Abstract
Immunoglobulin class switch recombination (CSR) occurs by an intrachromosomal deletion requiring generation of double-stranded DNA breaks (DSBs) in immunoglobulin switch region DNA. The initial steps of DSB formation have been elucidated: cytosine deamination by activation-induced cytidine deaminase (AID) and the generation of abasic sites by uracil-DNA glycosylase (UNG). We show that abasic sites are converted into single-strand breaks (SSBs) by apurinic/apyrimidinic endonucleases (APE1 and APE2). If SSBs are near to each other on opposite strands, they will generate DSBs; but if distal from each other, mismatch repair appears to be required to generate DSBs. The resulting S region DSBs occur at dC residues that are preferentially targeted by AID. We also investigate whether DNA polymerase β, which correctly repairs SSBs resulting from APE activity, attempts to repair the breaks during CSR. We find that although polymerase β does attempt to repair S region DNA breaks in switching B cells, the frequency of AID-instigated breaks appears to outnumber the SSBs repaired correctly by polymerase β, and thus some DSBs and mutations are generated. We also show that the S region DSBs are introduced and resolved during the G1 phase of the cell cycle.
Keywords: antibody class switch, DNA recombination, activation induced cytidine deaminase
1. Introduction
Activation of B lymphocytes by antigen and co-stimulatory signals, usually from T lymphocytes, initiates two processes of antibody diversification. Somatic hypermutation (SHM) introduces mutations in the variable region genes, which, in conjunction with antigen selection, increases antibody affinity. Class switch recombination (CSR) enables B cells to diversify the constant region and thereby the effector function of the antibody molecule, while maintaining the same antigen-binding domain.
CSR occurs by an intrachromosomal deletional recombination between switch (S) region sequences located upstream of the constant region genes. S regions consist of tandem repeats of short (20–80 bp) G-rich sequences, extending from 2 to 10 kb in length, and recombination can occur at any site within or near the S regions. CSR is thought to occur predominantly by a non-homologous end joining (NHEJ) mechanism, but it can also occur by an alternative end-joining mechanism that uses short microhomologies between the different S regions (Yan et al. 2007; Stavnezer et al. 2008).
Activation-induced cytidine deaminase (AID) is essential for both SHM and CSR (Muramatsu et al. 2000; Revy et al. 2000) and multiple reports indicate that its role is to deaminate dC bases within immunoglobulin (Ig) variable region genes and S regions (Di Noia & Neuberger 2002; Petersen-Mahrt et al. 2002; Rada et al. 2002; Bransteitter et al. 2003; Chaudhuri et al. 2003; Li et al. 2004; Longerich et al. 2006). DNA replication across the resulting dU residues introduces mutations that are characteristic of SHM, and that are also found in the regions surrounding S–S junctions. However, removal of the dU bases by UNG is required to introduce the DNA breaks necessary for CSR (Schrader et al. 2005). CSR is reduced by 95 per cent in B cells from mice deficient in the base excision repair (BER) enzyme uracil DNA glycosylase (UNG2; Rada et al. 2002). CSR is also severely reduced in patients who have deleterious mutations in UNG (Imai et al. 2003).
2. APE1 and APE2 create single-strand breaks in S regions at dC bases during CSR
As UNG creates abasic sites, we asked whether apurinic/apyrimidinic endonuclease APE1, the BER enzyme that can incise the phosphate backbone at abasic sites, is necessary for the creation of double-strand breaks (DSBs) in S regions, and thus for CSR (Guikema et al. 2007). In this same study, we investigated whether a homologous enzyme that has abasic endonuclease activity, APE2, might also contribute to CSR. We could not obtain ape1−/− B cells, even by foetal liver transfer, as ape1−/− mice die in utero prior to formation of haematopoietic cells. However, APE1 heterozygous mice have DNA repair defects, enabling us to examine the contribution of APE1 to CSR. APE2 is a more recently discovered AP endonuclease, with homology to APE1, but mice deficient in APE2 have no obvious repair defects, although they have a 50 per cent reduction in B and T cells relative to wild-type (WT) littermates (Tsuchimoto et al. 2001; Ide et al. 2004). The ape2 gene is located on the X chromosome, so we use male APE2-deficient mice (termed ape2Y/−).
To determine if APE1 or APE2 are present in switching B cells, we examined the regulation of these proteins in splenic B cells induced to switch by Western blot, and found that they are both present. APE1 was found to be constitutively expressed in the nucleus of switching B cells whereas APE2 is induced when splenic B cells are treated to induce CSR (Guikema et al. 2007).
