Abstract
Activation-induced (cytidine) deaminase (AID) efficiently introduces multiple and diversified deaminations in immunoglobulin (Ig) variable and switch regions. Here, we review studies of AID, and the APOBEC family member, APOBEC3G, demonstrating that both enzymes introduce multiple deaminations by processive action on single-stranded DNA and that deaminations occur stochastically at hot- and cold-spot targets. In a more detailed analysis of AID, we examine phosphorylation-null mutants, particularly, S38A and S43P. S43P mutant AID has been identified in a patient with hyper-IgM immunodeficiency syndrome. The phosphorylation-null mutants have essentially the same specific activity, processivity and ability to undergo transcription-dependent deamination compared with wild-type (WT) AID. Although the phosphorylation-null mutants still deaminate 5′-WRC hot spots, the mutant deamination spectra differ from WT AID. The mutants strongly prefer two motifs, 5′AGC and 5′GGC, which are disfavoured by WT AID. Differences in deamination specificities can be attributed primarily to the replacement of Ser rather than to the absence of phosphorylation. The 5′GGC motif occurs with exceptionally high frequency on the non-transcribed strand of human switch regions, IgG4 and IgE. The potential for S43P to catalyse large numbers of aberrant deaminations in switch region sequences suggests a possible relationship between non-canonical AID deamination specificity and a loss of antibody diversification.
Keywords: somatic hypermutation, class switch recombination, activation-induced (cytidine) deaminase phosphorylation, processivity, deamination spectrum, transcription
1. Introduction
The APOBEC family of polynucleotide cytidine deaminases is a diverse group of 11 enzymes with RNA or single-stranded (ss) DNA editing functions that play a role in mRNA regulation (APOBEC1), humoral immunity (activation-induced (cytidine) deaminase, AID) and innate immunity against retroviruses and retrotransposons (APOBEC3A–H; Harris & Liddament 2004; Pham et al. 2005; Chiu & Greene 2008). Some members remain with no known function (APOBEC2 and APOBEC4; Rogozin et al. 2005; Prochnow et al. 2007). Throughout the study of the APOBEC enzymes that are involved in immunity, two family members, AID and APOBEC3G, have taken centre stage.
Humans and mice have evolved to have a remarkable immune system that can produce an estimated 109 different variants of antibodies (Ab) in response to the invasion of foreign antigens (Milstein 1987). The creation of this massive number of Ab variants in B cells occurs through three different processes: V(D)J recombination; somatic hypermutation (SHM); and class switch recombination (CSR). The B-cell-specific protein, AID, is required to initiate both SHM and CSR. First, during V(D)J recombination, immunoglobulin (Ig) V (variable), D (diversity) and J (joining) gene segments randomly recombine to generate an initial repertoire of low-affinity IgM Ab with unique binding sites. Following recognition of a foreign antigen with low-affinity Ab, B cells are activated and undergo affinity maturation via SHM and CSR (see reviews, Di Noia & Neuberger 2007; Peled et al. 2008; Stavnezer et al. 2008).
Part of B-cell activation involves transcription of variable (V) and switch (S) regions in Ig loci (Maizels 1995; Peters & Storb 1996). Transcription is also necessary to provide AID with an ssDNA substrate. The AID deaminations preferentially occur at 5′-WRC hot spots (W=A or T; R=purine) and much less frequently occur at 5′-SYC cold spots (S=C or G; Y=pyrimidine; Pham et al. 2003; Bransteitter et al. 2004). Downstream action on AID deaminations by base excision repair or mismatch repair enzymes continues the pathway to antibody maturation (Peled et al. 2008; Stavnezer et al. 2008). SHM introduces an exceptionally high mutation rate, approximately 10−3–10−4 per base pair per cell division, within the V(D)J-rearranged Ig genes creating a more specific Ab repertoire (Rajewsky et al. 1987). CSR is a unique region-specific recombination event in which donor S regions, coding for an IgM Ab, recombine with acceptor S regions to produce IgG, IgA or IgE antibody isotypes (Stavnezer et al. 2008). The CSR event is initiated by DNA breaks incurred from DNA repair enzymes removing uracils (Chaudhuri et al. 2007; Stavnezer et al. 2008). AID is absolutely required to initiate SHM and CSR (Muramatsu et al. 2000), and humans and mice deficient for AID suffer from an immunodeficient hyper-IgM syndrome (HIGM2; Durandy et al. 2007).
APOBEC3G deamination is mainly involved in restricting HIV infection of T cells, although restriction of other retroviruses does occur (Suspene et al. 2006; Chiu & Greene 2008; Vartanian et al. 2008). APOBEC3G can restrict infections in HIV strains that lack the APOBEC3G antagonist, Vif (Sheehy et al. 2002). APOBEC3G deaminates C residues in ssDNA, with preferential deamination of 5′YCC motifs (Beale et al. 2004; Bishop et al. 2004). The ssDNA activity limits APOBEC3G deamination to only the first cDNA strand reverse transcribed (Suspene et al. 2004; Yu et al. 2004). As the reverse transcriptase copies past uracils during synthesis of the second DNA strand, G→A transitions will occur. These mutations may eliminate HIV infectivity by gene inactivation.
Although AID and APOBEC3G perform different cellular functions, both enzymes have evolved to efficiently introduce multiple and diversified deaminations on their respective ssDNA targets (Pham et al. 2003; Bransteitter et al. 2004; Suspene et al. 2004; Yu et al. 2004). Biochemical studies of AID and APOBEC3G have indicated that both enzymes act processively on ssDNA, and deaminations occur stochastically at the hot- and cold-spot targets (Pham et al. 2003, 2007; Bransteitter et al. 2004; Chelico et al. 2006, 2008). Here, we discuss how processive deaminations of AID and APOBEC3G relate to their actions in vivo with a more in-depth review of AID mutants affecting processivity and deamination specificity.
