Abstract
By temporarily deferring the repair of DNA lesions encountered during replication, the bypass of DNA damage is critical to the ability of cells to withstand genomic insults. Damage bypass can be achieved either by recombinational mechanisms that are generally accurate or by a process called translesion synthesis. Translesion synthesis involves replacing the stalled replicative polymerase with one of a number of specialized DNA polymerases whose active sites are able to tolerate a distorted or damaged DNA template. While this property allows the translesion polymerases to synthesize across damaged bases, it does so with the trade-off of an increased mutation rate. The deployment of these enzymes must therefore be carefully regulated. In addition to their important role in general DNA damage tolerance and mutagenesis, the translesion polymerases play a crucial role in converting the products of activation induced deaminase-catalysed cytidine deamination to mutations during immunoglobulin gene somatic hypermutation. In this paper, we specifically consider the control of translesion synthesis in the context of the timing of lesion bypass relative to replication fork progression and arrest at sites of DNA damage. We then examine how recent observations concerning the control of translesion synthesis might help refine our view of the mechanisms of immunoglobulin gene somatic hypermutation.
Keywords: translesion synthesis, immunoglobulin hypermutation, DNA polymerases, post-replication repair, mismatch repair
1. Introduction
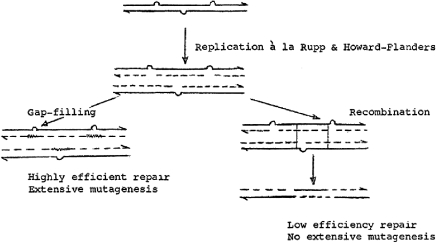
The majority of mutations in DNA arise in consequence of errors introduced during replication, and are frequently precipitated by DNA damage. It is just over 40 years since Evelyn Witkin showed that mutation in Escherichia coli is not an inevitable consequence of DNA damage, such as UV light. Instead, mutagenesis is determined by specific genetic loci and is, along with a number of other phenotypes, induced following exposure to UV light (Witkin 1967a,b). At about the same time, Dean Rupp and Paul Howard-Flanders demonstrated that E. coli defective in the ability to excise UV light-induced pyrimidine dimers could replicate with normal kinetics even when their genome contains up to 50 lesions. However, replication in these circumstances is associated with the formation of gaps in the newly synthesized DNA, at approximately the same spacing as the dimers, which are subsequently filled by a recombinational mechanism that gives rise to sister strand exchanges (Rupp & Howard-Flanders 1968; Rupp et al. 1971). Integration of these observations led to the first models for DNA damage bypass. These proposed that replication blocked at a site of UV damage restarted downstream of the lesion leaving a gap that is filled either by recombination or by an error-prone form of DNA synthesis across the lesion, now termed translesion synthesis (Radman 1970; figure 1). Although this early evidence suggested that lesion bypass was a post-replicative phenomenon, many subsequent models for lesion bypass in higher organisms have considered it as operating at stalled replication forks themselves as a mechanism to preserve fork integrity and progression (reviewed in Lehmann & Fuchs 2006).
Figure 1.
Miroslav Radman's (1970) model for lesion bypass in Escherichia coli from his paper on the SOS response circulated privately in 1970. The full text with a commentary by Bridges (2005) was recently published in DNA Repair. Reproduced with kind permission from Miro Radman.
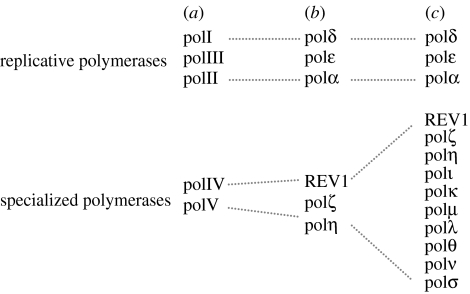
While mutagenic DNA damage bypass makes some kind of teleological sense in single-celled organisms in which it can be used as a mechanism to escape environmental bottlenecks, its preservation in higher organisms is more puzzling. Indeed, far from being lost, there has been a huge expansion in the number of specialized polymerases in vertebrates and there is clear evidence that these polymerases play a central role in DNA damage tolerance (figure 2).
Figure 2.
The expansion of specialized polymerases in vertebrate cells. Comparison of the currently known polymerases in (a) E. coli, (b) Saccharomyces cerevisiae and (c) humans.
The specialized translesion polymerases (reviewed in Prakash et al. 2005) are characterized by active sites that are tolerant of distortions in the DNA helix introduced by base damage or mismatches. While this property allows them to synthesize across damaged bases, it also renders them more likely to misincorporate on undamaged DNA. Use of these polymerases is therefore potentially mutagenic not only owing to the non- or mis-instructional nature of damaged bases, but also because their error rate is intrinsically higher. Their use must therefore be carefully regulated if the genomic mutation rate is not to be elevated to dangerous levels, particularly in multicellular organisms, in which the stability of the somatic genome is paramount.