We compared CSR in splenic B cells from ape1+/−, ape2Y/− and ape1+/− ape2−/− (termed double KO or DBL) mice cultured for 3 days with lipopolysaccharide (LPS) and specific combinations of cytokines that induce CSR to different isotypes, and analysed by flow cytometry for surface Ig expression. In each experiment, WT littermates from ape1+/− and ape2+/− cross-breedings were used as controls. We found that ape1+/− B cells switched slightly less well than WT cells in culture to all isotypes tested (figure 1). However, both ape2y/− and the DBL cells showed a greater reduction of CSR. Surprisingly, the DBL B cells switch as well as the ape2y−/− cells. These data indicate that both APE1 and APE2 contribute to CSR.
Figure 1.
CSR is reduced in splenic B cells from mice with reduced levels of APE1, APE2 or both APE1 and APE2. CFSE-stained B cells from WT littermates and ape1+/−, ape−/− or ape1+/− ape2−/− mice were cultured for 3 days with LPS+ interleukin (IL)-4 (IgG1 CSR), LPS+interferon-γ (IFNγ) (IgG2a CSR), LPS+dextran sulphate (IgG2b CSR), LPS+anti-IgD-dextran (anti-δ-dex)(IgG3 CSR), or with LPS+IL4, IL5, transforming growth factor (TGF)β, and anti-δ-dextran (IgA CSR). After culture, cells were fixed and stained for surface isotype expression and analysed by flow cytometry (Schrader et al. 1999). Shown are the average per cent switching relative to WT littermates, set at 100% (+s.e.m.), for the different isotypes. *p<0.05, **p<0.01 as determined by the one-sample t-test. Black bars, ape1+/−; dark grey bars, ape2Y/−; light grey bars, DBL. Originally published in Guikema et al. (2007).
Since APE1 deficiency is an embryonic lethal defect and ape2−/− lymphocytes were reported to have cell proliferation defects, we assessed proliferation of the B cells and also switching per cell division by staining splenic B cells with carboxyfluorescein diacetate succinimidyl ester (CFSE) prior to induction of CSR, and then following dilution of CFSE staining as cells divide. We found that there was no difference in the proliferation of any of the APE-deficient cells from WT cells in the B-cell cultures, and that CSR was reduced at all cell divisions in the APE2 and DBL cells (Guikema et al. 2007). We did observe a slight delay in G2/M phase in ape2−/− B cells, as previously reported (Ide et al. 2004), but it did not affect the number of cell divisions in our cultures.
As we hypothesize that APE1 and APE2 contribute to CSR by creating single-strand breaks (SSBs) at abasic sites in S regions, we expected to detect fewer DSBs in S regions by ligation-mediated polymerase chain reaction (LM-PCR). An example of an LM-PCR experiment performed with APE-deficient cells 2 days after the induction of CSR is shown in figure 2. As the DSBs are reduced in frequency in the double knockout cells but much less so in either the ape1+/− or ape2−/− cells, these data indicate that both APE1 and APE2 are involved in making DNA breaks during CSR. In similar experiments, we cloned and sequenced the Sμ and Sγ3 LM-PCR DNA fragments from WT cells to determine the sites of the DNA breaks (Schrader et al. 2005). We found that the blunt DSBs detected by LM-PCR in Sμ and Sγ3 in WT cells occur specifically at G:C bp in the WRC/GYW AID-targeting hotspot, consistent with their introduction by AID–UNG–APE (Guikema et al. 2007). Note that the few DSBs detected in AID and UNG-deficient B cells occur at random sites in the S region DNA (Schrader et al. 2005; table 1).
Figure 2.
Sμ DSBs are decreased in APE-deficient splenic B cells induced to switch. (a) LPS+IL4 and (b) LPS+anti-δ-dextran. LM-PCR was performed on threefold dilutions of DNA isolated from B cells stimulated to undergo CSR for 2 days under the indicated conditions. Primers specific for the 5′ and 3′ ends of Sμ were used in conjunction with a linker-specific primer. PCR products were blotted and hybridized to an internal Sμ-specific probe. GAPDH indicates amplification of an internal control for template input. Methods were previously described (Schrader et al. 2005). Originally published in Guikema et al. (2007).
Table 1.
