Abstract
This review focuses on the contribution of translesion DNA polymerases to immunoglobulin gene hypermutation, in particular on the roles of DNA polymerase eta (Polη) in the generation of mutations at A/T bases from the initial cytosine-targeted activation-induced cytidine deaminase (AID)-mediated deamination event, and of Polκ, an enzyme of the same polymerase family, used as a substitute when Polη is absent. The proposition that the UNG uracil glycosylase and the MSH2–MSH6 mismatch recognition complex are two competitive rather than alternative pathways in the processing of uracils generated by AID is further discussed.
Keywords: DNA polymerase eta (Polη), mismatch repair, translesion DNA synthesis
1. Introduction
Diversification of rearranged immunoglobulin genes is initiated by a locus-specific deamination process mediated by the enzyme activation-induced cytidine deaminase (AID), which produces uracils from cytosine deamination in the DNA encoding the immunoglobulin (Ig) variable regions (Muramatsu et al. 2000; Petersen-Mahrt et al. 2002). Such a nucleotide, albeit abnormal in DNA, is easily used as a template by replicative polymerases, and is thus susceptible to generate C to T and G to A mutations at the Ig locus. Three quarters of the mutations observed in memory B cells differ, however, from this simple mutation profile, implying that most mutations are generated by additional repair pathways that are sufficiently error-prone to further diversify this spectrum.
The seminal work of Rada et al. (2004) demonstrated that only two repair factors, uracil glycosylase and the MSH2–MSH6 complex, are involved in diversifying the outcome of the initial AID-mediated lesion, the mutation pattern in Ung×Msh2−/− mice, thus representing the footprint of AID deamination faithfully carried over replication. This work, as well as a more recent study (Liu et al. 2008), further established a quasi-quantitative correlation between the extent of cytosine deamination and the overall mutation load at the V gene loci, indicating that almost every deamination event is converted into a mutation, either at the deamination site or at a distance from it, a process that has to mobilize an error-prone DNA synthesis (Delbos et al. 2007; Weill & Reynaud 2008).
Such a process obviously requires repair pathways susceptible of generating errors at a very high rate. Accordingly, a base excision repair process resynthesizing a single base, even with low-fidelity polymerases, is unlikely to be involved. Two main pathways thus appear to be capable of generating such an error frequency: translesion synthesis opposed an abasic site that would be generated by the removal of the uracil base from DNA, and an MSH2/MSH6-driven short-patch error-prone DNA synthesis. This review focuses on the specific DNA polymerases involved in the context of the mutagenesis targeted around V genes. It should be mentioned that mutagenic processes that affect a second targeted region at the heavy chain locus, i.e. the switch regions, may slightly differ, and will not be discussed in this review. Effectively, hypermutation around the switch regions occurs as a bystander effect of a deamination activity whose biological outcome is the induction of double-strand breaks, and the high mutation load observed for this region in MSH2×UNG-deficient animals may suggest, although different interpretations are possible, a much higher frequency of UNG-mediated error-free repair during the normal isotype switch process (Xue et al. 2006).
2. Two confirmed partners of hypermutation among 11 non-replicative polymerases/transferases
Among the 11 non-replicative polymerases/transferases known to date, 10 have been studied for their possible contribution to hypermutation through the analysis of V gene mutations in activated B cells from polymerase-deficient animals. DNA polymerase nu (Polν) remains the only one not tested so far (table 1; Weill & Reynaud 2008). Whereas some contradictory data have been reported, a fair evaluation, taking into account notably the recent data published by the group of Patricia Gearhart for Polθ-deficient mice (Martomo et al. 2008), suggests that only two of them, Polη and Rev1, have been unambiguously demonstrated to be involved in hypermutation. Most convincingly, the impact of their inactivation on the Ig mutation pattern fits rather well with their known in vitro mutation signature. As discussed below, these two enzymes cannot account for the whole spectrum of mutations (in particular, most transversions at G/C bases), and there remains therefore another activity to be identified. DNA Polζ is a likely candidate, even though the drastic phenotype of the inactivation of its catalytic subunit, REV3L, has made so far any formal conclusion difficult. The complex functions of Polζ may therefore necessitate subtler mutagenesis, which would allow the dissociation of its catalytic activity from its other, so far unknown, functions, e.g. as an assembling or docking protein for other components of the replication/repair machinery, in order to address its specific contribution to Ig gene hypermutation.