2. Definition of processive and distributive enzymes
Processive enzymes catalyse multiple reactions in a single substrate-DNA encounter before dissociating to another molecule. By contrast, distributive enzymes catalyse one reaction per substrate-DNA encounter. The mechanisms of enzyme processivity can be summarized by two main types of movements, one- and three dimensional (Berg et al. 1981; von Hippel & Berg 1989; Halford & Marko 2004). DNA and RNA polymerases are classical one-dimensional processive enzymes. Common attributes are that the enzyme has a starting point, the primer/template junction, and then sequentially moves in one direction inserting nucleotides opposite template DNA. Numerous other enzymes use three-dimensional movement by facilitated diffusion, such as DNA glycosylases (Higley & Lloyd 1993; Bennett et al. 1995), human AP endonuclease (Carey & Strauss 1999), T4 endonuclease V (Dowd & Lloyd 1990), restriction endonucleases (Jack et al. 1982; Terry et al. 1985; Stanford et al. 2000) and AID and APOBEC3G (Pham et al. 2003, 2007; Chelico et al. 2006). These enzymes face a formidable challenge of finding ‘needle in a haystack’ substrate motifs in DNA after an arbitrary starting point is established by an initial random binding event. The three-dimensional processive mechanism increases the search efficiency by enabling the enzyme to employ a combination of both one-dimensional ‘sliding’ along the DNA and three-dimensional ‘hopping’ or ‘jumping’ mechanisms (von Hippel & Berg 1989; Stanford et al. 2000). The latter type of movement consists of consecutive microscopic dissociations and reassociations with the same substrate (von Hippel & Berg 1989; Halford & Marko 2004). A consequence of a three-dimensional search is that not each DNA base must be sampled so that the actions of the enzyme are inherently stochastic.
3. AID and APOBEC3G processively deaminate ssDNA in vitro
(a) Deamination spectra
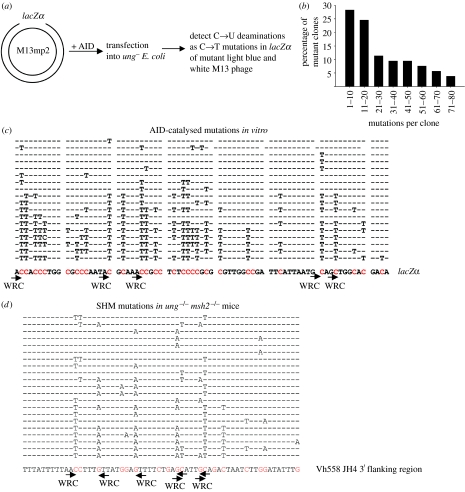
AID processivity was first uncovered in an in vitro assay using M13mp2 circular DNA substrate containing a lacZα reporter target sequence in a 365 nt ss gapped region (figure 1a). Following incubation with Sf9-expressed AID, deaminations are detected as C→T mutations occurring in DNA isolated from individual mutant phage progeny recovered from ung− Escherichia coli transfected with AID-treated DNA (Pham et al. 2003; Bransteitter et al. 2004). This in vitro system also demonstrated that AID preference for 5′-WRC hot spots and avoidance of 5′-SYC cold spots is an intrinsic biochemical property of the enzyme (Pham et al. 2003). Under conditions that limit M13 DNA targeting to a single AID molecule where the ratio of mutant phage (clear or light blue plaques) to wild-type (WT) phage (dark blue plaques) is less than 5 per cent, roughly two-thirds of the mutant clones had more than 10 mutations, with some clones containing 60–80 mutations (figure 1b). Since more than 95 per cent of the DNA substrates have no mutations, as confirmed by sequencing unmutated DNA isolated from blue plaques, the presence of multiple mutations in clones (figure 1c) clearly indicates that AID acts processively. Similarly, APOBEC3G also deaminates the lacZα target in a processive manner (P. Pham, L. Chelico & M. F. Goodman 2006, unpublished data).
Figure 1.
AID-catalysed processive deaminations in vitro demonstrate hallmark features of SHM in vivo. (a) A sketch of the assay used to detect C→T mutations resulting from AID deamination activity occurring on individual gapped M13 DNA substrates with the ss lacZα region. (b) The distribution of mutant clones containing various amounts of C→T mutations after 10 min incubation with AID. (c) DNA sequencing data obtained from a subset of 18 individual M13 DNA clones. Individual sequences show location of C→T mutations. Sequence context of a 74 nt fragment of the lacZα region is shown below clones. (a–c) is adapted from Pham et al. (2003). (d) SHM pattern in a 60 bp long fragment of Vh558 JH 3′ flanking region from ung−/− msh2−/− mice. Partial DNA sequencing data and representative clones are adapted from Rada et al. (2004), Supplemental Data. SHM mutations in these mice result from DNA replication over uracils from AID catalysis on the non-transcribed (C→T) and transcribed strands (G→A). Positions and orientations of WRC hot-spot motifs are shown by arrows.
AID processive deamination on ssDNA is highly stochastic resulting in heterogeneous multiple mutations in the lacZα region of individual clones (figure 1c). Although there are select 5′-WRC hot spots deaminated in the majority of clones, clearly, deaminating every 5′WRC target is not a requirement (figure 1c; Pham et al. 2003). The stochastic property of AID processive action and its preference for 5′-WRC hot-spot motifs in vivo are captured by the observed SHM mutations and clonal heterogeneity in mice deficient in both mismatch repair and base excision repair systems (ung−/−msh2−/−; figure 1d; Rada et al. 2004). In these mice, C→T and G→A SHM mutations are generated by DNA replication opposite uracils, from AID-catalysed deaminations, on both transcribed and non-transcribed strands of V and S region of Ig genes (Rada et al. 2004).
A strong positive charge concentrated at the N-terminus (+11) of AID seems to promote processive interactions with the negatively charged ssDNA (Pham et al. 2003; Bransteitter et al. 2004). By decreasing the +11 charge of WT AID to +7 in the mutant R35E/R36D, we found that there were approximately twofold less mutations per clone than WT AID (Bransteitter et al. 2004). This suggested that R35E/R36D mutant AID was less processive and could not remain bound to the same ssDNA long enough to catalyse as many processive deaminations as the WT. This was not due to a decrease in activity, but increased cycling between ssDNA substrates, as demonstrated by the accompanying approximately twofold increase in the number of mutant clones (Bransteitter et al. 2004).