Blunt DSBs in Sμ from WT splenic B cells occur preferentially at G:C bp and at WRC/GYW AID hotspots.
| Sμ | ||||
|---|---|---|---|---|
| nt at break | WT % (nbra) | aid−/− % (nbr) | ung−/− % (nbr) | sequenceb |
| G | 73.3(55) | 62.5(15) | 53(9) | 40.7 |
| C | 9.3 (7) | 4.2 (1) | 5.8 (1) | 16.1 |
| A | 8.0 (6) | 20.8 (5) | 35.3 (6) | 21.4 |
| T | 9.3 (7) | 12.5 (3) | 5.8 (1) | 21.8 |
| total | 75 breaks | 24 breaks | 17 breaks | 2000 nts |
| G+C | 82.6% | 66.7% | 58.8% | 56.8% |
| p valuec: | 0.001 | n.s.d | n.s. | |
| hotspots | 48.0% | 4.2% | 11.8% | 23.2%e |
| p valuec: | 0.002 | n.s.d | n.s. | |
Number of breaks.
The distribution of nucleotides in the genomic sequence analysed.
Significance of difference from random by Fisher's exact t-test.
not significant.
% of nucleotides in genomic sequence that are in G:C bp at the underlined nucleotide in WRC/GYW hotspots.
We examined the sites of blunt DSBs in the APE-deficient cells and found that the DSBs in the single-deficient ape1+/− or ape2−/− cells still show some preference for G:C bp in AID hotspots, although in each case the preference was lower than that in WT cells. However, DSBs in the DBL cells occur at random DNA sites, i.e. not significantly different from the nucleotide sequence of the Sμ region itself (Guikema et al. 2007). These data suggest that the few DNA breaks detected in the DBL B cells are mostly not at the site of AID-instigated lesions, and therefore are not due to an alternative BER pathway. The remaining slight, but statistically insignificant, preference for G:C bp in the DBL cells could be due to the haploid levels of APE1 activity. Thus, although APE2 is reported to have weaker endonuclease and stronger exonuclease activity than APE1 (Hadi et al. 2002), the finding that in the DBL cells the DSBs occur less frequently at G:C bp in AID hotspots than in either APE1 or APE2 singly deficient cells indicates that both APE1 and APE2 function as endonucleases during CSR. This is also supported by our finding that staggered DSBs in WT cells occur specifically at G:C bp (89%), but those in the DBL KO cells do not (50%; Guikema et al. 2007).
We interpret these data to indicate that in WT cells, AID instigates SSBs by the BER pathway, and then after SSBs are introduced, the DSBs required for CSR are created by end processing on either strand to the site of the initiating lesion on the other strand. If the SSBs on opposite strands are near each other, they can form a DSB spontaneously. However, if they are more distal, our data suggest that this processing is dependent on mismatch repair (MMR) proteins (Stavnezer and Schrader 2006; Guikema et al. 2007; Schrader et al. 2007). Figure 3 presents the model for how MMR could convert SSBs to DSBs. This model is supported by a large amount of data (Stavnezer & Schrader 2006; Schrader et al. 2007). We find that the sites of staggered DSBs, which are detected by using T4 DNA polymerase (Pol) to fill in or chew back from staggered ends prior to ligating on the linker primers, also occur at G:C bp (Schrader et al. 2005; Guikema et al. 2007). End processing of the staggered DSBs in vivo by fill-in DNA synthesis or Ercc1-XPF could create blunt DSBs ending at G:C bp in AID hotspots.
Figure 3.
Model for role of MMR in CSR: to convert SSBs to DSBs. AID is hypothesized to introduce several dU bases into S regions during one cell cycle. Some of the dU residues are excised by UNG, and some of the abasic sites are nicked by APE. The U:G mismatches that remain would be substrates for Msh2–Msh6. Msh2–Msh6, along with Mlh1–Pms2, recruit Exo1 (and accessory proteins) to a nearby 5′ nick, from where exonuclease (Exo)1 begins to excise towards the mismatch, creating a DSB with a 5′ single-strand overhang. The overhang can be filled in by a DNA Pol, usually a translesion polymerase due to the presence of abasic sites. Alternatively, the overhang is removed by a 5′ flap endonuclease (Fen1) or by Exo1. Short overhangs can be used during end joining, generating microhomologies at the junction. 3′ overhangs can be excised by Ercc1-XPF.
