DNA, and the efficiency of the enzyme with different substrates depends on reaction conditions (Kavli et al. 2002). SMUG1 was thought to be present only in vertebrates, but, in fact, it is also found in some bacteria. Interestingly, bacteria contain either the UNG-type enzyme or the SMUG1-type enzyme, usually not both (Pettersen et al. 2007). The activity of SMUG1 on the double-stranded substrate is strongly stimulated by AP-endonuclease APE1. Whereas UNG2 is highly specific for uracil, SMUG1 also efficiently removes 5-hydroxymethyluracil (HmU) and it may be the major enzyme correcting this lesion (Kavli et al. 2007). Studies on Ung knockout, Smug1 siRNA knockdown and Ung knockout/Smug1 knockdown mouse cells have indicated that Smug1 and Ung2 are both required for the prevention of mutations and that their functions are not redundant (An et al. 2005). Human SMUG1 binds tightly to AP sites and inhibits cleavage by AP-endonucleases. When expressed in Escherichia coli cells, it is unable to repair U:G mismatches induced by AID, inhibits proliferation and cannot reduce mutation rates, unlike UNG2 which alleviates the effects of AID. This is probably a reflection of the low catalytic turnover of SMUG1 compared with UNG-type enzymes. These results indicate that SMUG1 probably has its main function in non-proliferating or proliferating cells outside the S-phase. Unlike UNG2, SMUG1 makes contact with both DNA strands. It penetrates the double helix with a wedge motif that binds tightly to the abasic site. Interestingly, mutations in this motif decrease binding and increase catalytic efficiency several fold. Presumably, it is the role of this enzyme to carry out slow repair of U:G mismatches in DNA that is not undergoing replication, and, in this process, it may bind tightly to the AP site to protect it from further damage until the next player in the repair process arrives (Pettersen et al. 2007).
(c) T/U mismatch DNA glycosylase
In spite of its name, T/U mismatch DNA glycosylase (TDG) has a strong preference for uracil over thymine. TDG is an intriguing protein that, similar to SMUG1, has a low turnover number and strong binding to AP sites, and its activity is stimulated by APE1. As with SMUG1, the binding of the glycosylase to the AP site inhibits cleavage by the downstream AP endonuclease (Waters et al. 1999). Interestingly, the catalytic efficiency of the protein is increased by SUMOylation (Hardeland et al. 2002). It also has a strong preference for U:G mismatches. Unlike SMUG1, it is strictly cell-cycle regulated. However, it is regulated opposite to UNG2 by displaying the highest expression in the G1-phase and the lowest in the S-phase (Hardeland et al. 2007). While TDG has not been assumed to have an important function in uracil repair compared with the ‘leading’ enzymes UNG2 and SMUG1, this issue is far from settled and not based on good experimental evidence. The interesting expression pattern in the cell cycle and its substrate preference would predict a role in U:G repair outside the S-phase. How this role is shared with SMUG1 and UNG2 remains unclear.
(d) Uracil-DNA glycosylase MBD4
This glycosylase has the capacity to remove uracil and thymine resulting from deamination of CpG and methylated CpG, respectively (Hendrich et al. 1999). It was discovered as a protein that binds to methylated DNA (Hendrich & Bird 1998). Many of these properties resemble those of TDG. Unlike other uracil-DNA glycosylases, MBD4 also interacts directly with MLH1, suggesting a role in mismatch repair (Bellacosa 2001). Overexpression of a truncated form of MBD4 in an MSH6-defective human colon carcinoma cell line with microsatellite instability increases structural chromosomal rearrangements, including multiple reciprocal translocations, after irradiation. This may suggest a wider role for MBD4 in DNA damage response and maintenance of chromosomal stability (Abdel-Rahman et al. 2008). It may seem unlikely that it is the glycosylase function of MBD4 that is responsible for this type of structural instability.
4. Concluding remarks
It was thought for a long time that uracil in DNA is solely a lesion resulting from incorporation of dUMP during replication and spontaneous cytosine deamination. It is now clear that nature uses uracil in DNA in highly sophisticated mechanisms of adaptive immunity and innate responses to viral infection. This has greatly increased the interest in the roles of this class of enzymes. In most cells, however, genomic uracil remains a mutagenic lesion that must be accurately repaired by BER. The presence of several distinct uracil-DNA glycosylases indicates the importance of proper processing of DNA uracil. The individual functions of these enzymes are still not fully understood. The best established roles are those of UNG2 in the post-replicative repair of U:A pairs in all replicating cells and in SHM and CSR in B cells. Here, the other uracil-DNA glycosylases, SMUG1, TDG and MBD4, apparently have no role. They rather function in general repair of U:G mismatches (or HmU for SMUG1), possibly in different parts of the cell cycle, in different sequence contexts and, perhaps, in different subnuclear compartments. The available evidence from mammalian cells suggests that UNG2 also has a general role in U:G repair. Finally, there may be species differences with respect to the relative contribution of different uracil-DNA glycosylases in U:G repair and mutation prevention. This, as well as the overall regulation of BER, remains to be clarified.
