Abstract
The intrinsic antiretroviral factor APOBEC3G (A3G) is highly active against HIV-1 and other retroviruses. In different cell types, A3G is expressed in high-molecular-mass (HMM) RNA–protein complexes or low-molecular-mass (LMM) forms displaying different biological activities. In resting CD4 T cells, a LMM form of A3G potently restricts HIV-1 infection soon after virion entry. However, when T cells are activated, LMM A3G is recruited into HMM complexes that include Staufen-containing RNA granules. These complexes are probably nucleated by the induced expression of Alu/hY retroelement RNAs that accompany T-cell activation. HMM A3G sequesters these retroelement RNAs away from the nuclear long interspersed nuclear element-derived enzymes required for Alu/hY retrotransposition. Human immunodeficiency virus (HIV) exploits this ‘window of opportunity’ provided by the loss of LMM A3G in activated CD4 T cells to productively infect these cells. During HIV virion formation, newly synthesized LMM A3G is preferentially encapsidated but only under conditions where Vif is absent and thus not able to target A3G for proteasome-mediated degradation. Together, these findings highlight the discrete functions of the different forms of A3G. LMM A3G opposes the external threat posed by exogenous retroviruses, while HMM A3G complexes oppose the internal threat posed by the retrotransposition of select types of retroelements.
Keywords: APOBEC3G, HIV-1, Vif, Alu, retrotransposition, RNA granules
1. Retroviruses and cytidine deaminases
Biological interactions with retroviruses and related endogenous retroelements have played important roles in human evolution. These interactions are dynamic and reciprocal. Humans and retroviruses have evolved mechanisms to counteract the other's biological activities. Since the identification of human immunodeficiency virus (HIV) in 1983, this pathogenic retrovirus has become a prototype for studies of many aspects of retroviral biology, including virus–host interactions.
HIV-1 is a member of the primate lentivirus family of retroviruses and is the product of a cross-species (zoonotic) transmission event of related but distinct lentiviruses (simian immunodeficiency viruses or SIVs) that naturally infect non-human primates in sub-Saharan Africa. SIVcpz in chimpanzees (Pan troglodytes) is the precursor of pandemic HIV-1. Similarly, SIVsm from sooty mangabeys (Cercocebus atys) is the immediate source of HIV-2. Other lentiviruses, such as SIVagm from African green monkeys, have not been transmitted to humans. It is clear that species-specific barriers to infection oppose successful establishment of various zoonotic retroviral infections. These barriers, which are now becoming better understood, provide an important resource for better understanding the evolutionary conflict that continues to occur between pathogenic retroviruses and their cellular hosts.
2. The HIV-1 Vif phenotype
In contrast to simple retroviruses encoding only Gag, Pol and Env gene products, HIV-1 encodes six additional auxiliary proteins (Tat, Rev, Nef, Vif, Vpr and Vpu) that orchestrate the pathogenic interplay of HIV-1 with its human host. While the major functions of Vpr, Vpu, Tat, Rev and Nef were largely unravelled within a few years after the HIV-1 genome was sequenced, the mechanism underlying the function of Vif remained shrouded in mystery for almost two decades.
Vif corresponds to a basic 23 kDa phosphoprotein that is expressed late in the retroviral life cycle and is highly conserved among all of the primate lentiviruses with the notable exception of the equine infectious anaemia virus. The function of Vif is tightly linked to the nature of the virus-producing cell (Gabuzda et al. 1992). Several T-cell lines (e.g. Jurkat and SupT1) and non-haemopoietic cell lines (e.g. HeLa, 293T and COS) produce infectious HIV-1 virions in the absence of Vif (Δvif HIV-1) and are termed ‘permissive’. Conversely, Δvif HIV-1 virions derived from non-permissive cells, including primary CD4 T cells and macrophages, the natural targets of HIV-1 infection, are non-infectious (Sova & Volsky 1993; von Schwedler et al. 1993). However, the molecular basis for these interesting cell-dependent differences remained unclear for many years.
3. Investigating Vif action reveals APOBEC3G as an innate antiviral factor
The identification of human APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G or A3G) in non-permissive cells as a potent innate inhibitor of retroviruses emerged from studies investigating HIV-1 Vif action.
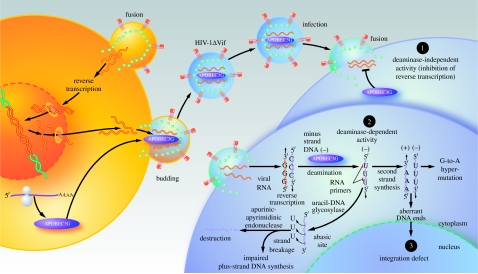
A key insight into how Vif allowed wild-type HIV-1 to readily spread in cultures of non-permissive cells was made when heterokaryons of permissive and non-permissive cells were produced and infected with Δvif HIV-1 (Madani & Kabat 1998; Simon et al. 1998). Analysis of the progeny virions emanating from these heterokaryons revealed that they were non-infectious, which suggested that Vif somehow overcomes the inhibitory effect of one or more factors produced by non-permissive cells. Using subtractive hybridization techniques for analysis of two closely related cells differing in permissivity, Sheehy et al. (2002) succeeded in identifying A3G, a known cytidine deaminase, as the host antiviral factor produced by non-permissive cells and thwarted by Vif. Virions budding from non-permissive cells were found to contain A3G, providing an explanation for how viral replication might be altered during the next round of viral spread (figure 1).
Figure 1.
Sequential strategies for the restriction of HIV-1 by virion-incorporated A3G. In the absence of Vif, A3G in the cytoplasm of virus-producing cells is effectively incorporated into budding virions and carried forward into the next target cells, where it can act as a potent inhibitor of HIV-1 replication. These inhibitory effects involve both deaminase-independent and -dependent antiviral actions. A3G bound to HIV-1 RNA may physically impede reverse transcriptase movement on the viral RNA template, resulting in a deaminase-independent block in reverse transcription (1). However, this inhibition is frequently incomplete, and minus-strand viral DNA is generated. A3G mediates extensive deamination of dC residues in this newly synthesized single-stranded viral DNA. This action of A3G effectively halts HIV replication because of the resulting dG-to-dA changes in the viral plus strand or because the uracil-containing minus strand is destroyed by the actions of uracil DNA glycosylase and apurinic–apyrimidinic endonuclease (2). Additionally, diminished chromosomal integration of the double-stranded viral DNA may occur due to defects in tRNALys3 primer cleavage, leading to the formation of viral DNA with aberrant ends (3).
4. APOBEC3G and other members of the cytidine deaminase family
A3G (384 amino acids, 46 kDa) belongs to a much larger family of cytidine deaminases that share a conserved zinc-binding motif (Cys/His)-Xaa-Glu-Xaa23–28-Pro-Cys-Xaa2–4-Cys (Jarmuz et al. 2002). These enzymes mediate hydrolytic deamination at the C4 position of the C (or dC) base, converting C to U (or dC to dU). These changes are often referred to as RNA or DNA editing (Teng et al. 1993; Harris et al. 2002). In humans, the APOBEC enzyme family includes activation-induced deaminase (AID), APOBEC1, APOBEC2, APOBEC3A–H and APOBEC4 (Jarmuz et al. 2002; Conticello et al. 2005; Rogozin et al. 2005; OhAinle et al. 2006).
APOBEC1 is primarily expressed in gastrointestinal tissue (Teng et al. 1993). As the central component of an RNA editosome complex, it edits the apolipoprotein B mRNA transcript at cytosine6666 converting a glutamine at this position to an in-frame stop codon giving rise to a truncated version of the apoB protein (Teng et al. 1993; Yamanaka et al. 1995; Mehta et al. 2000). The longer apoB100 and the shorter apoB48 proteins differ in their effects on cholesterol metabolism.
AID is selectively expressed in germinal centre B cells (Muramatsu et al. 1999), where it catalyses deamination at the DNA level, and thus promotes immunoglobulin gene diversification via somatic hypermutation and class switch recombination (Muramatsu et al. 2000).
The highly related APOBEC3 genes (APOBEC3A–H) (7, 10, 12 and 125) are located in a single cluster on human chromosome 22 at q13.2. Four of the APOBEC3 proteins (A3B, A3DE, A3F and A3G) have duplicated cytidine deaminase domains (CDs) while A3A, A3C and A3H have only one CD (Jarmuz et al. 2002; Conticello et al. 2005; Dang et al. 2006; OhAinle et al. 2006). Phylogenetic analyses suggest that the primordial APOBEC3 contained only one CD, which might have evolved from AID or APOBEC2 during the onset of vertebrate speciation (Conticello et al. 2005).