2. Mechanisms that control translesion synthesis in yeast and vertebrates
Following soon after the initial observations in E. coli, screens in the budding yeast Saccharomyces cerevisiae also revealed genetic loci required for DNA damage-induced mutagenesis (Lawrence & Christensen 1976; Lemontt 1971). Indeed, much of our current understanding of how translesion synthesis is controlled in eukaryotes has come from this organism. The genes required for mutagenesis in S. cerevisiae fall into the RAD6 epistasis group, which includes three translesion polymerases, REV1, Polζ (REV3/REV7) and Polη (RAD30; Lemontt 1971; Nelson et al. 1996a,b; McDonald et al. 1997; Johnson et al. 1999). The use of these enzymes in lesion bypass is controlled by the monoubiquitination of the DNA sliding clamp proliferating cell nuclear antigen (PCNA; Hoege et al. 2002). Monoubiquitination of PCNA increases the affinity of the specialized polymerases for the clamp through interactions between ubiquitin and UBM or UBZ ubiquitin-binding domains embedded within the polymerases themselves (Bienko et al. 2005). The monoubiquitination of PCNA is carried out by the RAD6/RAD18 heterodimer, which appears in turn to be recruited by regions of single-stranded DNA (Bailly et al. 1997; Davies et al. 2008) at stalled replication forks. In S. cerevisiae, this modification is a requirement for both translesion synthesis and an error-free pathway of lesion bypass defined by the RAD5 helicase (Hoege et al. 2002; Blastyak et al. 2007).
Understanding of DNA damage bypass pathways in vertebrates is now catching up with much being learned from the use of cell line models, in particular the chicken cell line DT40 (Buerstedde & Takeda 1991).
While the ubiquitination of PCNA is important for the control of translesion synthesis in DT40 (Arakawa et al. 2006; Simpson et al. 2006), its role is not as central as in budding yeast (Okada et al. 2002; Ross et al. 2005; Simpson & Sale 2005). Recent evidence has suggested that this may be, at least in part, due to greater prominence of a non-catalytic role of the Y-family polymerase REV1. Studies in yeast had already established that much of the contribution made by REV1 to translesion synthesis does not result from its rather restricted enzymatic activity (Nelson et al. 2000), REV1 being capable only of deoxycytidyl transfer opposite template G and a limited number of lesions (Nelson et al. 1996a; Zhang et al. 2002). Nonetheless, in yeast, the function of REV1 in damage tolerance is genetically dependent on RAD6 (Lawrence & Christensen 1976). By contrast, mutants of DT40 that lack both REV1 and the ability to ubiquitinate PCNA are much more sensitive to DNA damage than either single mutant, showing that PCNA ubiquitination and REV1 have substantially non-overlapping roles (Ross et al. 2005; Arakawa et al. 2006).
The C terminus of REV1 comprises two distinct regions. The first, found in the last 100 amino acids, interacts with each of the other translesion polymerases known to be involved in DNA damage bypass (Murakumo et al. 2001; Guo et al. 2003; Ohashi et al. 2004; Tissier et al. 2004). Adjacent to this is a second region that we demonstrated to mediate an interaction with PCNA (Ross et al. 2005) but which also contains the UBM ubiquitin-binding domains (Bienko et al. 2005). Although the precise relationship between these interactions remains to be established, this arrangement suggests that the C terminus of REV1 might also function, like ubiquitin, as an adaptor between the stalled replication fork and an incoming translesion polymerase.
3. The timing of translesion synthesis
Although the original proposal that translesion synthesis provides a mechanism for filling post-replicative gaps has recently been revisited (Lehmann & Fuchs 2006; Lopes et al. 2006; Waters & Walker 2006), much attention has been focused on the potential of these mechanisms to alleviate replication arrest ‘on-the-fly’ (i.e. at the fork, rather than after it). While these two modes of action are of course not mutually exclusive, there has, over the years, been a degree of controversy and confusion over when translesion synthesis operates.
The indication that PCNA ubiquitination and the non-catalytic function of the C terminus of REV1 might act to coordinate translesion synthesis in different pathways prompted us to consider whether such pathways might be temporally separated relative to replication fork arrest. To address this, we employed two complementary assays in DT40 cells that measure fork progression and post-replicative gap filling, respectively (Edmunds et al. 2008). Fork progression was monitored by measuring replication tracts in DNA fibres from cells that had been pulsed with halogenated nucleotides. This assay reveals the total length of DNA synthesized in a given time. By using two different labels, the length of DNA synthesized before and after DNA damage can be measured allowing the contribution of individual factors to maintaining fork progression on damaged DNA to be assessed. This approach yielded the surprising result that while REV1 was required to maintain wild-type levels of fork progression on DNA damaged with either UV light or 4-nitroquinoline-1-oxide, the ability to ubiquitinate PCNA was dispensable. However, assessment of post-replicative gap filling by alkaline sucrose velocity sedimentation showed a clear requirement for PCNA ubiquitination, as extensively documented previously in both yeast and vertebrate cells (Prakash 1981; Yamashita et al. 2002; Tateishi et al. 2003; Haracska et al. 2004).
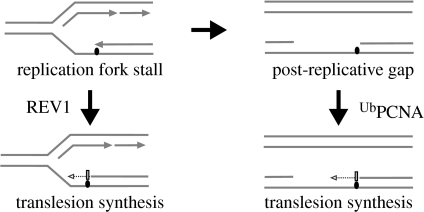
Together, these data suggested that REV1 and PCNA ubiquitination do indeed define distinct pathways for recruiting translesion synthesis and that these operate at different times relative to replication fork arrest (figure 3). Although this observation is surprising in the context of much of the recent thinking in the field, which has frequently envisaged PCNA ubiquitination as a mechanism for polymerase switching at stalled replication forks, recent experiments have suggested that practically all translesion synthesis is post-replicative in S. cerevisiae (Lopes et al. 2006). This may help to explain why, despite the C terminus of yeast REV1 making many of the same contacts as its vertebrate counterpart, lesion bypass in this organism is apparently so dependent on PCNA ubiquitination.
Figure 3.