(b) A tale of two cytosines
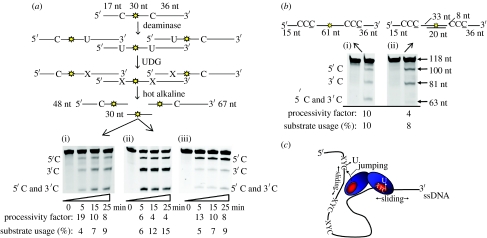
We developed a deamination assay to quantify the processive actions of AID and APOBEC3G in vitro (figure 2a; Chelico et al. 2006; Pham et al. 2007). For this assay, two deamination targets, either 5′AGC (AID) or 5′CCC (APOBEC3G), are placed within an ssDNA sequence. A fluorescein label located between the two deamination targets is used to detect all possible deaminations (figure 2a). In these experiments, less than 15 per cent of the substrate is used to ensure single-hit kinetics where the large majority ssDNA substrates are acted upon by only one enzyme (Creighton et al. 1995). The substrate can be deaminated at either the motif nearer the 5′-end (5′C), the motif nearer the 3′-end (3′C), or deaminated at both motifs (5′C and 3′C) (figure 2a).
Figure 2.
AID and APOBEC3G processively deaminate cytosines by sliding and jumping. (a) A sketch of the processivity assay. An 85 nucleotide (nt) ssDNA substrate with two 5′AGC or 5′CCC motifs for AID or APOBEC3G, respectively, with a central fluorescein reporter molecule (yellow star) is used to measure single and/or multiple deaminations occurring on an individual ssDNA substrate. AID and APOBEC3G can generate three types of products, a single deamination of the target C motif located either nearer the 5′-end (5′C) or nearer the 3′-end (3′C), or a double deamination occurring in both the 5′C and 3′C (5′C and 3′C) hot-spot motifs on the same DNA molecule. All three possible deamination products are detected by polyacrylamide gel electrophoresis following treatment of the ssDNA with UDG (uracil DNA glycosylase) and hot alkaline. ‘X’ denotes an abasic site. The integrated gel band intensities were used to determine the fraction of correlated double deaminations, generated by the processive action of a single AID or APOBEC3G (experimental details are contained in Chelico et al. 2006). AID deamination of the substrate is shown in (i). APOBEC3G deamination was tested in the absence (ii) or the presence (iii) of 5 mM MgCl2. Processivity factors and substrate usages are shown below the figures. (b) APOBEC3G deamination, without MgCl2, of a 118 nt ssDNA in the absence (i) or the presence (ii) of a 20 nt dsDNA region between the two 5′-CCC motifs. The internal fluorescein label is used to detect all possible deamination outcomes. The 5′C and 3′C deaminations are detected as the appearance of labelled 100- and 81 nt fragments, respectively; a double deamination results in a 63 nt labelled fragment (5′C and 3′C). Processivity factors and substrate usages are shown below the figures. Adapted from Chelico et al. (2008). (c) A sketch depicting the AID and APOBEC3G processive sliding and jumping mechanism on ssDNA. AID and APOBEC3G are depicted as a dimer; however, multiple oligomeric forms of APOBEC3G exist depending on the ssDNA substrate and salt concentration (Chelico et al. 2008). The placement of the active site (red circle) is schematically shown. AID and APOBEC3G appear to bind ssDNA randomly and move bidirectionally. Movement can either be sliding within a localized region or jumping to a distal part of the molecule without dissociating into the bulk solution. Model adapted from Chelico et al. (2006).
The deaminations are processive if the amount of double deaminations (5′C and 3′C) quantified from integrated gel band intensities is greater than the predicted double deaminations that would occur from two independent events (see Chelico et al. 2006). The value of this ratio is the processivity factor. A processivity factor significantly greater than 1 means that most double deaminations are correlated, i.e. occur from a single enzyme acting at both C targets. As expected, both AID and APOBEC3G deaminate ssDNA processively (processivity factor of at least 4; figure 2a(i)–(iii)). As a larger percentage of the substrate is deaminated, there is a reduction in the processivity factor that reflects the increased probability that a substrate can be acted upon independently by two enzymes (figure 2a(i)–(iii)). As indicated by the equal band intensities for both C targets, AID catalyses deaminations in an unbiased manner (figure 2a(i)).
APOBEC3G catalysis is exquisitely sensitive to the salt concentration (Chelico et al. 2008). For APOBEC3G, unbiased deaminations are catalysed with low salt (figure 2a(ii)) and directionally biased deaminations with high salt (5 mM MgCl2; figure 2a(iii)). In this unique mechanism, APOBEC3G achieves directionally biased deaminations without an energy source, i.e. this is an intrinsic property of the enzyme. Our current model has correlated monomers, which mainly form on DNA at low salt, with catalysis of unbiased deaminations and oligomers, such as dimers and tetramers, which form on DNA at high salt, with catalysis of directionally biased deaminations (Chelico et al. 2008). There is a salt-dependent inhibition of the intrinsic C deamination rate towards the 3′-ssDNA region, which offers a molecular basis for how oligomers impose a 5′-ssDNA end deamination bias (Chelico et al. 2008). For both unbiased and biased deaminations, there is a 30 nt ‘dead’ zone at the 3′-ssDNA end where deaminations are at least 10-fold less than elsewhere on the substrate (Chelico et al. 2008).