APOBEC2, a single-domain cytidine deaminase, is principally expressed in the cardiac and skeletal muscle. Despite rather extensive study, its function in muscle biology remains unknown. APOBEC2 is the closest known paralog of APOBEC3 for which a high-resolution structure is available (Prochnow et al. 2007). However, this deaminase lacks enzymatic activity and its precise function in cells is also unknown.
When expressed in Escherichia coli, A3G, AID and even APOBEC1 catalyse the deamination of dC residues in single-stranded DNA (Harris et al. 2002; Beale et al. 2004), suggesting that single-stranded DNA is the favoured substrate for A3G. This finding provides an explanation for how viral replication might be blocked in the next target cell via the inherent deaminase activity of A3G acting on the nascent reverse transcribed cDNAs produced by the virus (figure 1).
5. Vif circumvents antiviral activity of APOBEC3G
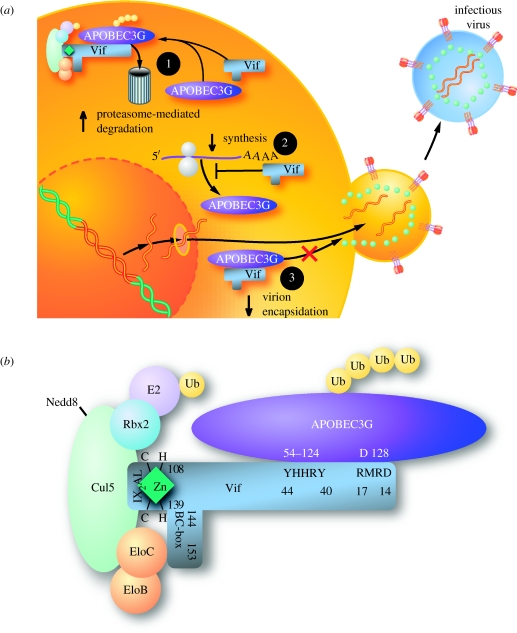
Human A3G expressed in non-permissive cells poses a significant threat to the replication and spread of HIV-1. Vif counters this threat by directly interacting with A3G and linking A3G to an efficient E3 ubiquitin ligase complex that mediates polyubiquitylation of both A3G and Vif. These events promote accelerated degradation of these proteins by the 26S proteasome (Conticello et al. 2003; Marin et al. 2003; Sheehy et al. 2003; Stopak et al. 2003). Vif also partially impairs the translation of A3G mRNA, although the mechanism remains undefined (Stopak et al. 2003). Nevertheless, the combined effects of accelerated degradation and diminished synthesis result in the nearly complete depletion of intracellular A3G in the producer cell during wild-type HIV-1 infection. These actions of Vif prevent virion encapsidation of A3G, thereby ensuring high infectivity of the progeny virions (Mariani et al. 2003; Stopak et al. 2003; figure 2a).
Figure 2.
The interplay of Vif and APOBEC3G. (a) Neutralization of A3G in virus-producing cells by HIV-1 Vif. Vif defeats the antiviral activity of A3G principally by both binding to A3G and recruiting an E3 ligase complex that mediates polyubiquitylation (Ubn) of A3G and its accelerated degradation in 26S proteasomes (1). Vif also partially impairs the translation of A3G mRNA (2). These dual effects of Vif effectively deplete A3G in the virus-producing cells and, thus, A3G is not available for incorporation into virions budding from these cells. Other auxiliary functions of Vif have been proposed, including physical exclusion of A3G from virion encapsidation in the absence of degradation, perhaps due to sequestration of A3G away from the sites of viral assembly/budding (3). (b) Model of the Vif–cullin5–elongin BC complex. Vif employs multiple protein interaction domains to orchestrate A3G degradation. The N-terminal region of Vif binds to an N-terminal region of A3G (amino acids 54–124). The SLQ(Y/F)LA motif (amino acids 144–150) of Vif mediates binding to the elongin C (EloC) component of the E3 ligase complex. Finally, a novel zinc-binding motif, (HCCH, amino acids 108–139) within Vif, containing two conserved cysteines, mediates a second interaction with the cullin 5 (Cul5) component. The Cul5–Vif E3 ubiquitin ligase binds A3G and brings it into close proximity with the E2 ubiquitin-conjugating enzyme.
In the absence of Vif, A3G effectively incorporates into budding HIV-1 virions (Mariani et al. 2003; Stopak et al. 2003) by interacting with the nucleocapsid region of the Gag polyprotein (Alce & Popik 2004; Cen et al. 2004; Luo et al. 2004). This interaction is further strengthened by A3G's propensity to bind single-stranded nucleic acids, particularly viral RNA at the plasma membrane site of virion budding (Schafer et al. 2004; Svarovskaia et al. 2004; Zennou et al. 2004; Khan et al. 2005; Burnett & Spearman 2007; Khan et al. 2007; Soros et al. 2007). The incorporation of only seven molecules of A3G into Δvif HIV virions produced by human peripheral blood mononuclear cells is sufficient to greatly impair HIV-1 replication (Xu et al. 2007). Thus, the A3G–Vif–E3 ligase axis is a compelling new target for the development of novel antiviral drugs that could re-enable the potent defensive properties of A3G.
6. Further insights into Vif action
Deletional mutagenesis coupled with co-immunoprecipitation analyses reveals that the N-terminal region of Vif binds to the N-terminal region of A3G (amino acids 54–124; Conticello et al. 2003; Marin et al. 2003; Simon et al. 2005; Wichroski et al. 2005; figure 2). Further mechanistic insights have emerged from cross-species studies. For example, Vif from SIVagm effectively triggers the degradation of African green monkey A3G but fails to neutralize either human or chimpanzee A3G. Similarly, HIV-1 Vif cannot induce degradation of African green monkey or rhesus macaque A3G. These species-specific effects appear to be governed by a single amino acid (residue 128) in A3G (Mariani et al. 2003; Bogerd et al. 2004; Mangeat et al. 2004; Schrofelbauer et al. 2004; Xu et al. 2004) and amino acids 14–17 (DRMR) in Vif (Schrofelbauer et al. 2006). These species-specific limitations in Vif activity probably form an important barrier that minimizes the frequency of zoonotic transmission of many primate lentiviruses. The fact that the Vif gene products of SIVcpz and SIVsm, the precursors of HIV-1 and 2, respectively, degrade human A3G (Mariani et al. 2003; Gaddis et al. 2004; Xu et al. 2004) provides a compelling explanation for how these viruses successfully spawned the HIV-1 and 2 epidemics in humans.
The regions in Vif critical for interacting with the E3 ligase complex have also been mapped by mutagenesis (figure 2b). The SLQ(Y/F)LA motif (amino acids 144–150) of Vif interacts directly with elongin C (Marin et al. 2003; Yu et al. 2003, 2004c; Mehle et al. 2004a,b). A cryptic zinc-coordination motif, His-Xaa5-Cys-Xaa17–18-Cys-Xaa3–5-His (HCCH, amino acids 108–139), connects Vif to cullin 5 (Luo et al. 2005; Mehle et al. 2006; Xiao et al. 2006). Through these interactions, Vif recruits a ubiquitin ligase (E3) complex comprising elongin C, elongin B, cullin 5, Nedd8 and Rbx1 (Yu et al. 2003, 2004c) that mediates the polyubiquitylation and triggers degradation of A3G (Conticello et al. 2003; Marin et al. 2003; Sheehy et al. 2003; Stopak et al. 2003). Mutation of the SLQ or the HCCH motif in Vif, or overexpression of cullin 5 mutants that fail to engage Nedd8 or Rbx1 renders Vif incapable of degrading A3G and leads to greater A3G antiviral activity (Mehle et al. 2004a,b, 2006; Yu et al. 2003, 2004c). Thus, the Vif:cullin 5 and Vif:elongin C interaction sites are additional potential targets for antiviral drug design.
Although Vif-induced degradation of A3G is important in overcoming the antiviral effects of A3G, non-degradative mechanisms of Vif action have also been proposed. Vif may physically exclude A3G from sites of viral assembly/budding or inhibit A3G encapsidation by competing for binding to viral components, such as the nucleocapsid or viral genomic RNA (Mariani et al. 2003; Kao et al. 2004; Opi et al. 2007; figure 2a).