Table 1.
Non-replicative DNA polymerases/transferases: impact of their inactivation on Ig gene hypermutation in the mouse.
| polymerase family | polymerase name | role in hypermutation | specific comments |
|---|---|---|---|
| A | Pol theta (θ) | no | conflicting data |
| Pol nu (ν) | not tested | ||
| B | Pol zeta (ζ) | not settled | embryonic lethal phenotype |
| X | Pol beta (β) | no | |
| Pol lambda (λ) | no | ||
| Pol mu (μ) | no | ||
| Tdt | no | ||
| Y | Pol eta (η) | yes | |
| Pol iota (ι) | no | ||
| Pol kappa (κ) | no | back-up of Pol eta in Pol eta-deficient animals | |
| Rev1 | yes | strand-biased contribution |
3. Polη as the sole contributor of A/T mutagenesis: a main player with substitutes
In 1998, the first studies of mismatch repair (MMR)-deficient mice uncovered a role of MSH2 in the generation of mutations at A/T bases, while, surprisingly, the absence of the effector part of the MMR complex (PMS2 and MLH1) had, at most, a simple quantitative impact (Frey et al. 1998; Phung et al. 1998, 1999; Rada et al. 1998; Ehrenstein et al. 2001). This dichotomy was documented further by the analysis of mice deficient for MSH6 and Exo1, the exonuclease involved in the excision of the patch containing the mismatch from an existing nick in DNA (Wiesendanger et al. 2000; Bardwell et al. 2004; Martomo et al. 2004), thus uncovering an unusual dissociation between the MMR partners and restricting the implication in the hypermutation mechanism to the MSH2–MSH6 mismatch binding part of the complex (Reynaud et al. 1999).
The second activity characterized as being involved in A/T mutagenesis was Polη, a then recently discovered translesion DNA synthesis (TLS) polymerase, through the analysis of patients with the variant form of the xeroderma pigmentosum syndrome (XPV), a genetic disease corresponding to the dysfunction of this enzyme (Zeng et al. 2001; Faili et al. 2004). However, there was a consistent residual A/T mutagenesis in both cases, particularly for XPV patients whose genetic defect is obviously variable, suggesting that the generation of A/T mutations might be more complex than a sole MSH2–MSH6 pathway recruiting Polη.
Inactivation of Polη in the mouse confirmed its major impact on A/T mutagenesis (Delbos et al. 2005; Martomo et al. 2005). However, analysis of the residual A/T mutagenesis in both Polη- and MSH2-deficient contexts uncovered a rather different mutation profile. In the Polη−/− background, these mutations evoked the signature of another polymerase, Polκ, which has an unusual error specificity resulting predominantly in T to G transversions (Ohashi et al. 2000; Zhang et al. 2000). The A/T pattern in Msh2−/− animals was by contrast compatible with the involvement of Polη in an UNG-mediated short-patch repair process with lower mutational efficiency (table 2; Delbos et al. 2005). The mutation pattern of Polη×MSH2-deficient animals, which showed a frequency of A/T mutations corresponding to the PCR background, confirmed this proposition. Such a process may only take place in the absence of MSH2, being competed out in the normal situation (see model below). This led us to propose that Polη is the only contributor of A/T mutations in the mouse (Delbos et al. 2007).
Table 2.
Mutation pattern of JH4 intronic sequences from Peyer's patch B cells with polymerase/repair-deficient backgrounds.
| within A/T mutations | ||||
|---|---|---|---|---|
| transitions | transversions | |||
| genetic background | G/C versus A/T mutations (percentage of total mutations) | A to GT to C | A to TT to A | A to CT to G |
| wild-type | 49 : 51 | 50 | 27 | 23 |
| Polη−/− | 84 : 16 | 19 | 25 | 56 |
| Msh2−/− | 90 : 10 | 36 | 38 | 27 |
| Msh2×Polη−/− | 99 : 1 | — | — | — |
Data from Delbos et al. (2007).
The back-up role proposed for Polκ was further confirmed by the analysis of Polη×Polκ double-deficient mice in which A/T mutagenesis was further reduced, although, in contrast to the previous situation, some A/T mutations were still detectable (Faili et al. submitted; table 1). It therefore suggests that, as long as the MSH2–MSH6 complex is present, other polymerases may still be recruited. However, it is difficult to discern a specific polymerase signature in this genetic configuration, due to the low number of A/T mutations collected even from large mutation databases. The last member of the Y family, Polι, could obviously be considered a logical candidate for this third line recruitment.