(c) The stochastic nature of three-dimensional processivity
The data generated from these processivity experiments (figure 2a(i)) can also be used to calculate the probability that if AID deaminates at the 5′-end target it will move on to deaminate the 3′-end target and vice versa (Chelico et al. 2006; Pham et al. 2008). This serves as a measure of AID's correlated deamination efficiency. Although the deaminations are processive (processivity factor of at least 8; figure 2a), the stochastic nature of AID deaminations predicts that correlated deamination of both C targets will not always occur at every AID–target C encounter (figure 1c). Accordingly, the correlated deamination efficiency of AID deamination at both C targets is only 30 per cent. This means that upon binding to DNA, 70 per cent of the time AID deaminates just one of the C targets and either bypasses the other target site or dissociates into the bulk solution.
The efficiency for APOBEC3G processivity depends on whether unbiased or directionally biased deaminations take place. Similar to AID, the correlated deamination efficiency of APOBEC3G unbiased deamination is 40 per cent (figure 2a(ii)). However, the directionally biased deamination mode has two different correlated deamination efficiencies for the 3′→5′ and 5′→3′ directions (figure 2a(iii)). The probability that APOBEC3G deaminates the 3′C and moves on to deaminate the 5′C is 60 per cent. The reverse probability, that deamination at the 5′C produces a correlated deamination with the 3′C is much less, 25 per cent, and illustrates the biased nature of APOBEC3G. Still, the biased deamination mode remains stochastic because only 60 per cent of the deaminations proceeding from 3′→5′ are correlated, i.e. this value never reaches 100 per cent.
(d) Contributions of sliding and jumping
We have confirmed that the three-dimensional scanning mechanism of AID and APOBEC3G involves both jumping and sliding (Bransteitter et al. 2004; Chelico et al. 2006, 2008). For APOBEC3G, we demonstrate jumping by using a short double-stranded (ds)DNA region as a block between two C targets (figure 2b). APOBEC3G binds to the dsDNA region with approximately fivefold less affinity than to ssDNA, making it unlikely that sliding across the dsDNA can occur (Chelico et al. 2008). Accordingly, the dsDNA block decreases the processivity factor, from that of completely ssDNA, by a factor of approximately 2, although processive deaminations still occur (processivity factor of 4, figure 2b). This suggests that the jumping motion of APOBEC3G contributes to a significant portion of the processivity (Chelico et al. 2006, 2008). Deamination spectra of AID show large gaps between clustered deamination sites suggesting that the enzyme can jump and slide (Pham et al. 2003, 2008; Bransteitter et al. 2004). Sliding seems to permit deamination of closely spaced residues whereas jumping would facilitate reaching multiple regions of an ssDNA substrate (figure 2c).
4. AID processive deamination during transcription does not require Ser38 phosphorylation and RPA
SHM in B cells requires active transcription of the V-region (Maizels 1995; Peters & Storb 1996). Since AID does not deaminate dsDNA, active transcription is needed to generate the ssDNA substrate for AID deamination activity. AID-catalysed deaminations on transcribed dsDNA must be highly dynamic to follow along with the moving transcription bubble. Nevertheless, AID retains its key biochemical properties on transcribed dsDNA substrates: a preference for 5′-WRC hot-spot motifs and diverse clonal mutagenic distribution (Bransteitter et al. 2004; Pham et al. 2008). AID, purified from mouse B cells, has been shown to be phosphorylated on Ser38, Tyr184 and possibly Thr27 (Basu et al. 2005; McBride et al. 2006; Pasqualucci et al. 2006). Although phosphorylation does not appear to be B-cell specific (McBride et al. 2006), AID phosphorylation at Ser38 was suggested to be the modification targeting AID to transcribed dsDNA through interactions with replication protein A (RPA) (Chaudhuri et al. 2004; Basu et al. 2005). On the other hand, recombinant AID, expressed in E. coli or Sf9 insect cells, has been shown to deaminate transcribed dsDNA in the absence of RPA (Pham et al. 2003, 2008; Sohail et al. 2003; Bransteitter et al. 2004; Shen & Storb 2004; Besmer et al. 2006). Since the phosphorylation mutant S38A has differing levels of CSR inhibition in vivo (Basu et al. 2005, 2007; Pasqualucci et al. 2006; Shinkura et al. 2007), we have carried out a comparative biochemical analysis using WT AID, S38A and additional phosphorylation defective mutants, to examine the alternative roles of phosphorylation that could account for a loss of AID CSR activity in vivo.
We have found that AID expressed in Sf9 insect cells can be phosphorylated on Ser 38 and Thr27 as well as two additional phosphorylation sites, Ser41 and Ser43 (Pham et al. 2008). Analysis of this mass spectrometry data suggests that only one residue can be phosphorylated on AID at a time (Pham et al. 2008). Phosphorylated AID has been determined to be only a few per cent or at maximum 15 per cent of the total population (McBride et al. 2006; Pham et al. 2008). For our comparative biochemical study, mutations were made by replacing Ser 38, 41 or 43 with either Ala (phosphorylation null) or Asp (phosphorylation mimic) (Pham et al. 2008). An additional mutation of Ser43 to Pro was made to characterize the phosphorylation-null HIGM-2 mutation found in a patient (Zhu et al. 2003).
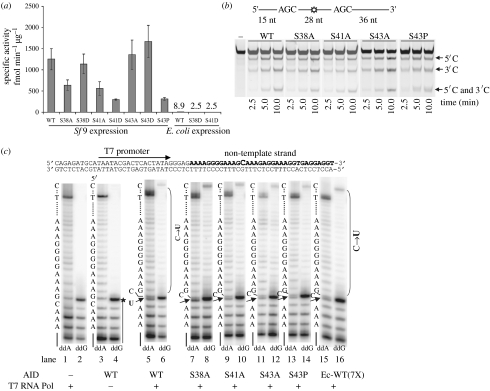
We found that a lack of phosphorylation or that mimicking phosphorylation did not change AID-specific activity more than a factor of 3 (figure 3a). AID produced from E. coli, which is not phosphorylated, is also active although its specific activity is approximately 50- to 100-fold lower compared with Sf9-expressed AID (figure 3a). We do not attribute the lower specific activity of E. coli AID to a lack of phosphorylation because no Sf9 insect cell phosphorylation-null mutant has an equivalent loss in activity (figure 3a). Mutations at phosphorylation sites had no effect on the processivity (figure 3b) or the stochastic nature of AID scanning, i.e. all phosphorylation mutants had a correlated deamination efficiency of approximately 30 per cent (Pham et al. 2008). Phosphorylation mutants of AID appear to have normal deamination activity on dsDNA undergoing transcription, in the absence of RPA (figure 3c; Pham et al. 2008). E. coli AID can also deaminate ssDNA during active transcription (figure 3c), although sevenfold excess enzyme was added to reach an activity level equivalent to Sf9 AID (figure 3a). AID phosphorylation is surely important in vivo, probably facilitating nuclear entry since phosphorylated AID is enriched in chromatic fractions (McBride et al. 2006). Yet, there appears to be no effect of phosphorylation on AID deamination activity on either ssDNA or dsDNA undergoing transcription.