7. Regulation of cytidine deaminases and cancer
How is the potentially promiscuous mutagenic activity of the cytidine deaminases controlled, particularly during cell division when this cytoplasmic enzyme could readily access nuclear DNA? In this regard, forced expression of APOBEC1 as a transgene in the livers of mice consistently leads to hepatic dysplasia and hepatocellular carcinoma, possibly due to promiscuous RNA editing of tumour suppressors or oncogenes (Yamanaka et al. 1995). High-level expression of AID is similarly linked with the development of various large B-cell and non-Hodgkin's lymphomas (Revy et al. 2000). The constitutive and ubiquitous transgenic expression of AID in mice similarly results in the development of various cancers, specifically T-cell lymphomas, micro-adenomas and lung adenocarcinomas (Okazaki et al. 2003). The associations of APOBEC1 and AID with cancer emphasize how tight intracellular regulation of these cytidine deaminases is probably required to minimize the chances of mutations in genomic DNA leading to cellular transformation.
8. Assembly into large RNA–protein complexes inhibits APOBEC3G enzymatic activity
How is the potentially mutagenic activity of the A3G enzymes negatively regulated? One possibility was suggested immediately by subcellular localization studies, which indicated that A3G is strongly retained in the cytoplasm (Mangeat et al. 2003; Muckenfuss et al. 2006; OhAinle et al. 2006; Wichroski et al. 2006; Gallois-Montbrun et al. 2007). While such sequestration would limit promiscuous editing of genomic DNA during many phases of the cell cycle, nuclear access could occur during mitosis when nuclear membranes break down. Another possibility involving negative regulation by complex assembly was suggested by functional clues from APOBEC1 and AID. The enzymatic activity of APOBEC1 is regulated by its assembly with an additional factor termed ACF (APOBEC1 complementing factor), leading to the formation of a multicomponent enzyme complex. AID has no measurable deaminase activity unless pretreated with RNase to remove inhibitory RNAs bound to AID. Indeed, endogenous A3G expressed in the cytoplasm of H9 T-cell lines and mitogen-activated CD4 T cells is assembled in 5–15 MDa high-molecular-mass (HMM) ribonucleoprotein (RNP) complexes, and the deaminase activity of A3G is greatly inhibited in these complexes. Interestingly, these HMM A3G complexes can be artificially converted to an enzymatically active low-molecular-mass (LMM) form by treatment with RNase A, suggesting that one or more cellular RNAs play an important role in the assembly of HMM A3G complexes (Chiu et al. 2005).
9. RNP complexes in HMM A3G complex assembly
The HMM A3G RNP complexes contain at least 95 different proteins, as determined by tandem affinity purification and mass spectrometry (Chiu et al. 2006; Kozak et al. 2006; Gallois-Montbrun et al. 2007). Numerous cellular RNA-binding proteins with diverse roles in RNA function, metabolism and fate determination are found in these HMM A3G complexes, and their participation in the complex occurs in an RNA-dependent manner (Chiu et al. 2006; Kozak et al. 2006; Gallois-Montbrun et al. 2007). Careful analysis suggests that these components fall into at least three previously defined multi-subunit RNP complexes in human cells: (i) Staufen-containing, polysome-associated RNA granules, (ii) Ro RNPs, and (iii) select components of prespliceosomes plus reservoirs for transcriptional regulators (Chiu et al. 2006). Of note, the protein cofactors in the latter class are quite multifunctional, and many participate in Staufen-containing RNA granules and function as cytoplasmic regulators of translation.
Staufen-containing RNA granules correspond to more than 10 MDa macromolecular RNP complexes comprising ribosomal subunits, scaffold proteins, translation machinery, RNA-binding proteins, helicases and various decay enzymes (Kanai et al. 2004; Villace et al. 2004; Anderson & Kedersha 2006; Kiebler & Bassell 2006). Ro RNPs are the major RNP autoantigens recognized by sera from patients with various connective tissue diseases. In human cells, Ro RNPs contain one of the four human small Y (hY) RNAs (hY1, hY3, hY4 and hY5) and two core proteins (60 kDa Ro and 50 kDa La; Fabini et al. 2000). Almost all of the proteins that participate in the formation of Staufen-containing RNA granules and Ro RNPs are readily detectable in the purified HMM A3G RNP complexes.
An association of A3G with exogenously expressed components of stress granules (SGs) and processing bodies (PBs) has been reported (Kozak et al. 2006; Wichroski et al. 2006; Gallois-Montbrun et al. 2007). The fact that Staufen RNA granules, SGs and PBs represent a continuum of granular-like structures in which cargos may be transferred back and forth could contribute to these results. These findings imply that the dynamic spatial organizations between RNA granules and related cytoplasmic complexes could affect the status of A3G complexes and control A3G's antiviral activity.
Intriguingly, sequencing of the most prominent RNA components in HMM A3G complexes identified human Alu and small hY (hY1–5) endogenous retroelement RNAs (Chiu et al. 2006). In the presence of A3G, Alu and hY RNAs are selectively recruited and enriched in Staufen RNA granules and Ro RNPs, respectively, suggesting a potential physiological function for these complexes (Chiu et al. 2006). Specifically, endogenous non-autonomous retroelements (i.e. Alu and hY RNAs) are probably the natural cellular targets of A3G.
10. Intravirion APOBEC3G complexes: an unusual virus–host interaction
The fact that cellular A3G principally resides in 5–15 MDa HMM RNP complexes in activated, virus-producing T cells prompted analysis to determine which form of A3G is actually incorporated into the budding virions. Pulse-chase radiolabelling and size-fractionation studies revealed that cellular A3G rapidly assembles (less than 30 min) into HMM complexes (Soros et al. 2007), and as a consequence only small quantities of newly synthesized LMM A3G occur in cells. However, pulse-chase studies further revealed that virion A3G is mainly recruited from the cellular pool of newly synthesized enzymes (Soros et al. 2007). Interaction of A3G with HIV genomic RNA in the virion core leads to the assembly of large intravirion A3G complexes and unexpectedly to inhibition of its enzymatic activity (Soros et al. 2007). This process is analogous to the inactivation of cellular A3G activity when it engages cellular RNAs in the HMM RNA–protein complexes (Chiu et al. 2005).
How is the enzyme ultimately activated? Intriguingly, the answer appears to involve reverse transcription and the action of HIV-1 RNase H. As the minus-strand cDNA is synthesized during HIV reverse transcription, the genomic RNA template is degraded by the RNase H activity of the reverse transcriptase, generating the minus-strand DNA substrate of A3G and also removing the inhibitory RNA bound to A3G and thereby activating its deaminase activity (Soros et al. 2007). These findings highlight a most unusual virus–host interaction, in which initiation of the antiviral enzymatic activity of A3G is contingent on the prior action of an essential viral enzyme.
11. Mechanisms of APOBEC3G antiviral and anti-retroelement action
The antiviral mechanism of A3G has been attributed to two fundamental properties: its ability to bind single-stranded RNA and its inherent deaminase activity on single-stranded DNA substrates. These two properties map to two A3G CDs. The N-terminal CD1 mediates RNA binding and virion encapsidation (Navarro et al. 2005; Newman et al. 2005; Iwatani et al. 2006). The C-terminal CD2 confers deaminase activity (Hache et al. 2005; Navarro et al. 2005; Newman et al. 2005; Iwatani et al. 2006) and sequence specificity for modification of the single-stranded DNA substrate (Harris et al. 2003; Zhang et al. 2003; Bishop et al. 2004; Liddament et al. 2004; Wiegand et al. 2004).
12. Deaminase-independent antiviral activity of APOBEC3G
RNA binding appears to play a major role in rendering ΔVif HIV-1 virions non-infectious. Specifically, A3G analogues containing inactivated C-terminal CD2 can still substantially reduce the infectivity of Δvif HIV-1 (Newman et al. 2005; Bishop et al. 2006). The RNA-binding activity of the N-terminal CD1 appears critically involved in this non-enzymatic form of inhibition, which probably involves binding to HIV-1 RNA in the virion core and physical impairment of reverse transcriptase activity (figure 1). Additionally, A3G in Δvif HIV-1 virions reduces the ability of tRNALys3 primers to initiate reverse transcription by 50 per cent or more, again providing a mechanism for a block at the level of reverse transcription (Guo et al. 2006). A3G might also cause defects in tRNALys3 cleavage during plus-strand DNA transfer, leading to the formation of aberrant viral DNA ends that could interfere with subsequent chromosomal integration (Mbisa et al. 2007). Of note, the N-terminal linker region of A3G has been implicated as a docking site for the C-terminal domain of HIV-1 integrase. The association of A3G with components of the preintegration complex, such as integrase, might negatively influence nuclear import of the complex, thereby further impairing viral DNA integration (Luo et al. 2007).