3. Inhibitory role for DNA Polymerase β during CSR
Since APE creates SSBs during CSR, we asked if the next enzyme in the BER pathway, DNA Polβ, is involved in CSR. Polβ has two activities important for error-free BER. First is addition of a single correct nucleotide at the site of the lesion, which would add a dC to the 3′ end at the nick created by AID–UNG–APE. Second is lyase activity, which excises the deoxyribose phosphate (dRP) residue at the 5′ end of the nicked strand, remaining after APE nicks the phosphate backbone. This appears to be the rate-limiting step during BER (Srivastava et al. 1998), and thus is thought to occur after a dC is added to the 3′ end. Subsequently, DNA ligase 1 or DNA ligase III-XRCC1 completes repair by sealing the nick. Correct repair of the SSBs would inhibit CSR. Hence, an intriguing question arises as to how the S region nicks are spared from faithful BER so that they can be converted into DSBs to provide the essential intermediates for CSR. One appealing hypothesis is that BER components downstream of UNG and APE might be downregulated in cells undergoing CSR or specifically prevented from accessing S region lesions. Indeed, the recent finding that the amount of Polβ is inversely correlated with the frequency of SHM in subclones of the human Burkitt lymphoma cell line BL2 makes this hypothesis even more attractive (Poltoratsky et al. 2007). Alternatively, it is possible that the introduction of numerous S region lesions overwhelms the BER machinery, although BER activity is not inhibited during CSR.
To gain some insight into the potential regulation of Polβ activity in switching B cells, we examined levels of Polβ protein in mouse splenic B cells induced to undergo CSR. B cells from WT and AID-deficient mice were treated with LPS plus interleukin (IL)4 or with LPS plus interferon (IFN)γ for various time periods, and Western blots of nuclear and cytoplasmic extracts from the cultured B cells were prepared. Polβ accumulated in the nuclei of cells undergoing CSR, and cytoplasmic Polβ was coincidently reduced, suggesting that Polβ was redistributed from the cytoplasm to the nucleus in switching B cells (Wu & Stavnezer 2007). Polβ nuclear translocation was not AID dependent, as Polβ underwent similar translocation in AID-deficient B cells. This translocation might be due to the requirement for BER to repair the large amount of oxidative DNA damage occurring in rapidly proliferating B cells (Ito et al. 2007).
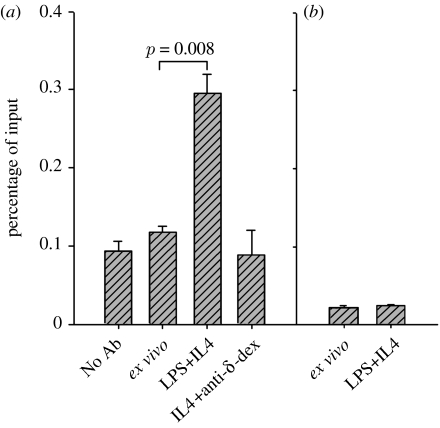
We considered the possibility that Polβ might be prevented from accessing S regions in switching B cells. To address this, we used chromatin immunoprecipitation (ChIP) to detect the association of Polβ with the Sμ region. To obtain quantitative results, ChIP was analysed by real-time PCR (figure 4). Stimulation with LPS plus IL4 for 3 days resulted in a 2.6-fold enrichment of Polβ association with the Sμ region compared with ex vivo B cells, whereas no enrichment above background was observed upon treatment with IL4 plus anti-IgD conjugated to dextran (anti-δ-dex), a treatment that induces B-cell proliferation but not CSR. Figure 4 also shows that Polβ does not associate with the Cμ gene in either ex vivo or LPS plus IL4 activated B cells, consistent with previous data showing that AID-dependent DSBs are found in S regions, but not in the Cμ gene (Catalan et al. 2003; Rush et al. 2004; Schrader et al. 2005). Altogether, the results clearly indicate that Polβ localizes to nuclei and binds the Sμ region DNA during CSR in cultured B cells.
Figure 4.
DNA Polβ binds to the IgSμ region but not the Cμ gene, in B cells undergoing CSR. (a) Sμ and (b) Cμ. Chromatin immunoprecipitation (ChIP) of extracts from WT splenic B cells, ex vivo or cultured as indicated, analysed by quantitative PCR. IP data are plotted relative to input DNA signal (corrected for the relative amounts of the sample analysed). The significance of the difference between ex vivo and cells activated with LPS+IL4 was calculated by the paired t-test. Originally published in Guikema et al. (2007).