Figure 3.
AID phosphorylation-null and phosphorylation-mimicking mutants have similar activity on ssDNA, processivity and the ability to deaminate dsDNA undergoing transcription. (a) Specific activities of Sf9-expressed WT and mutant AID are similar, within a factor of 3. E. coli-expressed WT and mutant AID have significantly lower specific activities (50- to 100-fold less) compared with AID-expressed Sf9. (b) Phosphorylation mutants S38A, S41A, S43A and S43P processively deaminate two 5′-WRC motifs, as evidenced by the presence of a double deamination band (5′C and 3′C) of comparable intensity to the WT AID. (c) WT and mutant AID deamination activity on linear dsDNA undergoing transcription by T7 RNA polymerase in the absence of RPA. C→U deamination by WT and mutant AID is detected by the presence of an intense termination band just below the template C band (indicated with a filled arrow) when ddATP is used in the place of dATP in the polymerase extension reactions (ddA, lanes 5, 7, 9, 11, 13 and 15). Consistent with the conversion of C→U in ddA lanes is the presence of bands resolving past the C template site (bracketed region) when ddGTP is used instead of dGTP (ddG, lanes 6, 8, 10, 12 and 14). Deamination occurs with similar efficiency for WT and mutant AID proteins expressed in Sf9 cells (lanes 5–14). Similar level of deamination is also observed for E. coli-expressed WT AID when added at sevenfold higher concentration than Sf9 WT AID (Ec-WT (7X), lanes 15 and 16). No deamination is detected on the dsDNA substrate in the absence of either AID or T7 RNA polymerase (lanes 1–4). The presence of intact C template is marked by the absence of a band just below the C template position in the ddA lanes (lanes 1 and 3) and by the absence of nucleotide incorporation past the C template site in the ddG lanes (lanes 2 and 4). A sketch of the dsDNA template is shown on the top. Adapted from Pham et al. (2008).
5. Amino acid residues affecting AID deamination specificity
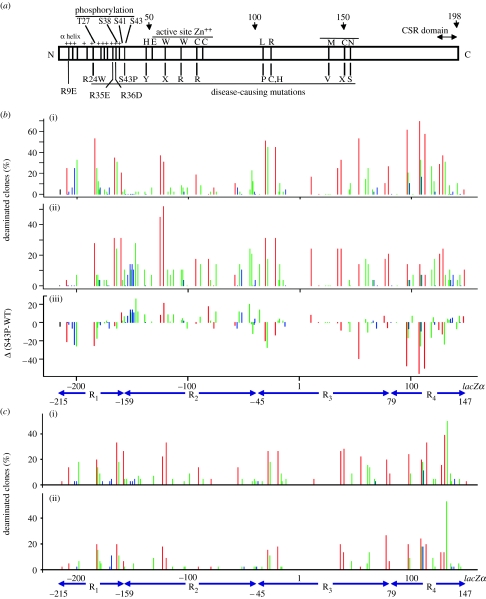
That a lack of AID phosphorylation did not change deamination activity leaves open the question of why phosphorylation-null mutants S38A and S43P have decreased and defective CSR in vivo, respectively. We tested the phosphorylation-null mutants using a lacZα reporter assay to determine whether there was a change in their deamination spectra from WT AID. The location of the phosphorylation-null mutations with respect to AID domain architecture is shown in figure 4a. All mutants demonstrated differences in the preferred trinucleotide sequence context from WT AID and the differences were similar for all mutants (Pham et al. 2008). The lacZα reporter assay also confirmed that the phosphorylation-null mutants act processively, i.e. there were at least 10 mutations per clone and only 10 per cent of the substrates were deaminated (Pham et al. 2008). Since dephosphorylation of WT AID does not change the deamination specificity (Pham et al. 2008), the different specificities of the phosphorylation-null mutants can be attributed to the replacement of Ser to Ala or Pro rather than an absence of phosphorylation. Here, we examine S43P as an example (figure 4b).
Figure 4.
Deamination spectra of WT AID and S43P, R35E and R35E/R36D mutant AID. (a) Schematic of AID domain structure with notations for disease-causing mutations and mutations constructed for biochemical study. (b,c) The gapped M13 DNA construct was incubated with WT or mutant AID for 2.5 min and each deamination spectrum was compiled from the sequencing of approximately 50 mutant phage clones. Bars represent the percentage of mutated clones containing a mutation at that position on the lacZα reporter sequence. Red bars, C deaminations in 5′-WRC hot-spot motifs; blue bars, deaminations in 5′-SYC cold-spot motifs; green bars, deamination of intermediate motifs (neither 5′WRC nor 5′SYC). The difference spectrum, (b(iii)) S43P versus WT, enables a side-by-side comparison of deamination frequencies at each site deaminated by (b(ii)) S43P and (b(i)) WT AID. The difference spectrum is the WT AID frequencies subtracted from the S43P AID frequencies. Bars below zero represent excess deaminations for WT AID and bars above zero denote excess S43P mutant AID deaminations (c(i) R35E and (ii) R35E/R36D). (a,c) is adapted from Bransteitter et al. (2004) and (b) adapted from Pham et al. (2008).