The role of non-enzymatic mechanisms of A3G antiviral function has been evaluated in other systems. For example, A3G also mediates deaminase-independent antiviral activity against human T-cell leukaemia virus type-1 (HTLV-1; Mahieux et al. 2005; Sasada et al. 2005) and hepatitis B virus (HBV; Rosler et al. 2004; Turelli et al. 2004a). Intriguingly, some retroviruses, such as HTLV-I, murine leukaemia virus (MLV) and Mason–Pfizer monkey virus, have evolved mechanisms to escape human A3G, mouse APOBEC3 (mA3) or rhesus A3G, respectively, by targeting the RNA-binding properties of these antiviral proteins (Kobayashi et al. 2004; Abudu et al. 2006; Derse et al. 2007; Doehle et al. 2005b, 2006).
13. Deaminase-dependent antiviral activity of APOBEC3G
If RNA binding solely accounts for the antiviral activity of A3G, why would the deaminase function be conserved? Although we cannot exclude the existence of other physiological functions, we believe that the conserved deaminase property of A3G provides an even more potent ‘second antiviral punch’ (figure 1).
During reverse transcription, the C-terminal CD2 of virion-incorporated A3G catalyses extensive dC-to-dU deamination, preferentially at the 3′-dC of the 5′-CC dinucleotides (Harris et al. 2003; Mangeat et al. 2003; Zhang et al. 2003; Bishop et al. 2004; Liddament et al. 2004; Wiegand et al. 2004) in the newly synthesized minus-strand viral DNA (Yu et al. 2004b; Chelico et al. 2006; Suspene et al. 2006). Some of the uracils generated by A3G may be excised by uracil DNA glycosylase, leading to abasic sites that could cause DNA degradation in the presence of apurinic–apyrimidinic endonucleases (Schrofelbauer et al. 2005; Yang et al. 2007a). A few viral minus strands appear to survive this attack and serve as templates for plus-strand synthesis, where the dU promotes dA misincorporation. The resultant dG-to-dA mutations further negate HIV-1 replication by altering viral open reading frames and introducing inappropriate translation termination codons (Harris et al. 2003; Lecossier et al. 2003; Mangeat et al. 2003; Zhang et al. 2003; figure 3). Additionally, the accumulation of dUs in minus-strand DNA may lead to decreased plus-strand synthesis due to aberrant initiation (Klarmann et al. 2003; figure 1).
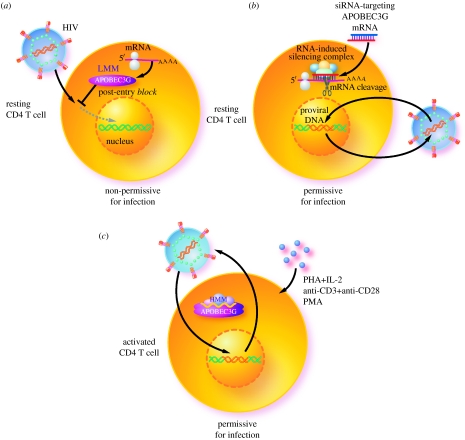
Figure 3.
(a) LMM A3G restricts HIV-1 after entry into resting peripheral blood CD4 T cells. Human A3G exists exclusively in LMM forms in peripheral blood-derived resting CD4 T cells and functions as a post-entry restriction factor to block replication of incoming HIV-1 viral particles. (b) RNA interference-mediated depletion of LMM A3G in resting CD4 T cells is sufficient to render these cells permissive for HIV-1 infection. (c) This restricting activity is forfeited when A3G is recruited into the HMM A3G complex upon CD4 T-cell activation by various mitogens (anti-CD3/CD28 and phorbol myristate acetate (PMA)) and cytokines (IL-2, IL-7 and IL-15).
These enzyme-dependent effects of A3G produce antiviral responses against many retroviruses in addition to HIV-1. These include SIV and equine infectious anaemia virus, and even distantly related retroviruses, such as MLV and foamy viruses (Harris et al. 2003; Mangeat et al. 2003; Mariani et al. 2003; Lochelt et al. 2005; Russell et al. 2005; Delebecque et al. 2006). Similarly, G-to-A hypermutation, albeit at low levels, has also been detected in HBV (Rosler et al. 2004; Suspene et al. 2005; Bonvin et al. 2006) and HTLV-1 (Mahieux et al. 2005) genomes produced in cells expressing A3G.
These studies and those interrogating intravirion A3G imply sequential strategies for virion-incorporated A3G to restrict HIV-1 (figure 1). Initially, the enzymatically latent form of A3G bound to HIV-1 RNA may impair generation of minus-strand DNA by physically impeding reverse transcriptase on its viral RNA template. However, this physical block may be incomplete and minus-strand viral DNA may be generated. Subsequently, A3G deaminase activity is restored when RNase H degrades the viral RNA and leaves the single-stranded DNA template intact for plus-strand synthesis, allowing extensive deamination of the minus-strand DNA. The relative effectiveness of these two antiviral actions in different cellular environments could explain why HBV replication is inhibited by deaminase-dependent actions of A3G in HepG2 cells but by deaminase-independent action in Huh7 cells (Rosler et al. 2004; Turelli et al. 2004a; Suspene et al. 2005; Bonvin et al. 2006).
14. Post-entry restriction of exogenous viral infection by LMM APOBEC3G
The finding of different forms of A3G has shed light on a long-standing mystery in HIV-1 biology: why resting CD4 T cells in lymphoid tissue are permissive to HIV-1 infection, while CD4 T cells circulating in the peripheral blood are not, even though A3G exists in both cell types. The answer is that A3G is expressed in two very different forms in these two populations of CD4 T cells. Circulating resting CD4 T cells have LMM A3G (Chiu et al. 2005) and are refractory to HIV-1 infection, at least in part, owing to an early post-entry restriction block at or immediately after the reverse transcription step (figure 3a). In sharp contrast, in lymphoid tissue-resident resting CD4 T cells, which display increased permissiveness to HIV-1 infection, A3G is predominantly found in HMM complexes (Kreisberg et al. 2006). RNA interference-mediated depletion of LMM A3G in resting CD4 T cells is sufficient to render these cells permissive for HIV-1 infection (Chiu et al. 2005; figure 3b). These results provide the first evidence that LMM A3G can function as a potent post-entry restriction factor that inhibits the replication of incoming HIV-1 virions in target cells independently of its prior incorporation into virions. However, this restricting activity is forfeited when A3G is recruited into the HMM A3G complex upon CD4 T-cell activation by various mitogens (anti-CD3/CD28 and phorbol myristate acetate) and cytokines (interleukins (IL)-2 and -15) (Chiu et al. 2005; Kreisberg et al. 2006; Stopak et al. 2007; figure 3c).
LMM A3G also governs the resistance of cells of monocyte lineage to infection with HIV-1 (Chiu et al. 2005; Pion et al. 2006; Ellery et al. 2007; Peng et al. 2007). Specifically, freshly isolated monocytes are refractory to infection and have LMM A3G (Chiu et al. 2005). Differentiation of these cells into macrophages promotes assembly of the HMM A3G complex (Chiu et al. 2005), a transition that correlates with a sharp increase in permissiveness to HIV-1 infection. HMM A3G complexes also are observed in the CD16+ subset of monocytes, which are more permissive to HIV infection than other monocytes (Ellery et al. 2007). Maturation of dendritic cells is associated with a sharp increase in A3G expression and the additional expression of LMM A3G as well as less permissiveness to HIV infection (Pion et al. 2006; Stopak et al. 2007).