A similar polymerase back-up has been proposed to explain the increased mutagenicity of UV light observed in XPV patients, the polymerase acting as a substitute being unable, in contrast to Polη, to bypass the UV-induced cyclobutane thymidine dimers in an error-free fashion. The contribution of Polι to this mutagenic back-up has recently been documented in vitro in UV-irradiated cells deficient for either or both of these two polymerases (Dumstorf et al. 2006; Gueranger et al. 2008). The situation in hypermutation appears somehow as a mirror image; the Polκ polymerase used as a back-up is less mutagenic than the default one, explaining why, in spite of the similar bias for A/T mutations of these two enzymes, the A/T mutagenesis and consequently the total mutation load drops in Polη-deficient B cells (figure 1). This symmetrical situation illustrates well the ‘paradox’ of TLS polymerase usage in the immune system, contrasting their hierarchical selection for error-free repair during the bypass of DNA damages with their recruitment for error-prone repair during hypermutation (Weill & Reynaud 2008).
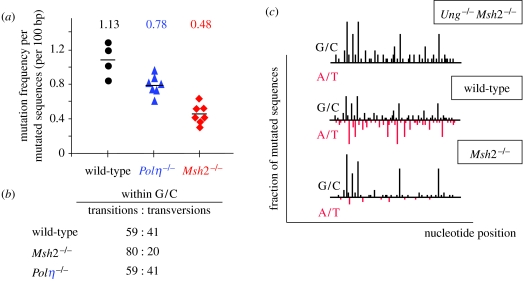
Figure 1.
Profound alteration of the mutation profile of MSH2-deficient B cells. (a) A two- to three-fold reduction in hypermutation in MSH2-deficient B cells. Mutation frequency of JH4 intronic sequences from Peyer's patch PNAhigh B cells was determined for individual mice. (b) A bias for G/C transitions in JH4 sequences from Peyer's patch B cells in MSH2-deficient animals. (c) A schematic of the distribution of mutations along the JH4 intronic sequence of Peyer's patch PNAhigh B cells, with G/C mutations plotted above the line representing the JH4 sequence and A/T mutations below. This conceptual representation illustrates the similar quantitative levels of G/C mutagenesis at hotspot positions between the Msh2×Ung−/− and the Msh2−/− backgrounds, while being greatly diminished at other G/C bases. G/C mutagenesis is overall lower in the wild-type context, in which approximately half of the deaminated cytosines are repaired to generate mutations at A/T bases. Mutation data taken from Rada et al. (2004) and Delbos et al. (2007).
Albeit being mainly error-prone opposite T nucleotides, Polη may also have a low mutagenicity at G/C bases, thus accounting for the residual 3–4% transversions at G/C bases observed in the UNG-deficient background (and for some G/C transitions that cannot obviously be distinguished from the uracils carried over replication; Rada et al. 2002; Di Noia et al. 2006).
4. Profound alteration of the mutation profile in MSH2-deficient mice
MMR and UNG-mediated pathways have been proposed as two alternative modes of processing of uracils during the somatic mutation process (Rada et al. 2004). There is, however, a clear asymmetry in these processes. UNG deficiency results in a small increase in mutagenesis at G/C bases (55–60 versus 45–50% in wild-type animals), which consists mostly in transitions (Rada et al. 2002). By contrast, A/T mutagenesis is not affected, establishing the complete independence of mismatch handling from any uracil removal activity. Most notably, there is no alteration of the overall targeting at either G/C or A/T bases.