Although both S43P and WT AID still prefer to deaminate 5′-WRC hot-spot motifs and disfavour deaminating 5′-SYC cold-spot motifs, they show differences in the deamination frequency of intermediate motifs (figure 4b). A statistical comparison of WT and mutant S43P AID spatial deamination patterns was made by dividing the 365 nucleotide (nt) ssDNA gapped region into four subregions: R1 (nucleotide positions −215 to −159); R2 (−158 to −45); R3 (−44 to 79); and R4 (80–147). In R1, R2 or R4, there are large differences between mutant S43P and WT AID (figure 4b). Disparities between S43P and WT AID are quantified with a difference spectrum constructed by subtracting the frequency of deaminated clones in the WT AID spectra from that of S43P AID at each C site in the 365 nt gap (figure 4b(iii)). Excess deaminations for WT AID extend below 0 and excess S43P mutant AID deaminations extend above 0.
Upon examination of the lacZα sequence, it becomes clear that the large disparity in the WT and S43P AID spectra is caused by differences in the distribution of the intermediate triplet motifs, 5′CGC and 5′GGC. For example, in R2, S43P deamination is favoured over WT AID deaminations, and the intermediate motifs 5′CGC and 5′GGC occur five and four times, respectively. These two intermediate motifs, as well as 5′AGC, have significantly higher mutability indexes than WT AID for S43P, as well as S38A (table 1). The deamination preference may be relevant to the loss of CSR in a patient carrying the S43P mutant AID allele, since a compilation of individual trinucleotide motifs in Ig switch regions shows that they are rich in the 5′AGC and 5′GGC motifs (table 2). Non-canonical deaminations by S43P mutant AID may lead to excess dsDNA breaks and aberrant recombination.
Table 1.
Three nucleotide motif mutability index (MI)a for WT and mutant AID (S38A and S43P) at 2.5 min incubation with lacZα gapped region.
| MI | ||||
|---|---|---|---|---|
| motif | WT | S38A | S43P | pb |
| hot spots | ||||
| AAC | 2.5 | 1.5 | 1.6 | 0.41 |
| AGC | 1.3 | 2.1 | 2.0 | 0.06 |
| TAC | 3.4 | 2.0 | 2.2 | 0.22 |
| TGC | 2.6 | 2.2 | 2.6 | 0.99 |
| MI average (WRC) | 2.5 | 1.9 | 2.1 | |
| cold spots | ||||
| CCC | 0.28 | 0.52 | 0.44 | 0.65 |
| CTC | 0.45 | 0.76 | 0.53 | 0.99 |
| GCC | 0.05 | 0.18 | 0.05 | 0.07 |
| GTC | 0.32 | 0.67 | 0.00 | 0.99 |
| MI average (SYC) | 0.28 | 0.53 | 0.26 | |
| intermediates | ||||
| ACC | 1.2 | 0.32 | 0.35 | 0.02 |
| ATC | 1.0 | 0.63 | 0.71 | 0.97 |
| CAC | 1.4 | 0.91 | 0.96 | 0.31 |
| CGC | 0.5 | 1.3 | 1.4 | 0.004 |
| GAC | 0.24 | 0.5 | 0.35 | 0.79 |
| GGC | 0.23 | 0.78 | 1.0 | 0.0001 |
| TCC | 0.48 | 1.1 | 1.1 | 0.75 |
| TTC | 0.53 | 0.71 | 0.71 | 0.46 |
| MI average | 0.70 | 0.78 | 0.82 | |
The MI is defined as the number of times a given trinucleotide motif within a segment of DNA contains a mutation, divided by the number of times the oligonucleotide would be expected to be mutated for a mechanism with no sequence bias.
The p-values were calculated by the paired Wilcoxon test (see Pham et al. 2008). Adapted from Pham et al. (2008).
Table 2.
Distribution of trinucleotide motifs in Ig switch regionsa.
| number of motifs | |||
|---|---|---|---|
| motif | IgA | IgE | IgG |
| hot spots | |||
| AAC | 6 | 37 | 24 |
| AGC | 79 | 90 | 148 |
| TAC | 4 | 10 | 7 |
| TGC | 7 | 30 | 83 |
| cold spots | |||
| CCC | 0 | 49 | 122 |
| CTC | 1 | 31 | 106 |
| GCC | 2 | 54 | 135 |
| GTC | 5 | 17 | 44 |
| intermediates | |||
| ACC | 4 | 42 | 74 |
| ATC | 2 | 20 | 21 |
| CAC | 2 | 34 | 97 |
| CGC | 3 | 11 | 30 |
| GAC | 15 | 62 | 79 |
| GGC | 65 | 126 | 196 |
| TCC | 2 | 30 | 108 |
| TTC | 0 | 22 | 57 |
The number of trinucleotide motifs was calculated for the non-transcribed strand of IgA (mouse germline IgA switch region, accession number gi:194415), IgE (human IgE switch region, accession number gi:32983) and IgG (human Ig gamma-4 switch region, accession number gi:31510). Adapted from Pham et al. (2008).
Altering the overall charge of AID by mutating positive Arg residues to negatively charged Asp or Glu (figure 4a) also changes the preferred trinucleotide motifs compared to WT AID (Bransteitter et al. 2004). The mutant R35E and the double mutant R35E/R36D decrease the net positive charge of AID, which not only decreases AID processivity (see §3a) but also promotes deamination of an intermediate motif (figure 4c). The deamination of 5′WRC sites is not changed in the single mutant R35E, but an intermediate trinucleotide motif, 5′CAC130, not targeted in the WT AID spectrum, becomes a dominant deamination site (figure 4c(i), subregion R4). For the double mutant R35E/R36D, not only is 5′CAC130 targeted for deamination but deamination of a nearby 5′-WRC motif (site 128) is attenuated fourfold (figure 4c(ii), subregion R4). Unlike the phosphorylation-null mutants, replacing positively charged amino acid residues with negative amino acid residues affects both the trinucleotide motif preference and processivity.