Interestingly, the post-entry antiviral action of cellular LMM A3G does not depend strictly on editing. Instead, it involves significant delays in the accumulation of HIV-1 late reverse transcription products (Chiu et al. 2005). As such, RNA binding by A3G is probably involved in the restriction of incoming HIV-1 by LMM A3G. However, G-to-A hypermutation has been noted in a minor subset of reverse transcripts from unstimulated peripheral blood mononuclear cells (Janini et al. 2001) and resting CD4 T cells (Chiu et al. 2005) infected with HIV-1. Although a recent study suggested that T cells contain an RNase-insensitive inhibitor of A3G deaminase activity (Thielen et al. 2007), LMM A3G possesses detectable enzymatic activity in vivo. Thus, A3G probably employs a dual strategy involving sequential non-enzymatic and enzymatic actions to achieve its potent post-entry antiviral effects.
15. HMM APOBEC3G opposes endogenous Alu retroelement retrotransposition
Identifying the RNA components of HMM A3G complexes (Chiu et al. 2006) has yielded important insights into their physiological functions and suggested that endogenous non-autonomous retroelements (i.e. Alu RNAs; Kazazian 2004) probably are the natural cellular targets of A3G.
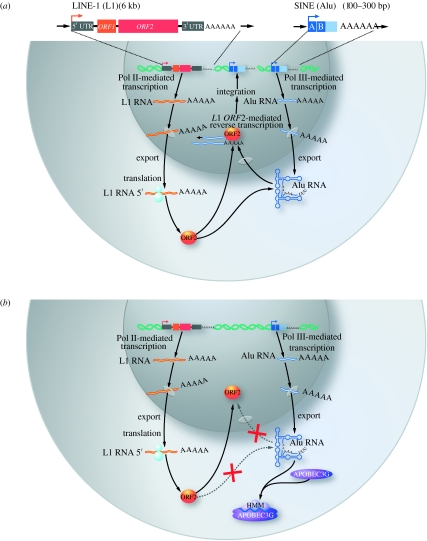
Alu elements, the most abundant short interspersed nucleoside elements, are a highly successful group of retroelements in humans, accounting for approximately 10 per cent of the human genome. They are amplified through retrotransposition, an intracellular process that involves cytoplasmic RNA intermediates, reverse transcription in the nucleus, and integration of the newly formed retroelement DNA at novel chromosomal sites. Alu elements encode no protein but can retrotranspose by ‘stealing’ the reverse transcriptase and endonuclease enzymes encoded by a set of autonomous retroelements termed long interspersed nuclear elements-1 (L-1; Dewannieux et al. 2003; Kazazian 2004; figure 4a).
Figure 4.
HMM A3G restricts Alu retrotransposition. (a) Alu retrotransposition mediated by long interspersed nuclear elements-1 (L1). Functional L1 elements are 6-kb-long autonomous retroelements that contain an internal RNA polymerase II (Pol II) promoter within their 5′-UTR, two open reading frames (ORF1 and ORF2), and a 3′-UTR. SINEs, including the most prominent and active member, Alu, are short RNA polymerase III (Pol III)-transcribed retroelements that contain an internal promoter but no protein coding capacity. Successful retrotransposition of Alu elements depends on their ability ‘to steal’ the reverse transcriptase/endonuclease enzymes encoded by L1 ORF2. (b) Non-enzymatic inhibitory mechanism for A3G to restrict Alu. Human A3G impairs the retrotransposition of Alu by sequestering Alu RNA transcripts in the cytoplasmic HMM complexes, especially Staufen-containing RNA granules, away from the nuclear L1 machinery, thereby interdicting the retrotransposition cycle.
When tested in an in vitro assay measuring Alu retrotransposition in living cells (Dewannieux et al. 2003), A3G greatly inhibited L1-dependent retrotransposition of Alu elements (Chiu et al. 2006; Hulme et al. 2007). Notably, A3G does not affect the retrotransposition of L1 (Turelli et al. 2004b; Esnault et al. 2005; Bogerd et al. 2006; Chen et al. 2006; Muckenfuss et al. 2006; Stenglein & Harris 2006). Catalytically inactive mutants of A3G also effectively inhibit Alu retrotransposition, suggesting a deaminase-independent mechanism for A3G action (Chiu et al. 2006; Hulme et al. 2007). Since A3G is primarily cytoplasmic, and Alu RNA is recruited to Staufen RNA granules in an A3G-dependent manner, A3G probably interrupts retrotransposition by sequestering transcribed Alu RNAs in the cytoplasm, denying Alu RNAs access to the nuclear enzymatic machinery of L1 (Chiu et al. 2006; figure 4b). Unfortunately, the assembly of HMM A3G complexes to combat Alu retrotransposition opens the door for HIV-1 infection as the post-entry restricting activity of LMM A3G is forfeited.
The assembly of HMM A3G complexes appears to require entry into the G1b phase of the cell cycle (Chiu et al. 2005), a stage characterized by RNA synthesis. Activation of CD4 T cells with phytohaemagglutinin and IL-2 induces high-level expression of selected endogenous retroelement, including Alu and hY RNAs. Conversely, almost all of the protein cofactors that participate in the HMM A3G complexes are constitutively expressed in resting CD4 T cells but are not assembled into complexes (Chiu et al. 2006). These findings raise the distinct possibility that the induced expression of Alu RNAs is the driving force for HMM A3G complex assembly. These findings also suggest that A3G directly binds to Alu and hY retroelement RNAs.
In sharp contrast to human APOBEC3 family genes (A3A–A3H) on chromosome 22, only a single APOBEC3 gene (mA3) is present on the syntenic chromosome 15 in rodents, suggesting that the APOBEC3 locus expanded after the genetic radiation of mice and humans. Phylogenetic analysis of primate A3G proteins reveals a high rate of positive selection (non-synonymous mutations) that predates the emergence and diversification of primate lentiviruses (Sawyer et al. 2004; Zhang & Webb 2004), strengthening the notion that the APOBEC3 gene family expanded to curtail the genomic instability caused by endogenous retroelements. Consistent with this notion, the activity of retrotransposons is at least 100-fold higher in mouse cells than in human cells (Maksakova et al. 2006).
16. APOBEC3 deaminases as inhibitors of endogenous retroelements
APOBEC3 proteins do, in fact, inhibit LTR retrotransposons. Human A3B, A3C, A3F, A3G and mA3 all effectively inhibit mouse IAP and MusD retroelements (Esnault et al. 2005; Bogerd et al. 2006; Chen et al. 2006), while A3F restricts a pseudo-ancestral human endogenous retrovirus (HERV-K) DNA sequence that was recently reconstructed based on the fossil record of ancient endogenous retroviruses (Lee & Bieniasz 2007). Reminiscent of the dual inhibitory effects of A3G in HIV-1 replication, APOBEC3 proteins exert dual restricting effects on these endogenous retroviruses, involving both a decrease in the number of transposed DNA copies and extensive editing of the transposed copies (Esnault et al. 2005, 2006). Interestingly, A3A inhibits IAP and MusD retrotransposition through a novel deamination-independent mechanism (Bogerd et al. 2006).
Human APOBEC3 proteins, including A3A, A3B, A3C and A3F, inhibit non-LTR retrotransposons, such as L1 retroelements (Bogerd et al. 2006; Chen et al. 2006; Muckenfuss et al. 2006; Stenglein & Harris 2006); A3A, A3B, and to a lesser extent, A3C also inhibit L1-mediated Alu retrotransposition (Bogerd et al. 2006). This finding could reflect the ability of these deaminases to enter the nucleus (Bogerd et al. 2006; Chen et al. 2006; Muckenfuss et al. 2006), where L1-mediated reverse transcription occurs. Human A3B and A3F may also directly interact with the L1 enzymatic machinery through a highly homologous region in these two deaminases (Stenglein & Harris 2006). The relatively high-level expression of APOBEC3 proteins in human testis and ovary (A3G, A3F and A3C) (Jarmuz et al. 2002; OhAinle et al. 2006) and embryonic stem cells (A3B) (Bogerd et al. 2006), where extensive genome demethylation prevails and retroelements are thought to be transcribed (Kazazian 2004; Maksakova et al. 2006), points to a physiologically relevant role for these deaminases in the protection of these cells from the potentially deleterious effects of endogenous retrotransposition.