REV1 and PCNA ubiquitination define temporally distinct mechanisms for controlling translesion synthesis. On encountering a lesion (black oval) that blocks the replicative DNA polymerases, translesion synthesis can be directly recruited (dotted line), thereby allowing the fork to continue. This recruitment requires the C terminus of REV1. Alternatively, the fork can arrest and restart downstream of the template lesion leaving a gap. This gap is flagged by PCNA ubiquitination, which also facilitates the recruitment of translesion polymerases. We suggest that the former mechanism might be favoured on the leading strand, the latter on the lagging strand where discontinuous Okazaki fragment synthesis will lead more readily to gap formation.
In the second part of this paper, we will consider whether these observations might help to refine our view of how and when mutagenic processing of activation induced deaminase (AID)-mediated DNA damage in the immunoglobulin loci takes place.
4. Deamination in the immunoglobulin locus: a simple lesion processed by multiple pathways
Three seemingly diverse processes (somatic hypermutation, gene conversion and class switch recombination) constitute an essential facet of the adaptive immune response by generating and refining a practically infinite range of antibody specificities. All three are initiated by the deamination of cytidine within the immunoglobulin loci by AID (Muramatsu et al. 2000; Revy et al. 2000; Arakawa et al. 2002; Harris et al. 2002). Since the previous Discussion Meeting on antibody diversification in 2000, a wealth of data has led to our current understanding of how a simple deamination event triggers the diverse and complex outcomes that create immunoglobulin diversity. The broader aspects of immunoglobulin diversification have been reviewed extensively elsewhere (Kenter 2005; Maizels 2005; Di Noia & Neuberger 2007). Here, we will concentrate on how deamination of dC leads to the full range of point mutations seen during immunoglobulin gene hypermutation.
Although AID works only on dC in DNA, generating uracil, mutations in the immunoglobulin genes in mouse and man are found at all four bases. Unlike general replication errors, which exhibit a marked bias to transition mutations, the spectrum of immunoglobulin hypermutation reveals preferential focusing to certain hotspots and exhibits a ratio of transitions to transversions much closer to 50 per cent (Betz et al. 1993). An important clue as to how dC deamination can lead to the full spectrum of mutations seen in the immunoglobulin genes comes from demonstrations that the genetic requirements for the formation of mutations at dG:dC base pairs and dA:dT base pairs are different. The first of these observations predated the discovery of AID. Mice deficient for a key component of the mismatch repair machinery, MSH2, exhibit a substantial reduction in mutations at dA:dT base pairs, while retaining practically normal levels of mutation at dG:dC bases (Frey et al. 1998; Rada et al. 1998). This observation led to the suggestion that somatic hypermutation is a two-phase process with phase I generating mutations at dG:dC and phase II at dA:dT (Rada et al. 1998; Neuberger et al. 2003).
5. Hypermutating cell lines and G:C mutation: models of abasic site bypass
Curiously, in all cell line models that exhibit constitutive or inducible immunoglobulin hypermutation, mutations are formed predominantly at dG:dC base pairs (Denepoux et al. 1997; Sale & Neuberger 1998; Harris et al. 2001). For example, in the Burkitt lymphoma line Ramos over 80 per cent of mutations are found at dG:dC base pairs (Sale & Neuberger 1998). In DT40 cells, which normally diversify their immunoglobulin genes predominantly by gene conversion, disruption of homologous recombination or removal of the V pseudogene donors results in almost exclusive (approx. 95%) mutations at dG:dC (Arakawa et al. 2004; Sale et al. 2001). Thus, while hypermutating variants of DT40 provide a genetically most attractive model system, the immunoglobulin mutagenesis in these lines probably reflects simply the direct consequence of replication across either dU, generating dC→dT transition mutations, or across abasic sites formed following excision of uracil by uracil DNA glycosylase (UNG; Di Noia & Neuberger 2002; Sale 2004). Nevertheless, hypermutation in cell lines appears to accurately reflect the ‘phase I’ mutagenesis seen in mice and humans. The reasons for this dG:dC bias, when the parental organism is perfectly capable of mutagenesis at dA:dT base pairs as well, remain unclear. However, it is tempting to speculate that it is related to a common feature or combination of features of transformed germinal centre B cells such as deregulated MYC or BCL-6 expression and loss of p53.
The contributions of PCNA ubiquitination and of REV1 to bypass of abasic sites in the immunoglobulin loci of DT40 broadly mirror their roles in DNA damage tolerance. Efficient mutagenesis requires both REV1 and the ubiquitination of PCNA but loss of both results in almost total abrogation of mutation (Arakawa et al. 2006). However, part of the reduction in mutation in rev1 DT40 cells may be due to loss of the deoxycytidyl transferase activity of REV1 since abasic sites are good substrates for this enzyme. Reconstitution of rev1 DT40 cells with human REV1 carrying catalytically inactivating point mutations demonstrated that REV1 was required for dC→dG transversion mutations on both strands of the immunoglobulin light chain locus in DT40. However, the decrease in dC→dG transversions was fully compensated for by an increase in dC→dA and dC→dT mutations (Ross & Sale 2006). Surprisingly, although disruption of the whole of REV1 results in decreased levels of point mutation, deletion of the C terminus does not. Furthermore, the frequency of dC→dG transversions is unaffected suggesting that the C terminus is not required to recruit the catalytic activity of REV1. However, an increase in dC→dA transversions on either strand, at the expense of dC→dT transitions, suggested that the C terminus of REV1 can nonetheless influence polymerase selection at abasic sites even when the catalytic activity of the protein is not employed.