To visualize the location of these specificity altering mutations on AID, we used the X-ray crystal structure of the closely related APOBEC family member, APOBEC2 (Prochnow et al. 2007). Residues of APOBEC2 equivalent to AID Arg35, Arg36 and Ser43 appear to be on solvent exposed surfaces but not near the zinc-binding deaminase motif. Our model that positively charged AID has affinity for the negatively charged DNA backbone (Bransteitter et al. 2004) suggests that multiple residues external to the active site play a role in ssDNA interactions. Altering these interactions appears to change the deamination specificity.
6. Conclusions
AID initiates SHM and CSR by deamination of C targets within the V- and S-regions of Ig genes creating G·U mispairs. Subsequent processing of the G·U mispairs by DNA replication and error-prone base excision repair and mismatch repair in B cells ensures the generation of highly diversified V-region SHM mutations or double-strand breaks in S regions needed for CSR. AID random processive deaminations in these regions appear to be important biologically. AID processive action during transcription would ensure that a significant number of deaminations occurred over a wide range of the Ig V- and S-regions. Apart from processivity, it appears that during CSR the deamination specificity for 5′-WRC sequences is also important for proper AID function. In vitro WT AID deamination spectra measured on ssDNA and on transcribed dsDNA agree well with SHM mutation spectra in humans (Pham et al. 2003; Bransteitter et al. 2004). Therefore, it seems possible that S43P mutant AID would create the same high levels of U at trinucleotide motifs, 5′AGC and 5′GGC, prevalent in S-region motifs in vivo. If aberrant recombination were to result from excess deamination in motifs disfavoured by WT AID then this might contribute to the lack of CSR in B cells expressing S43P AID (Zhu et al. 2003). A similar scenario may occur with S38A, with differing degrees of CSR inhibition (Basu et al. 2005; McBride et al. 2006; Pasqualucci et al. 2006; Shinkura et al. 2007), depending on the extent of the atypical deamination spectra (Pham et al. 2008).
In vivo, APOBEC3G must act during reverse transcription of the HIV genome to deaminate the nascent ss-cDNA strand before it becomes part of the dsDNA proviral genome (Yu et al. 2004). If APOBEC3G deaminations do not cause sufficient ‘hypermutation’ to inactivate the HIV virus, the mutations may instead make a contribution to the evolution of drug resistance (Mulder et al. 2008; Pillai et al. 2008). For this reason, APOBEC3G processive behaviour is advantageous to introduce the maximum number of deaminations in this limited time. Attributed to what we have demonstrated in vitro for APOBEC3G directionally biased deaminations (Chelico et al. 2006, 2008), there appears to be localized regions of polarized deaminations identified in select hypermutated HIV patient genomes (Suspene et al. 2006).
We have examined the scanning and deamination mechanisms of AID and APOBEC3G. Both AID and APOBEC3G employ sliding and jumping modes to processively deaminate multiple C residues in a single ssDNA encounter. For each enzyme, their processive scanning mechanisms appear to be solely attributable to intrinsic biochemical properties of the enzymes and do not require protein or nucleotide cofactors. The in vitro characteristics of AID and APOBEC3G deamination mimic what has been demonstrated in vivo and suggest that the differences in their stochastic processive scanning mechanisms may have evolved to be optimal for their in vivo function.
Acknowledgments
This work was supported by the National Institutes of Health grants ESO13192 and R37GM21422.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘DNA deamination in immunity, virology and cancer’.
References
- Basu U., Chaudhuri J., Alpert C., Dutt S., Ranganath S., Li G., Schrum J.P., Manis J.P., Alt F.W. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. doi:10.1038/nature04255 [DOI] [PubMed] [Google Scholar]
- Basu U., Chaudhuri J., Phan R.T., Datta A., Alt F.W. Regulation of activation induced deaminase via phosphorylation. Adv. Exp. Med. Biol. 2007;596:129–137. doi: 10.1007/0-387-46530-8_11. doi:10.1007/0-387-46530-8_11 [DOI] [PubMed] [Google Scholar]
- Beale R.C., Petersen-Mahrt S.K., Watt I.N., Harris R.S., Rada C., Neuberger M.S. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. doi:10.1016/j.jmb.2004.01.046 [DOI] [PubMed] [Google Scholar]
- Bennett S.E., Sanderson R.J., Mosbaugh D.W. Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosylase is affected by NaCl concentration. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. doi:10.1021/bi00018a014 [DOI] [PubMed] [Google Scholar]
- Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. doi:10.1021/bi00527a028 [DOI] [PubMed] [Google Scholar]
- Besmer E., Market E., Papavasiliou F.N. The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol. Cell. Biol. 2006;26:4378–4385. doi: 10.1128/MCB.02375-05. doi:10.1128/MCB.02375-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Sheehy A.M., Davidson N.O., Cho S.J., Malim M.H. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. doi:10.1016/j.cub.2004.06.057 [DOI] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Calabrese P., Goodman M.F. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 2004;279:51 612–51 621. doi: 10.1074/jbc.M408135200. doi:10.1074/jbc.M408135200 [DOI] [PubMed] [Google Scholar]
- Carey D.C., Strauss P.R. Human apurinic/apyrimidinic endonuclease is processive. Biochemistry. 1999;38:16 553–16 560. doi: 10.1021/bi9907429. doi:10.1021/bi9907429 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Khuong C., Alt F.W. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. doi:10.1038/nature02821 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. doi:10.1016/s0065-2776(06)94006-1 [DOI] [PubMed] [Google Scholar]
- Chelico L., Pham P., Calabrese P., Goodman M.F. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. doi:10.1038/nsmb1086 [DOI] [PubMed] [Google Scholar]
- Chelico L., Sacho E.J., Erie D.A., Goodman M.F. A model for oligomeric regulation of APOBEC3G cytosine deaminase-dependent restriction of HIV. J. Biol. Chem. 2008;283:13 780–13 791. doi: 10.1074/jbc.M801004200. doi:10.1074/jbc.M801004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.L., Greene W.C. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. doi:10.1146/annurev.immunol.26.021607.090350 [DOI] [PubMed] [Google Scholar]