17. APOBEC3 deaminases as innate antiviral restriction factors
Considerable effort has been devoted to defining the function and spectrum of antiviral activities for each of the members of the extended human APOBEC3 family. Apart from A3G, several human APOBEC3 family members (e.g. A3F, A3B and A3DE) also inhibit HIV-1. A3F is coordinately expressed with A3G in primary cellular targets of HIV-1 (CD4 T cells and macrophages) (Bishop et al. 2004; Liddament et al. 2004; Wiegand et al. 2004). Similar to A3G, virion-incorporated A3F probably achieves its restrictive effect through a two-pronged sequential attack: initial impairment of viral reverse transcription followed by mutational inactivation of successfully formed reverse transcripts (Bishop et al. 2006; Holmes et al. 2007; Yang et al. 2007b). LMM forms of A3F also probably mediate post-entry restriction of HIV-1. The dG-to-dA hypermutations matching the recognized sequence preferences for both A3G (CC) and A3F (TC) are detectable in a minor subset of slowly formed reverse transcripts isolated from resting CD4 T cells infected with HIV-1 (Chiu et al. 2005). Furthermore, siRNA-mediated downregulation of A3F expression in dendritic cells increases the permissiveness of these cells to HIV-1 infection (Pion et al. 2006). Human A3B also has moderate activity against HIV-1 when packaged into virions and is quite resistant to Vif-induced degradation (Bishop et al. 2004; Doehle et al. 2005a). Human A3DE is the most recent APOBEC3 protein found to suppress HIV-1 infection. It deaminates HIV-1 minus-strand DNA preferentially at an AC dinucleotide motif that is distinct from that of other APOBEC3 members, but signatures of this type of mutation are evident in clinical HIV-1 isolates (Dang et al. 2006).
HIV is not the sole viral target of the APOBEC3 gene products. Indeed, human A3G actively suppresses the spread of HIV-1, SIV and MLV; A3F, A3B and A3DE inhibit the replication of HIV-1 and SIV, but not MLV (Bishop et al. 2004; Doehle et al. 2005a; Dang et al. 2006). Human A3C blocks SIV and HIV-1, although the anti-HIV effects are quite modest (Yu et al. 2004a). Within the nucleus, A3A blocks the replication of adeno-associated virus, which replicates as a single-stranded DNA in the nuclei of infected cells (Chen et al. 2006). In close parallel to the post-entry restricting function of A3G and A3F against HIV-1, the deaminase-independent actions of A3G, A3F, A3B and A3C sharply interfere with the HBV replication in cotransfected hepatoma cell lines. Mouse protein mA3 also appears to restrict mouse mammary tumour virus, as highlighted by the increased vulnerability of mA3 knockout mice to MMTV infection (Okeoma et al. 2007). This fascinating spectrum of antiviral activities for the APOBEC3 family of enzymes is certainly consistent with the impressive expansion and rapid evolution of the primate A3 proteins.
18. Conclusions
Studies of A3G biology highlight the multifunctional properties of A3G as an antiviral factor. Using its inherent deaminase activity or deaminase-independent RNA-binding anti-viral mechanisms, A3G acts at distinct steps along the retroviral life cycle. The ability of LMM A3G to counteract HIV-1 at the post-entry level and to inhibit Alu retrotransposition in its HMM form further highlights the economical use of this enzyme in the host to minimize the threat posed by both exogenous retroviruses and endogenous retroviral elements. The disproportionate activities, expression profiles, and even the intracellular localizations of the various APOBEC3 members (please refer to Chiu & Greene (2008) for a comprehensive review) further support the notion that members of this gene family may have evolved to function as important guardians of the integrity of the human genome that counter the adverse effects of both exogenous retroviruses and endogenous retroelements.
Acknowledgments
We wish to acknowledge the outstanding contributions from the various laboratories in the APOBEC field that we were unable to cite owing to space limitations. Research in our laboratory is supported by funding from the National Institutes of Health grants R01 AI065329-01 (to W.C.G.) and RR18928-01; the San Francisco Women's HIV Interdisciplinary Network (National Institutes of Health grant P01 HD40543 to W.C.G.); American Foundation for AIDS Research grant 10652535-RFHF (to Y.-L.C.); University of California San Francisco–Gladstone Institute of Virology and Immunology Center for AIDS Research grant AI0277635P30GY. We thank John C. W. Carroll for graphics arts, Gary Howard and Stephen Ordway for editorial assistance and Robin Givens for administrative support. This review was adapted in part from a recent paper published in Annual Review of Immunology vol. 26 (c) 2008 including the use of select figures with permission. doi:10.1146/annurev.immunol.26.021607.090350.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘DNA deamination in immunity, virology and cancer’.
References
- Abudu A., Takaori-Kondo A., Izumi T., Shirakawa K., Kobayashi M., Sasada A., Fukunaga K., Uchiyama T. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 2006;16:1565–1570. doi: 10.1016/j.cub.2006.06.055. doi:10.1016/j.cub.2006.06.055 [DOI] [PubMed] [Google Scholar]
- Alce T.M., Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279:34 083–34 086. doi: 10.1074/jbc.C400235200. doi:10.1074/jbc.C400235200 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. doi:10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale R.C., Petersen-Mahrt S.K., Watt I.N., Harris R.S., Rada C., Neuberger M.S. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. doi:10.1016/j.jmb.2004.01.046 [DOI] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Sheehy A.M., Davidson N.O., Cho S.J., Malim M.H. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. doi:10.1016/j.cub.2004.06.057 [DOI] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Malim M.H. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. doi:10.1128/JVI.00839-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Doehle B.P., Wiegand H.L., Cullen B.R. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl Acad. Sci. USA. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. doi:10.1073/pnas.0307713101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Wiegand H.L., Doehle B.P., Lueders K.K., Cullen B.R. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. doi:10.1093/nar/gkj416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin M., et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. doi:10.1002/hep.21187 [DOI] [PubMed] [Google Scholar]
- Burnett A., Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. doi:10.1128/JVI.02237-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S., Guo F., Niu M., Saadatmand J., Deflassieux J., Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 2004;279:33 177–33 184. doi: 10.1074/jbc.M402062200. doi:10.1074/jbc.M402062200 [DOI] [PubMed] [Google Scholar]
- Chelico L., Pham P., Calabrese P., Goodman M.F. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. doi:10.1038/nsmb1086 [DOI] [PubMed] [Google Scholar]
- Chen H., Lilley C.E., Yu Q., Lee D.V., Chou J., Narvaiza I., Landau N.R., Weitzman M.D. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. doi:10.1016/j.cub.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Greene W.C. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. doi:10.1146/annurev.immunol.26.021607.090350 [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Soros V.B., Kreisberg J.F., Stopak K., Yonemoto W., Greene W.C. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. doi:10.1038/nature03493 [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Witkowska H.E., Hall S.C., Santiago M., Soros V.B., Esnault C., Heidmann T., Greene W.C. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:15 588–15 593. doi: 10.1073/pnas.0604524103. doi:10.1073/pnas.0604524103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello S.G., Harris R.S., Neuberger M.S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. doi:10.1016/j.cub.2003.10.034 [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Thomas C.J., Petersen-Mahrt S.K., Neuberger M.S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. doi:10.1093/molbev/msi026 [DOI] [PubMed] [Google Scholar]
- Dang Y., Wang X., Esselman W.J., Zheng Y.H. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 2006;80:10 522–10 533. doi: 10.1128/JVI.01123-06. doi:10.1128/JVI.01123-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delebecque F., et al. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 2006;80:605–614. doi: 10.1128/JVI.80.2.605-614.2006. doi:10.1128/JVI.80.2.605-614.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Hill S.A., Princler G., Lloyd P., Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl Acad. Sci. USA. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. doi:10.1073/pnas.0609444104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. doi:10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Doehle B.P., Schafer A., Cullen B.R. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005a;339:281–288. doi: 10.1016/j.virol.2005.06.005. doi:10.1016/j.virol.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Doehle B.P., Schafer A., Wiegand H.L., Bogerd H.P., Cullen B.R. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 2005b;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. doi:10.1128/JVI.79.13.8201-8207.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle B.P., Bogerd H.P., Wiegand H.L., Jouvenet N., Bieniasz P.D., Hunter E., Cullen B.R. The betaretrovirus Mason–Pfizer monkey virus selectively excludes simian APOBEC3G from virion particles. J. Virol. 2006;80:12 102–12 108. doi: 10.1128/JVI.01600-06. doi:10.1128/JVI.01600-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellery P.J., et al. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J. Immunol. 2007;178:6581–6589. doi: 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A.J., Heidmann T., Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. doi:10.1038/nature03238 [DOI] [PubMed] [Google Scholar]