As Polβ-deficient mice die just before birth, in order to generate mice with polβ−/− B cells, 2×106 foetal liver cells from polβ−/− and from polβ+/+ day 18.5 post-coitus foetuses were injected intravenously into sublethally irradiated recipient mice, as previously described (Esposito et al. 2000). Since the recipient cells bear CD45.1 and the donor cells bear CD45.2, successful reconstitution could be verified by fluorescence-activated cell sorting (FACS) analysis with antibodies recognizing CD45.1 and CD45.2. The recipient mice were sacrificed six weeks after foetal liver cell injection; FACS analysis revealed that their splenic B cells were almost exclusively CD45.2+(95–99%), indicating successful transfer and reconstitution. Lack of Polβ protein in splenic B cells in recipients that received polβ−/− foetal liver cells was confirmed by Western blot analysis (Wu & Stavnezer 2007). Analysis of splenic B-cell subsets showed that the proportion of immature, marginal zone and follicular Bcells was similar between the polβ+/+ and polβ−/− spleens (Wu & Stavnezer 2007). By CFSE analysis, DNA content analysis and by measuring [3H]-thymidine incorporation, we found no differences in cell divisions or proliferation between WT and KO splenic B cells (Wu & Stavnezer 2007).
If Polβ repairs SSBs during CSR, its deletion might result in an increase of CSR. We examined CSR to several isotypes, and found that switching to IgG2a is increased in the polβ−/− B cells, but no other isotype is significantly affected (figure 5). As isotype specificity is regulated by germ-line (GL) transcription of the unrearranged Sx-Cx segment, we asked if this specific stimulation of IgG2a CSR might be due to increased levels of GL γ2a transcripts in polβ−/− cells, but found they are not increased (Wu & Stavnezer 2007). Since Polβ has no known involvement in other cellular pathways except BER, it is unlikely that Polβ deficiency alters the signal transduction pathway specifically for IgG2a induction. S regions consist of tandem repeats that are unique to each isotype, although all contain numerous targets for AID: the hotspot motif WRC/GYW, where W=A or T, R=A or G, Y=C or T (Pham et al. 2003; Bransteitter et al. 2004; Yu et al. 2004). We considered the possibility that IgG2a CSR might be inhibited by Polβ due to the fact that there are fewer AID hotspot targets in Sγ2a than in other S regions, and thus it was possible that SSBs might be limiting for IgG2a CSR but not for other isotypes. To test this hypothesis, we developed suboptimal conditions for CSR for each isotype, by reducing the concentration of LPS and cytokines in culture, and examined CSR under these conditions in polβ−/− cells and WT controls. We reasoned that under suboptimal conditions, DNA breaks might be limiting and thus Polβ might inhibit CSR to other isotypes in addition to IgG2a. Under suboptimal conditions, lower levels of AID protein and GL γ1 and γ3 transcripts are induced in both WT and polβ−/− cells. As expected, suboptimal conditions results in decreased CSR efficiency, but polβ−/− cells switch relatively better than polβ+/+ cells to IgG2b, IgG3 and IgG2a (Wu & Stavnezer 2007). However, CSR to IgG1 and IgA still does not differ between WT and Polβ-deficient cells. This might be due to the fact that Sγ1 and Sα sequences have more of the hottest of the AID hotspots (AGCT) than any other S region except for Sμ (Wu & Stavnezer 2007).
Figure 5.
DNA Polβ-deficient B cells show moderate increase in CSR to IgG2a. (a) Representative FACS analysis for surface Ig isotype expression in transferred CD45.2+ cells ((i) IgG1, (ii) IgG2a, (iii) IgG2b, (iv) IgG3 and (v) IgA). WT and polβ−/− splenic B cells were stimulated to switch to the indicated isotypes (optimal conditions) for 4 days, then stained with anti-CD45.2 and antibodies specific for the indicated isotypes. Stained cells were analysed by flow cytometry. (b) Percentage of switching to the indicated isotypes in polβ−/− cells relative to WT cells. Mean of 4 experiments +s.e.m. is shown. Difference between WT and polβ−/− for IgG2a is significant (p<0.01). Originally published in ref Wu & Stavnezer (2007).
If Polβ possesses the ability to repair SSBs in S regions, as we hypothesize, its deletion should result in the accumulation of SSBs and consequently an increase in DSBs. We used LM-PCR to detect the DSBs in Sμ and Sγ3 regions from polβ−/− and WT B cells. Splenic B cells were activated to switch for 2 days, and genomic DNA was prepared for LM-PCR. In agreement with previous findings (Schrader et al. 2005), abundant DSBs were detected in WT cells at this time point, with very few breaks detectable in AID-deficient cells treated identically. Remarkably, 2.3-fold more Sμ DSBs were detected in polβ−/− cells than in WT cells, and a threefold increase in DSBs was observed in the acceptor Sγ3 region in polβ−/− cells (figure 6; Wu & Stavnezer 2007). To ascertain whether the increased DSBs in S regions of polβ−/− cells are relevant to CSR, and not due to a non-specific increase in DSBs, we assayed DSBs at the Cμ region. Very few breaks in the Cμ gene were detected, and no increase was detected in polβ−/− cells (Wu & Stavnezer 2007). These results clearly demonstrate that Polβ is able to repair DSBs induced in Ig S regions during CSR, as its absence leads to increased S region DSBs. The sequence specificity of the DSB sites in Sμ are similar between polβ+/+ and polβ−/− B cells (Wu & Stavnezer 2007).