- Creighton S., Bloom L.B., Goodman M.F. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. doi:10.1016/0076-6879(95)62021-4 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. doi:10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Dowd D.R., Lloyd R.S. Biological significance of facilitated diffusion in protein–DNA interactions. Applications to T4 endonuclease V-initiated DNA repair. J. Biol. Chem. 1990;265:3424–3431. [PubMed] [Google Scholar]
- Durandy A., Taubenheim N., Peron S., Fischer A. Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv. Immunol. 2007;94:275–306. doi: 10.1016/S0065-2776(06)94009-7. doi:10.1016/s0065-2776(06)94009-7 [DOI] [PubMed] [Google Scholar]
- Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. doi:10.1093/nar/gkh624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Liddament M.T. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. doi:10.1038/nri1489 [DOI] [PubMed] [Google Scholar]
- Higley M., Lloyd R.S. Processivity of uracil DNA glycosylase. Mutat. Res. 1993;294:109–116. doi: 10.1016/0921-8777(93)90019-d. [DOI] [PubMed] [Google Scholar]
- Jack W.E., Terry B.J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc. Natl Acad. Sci. USA. 1982;79:4010–4014. doi: 10.1073/pnas.79.13.4010. doi:10.1073/pnas.79.13.4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Somatic hypermutation: how many mechanisms diversify V region sequences? Cell. 1995;83:9–12. doi: 10.1016/0092-8674(95)90227-9. doi:10.1016/0092-8674(95)90227-9 [DOI] [PubMed] [Google Scholar]
- McBride K.M., Gazumyan A., Woo E.M., Barreto V.M., Robbiani D.F., Chait B.T., Nussenzweig M.C. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl Acad. Sci. USA. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. doi:10.1073/pnas.0603272103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C. Diversity and the genesis of high affinity antibodies. Biochem. Soc. Trans. 1987;15:779–787. doi: 10.1042/bst0150779. [DOI] [PubMed] [Google Scholar]
- Mulder L.C., Harari A., Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl Acad. Sci. USA. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. doi:10.1073/pnas.0710190105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. doi:10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Kitaura Y., Gu H., Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl Acad. Sci. USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. doi:10.1073/pnas.0509969103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled J.U., Kuang F.L., Iglesias-Ussel M.D., Roa S., Kalis S.L., Goodman M.F., Scharff M.D. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. doi:10.1146/annurev.immunol.26.021607.090236 [DOI] [PubMed] [Google Scholar]
- Peters A., Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. doi:10.1016/S1074-7613(00)80298-8 [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. doi:10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Goodman M.F. Reward versus risk: DNA cytidine deaminases triggering immunity and disease. Biochemistry. 2005;44:2703–2715. doi: 10.1021/bi047481+. doi:10.1021/bi047481+ [DOI] [PubMed] [Google Scholar]
- Pham P., Chelico L., Goodman M.F. DNA deaminases AID and APOBEC3G act processively on single-stranded DNA. DNA Repair (Amst.) 2007;6:689–692. doi: 10.1016/j.dnarep.2007.01.001. doi:10.1016/j.dnarep.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Pham P., Smolka M.B., Calabrese P., Landolph A., Zhang K., Zhou H., Goodman M.F. Impact of phosphorylation and phosphorylation-null mutants on the activity and deamination specificity of activation-induced cytidine deaminase. J. Biol. Chem. 2008;283:17 428–17 439. doi: 10.1074/jbc.M802121200. doi:10.1074/jbc.M802121200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S.K., Wong J.K., Barbour J.D. Turning up the volume on mutational pressure: is more of a good thing always better? (A case study of HIV-1 Vif and APOBEC3) Retrovirology. 2008;5:26. doi: 10.1186/1742-4690-5-26. doi:10.1186/1742-4690-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. doi:10.1038/nature05492 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., Neuberger M.S. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. doi:10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Rajewsky K., Forster I., Cumano A. Evolutionary and somatic selection of the antibody repertoire in the mouse. Science. 1987;238:1088–1094. doi: 10.1126/science.3317826. doi:10.1126/science.3317826 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Basu M.K., Jordan I.K., Pavlov Y.I., Koonin E.V. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. doi:10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Shen H.M., Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc. Natl Acad. Sci. USA. 2004;101:12 997–13 002. doi: 10.1073/pnas.0404974101. doi:10.1073/pnas.0404974101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R., Okazaki I.M., Muto T., Begum N.A., Honjo T. Regulation of AID function in vivo. Adv. Exp. Med. Biol. 2007;596:71–81. doi: 10.1007/0-387-46530-8_7. doi:10.1007/0-387-46530-8_7 [DOI] [PubMed] [Google Scholar]
- Sohail A., Klapacz J., Samaranayake M., Ullah A., Bhagwhat A.S. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucl. Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. doi:10.1093/nar/gkg464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford N.P., Szczelkun M.D., Marko J.F., Halford S.E. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. doi: 10.1093/emboj/19.23.6546. doi:10.1093/emboj/19.23.6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., Schrader C.E. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. doi:10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R., Sommer P., Henry M., Ferris S., Guetard D., Pochet S., Chester A., Navaratnam N., Wain-Hobson S., Vartanian J.P. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. doi:10.1093/nar/gkh554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R., Rusniok C., Vartanian J.P., Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. doi:10.1093/nar/gkl555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry B.J., Jack W.E., Modrich P. Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J. Biol. Chem. 1985;260:13 130–13 137. [PubMed] [Google Scholar]
- Vartanian J.P., Guetard D., Henry M., Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. doi:10.1126/science.1153201 [DOI] [PubMed] [Google Scholar]
- von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- Yu Q., Konig R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. doi:10.1038/nsmb758 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Nonoyama S., Morio T., Muramatsu M., Honjo T., Mizutani S. Type two hyper-IgM syndrome caused by mutation in activation-induced cytidine deaminase. J. Med. Dent. Sci. 2003;50:41–46. [PubMed] [Google Scholar]