- Esnault C., Millet J., Schwartz O., Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. doi:10.1093/nar/gkl054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabini G., Rutjes S.A., Zimmermann C., Pruijn G.J., Steiner G. Analysis of the molecular composition of Ro ribonucleoprotein complexes. Identification of novel Y RNA-binding proteins. Eur. J. Biochem. 2000;267:2778–2789. doi: 10.1046/j.1432-1327.2000.01298.x. doi:10.1046/j.1432-1327.2000.01298.x [DOI] [PubMed] [Google Scholar]
- Gabuzda D.H., Lawrence K., Langhoff E., Terwilliger E., Dorfman T., Haseltine W.A., Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+T lymphocytes. J. Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddis N.C., et al. Further investigation of simian immunodeficiency virus Vif function in human cells. J. Virol. 2004;78:12 041–12 046. doi: 10.1128/JVI.78.21.12041-12046.2004. doi:10.1128/JVI.78.21.12041-12046.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S., Kramer B., Swanson C.M., Byers H., Lynham S., Ward M., Malim M.H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. doi:10.1128/JVI.02287-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 2006;80:11 710–11 722. doi: 10.1128/JVI.01038-06. doi:10.1128/JVI.01038-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G., Liddament M.T., Harris R.S. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 2005;280:10 920–10 924. doi: 10.1074/jbc.M500382200. doi:10.1074/jbc.M500382200 [DOI] [PubMed] [Google Scholar]
- Harris R.S., Petersen-Mahrt S.K., Neuberger M.S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. doi:10.1016/S1097-2765(02)00742-6 [DOI] [PubMed] [Google Scholar]
- Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malim M.H. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. doi:10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Holmes R.K., Koning F.A., Bishop K.N., Malim M.H. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. doi:10.1074/jbc.M607298200 [DOI] [PubMed] [Google Scholar]
- Hulme A.E., Bogerd H.P., Cullen B.R., Moran J.V. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. doi:10.1016/j.gene.2006.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y., Takeuchi H., Strebel K., Levin J.G. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. doi:10.1128/JVI.02680-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janini M., Rogers M., Birx D.R., McCutchan F.E. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 2001;75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. doi:10.1128/JVI.75.17.7973-7986.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. doi:10.1006/geno.2002.6718 [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. doi:10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Kao S., Miyagi E., Khan M.A., Takeuchi H., Opi S., Goila-Gaur R., Strebel K. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1:27. doi: 10.1186/1742-4690-1-27. doi:10.1186/1742-4690-1-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H.H.J. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. doi:10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Khan M.A., et al. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. doi:10.1128/JVI.79.9.5870-5874.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A., Goila-Gaur R., Opi S., Miyagi E., Takeuchi H., Kao S., Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. doi:10.1186/1742-4690-4-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M.A., Bassell G.J. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. doi:10.1016/j.neuron.2006.08.021 [DOI] [PubMed] [Google Scholar]
- Klarmann G.J., Chen X., North T.W., Preston B.D. Incorporation of uracil into minus strand DNA affects the specificity of plus strand synthesis initiation during lentiviral reverse transcription. J. Biol. Chem. 2003;278:7902–7909. doi: 10.1074/jbc.M207223200. doi:10.1074/jbc.M207223200 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Takaori-Kondo A., Shindo K., Abudu A., Fukunaga K., Uchiyama T. APOBEC3G targets specific virus species. J. Virol. 2004;78:8238–8244. doi: 10.1128/JVI.78.15.8238-8244.2004. doi:10.1128/JVI.78.15.8238-8244.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak S.L., Marin M., Rose K.M., Bystrom C., Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281:29 105–29 119. doi: 10.1074/jbc.M601901200. doi:10.1074/jbc.M601901200 [DOI] [PubMed] [Google Scholar]
- Kreisberg J.F., Yonemoto W., Greene W.C. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 2006;203:865–870. doi: 10.1084/jem.20051856. doi:10.1084/jem.20051856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D., Bouchonnet F., Clavel F., Hance A.J. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. doi:10.1126/science.1083338 [DOI] [PubMed] [Google Scholar]
- Lee Y.N., Bieniasz P.D. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. doi:10.1371/journal.ppat.0030010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament M.T., Brown W.L., Schumacher A.J., Harris R.S. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. doi:10.1016/j.cub.2004.06.050 [DOI] [PubMed] [Google Scholar]
- Lochelt M., et al. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl Acad. Sci. USA. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. doi:10.1073/pnas.0501445102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Liu B., Xiao Z., Yu Y., Yu X., Gorelick R., Yu X.F. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 2004;78:11 841–11 852. doi: 10.1128/JVI.78.21.11841-11852.2004. doi:10.1128/JVI.78.21.11841-11852.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Xiao Z., Ehrlich E., Yu Y., Liu B., Zheng S., Yu X.F. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl Acad. Sci. USA. 2005;102:11 444–11 449. doi: 10.1073/pnas.0502440102. doi:10.1073/pnas.0502440102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Wang T., Liu B., Tian C., Xiao Z., Kappes J., Yu X.F. Cytidine deaminases APOBEC3G and APOBEC3F interact with HIV-1 integrase and inhibit proviral DNA formation. J. Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. doi:10.1128/JVI.02584-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N., Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 1998;72:10 251–10 255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieux R., Suspene R., Delebecque F., Henry M., Schwartz O., Wain-Hobson S., Vartanian J.P. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J. Gen. Virol. 2005;86:2489–2494. doi: 10.1099/vir.0.80973-0. doi:10.1099/vir.0.80973-0 [DOI] [PubMed] [Google Scholar]
- Maksakova I.A., Romanish M.T., Gagnier L., Dunn C.A., van de Lagemaat L.N., Mager D.L. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. doi:10.1371/journal.pgen.0020002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. doi:10.1038/nature01709 [DOI] [PubMed] [Google Scholar]
- Mangeat B., Turelli P., Liao S., Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 2004;279:14 481–14 483. doi: 10.1074/jbc.C400060200. doi:10.1074/jbc.C400060200 [DOI] [PubMed] [Google Scholar]
- Mariani R., Chen D., Schrofelbauer B., Navarro F., Konig R., Bollman B., Munk C., Nymark-McMahon H., Landau N.R. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. doi:10.1016/S0092-8674(03)00515-4 [DOI] [PubMed] [Google Scholar]
- Marin M., Rose K.M., Kozak S.L., Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. doi:10.1038/nm946 [DOI] [PubMed] [Google Scholar]
- Mbisa J.L., et al. HIV-1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. doi:10.1128/JVI.00272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A., Goncalves J., Santa-Marta M., McPike M., Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004a;18:2861–2866. doi: 10.1101/gad.1249904. doi:10.1101/gad.1249904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A., Strack B., Ancuta P., Zhang C., McPike M., Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004b;279:7792–7798. doi: 10.1074/jbc.M313093200. doi:10.1074/jbc.M313093200 [DOI] [PubMed] [Google Scholar]
- Mehle A., Thomas E.R., Rajendran K.S., Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 2006;281:17 259–17 265. doi: 10.1074/jbc.M602413200. doi:10.1074/jbc.M602413200 [DOI] [PubMed] [Google Scholar]
- Mehta A., Kinter M.T., Sherman N.E., Driscoll D.M. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell Biol. 2000;20:1846–1854. doi: 10.1128/mcb.20.5.1846-1854.2000. doi:10.1128/MCB.20.5.1846-1854.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H., Hamdorf M., Held U., Perkovic M., Lower J., Cichutek K., Flory E., Schumann G.G., Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281:22 161–22 172. doi: 10.1074/jbc.M601716200. doi:10.1074/jbc.M601716200 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18 470–18 476. doi: 10.1074/jbc.274.26.18470. doi:10.1074/jbc.274.26.18470 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. doi:10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Navarro F., Bollman B., Chen H., Konig R., Yu Q., Chiles K., Landau N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. doi:10.1016/j.virol.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Newman E.N., Holmes R.K., Craig H.M., Klein K.C., Lingappa J.R., Malim M.H., Sheehy A.M. Antiviral Function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. doi:10.1016/j.cub.2004.12.068 [DOI] [PubMed] [Google Scholar]