Figure 6.
LM-PCR demonstrates increased DSBs in (a) Sμ and (b) Sγ3 regions from polβ−/− cells relative to WT cells. Cells induced to switch for 2 days under the optimal conditions indicated. Methods similar to figure 2, except an Sγ3 specific primer and probe was used as indicated (Wu & Stavnezer 2007). Originally published in Wu & Stavnezer (2007).
Mutations observed in the segment 5′ to unrearranged (GL) Sμ segments have been regarded to reflect AID targeting to S regions (Petersen et al. 2001; Nagaoka et al. 2002; Schrader et al. 2003). If Polβ participates in repairing SSBs introduced by the AID–UNG–APE pathway, one would predict that Polβ deficiency should result in increased mutations in the GL Sμ segment, and that is indeed what we found. There was a twofold increase in mutations in both the GL 5′ Sμ segment and the recombined Sμ (with Sγ3) segment from activated polβ−/− B cells compared with WT B cells (Wu & Stavnezer 2007). The data are highly significant (p<0.001). This increase is remarkably similar to the increase that we measured for DSBs by the LM-PCR assay. These data unambiguously indicate that Polβ is able to correctly repair AID-initiated SSBs in S regions during CSR, thereby reducing DSBs and S region mutations.
In conclusion, we reason that if B cells were to downregulate BER during CSR, this could be deleterious, given the great amount of reactive oxygen species produced during B-cell activation and proliferation (Fedyk & Phipps 1994; Ito et al. 2004). Therefore, it seems plausible that instead a mechanism is adopted that endows S regions with such numerous AID targets that the ability of BER to repair them is overwhelmed, rather than abrogating overall BER ability and thus jeopardizing the integrity of the B-cell genome. Although it has been suggested that the number of AID-instigated lesions is small in comparison to the number of lesions sustained in the overall cellular genome every day, it is possible that the high local concentration of dU residues along with the great number of oxidized bases in cells induced to switch can overwhelm BER. The finding that artificially introduced I-SceI sites in Sμ and Sγ1 regions mediate CSR to IgG1 suggests that only a single DSB in the donor and acceptor S region is sufficient for CSR (Zarrin et al. 2007). However, introduction of numerous dU bases might be required in order to obtain DSBs in the donor and acceptor S regions simultaneously. In fact, examination of mutations in ung−/− msh2−/− mice demonstrated that AID introduces many more lesions into the Sμ region than the numbers of resulting mutations found in WT cells, most likely due to their being correctly repaired in WT mice (Xue et al. 2006). These considerations and our experimental data suggest that Polβ functions normally during CSR to repair AID-initiated DNA lesions, but that the numerous AID lesions overwhelm it, and thus some breaks remain unrepaired.