6. Genetic requirements for immunoglobulin hypermutation in mice and humans
Hypermutation in mice and humans is more complex than the simple bypass of AID-induced damage seen in DT40 as mutations at dA:dT base pairs are generally more numerous than those at dG:dC. Furthermore, genetic analyses in mice have so far revealed a far greater degree of redundancy between the different pathways that process dU than is evident in DT40, making interpretation of the contribution of individual components more problematic.
Analysis of hypermutation in rev1−/− mice also reveals a clear contribution of the catalytic activity of the protein in bypassing abasic sites. However, in contrast to DT40, this activity appears to be strongly strand biased with the decrease in dC→dG transversions being seen only on the coding (non-transcribed) strand (Jansen et al. 2006). The overall mutation burden was the same as in control animals, the loss of dC→dG transversions being compensated for by an increase in dC→dA transversions and in mutations at dA:dT base pairs. One possible source of the redundancy in mutagenesis at dG:dC is DNA polymerase θ, a member of the A-family of DNA polymerases, which is also capable of direct catalytic bypass of abasic sites, strongly favouring A and disfavouring C in vitro (Seki et al. 2004). Polθ has been shown to play a role in the formation of mutations at dG:dC during immunoglobulin hypermutation in mouse (Masuda et al. 2005; Zan et al. 2005), and while it is tempting to speculate, based on their in vitro activities, that Polθ and REV1 play compensatory roles to each other, this remains to be tested. Furthermore, very little is yet known about the mechanisms that control Polθ.
Mutagenesis at dA:dT cannot, of course, be explained by direct misincorporation opposite dU or an abasic site. Unlike mutagenesis at dG:dC, which requires uracil excision, mutagenesis at dA:dT depends heavily on mismatch recognition of dU:dG mispairs. First noted in Msh2−/− mice (Frey et al. 1998; Rada et al. 1998), a similar loss of dA:dT mutagenesis has been documented in mice deficient for MSH6 (Wiesendanger et al. 2000; Martomo et al. 2004) and EXO1 (Bardwell et al. 2004). Curiously, mice deficient in PMS2 and MLH1, proteins required to couple mismatch recognition to excision (reviewed in Jiricny 2006), do not exhibit as striking a phenotype in terms of hypermutation (Frey et al. 1998; Kim et al. 1999; Phung et al. 1999; Ehrenstein et al. 2001). This suggests that either other factors can substitute for PMS2 and MLH1 in this context or the requirement for MSH2/6 and EXO1 in dA:dT mutagenesis does not reflect their canonical role in mismatch repair. Importantly, the dependence of dA:dT mutagenesis on mismatch repair factors is not complete and it is clear that a proportion of dA:dT mutations are also triggered by excision of dU by UNG (Rada et al. 2004).
Two further genetic requirements help provide a very strong framework for understanding mutagenesis at dA:dT base pairs. Numerous lines of evidence point to a central role for DNA polymerase η (Zeng et al. 2001; Delbos et al. 2005). Indeed, recent data have suggested that it is the sole polymerase responsible (Delbos et al. 2007). Additionally, Langerak et al. (2007) have also very recently demonstrated a pivotal requirement for PCNA ubiquitination. Polη interacts with ubiquitinated PCNA via its C terminal UBZ domain (Bienko et al. 2005) and the mutation spectrum of Polη on undamaged DNA is similar to the spectrum of dA:dT mutations seen during immunoglobulin hypermutation (Pavlov et al. 2002) suggesting that, unlike mutations at dG:dC (which arise due to bypass of a non-instructional lesion), mutations at dA:dT arise in consequence of misincorporation during DNA patch synthesis by Polη. Together, these observations support a model in which PCNA ubiquitination recruits Polη to the immunoglobulin loci. As in the rev1−/− mice, redundancy of mutagenic mechanisms exists and a compensatory increase in mutation at dG:dC is seen in both the pcnaK164R and polη−/− animals. Interestingly, the frequency of dC→dG transversions on the coding strand, which as discussed above are highly dependent on the catalytic activity of REV1, was unaffected by disruption of PCNA ubiquitination, confirming that the recruitment of the catalytic activity of REV1 is independent of the ability of REV1 to bind ubiquitin on PCNA.
7. Can immunoglobulin hypermutation be understood in terms of the timing of translesion synthesis?
In this last section we examine how, despite some important differences between studies of damage tolerance in DT40 and hypermutation in mice, some of the concepts uncovered in DT40 might be useful in improving our understanding of immunoglobulin hypermutation.
Substantial evidence points to AID activity being linked to transcription (reviewed in Di Noia & Neuberger 2007), the transcription bubble providing the single-stranded DNA upon which AID has biochemically been shown to act (Bransteitter et al. 2003; Chaudhuri et al. 2003; Dickerson et al. 2003). Transcription of highly active loci such as the immunoglobulin genes in germinal centre B cells is likely to be only transiently interrupted by the passage of a replication fork. Thus, the deamination of dC will take place almost continuously both before and after replication of the immunoglobulin V genes.