By contrast, there is a profound alteration of the mutation profile in the MSH2-deficient context, which is not restricted to the sole reduction in A/T mutagenesis. These alterations include a lower mutation frequency, an increased proportion of transitions among G/C mutations and, most remarkably, a modified mutation profile at the V locus with a marked targeting at a few hotspot positions (figure 1; Frey et al. 1998; Phung et al. 1998; Rada et al. 1998). No comprehensive explanation had been put forward for this complex phenotype, which was also observed in MSH6- and Exo1-deficient animals (Bardwell et al. 2004; Martomo et al. 2004). Considering the mutation profile of Ung×Msh2−/− mice as the reference pattern of AID-mediated deaminations, we have therefore proposed that these alterations can be collectively explained by an increased error-free repair mediated by UNG when the MSH2–MSH6 pathway is not functional, with a fraction of uracils escaping this repair, notably at specific hotspot motifs (Delbos et al. 2007). A schematic of the mutation profile, comparing the wild-type, Msh2−/− and double Msh2×Ung−/− genetic backgrounds, is shown in figure 1c. This scheme highlights the contrasting impact of mutations in the MSH2-deficient context at a few hotspot positions (within a WGCW symmetrical context for most of them; Martomo et al. 2004), compared with the rest of the sequence. Most notably, the mutation frequency at these G/C hotspots is close to their deamination frequency, reflecting their conversion of deamination into mutation at the same position. This hotspot mutation frequency is by contrast approximately twofold lower in wild-type animals, thus suggesting that approximately half of the deaminations produced are converted by the MSH2–MSH6 pathway into mutations at nearby A/T bases. In the MSH2-deficient context, these hotspots are thus almost quantitatively prevented from being handled by UNG, while the rest of the sequence harbours a much higher level of error-free repair compared with the Ung×Msh2−/− profile. How and why are these hotspots protected remains unclear, but one could envision an increased stability of AID at a few deaminated sites, which would require the MSH2–MSH6 complex for efficiently dislodging it (Delbos et al. 2007).
5. Strand bias in hypermutation
It is often stated that AID targets both DNA strands equally in vivo (Franklin & Blanden 2008). However, considering again the AID targeting revealed by the mutation pattern of UNG–MSH2-deficient mice, the ratio of C to G mutations is in fact 60 : 40, i.e. slightly biased towards the non-transcribed (or coding) strand. This strand bias appears to be conserved by the UNG pathway (e.g. in Msh2−/− animals), but mutations at C and G are equalized in MSH2-proficient genetic backgrounds and translated into an A over T bias (table 3; Weill & Reynaud 2008).
Table 3.
The C over G targeting bias generated by AID at the V gene locus is eliminated by the MSH2–MSH6 pathway to generate an A to T mutation bias.
| genetic background | percentage of total mutations | |||
|---|---|---|---|---|
| C | G | A | T | |
| C over G bias (60 : 40) | ||||
| Ung−/−×Msh2−/−; Rada et al. (2002) | 61 | 38 | 1 | 0 |
| Msh2−/−; Rada et al. (2004) | 54 | 36 | 6 | 4 |
| Polη−/−×Msh2−/−; Rada et al. (2004) | 62 | 37 | 0 | 1 |
| no C/G bias–A over T bias (70 : 30) | ||||
| wild-type; Rada et al. (2004) | 25 | 25 | 34 | 16 |
| Ung−/−; Delbos et al. (2007) | 29 | 29 | 29 | 13 |
| Polη−/−; Rada et al. (2004) | 41 | 43 | 11 | 6 |
Polη is intrinsically two to three times more error-prone at copying Ts than As, but these figures have been obtained from an in vitro synthesis of several hundred bases (Pavlov et al. 2002), which may not exactly correspond to the in vivo situation. The strand bias at As over Ts may thus come from an imbalanced repair that combines several steps of unequal strand processing: an increased repair on the coding strand resulting in the equalization of C/G mutagenesis; a longer patch synthesis when copying the non-coding versus the coding strand (i.e. according to a DNA polymerase synthesis proceeding along the direction of elongation of the RNA polymerase complex versus backwards); the high error rate of Polη when copying Ts. This last feature appears to correspond not only to the specificity of Polη, but also, and as observed experimentally, to that of Polκ (table 3). This model would altogether favour an MSH2–MSH6 repair process operating on the U-containing strand rather than opposed to it.
This issue has recently been addressed by the analysis of a transgenic mutation substrate in which a single C residue was embedded in a long stretch of As and Ts (Unniraman & Schatz 2007). These authors concluded that only the coding strand was targeted for error-prone repair, thus proposing that the A/T bias would come from the specific mutagenic properties of Polη synthesizing only one DNA strand. However, the mutation data obtained in this study showed a strong bias for T mutations that do not correspond to the normal mutation pattern of V genes for which mutagenesis at As dominates, which makes this elegant approach somewhat inconclusive at this point.