4. S region DNA breaks are introduced and repaired during G1 phase of the cell cycle
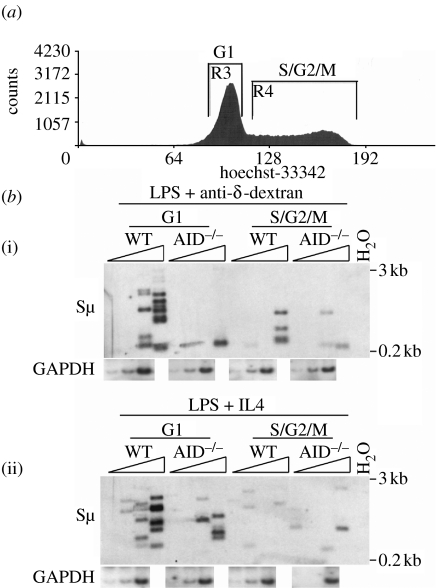
As described above (figure 3), our data indicate that MMR converts distal SSBs on opposite DNA strands to DSBs during CSR. The model posits that the MMR heterodimer Msh2–Msh6 recognizes dU:dG mismatches, this recruits Mlh1–Pms2, and then exonuclease 1 (Exo1) is recruited to the nearest SSB introduced by APE acting at AID–UNG lesions (Stavnezer & Schrader 2006). Exo1 activity then results in conversion of the SSB to a DSB. Alternatively, it has been proposed that DNA replication could convert AID-initiated SSBs into DSBs when the elongating DNA strand reaches a nick on the template strand. To address this alternative possibility, we examined the cell cycle regulation of Sμ DSBs (Schrader et al. 2007). Splenic B cells were activated for 2 days with LPS and IL4 or LPS and anti-δ-dex, stained with Hoechst 33342 and then sorted into G1 and S/G2/M fractions on the basis of DNA content (figure 7a). Dead cells were excluded by 7-AAD staining. At 48 hours there are no detectable undivided cells in these cultures. As shown in figure 7b, AID-dependent DSBs were almost exclusively detected in the G1 fraction, indicating that DSBs are created and resolved during G1 phase, and were not created during S phase. We then examined whether UNG, APE1 and APE2 are all present during G1 phase, by Western blotting of extracts from cells sorted into G1, S and G2/M phases. We found that all three of these proteins are as abundant in G1 phase as the other cell cycle phases (Schrader et al. 2007; J. E. J. Guikema 2008, unpublished data). We also found that UNG activity was equal in splenic B cells in G1 and S phase (Schrader et al. 2007).
Figure 7.
AID-dependent Sμ DSBs are detected in G1, but not in S/G2/M phase cells. (a) Splenic B cells were activated for 2 days, stained with Hoechst 33342, and sorted into the indicated two populations, as shown. (b(i)(ii)) LM-PCR was performed on DNA from viable sorted activated cells. Threefold dilutions of 7200 cell equivalents were amplified. PCR amplification of the GAPDH gene (except for the highest input) is shown. The figure is representative of two experiments. Adapted with permission from Schrader et al. (2007). Copyright 2007, The American Association of Immunologists, Inc.
These data indicate that the AID–UNG–APE pathway leads to DSBs in Sμ during G1 phase, and that these DSBs are resolved during the G1 phase, demonstrating that AID-dependent DSBs are not due to replication across SSBs during S phase. These results agree with a previous report showing that γH2AX/Nbs1 foci are detected during G1/early S phase in splenic B cells activated to switch (Petersen et al. 2001). The lack of DSBs during S phase would prevent the DSBs from blocking replication and reduce the likelihood of aberrant recombination. Also, this finding is consistent with the results of others showing that CSR occurs by NHEJ (Casellas et al. 1998; Manis et al. 1998; Yan et al. 2007), rather than by homologous recombination, which requires a homologous chromatid to serve as a template for repair. It will be very interesting to determine how the cell-cycle regulation of S region DSBs is enforced.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘DNA deamination in immunity, virology and cancer’.
References
- Bransteitter R., Pham P., Scharff M.D., Goodman M.F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl Acad. Sci. USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. doi:10.1073/pnas.0730835100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Calabrese P., Goodman M.F. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 2004;279:51 612–51 621. doi: 10.1074/jbc.M408135200. doi:10.1074/jbc.M408135200 [DOI] [PubMed] [Google Scholar]
- Casellas R., et al. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. doi:10.1093/emboj/17.8.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan N., Selz F., Imai K., Revy P., Fischer A., Durandy A. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J. Immunol. 2003;171:2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. doi:10.1038/nature01574 [DOI] [PubMed] [Google Scholar]
- Di Noia J., Neuberger M.S. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. doi:10.1038/nature00981 [DOI] [PubMed] [Google Scholar]
- Esposito G., Texido G., Betz U.A., Gu H., Muller W., Klein U., Rajewsky K. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc. Natl Acad. Sci. USA. 2000;97:1166–1171. doi: 10.1073/pnas.97.3.1166. doi:10.1073/pnas.97.3.1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedyk E.R., Phipps R.P. Reactive oxygen species and not lipoxygenase products are required for mouse B-lymphocyte activation and differentiation. Int. J. Immunopharmacol. 1994;16:533–546. doi: 10.1016/0192-0561(94)90105-8. doi:10.1016/0192-0561(94)90105-8 [DOI] [PubMed] [Google Scholar]
- Guikema J.E., Linehan E.K., Tsuchimoto D., Nakabeppu Y., Strauss P.R., Stavnezer J., Schrader C.E. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. doi:10.1084/jem.20071289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi M.Z., Ginalski K., Nguyen L.H., Wilson D.M., III Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol. 2002;316:853–866. doi: 10.1006/jmbi.2001.5382. doi:10.1006/jmbi.2001.5382 [DOI] [PubMed] [Google Scholar]