Acknowledgments
This work has been supported by the European Community (Integrated Project DNA repair, grant no. LSHG-CT-2005-512113), the Norwegian Cancer Association, the Cancer Fund at St Olav's Hospital, the Svanhild and Arne Must Fund for Medical Research, the National Programme for Research in Functional Genomics in Norway (FUGE) and the Norwegian Research Council.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘DNA deamination in immunity, virology and cancer’.
References
- Abdel-Rahman W.M., Knuutila S., Peltomaki P., Harrison D.J., Bader S.A. Truncation of MBD4 predisposes to reciprocal chromosomal translocations and alters the response to therapeutic agents in colon cancer cells. DNA Repair. 2008;7:321–328. doi: 10.1016/j.dnarep.2007.11.009. doi:10.1016/j.dnarep.2007.11.009 [DOI] [PubMed] [Google Scholar]
- Akbari M., et al. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004;32:5486–5498. doi: 10.1093/nar/gkh872. doi:10.1093/nar/gkh872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari M., Visnes T., Krokan H.E., Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair. 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. doi:10.1016/j.dnarep.2008.01.002 [DOI] [PubMed] [Google Scholar]
- An Q., Robins P., Lindahl T., Barnes D.E. C→T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. doi:10.1038/sj.emboj.7600689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S., Ericsson M., Dai H.Y., Pena-Diaz J., Slupphaug G., Nilsen H., Aarset H., Krokan H.E. Monoclonal B-cell hyperplasia and leukocyte imbalance precede development of B-cell malignancies in uracil-DNA glycosylase deficient mice. DNA Repair. 2005a;4:1432–1441. doi: 10.1016/j.dnarep.2005.08.004. doi:10.1016/j.dnarep.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Andersen S., Heine T., Sneve R., Konig I., Krokan H.E., Epe B., Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005b;26:547–555. doi: 10.1093/carcin/bgh347. doi:10.1093/carcin/bgh347 [DOI] [PubMed] [Google Scholar]
- Bellacosa A. Role of MED1 (MBD4) gene in DNA repair and human cancer. J. Cell Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. doi:10.1002/jcp.1064 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. doi:10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Eftedal I., Guddal P.H., Slupphaug G., Volden G., Krokan H.E. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993;21:2095–2101. doi: 10.1093/nar/21.9.2095. doi:10.1093/nar/21.9.2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen L., et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. doi:10.1038/sj.emboj.7601958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U., Steinacher R., Jiricny J., Schär P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. doi:10.1093/emboj/21.6.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U., Kunz C., Focke F., Szadkowski M., Schär P. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 2007;35:3859–3867. doi: 10.1093/nar/gkm337. doi:10.1093/nar/gkm337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B., Hardeland U., Ng H.H., Jiricny J., Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. doi:10.1038/45843 [DOI] [PubMed] [Google Scholar]
- Imai K., et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. doi:10.1038/ni974 [DOI] [PubMed] [Google Scholar]
- Kavli B., et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39 926–39 936. doi: 10.1074/jbc.M207107200. doi:10.1074/jbc.M207107200 [DOI] [PubMed] [Google Scholar]
- Kavli B., Andersen S., Otterlei M., Liabakk N.B., Imai K., Fischer A., Durandy A., Krokan H.E., Slupphaug G. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J. Exp. Med. 2005;201:2011–2021. doi: 10.1084/jem.20050042. doi:10.1084/jem.20050042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavli B., Otterlei M., Slupphaug G., Krokan H.E. Uracil in DNA—general mutagen, but normal intermediate in acquired immunity. DNA Repair. 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. doi:10.1016/j.dnarep.2006.10.014 [DOI] [PubMed] [Google Scholar]
- Krokan H.E., Standal R., Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan H.E., Drabløs F., Slupphaug G. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. doi:10.1038/sj.onc.1205996 [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. doi:10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., Schatz D.G. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. doi:10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18 470–18 476. doi: 10.1074/jbc.274.26.18470. doi:10.1074/jbc.274.26.18470 [DOI] [PubMed] [Google Scholar]
- Nilsen H., et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. doi:10.1016/S1097-2765(00)80271-3 [DOI] [PubMed] [Google Scholar]
- Nilsen H., Haushalter K.A., Robins P., Barnes D.E., Verdine G.L., Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. doi:10.1093/emboj/20.15.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H., Stamp G., Andersen S., Hrivnak G., Krokan H.E., Lindahl T., Barnes D.E. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. doi:10.1038/sj.onc.1206860 [DOI] [PubMed] [Google Scholar]
- Otterlei M., et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. doi:10.1093/emboj/18.13.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen H.S., Sundheim O., Gilljam K.M., Slupphaug G., Krokan H.E., Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. doi:10.1093/nar/gkm372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. doi:10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Slupphaug G., Eftedal I., Kavli B., Bharati S., Helle N.M., Haug T., Levine D.W., Krokan H.E. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. doi:10.1021/bi00001a016 [DOI] [PubMed] [Google Scholar]
- Slupphaug G., Kavli B., Krokan H.E. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Sousa M.M., Krokan H.E., Slupphaug G. DNA-uracil and human pathology. Mol. Aspects Med. 2007;28:276–306. doi: 10.1016/j.mam.2007.04.006. doi:10.1016/j.mam.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Waters T.R., Gallinari P., Jiricny J., Swann P.F. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. doi:10.1074/jbc.274.1.67 [DOI] [PubMed] [Google Scholar]