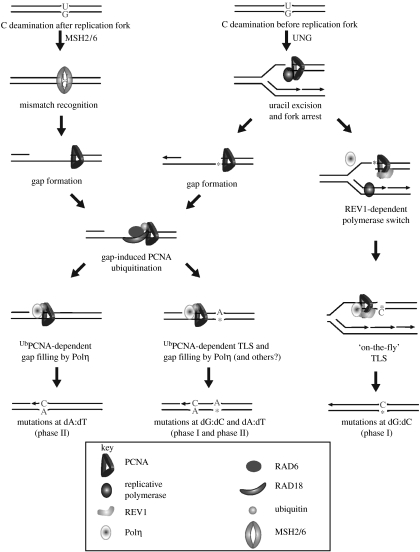
Uracil excision in the context of base excision repair is rapidly followed by elimination of the resulting abasic site by AP endonuclease and patch repair (reviewed in Kavli et al. 2007). However, for mutagenesis, the abasic site must be replicated before it is acted on by AP endonuclease. This is most likely to be achieved when U is encountered by a replication fork and is removed by UNG2 physically recruited to PCNA (Otterlei et al. 1999; Warbrick 2000). The resulting abasic site poses a block to the replicative polymerases leaving the replication machinery with two choices, to directly bypass the lesion, maintaining fork progression, or to arrest leaving a gap and deferring bypass of the lesion until later (figure 4).
Figure 4.
A model for how the timing of AID-mediated deamination and of uracil processing might dictate the mutagenic outcomes during immunoglobulin diversification. Deamination before and after passage of the replication fork triggers different mutagenic responses. Before arrival of the fork, U is recognized by UNG2 and removed to form an abasic site. This abasic site triggers either REV1-mediated ‘on-the-fly’ lesion bypass and mutagenesis at dG:dC or it triggers replication arrest and the formation of a post-replicative gap (see also figure 3). The lesion containing gap triggers PCNA ubiquitination and is subsequently bypassed by DNA polymerase η resulting in mutations at both dG:dC from bypass of the abasic site and at dA:dT from misincorporation during patch synthesis. C deamination after replication, but still with S phase, results in mismatch recognition and excision of the dU-containing strand. The resulting gap triggers PCNA ubiquitination and recruitment of DNA polymerase η with mutations arising from misincorporation during patch synthesis. TLS, translesion synthesis.
By analogy with bypass of UV damage in DT40, bypass of the abasic site at the fork should require REV1, while bypass at a post-replicative gap should require ubiquitination of PCNA (figure 4). It is not clear what would determine the division of labour between these two mechanisms, but an obvious possibility is whether the deamination has occurred on the leading or lagging strand. Owing to the inherently discontinuous nature of lagging strand synthesis, gap formation may be favoured on the lagging strand, while a mechanism to maintain fork progression would be favoured on the leading strand. Current evidence suggests that replication in the immunoglobulin loci of mature B cells proceeds from downstream of the C region, back towards the V region (Zhou et al. 2002; Norio et al. 2005). Thus, the (coding) non-transcribed strand of the V gene would also be the leading strand. Our model suggests that the use of REV1 for abasic site bypass might be favoured on this strand, a notion consistent with the observed strand bias of dC→dG mutagenesis seen in rev1−/− mice (Jansen et al. 2006). The analysis of hypermutation in rev1−/−/pcnaK164R doubly mutant mice will certainly be illuminating for testing the extent to which these two mechanisms compensate for each other and for uncovering other modes of translesion synthesis control.
The strong dependence of mutagenesis at dA:dT on PCNA ubiquitination places it as a post-replicative event in our model. Although it has recently been proposed that dA:dT mutagenesis takes place in G1 (Franklin & Blanden 2008), this is not consistent with the observations that show a requirement for replication forks in order for PCNA ubiquitination to be detectable in both budding yeast and human cells (Kannouche et al. 2004; Davies et al. 2008). Furthermore, we propose that the two pathways that result in dA:dT mutations (dU excision and recognition of dU:dG by MSH2/6; Rada et al. 2004) converge on a single-strand gap formed or remaining after bulk replication has been completed (figure 4). This model suggests that a key difference between the gaps created in each pathway is whether or not they contain an abasic site and, therefore, whether or not they potentially give rise to linked mutations at both dG:dC and dA:dT. However, it is debateable whether such linkage would be convincingly detectable as the proportion of mutations generated by this pathway is probably too small (Rada et al. 2004) and the range of gap lengths, and hence the range over which linkage might occur, is potentially quite large. Original experimental estimates for post-replicative gaps formed after UV irradiation suggested a length of approximately 800 bp (Lehmann 1972). However, the limit of the hypermutation domain in the immunoglobulin loci suggests rather shorter distances of the order of 30–100 nucleotides (Di Noia & Neuberger 2007).
Although dU excision can trigger mutation at dA:dT pairs, most of these mutations occur independently of UNG, instead relying on recognition of dU:dG mismatches by the MSH2/MSH6 heterodimer (Rada et al. 2004). This involvement of mismatch repair raises a paradox. Mismatch repair usually functions to eliminate base misincorporations during replication and is therefore antimutagenic. However, in the context of AID-induced dU:dG mismatches, recognition by MSH2/6 is clearly mutagenic. Why this is the case remains unclear but is likely to be related to the recruitment of PCNA ubiquitination. The involvement of EXO1 in the generation of mutations at dA:dT suggests that mismatch recognition is likely to result in the exonucleolytic degradation of one strand to create a gap. Why might gaps created by recognition of dU:dG trigger PCNA ubiquitination? Recruitment of PCNA ubiquitination by mismatch repair is not without precedent. Observations from Serge Boiteux's group suggest that PCNA ubiquitination and Polη are recruited to gaps created by the post-replicative recognition of 8-oxoG containing mismatches in yeast, in this context resulting in error-free dCMP incorporation opposite to the lesion by Polη rather than inaccurate dA incorporation by Polδ (de Padula et al. 2004).