- Ide Y., Tsuchimoto D., Tominaga Y., Nakashima M., Watanabe T., Sakumi K., Ohno M., Nakabeppu Y. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood. 2004;104:4097–4103. doi: 10.1182/blood-2004-04-1476. doi:10.1182/blood-2004-04-1476 [DOI] [PubMed] [Google Scholar]
- Imai K., et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. doi:10.1038/ni974 [DOI] [PubMed] [Google Scholar]
- Ito K., et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. doi:10.1038/nature02989 [DOI] [PubMed] [Google Scholar]
- Ito K., et al. Regulation of reactive oxygen species by Atm is essential for proper response to DNA double-strand breaks in lymphocytes. J. Immunol. 2007;178:103–110. doi: 10.4049/jimmunol.178.1.103. [DOI] [PubMed] [Google Scholar]
- Li Z., Woo C.J., Iglesias-Ussel M.D., Ronai D., Scharff M.D. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. doi:10.1101/gad.1161904 [DOI] [PubMed] [Google Scholar]
- Longerich S., Basu U., Alt F., Storb U. AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. doi:10.1016/j.coi.2006.01.008 [DOI] [PubMed] [Google Scholar]
- Manis J.P., Gu Y., Lansford R., Sonoda E., Ferrini R., Davidson L., Rajewsky K., Alt F.W. Ku70 is required for late B cell development and immunoglobulin heavy chain switching. J. Exp. Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. doi:10.1084/jem.187.12.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. doi:10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nagaoka H., Muramatsu M., Yamamura N., Kinoshita K., Honjo T. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Sm region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J. Exp. Med. 2002;195:529–534. doi: 10.1084/jem.20012144. doi:10.1084/jem.20012144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S., et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. doi:10.1038/414660a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–104. doi: 10.1038/nature00862. doi:10.1038/nature00862 [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. doi:10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Poltoratsky V., Prasad R., Horton J.K., Wilson S.H. Down-regulation of DNA polymerase beta accompanies somatic hypermutation in human BL2 cell lines. DNA Repair (Amst.) 2007;6:244–253. doi: 10.1016/j.dnarep.2006.10.003. doi:10.1016/j.dnarep.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. doi:10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Revy P., et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. doi:10.1016/S0092-8674(00)00079-9 [DOI] [PubMed] [Google Scholar]
- Rush J.S., Fugmann S.D., Schatz D.G. Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to S mu in Ig class switch recombination. Int. Immunol. 2004;16:549–557. doi: 10.1093/intimm/dxh057. doi:10.1093/intimm/dxh057 [DOI] [PubMed] [Google Scholar]
- Schrader C.E., Edelmann W., Kucherlapati R., Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. doi:10.1084/jem.190.3.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C.E., Bradley S.P., Vardo J., Mochegova S.N., Flanagan E., Stavnezer J. Mutations occur in the Ig Sμ region but rarely in Sγ regions prior to class switch recombination. EMBO J. 2003;22:5893–5903. doi: 10.1093/emboj/cdg550. doi:10.1093/emboj/cdg550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C.E., Linehan E.K., Mochegova S.N., Woodland R.T., Stavnezer J. Inducible DNA breaks in Ig S regions are dependent upon AID and UNG. J. Exp. Med. 2005;202:561–568. doi: 10.1084/jem.20050872. doi:10.1084/jem.20050872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C.E., Guikema J.E., Linehan E.K., Selsing E., Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J. Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- Srivastava D.K., Berg B.J., Prasad R., Molina J.T., Beard W.A., Tomkinson A.E., Wilson S.H. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998;273:21 203–21 209. doi: 10.1074/jbc.273.33.21203. doi:10.1074/jbc.273.33.21203 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Schrader C.E. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 2006;22:23–28. doi: 10.1016/j.tig.2005.11.002. doi:10.1016/j.tig.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E.J., Schrader C.E. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. doi:10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. doi:10.1093/nar/29.11.2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. doi:10.1084/jem.20070285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K., Rada C., Neuberger M.S. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. doi:10.1084/jem.20061067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. doi:10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
- Yu K., Huang F.T., Lieber M.R. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. doi:10.1074/jbc.M311616200 [DOI] [PubMed] [Google Scholar]
- Zarrin A.A., Del Vecchio C., Tseng E., Gleason M., Zarin P., Tian M., Alt F.W. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. doi:10.1126/science.1136386 [DOI] [PubMed] [Google Scholar]