- OhAinle M., Kerns J.A., Malik H.S., Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. doi:10.1128/JVI.80.8.3853-3862.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki I.M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., Honjo T. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. doi:10.1084/jem.20030275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma C.M., Lovsin N., Peterlin B.M., Ross S.R. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445:927–930. doi: 10.1038/nature05540. doi:10.1038/nature05540 [DOI] [PubMed] [Google Scholar]
- Opi S., Kao S., Goila-Gaur R., Khan M.A., Miyagi E., Takeuchi H., Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. doi:10.1128/JVI.02694-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Greenwell-Wild T., Nares S., Jin W., Lei K.J., Rangel Z.G., Munson P.J., Wahl S.M. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110:393–400. doi: 10.1182/blood-2006-10-051763. doi:10.1182/blood-2006-10-051763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pion M., Granelli-Piperno A., Mangeat B., Stalder R., Correa R., Steinman R.M., Piguet V. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J. Exp. Med. 2006;203:2887–2893. doi: 10.1084/jem.20061519. doi:10.1084/jem.20061519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. doi:10.1038/nature05492 [DOI] [PubMed] [Google Scholar]
- Revy P., et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. doi:10.1016/S0092-8674(00)00079-9 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Basu M.K., Jordan I.K., Pavlov Y.I., Koonin E.V. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- Rosler C., Kock J., Malim M.H., Blum H.E., von Weizsacker F. Comment on “Inhibition of hepatitis B virus replication by APOBEC3G”. Science. 2004;305:1403. doi: 10.1126/science.1101974. doi:10.1126/science.1100464 author reply 1403. [DOI] [PubMed] [Google Scholar]
- Russell R.A., Wiegand H.L., Moore M.D., Schafer A., McClure M.O., Cullen B.R. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. doi:10.1128/JVI.79.14.8724-8731.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasada A., Takaori-Kondo A., Shirakawa K., Kobayashi M., Abudu A., Hishizawa M., Imada K., Tanaka Y., Uchiyama T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005;2:32. doi: 10.1186/1742-4690-2-32. doi:10.1186/1742-4690-2-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S.L., Emerman M., Malik H.S. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. doi:10.1371/journal.pbio.0020275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A., Bogerd H.P., Cullen B.R. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. doi:10.1016/j.virol.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Schrofelbauer B., Chen D., Landau N.R. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl Acad. Sci. USA. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. doi:10.1073/pnas.0307132101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B., Yu Q., Zeitlin S.G., Landau N.R. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 2005;79:10 978–10 987. doi: 10.1128/JVI.79.17.10978-10987.2005. doi:10.1128/JVI.79.17.10978-10987.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B., Senger T., Manning G., Landau N.R. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. doi:10.1128/JVI.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. doi:10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Malim M.H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. doi:10.1038/nm945 [DOI] [PubMed] [Google Scholar]
- Simon J.H., Gaddis N.C., Fouchier R.A., Malim M.H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 1998;4:1397–1400. doi: 10.1038/3987. doi:10.1038/3987 [DOI] [PubMed] [Google Scholar]
- Simon V., Zennou V., Murray D., Huang Y., Ho D.D., Bieniasz P.D. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. doi:10.1371/journal.ppat.0010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros V.B., Yonemoto W., Greene W.C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. doi:10.1371/journal.ppat.0030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sova P., Volsky D.J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M.D., Harris R.S. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281:16 837–16 841. doi: 10.1074/jbc.M602367200. doi:10.1074/jbc.M602367200 [DOI] [PubMed] [Google Scholar]
- Stopak K., de Noronha C., Yonemoto W., Greene W.C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. doi:10.1016/S1097-2765(03)00353-8 [DOI] [PubMed] [Google Scholar]
- Stopak K.S., Chiu Y.L., Kropp J., Grant R.M., Greene W.C. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J. Biol. Chem. 2007;282:3539–3546. doi: 10.1074/jbc.M610138200. doi:10.1074/jbc.M610138200 [DOI] [PubMed] [Google Scholar]
- Suspene R., Guetard D., Henry M., Sommer P., Wain-Hobson S., Vartanian J.P. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. doi:10.1073/pnas.0408223102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R., Rusniok C., Vartanian J.P., Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. doi:10.1093/nar/gkl555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia E.S., Xu H., Mbisa J.L., Barr R., Gorelick R.J., Ono A., Freed E.O., Hu W.S., Pathak V.K. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 2004;279:35 822–35 828. doi: 10.1074/jbc.M405761200. doi:10.1074/jbc.M405761200 [DOI] [PubMed] [Google Scholar]
- Teng B., Burant C.F., Davidson N.O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. doi:10.1126/science.8511591 [DOI] [PubMed] [Google Scholar]
- Thielen B.K., Klein K.C., Walker L.W., Rieck M., Buckner J.H., Tomblingson G.W., Lingappa J.R. T cells contain an RNase-insensitive inhibitor of APOBEC3G deaminase activity. PLoS Pathog. 2007;3:1320–1334. doi: 10.1371/journal.ppat.0030135. doi:10.1371/journal.ppat.0030135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P., Mangeat B., Jost S., Vianin S., Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004a;303:1829. doi: 10.1126/science.1092066. doi:10.1126/science.1092066 [DOI] [PubMed] [Google Scholar]
- Turelli P., Vianin S., Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J. Biol. Chem. 2004b;279:43 371–43 373. doi: 10.1074/jbc.C400334200. doi:10.1074/jbc.C400334200 [DOI] [PubMed] [Google Scholar]
- Villace P., Marion R.M., Ortin J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. doi:10.1093/nar/gkh552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U., Song J., Aiken C., Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski M.J., Ichiyama K., Rana T.M. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J. Biol. Chem. 2005;280:8387–8396. doi: 10.1074/jbc.M408048200. doi:10.1074/jbc.M408048200 [DOI] [PubMed] [Google Scholar]
- Wichroski M.J., Robb G.B., Rana T.M. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. doi:10.1371/journal.ppat.0020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H.L., Doehle B.P., Bogerd H.P., Cullen B.R. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. doi:10.1038/sj.emboj.7600246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Ehrlich E., Yu Y., Luo K., Wang T., Tian C., Yu X.F. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology. 2006;349:290–299. doi: 10.1016/j.virol.2006.02.002. doi:10.1016/j.virol.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Xu H., Svarovskaia E.S., Barr R., Zhang Y., Khan M.A., Strebel K., Pathak V.K. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl Acad. Sci. USA. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. doi:10.1073/pnas.0400830101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Chertova E., Chen J., Ott D.E., Roser J.D., Hu W.S., Pathak V.K. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360:247–256. doi: 10.1016/j.virol.2006.10.036. doi:10.1016/j.virol.2006.10.036 [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Balestra M.E., Ferrell L.D., Fan J., Arnold K.S., Taylor S., Taylor J.M., Innerarity T.L. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl Acad. Sci. USA. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. doi:10.1073/pnas.92.18.8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Chen K., Zhang C., Huang S., Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 2007a;282:11 667–11 675. doi: 10.1074/jbc.M606864200. doi:10.1074/jbc.M606864200 [DOI] [PubMed] [Google Scholar]
- Yang Y., Guo F., Cen S., Kleiman L. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology. 2007b;365:92–100. doi: 10.1016/j.virol.2007.03.022. doi:10.1016/j.virol.2007.03.022 [DOI] [PubMed] [Google Scholar]
- Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X.F. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. doi:10.1126/science.1089591 [DOI] [PubMed] [Google Scholar]
- Yu Q., Chen D., Konig R., Mariani R., Unutmaz D., Landau N.R. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 2004a;279:53 379–53 386. doi: 10.1074/jbc.M408802200. doi:10.1074/jbc.M408802200 [DOI] [PubMed] [Google Scholar]
- Yu Q., Konig R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004b;11:435–442. doi: 10.1038/nsmb758. doi:10.1038/nsmb758 [DOI] [PubMed] [Google Scholar]
- Yu Y., Xiao Z., Ehrlich E.S., Yu X., Yu X.F. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004c;18:2867–2872. doi: 10.1101/gad.1250204. doi:10.1101/gad.1250204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V., Perez-Caballero D., Gottlinger H., Bieniasz P.D. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 2004;78:12 058–12 061. doi: 10.1128/JVI.78.21.12058-12061.2004. doi:10.1128/JVI.78.21.12058-12061.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Webb D.M. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 2004;13:1785–1791. doi: 10.1093/hmg/ddh183. doi:10.1093/hmg/ddh183 [DOI] [PubMed] [Google Scholar]
- Zhang H., Yang B., Pomerantz R.J., Zhang C., Arunachalam S.C., Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. doi:10.1038/nature01707 [DOI] [PMC free article] [PubMed] [Google Scholar]