A critical issue therefore is what determines whether or not PCNA ubiquitination is recruited to gaps formed following mismatch recognition. One possibility again relates to timing, this time of mismatch recognition. Canonical mismatch repair is likely to act almost immediately a base is misincorporated at the fork and therefore not result in a persistent gap. Thus, if dU:dG formation and its recognition happen after replication, rather than at the fork, the resulting gap may then be treated in much the same way as a gap created by a replication fork stall with downstream restart, with both resulting in PCNA ubiquitination (figure 4). Alternatively, there may be mechanisms that directly couple mismatch repair with PCNA ubiquitination and it is of note here that both MSH2 and MSH6 were identified in a macromolecular complex containing RAD18 and Polη (Yuasa et al. 2006).
The use of PCNA ubiquitination to flag persistent unreplicated gaps is an important strategy employed by cells to ensure the bypass of all lesions and the generation of a complete copy of the genome. In the context of immunoglobulin hypermutation, this strategy, normally reserved for difficult lesion-containing gaps, is subverted, via mismatch excision, by the unusual and high level of post-replicative damage created by AID.
Acknowledgments
We would like to thank Cristina Rada and Michael Neuberger for many stimulating discussions. Work in the laboratory is supported by the Medical Research Council and the Association for International Cancer Research.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘DNA deamination in immunity, virology and cancer’.
References
- Arakawa H., Hauschild J., Buerstedde J.M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. doi:10.1126/science.1067308 [DOI] [PubMed] [Google Scholar]
- Arakawa H., Saribasak H., Buerstedde J.M. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2:E179. doi: 10.1371/journal.pbio.0020179. doi:10.1371/journal.pbio.0020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H., Moldovan G.L., Saribasak H., Saribasak N.N., Jentsch S., Buerstedde J.M. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. doi:10.1371/journal.pbio.0040366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V., Lauder S., Prakash S., Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997;272:23 360–23 365. doi: 10.1074/jbc.272.37.23360. doi:10.1074/jbc.272.37.23360 [DOI] [PubMed] [Google Scholar]
- Bardwell P.D., Woo C.J., Wei K., Li Z., Martin A., Sack S.Z., Parris T., Edelmann W., Scharff M.D. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 2004;5:224–229. doi: 10.1038/ni1031. doi:10.1038/ni1031 [DOI] [PubMed] [Google Scholar]
- Betz A.G., Rada C., Pannell R., Milstein C., Neuberger M.S. Passenger transgenes reveal intrinsic specificity of the antibody hypermutation mechanism: clustering, polarity, and specific hot spots. Proc. Natl Acad. Sci. USA. 1993;90:2385–2388. doi: 10.1073/pnas.90.6.2385. doi:10.1073/pnas.90.6.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M., et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. doi:10.1126/science.1120615 [DOI] [PubMed] [Google Scholar]
- Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. doi:10.1016/j.molcel.2007.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Scharff M.D., Goodman M.F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. doi:10.1073/pnas.0730835100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges B.A. Error-prone DNA repair and translesion DNA synthesis. II: the inducible SOS hypothesis. DNA Repair (Amsterdam) 2005;4:725–726. doi: 10.1016/j.dnarep.2004.12.009. doi:10.1016/j.dnarep.2004.12.009 see also p. 739. [DOI] [PubMed] [Google Scholar]
- Buerstedde J.M., Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. doi:10.1016/0092-8674(91)90581-I [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. doi:10.1038/nature01574 [DOI] [PubMed] [Google Scholar]
- Davies A.A., Huttner D., Daigaku Y., Chen S., Ulrich H.D. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. doi:10.1016/j.molcel.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Padula M., Slezak G., Auffret van Der Kemp P., Boiteux S. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:5003–5010. doi: 10.1093/nar/gkh831. doi:10.1093/nar/gkh831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F., De Smet A., Faili A., Aoufouchi S., Weill J.C., Reynaud C.A. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. doi:10.1084/jem.20050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F., Aoufouchi S., Faili A., Weill J.C., Reynaud C.A. DNA polymerase η is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2007;204:17–23. doi: 10.1084/jem.20062131. doi:10.1084/jem.20062131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denepoux S., Razanajaona D., Blanchard D., Meffre G., Capra J.D., Banchereau J., Lebecque S. Induction of somatic mutation in a human B cell line in vitro. Immunity. 1997;6:35–46. doi: 10.1016/s1074-7613(00)80240-x. doi:10.1016/S1074-7613(00)80240-X [DOI] [PubMed] [Google Scholar]
- Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. doi:10.1084/jem.20030481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J., Neuberger M.S. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. doi:10.1038/nature00981 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. doi:10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Edmunds C.E., Simpson L.J., Sale J.E. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. doi:10.1016/j.molcel.2008.03.024 [DOI] [PubMed] [Google Scholar]
- Ehrenstein M.R., Rada C., Jones A.M., Milstein C., Neuberger M.S. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc. Natl Acad. Sci. USA. 2001;98:14 553–14 558. doi: 10.1073/pnas.241525998. doi:10.1073/pnas.241525998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A., Blanden R.V. The strand bias paradox of somatic hypermutation at immunoglobulin loci. Trends Immunol. 2008;29:167–172. doi: 10.1016/j.it.2008.01.008. doi:10.1016/j.it.2008.01.008 [DOI] [PubMed] [Google Scholar]
- Frey S., Bertocci B., Delbos F., Quint L., Weill J.C., Reynaud C.A. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. doi:10.1016/S1074-7613(00)80594-4 [DOI] [PubMed] [Google Scholar]
- Guo C., Fischhaber P.L., Luk-Paszyc M.J., Masuda Y., Zhou J., Kamiya K., Kisker C., Friedberg E.C. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. doi:10.1093/emboj/cdg626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Torres-Ramos C.A., Johnson R.E., Prakash S., Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. doi:10.1128/MCB.24.10.4267-4274.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Croom-Carter D.S., Rickinson A.B., Neuberger M.S. Epstein–Barr virus and the somatic hypermutation of immunoglobulin genes in Burkitt's lymphoma cells. J. Virol. 2001;75:10 488–10 492. doi: 10.1128/JVI.75.21.10488-10492.2001. doi:10.1128/JVI.75.21.10488-10492.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Sale J.E., Petersen-Mahrt S.K., Neuberger M.S. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. doi:10.1016/S0960-9822(02)00717-0 [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. doi:10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- Jansen J.G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. doi:10.1084/jem.20052227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. doi:10.1038/nrm1907 [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash S., Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 1999;274:15 975–15 977. doi: 10.1074/jbc.274.23.15975. doi:10.1074/jbc.274.23.15975 [DOI] [PubMed] [Google Scholar]
- Kannouche P.L., Wing J., Lehmann A.R. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. doi:10.1016/S1097-2765(04)00259-X [DOI] [PubMed] [Google Scholar]
- Kavli B., Otterlei M., Slupphaug G., Krokan H.E. Uracil in DNA-general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amsterdam) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. doi:10.1016/j.dnarep.2006.10.014 [DOI] [PubMed] [Google Scholar]
- Kenter A.L. Class switch recombination: an emerging mechanism. Curr. Top. Microbiol. Immunol. 2005;290:171–199. doi: 10.1007/3-540-26363-2_8. doi:10.1007/3-540-26363-2-8 [DOI] [PubMed] [Google Scholar]
- Kim N., Bozek G., Lo J.C., Storb U. Different mismatch repair deficiencies all have the same effects on somatic hypermutation: intact primary mechanism accompanied by secondary modifications. J. Exp. Med. 1999;190:21–30. doi: 10.1084/jem.190.1.21. doi:10.1084/jem.190.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P., Nygren A.O., Krijger P.H., van den Berk P.C., Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J. Exp. Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. doi:10.1084/jem.20070902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W., Christensen R. UV mutagenesis in radiation-sensitive strains of yeast. Genetics. 1976;82:207–232. doi: 10.1093/genetics/82.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A.R. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J. Mol. Biol. 1972;66:319–337. doi: 10.1016/0022-2836(72)90418-4. doi:10.1016/0022-2836(72)90418-4 [DOI] [PubMed] [Google Scholar]
- Lehmann A.R., Fuchs R.P. Gaps and forks in DNA replication: rediscovering old models. DNA Repair (Amsterdam) 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. doi:10.1016/j.dnarep.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Lemontt J.F. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. doi:10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Maizels N. Immunoglobulin gene diversification. Annu. Rev. Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. doi:10.1146/annurev.genet.39.073003.110544 [DOI] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Gearhart P.J. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 2004;200:61–68. doi: 10.1084/jem.20040691. doi:10.1084/jem.20040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., et al. DNA polymerase θ contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl Acad. Sci. USA. 2005;102:13 986–13 991. doi: 10.1073/pnas.0505636102. doi:10.1073/pnas.0505636102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Levine A.S., Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakumo Y., Ogura Y., Ishii H., Numata S., Ichihara M., Croce C.M., Fishel R., Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem. 2001;276:35 644–35 651. doi: 10.1074/jbc.M102051200. doi:10.1074/jbc.M102051200 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. doi:10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence C.W., Hinkle D.C. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996a;382:729–731. doi: 10.1038/382729a0. doi:10.1038/382729a0 [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence C.W., Hinkle D.C. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996b;272:1646–1649. doi: 10.1126/science.272.5268.1646. doi:10.1126/science.272.5268.1646 [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Gibbs P.E., Nowicka A.M., Hinkle D.C., Lawrence C.W. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol. Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. doi:10.1046/j.1365-2958.2000.01997.x [DOI] [PubMed] [Google Scholar]
- Neuberger M.S., Harris R.S., Di Noia J., Petersen-Mahrt S.K. Immunity through DNA deamination. Trends Biochem. Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. doi:10.1016/S0968-0004(03)00111-7 [DOI] [PubMed] [Google Scholar]
- Norio P., Kosiyatrakul S., Yang Q., Guan Z., Brown N.M., Thomas S., Riblet R., Schildkraut C.L. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. doi:10.1016/j.molcel.2005.10.029 [DOI] [PubMed] [Google Scholar]
- Ohashi E., Murakumo Y., Kanjo N., Akagi J., Masutani C., Hanaoka F., Ohmori H. Interaction of hRev1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. doi:10.1111/j.1356-9597.2004.00747.x [DOI] [PubMed] [Google Scholar]
- Okada T., Sonoda E., Yamashita Y.M., Koyoshi S., Tateishi S., Yamaizumi M., Takata M., Ogawa O., Takeda S. Involvement of vertebrate polκ in Rad18-independent postreplication repair of UV damage. J. Biol. Chem. 2002;277:48 690–48 695. doi: 10.1074/jbc.M207957200. doi:10.1074/jbc.M207957200 [DOI] [PubMed] [Google Scholar]
- Otterlei M., et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. doi:10.1093/emboj/18.13.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov Y.I., Rogozin I.B., Galkin A.P., Aksenova A.Y., Hanaoka F., Rada C., Kunkel T.A. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase eta during copying of a mouse immunoglobulin kappa light chain transgene. Proc. Natl Acad. Sci. USA. 2002;99:9954–9959. doi: 10.1073/pnas.152126799. doi:10.1073/pnas.152126799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung Q.H., Winter D.B., Alrefai R., Gearhart P.J. Hypermutation in Ig V genes from mice deficient in the MLH1 mismatch repair protein. J. Immunol. 1999;162:3121–3124. [PubMed] [Google Scholar]
- Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 1981;184:471–478. doi: 10.1007/BF00352525. doi:10.1007/BF00352525 [DOI] [PubMed] [Google Scholar]
- Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. doi:10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- Rada C., Ehrenstein M.R., Neuberger M.S., Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. doi:10.1016/S1074-7613(00)80595-6 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., Neuberger M.S. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. doi:10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Radman M. SOS replication: a distinct replication mechanism which is induced by DNA damaging treatments? DNA Repair. 1970;4:732–738. [PubMed] [Google Scholar]
- Revy P., et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. doi:10.1016/S0092-8674(00)00079-9 [DOI] [PubMed] [Google Scholar]
- Ross A.L., Sale J.E. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol. Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. doi:10.1016/j.molimm.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Ross A.L., Simpson L.J., Sale J.E. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. doi:10.1093/nar/gki279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W.D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. doi:10.1016/0022-2836(68)90445-2 [DOI] [PubMed] [Google Scholar]
- Rupp W.D., Wilde C.E., III, Reno D.L., Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. doi:10.1016/0022-2836(71)90204-X [DOI] [PubMed] [Google Scholar]
- Sale J.E. Immunoglobulin diversification in DT40: a model for vertebrate DNA damage tolerance. DNA Repair (Amsterdam) 2004;3:693–702. doi: 10.1016/j.dnarep.2004.03.042. doi:10.1016/j.dnarep.2004.03.042 [DOI] [PubMed] [Google Scholar]
- Sale J.E., Neuberger M.S. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. doi:10.1016/S1074-7613(00)80651-2 [DOI] [PubMed] [Google Scholar]
- Sale J.E., Calandrini D.M., Takata M., Takeda S., Neuberger M.S. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. doi:10.1038/35091100 [DOI] [PubMed] [Google Scholar]
- Seki M., Masutani C., Yang L.W., Schuffert A., Iwai S., Bahar I., Wood R.D. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. doi:10.1038/sj.emboj.7600424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L.J., Sale J.E. UBE2V2 (MMS2) is not required for effective immunoglobulin gene conversion or DNA damage tolerance in DT40. DNA Repair (Amsterdam) 2005;4:503–510. doi: 10.1016/j.dnarep.2004.12.002. doi:10.1016/j.dnarep.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Simpson L.J., Ross A.L., Szuts D., Alviani C.A., Oestergaard V.H., Patel K.J., Sale J.E. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. doi:10.1038/sj.embor.7400777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S., Niwa H., Miyazaki J., Fujimoto S., Inoue H., Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. doi:10.1128/MCB.23.2.474-481.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A., Kannouche P., Reck M.P., Lehmann A.R., Fuchs R.P., Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase poleta and REVl protein. DNA Repair (Amsterdam) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. doi:10.1016/j.dnarep.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. doi:10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-# [DOI] [PubMed] [Google Scholar]
- Waters L.S., Walker G.C. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl Acad. Sci. USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. doi:10.1073/pnas.0510167103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesendanger M., Kneitz B., Edelmann W., Scharff M.D. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6- deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. doi:10.1084/jem.191.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E.M. Mutation-proof and mutation-prone modes of survival in derivatives of Escherichia coli B differing in sensitivity to ultraviolet light. Brookhaven Symp. Biol. 1967a;20:17–55. [Google Scholar]
- Witkin E.M. The radiation sensitivity of Escherichia coli B: a hypothesis relating filament formation and prophage induction. Proc. Natl Acad. Sci. USA. 1967b;57:1275–1279. doi: 10.1073/pnas.57.5.1275. doi:10.1073/pnas.57.5.1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Okada T., Matsusaka T., Sonoda E., Zhao G.Y., Araki K., Tateishi S., Yamaizumi M., Takeda S. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. doi:10.1093/emboj/cdf534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa M.S., Masutani C., Hirano A., Cohn M.A., Yamaizumi M., Nakatani Y., Hanaoka F. A human DNA polymerase η complex containing Rad18, Rad6 and Rev1; proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells. 2006;11:731–744. doi: 10.1111/j.1365-2443.2006.00974.x. doi:10.1111/j.1365-2443.2006.00974.x [DOI] [PubMed] [Google Scholar]
- Zan H., Shima N., Xu Z., Al-Qahtani A., III, Evinger A.J., Zhong Y., Schimenti J.C., Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. doi:10.1038/sj.emboj.7600833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Winter D.B., Kasmer C., Kraemer K.H., Lehmann A.R., Gearhart P.J. DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2001;2:537–541. doi: 10.1038/88740. doi:10.1038/88740 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wu X., Rechkoblit O., Geacintov N.E., Taylor J.S., Wang Z. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 2002;30:1630–1638. doi: 10.1093/nar/30.7.1630. doi:10.1093/nar/30.7.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Ermakova O.V., Riblet R., Birshtein B.K., Schildkraut C.L. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. doi:10.1128/MCB.22.13.4876-4889.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]