6. Competitive rather than alternative repair pathways in hypermutation: a cell-cycle affair?
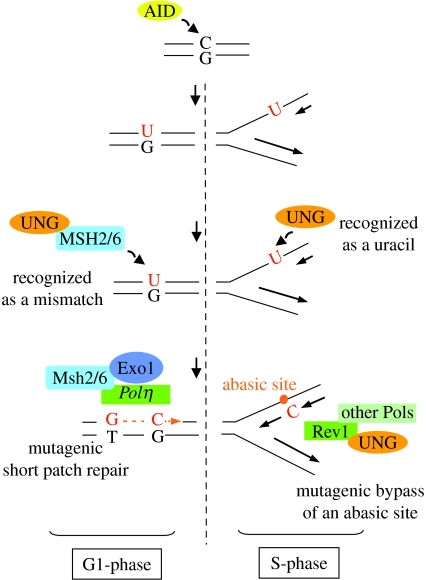
The model we wish to propose, in which many aspects remain speculative, aims at accommodating the relative independence of the MSH2–MSH6 pathway from UNG-mediated repair as observed in UNG-deficient animals, while the reverse situation, i.e. MSH2 or MSH6 deficiency, indicates that the MMR pathway strongly impinges on UNG activity (figure 2).
Figure 2.
Competitive repair pathways in hypermutation. See text for detailed comments.
One way to envision such a compartmentalization is a function that would rely on different phases of the cell cycle. AID expression is indeed not cell-cycle regulated, making it likely that cytosine deamination may take place at different phases of the cycle (Aoufouchi et al. 2008). By contrast, UNG is strongly upregulated in the late G1 and early S-phases, at which stage it can cope with misincorporated uracils during replication (Hagen et al. 2008). MMR would be normally considered as a post-replicative actor, but the restricted involvement of the sole MSH2–MSH6 and Exo1 partners precludes such a priori.
MSH2–MSH6 would thus recruit Polη (or its polymerase substitute) in G1, eliciting, after excision by Exo1 of a stretch of DNA containing the uracil, a DNA synthesis of sufficient length to generate approximately one distant mutation for each repaired uracil. The most elusive part of this process is the mode of DNA incision proximal to the uracil lesion, in the absence of the DNA discontinuities provided by replication and of the usual MLH1 and PMS2 effector partners. Effectively, no endonuclease activity that would specifically affect the A/T mutagenesis has been described so far. In the absence of MSH2–MSH6, the mismatches present in the late G1-phase would be handled by UNG, in an error-free or a low error-prone fashion, with a fraction of them escaping detection, notably at hotspot positions.
The action of UNG would be restricted to the S-phase, where it would mainly generate an abasic site without further repair, and these abasic sites would be bypassed by TLS polymerases acting, in contrast to Polη, in their classical damage tolerance function. The phenotype of Rev1 deficiency, with a major impact on C to G mutations and a minor one on G to C, is the perfect proof for the TLS role of Rev1, its unique signature of cytidine incorporation demonstrating a polymerization opposite an abasic site (Jansen et al. 2006). Its unequal strand contribution is so far unexplained, suggesting a strand-biased recruitment that may possibly correspond to a leading/lagging strand dichotomy of the TLS process.
The limited effect of Rev1 deficiency on G/C mutations obviously requires the implication of other TLS polymerases at the replication fork, generating notably G to T and C to A transversions. Some G/C transitions could be generated by TLS as well, according to the ‘A’ rule, a default pathway by which As are inserted opposite to non-instructive lesions (Goodman et al. 1993). The conditional inactivation of REV3L in mature B cells induces a large reduction in the overall mutation load that affects all types of mutations similarly, which has been interpreted as an indirect consequence of the reduced viability of centroblasts lacking REV3L activity (D. Schenten & K. Rajewsky 2008, personal communication). An alternative explanation would be that Polζ participates in the formation of all types of mutations, not only in the bypass of abasic sites, but also in the extension of the Polη patch synthesis. Such a global role remains possible, and it should be mentioned that our knowledge of the biochemical function of Polζ as a TLS extender enzyme relies only on experiments performed with the yeast protein, a quite different enzyme with half the size of mammalian REV3L (Gan et al. 2008).
Translesion bypass is recruited upon stalling of the replication fork, which induces the monoubiquitination of the proliferating cell nuclear antigen (PCNA) DNA sliding clamp, which, in turn, promotes its interaction with specific motifs of TLS polymerases (Kannouche et al. 2004). Unexpectedly, inactivation of the monoubiquitination site of PCNA (a Lys164 to Arg mutation) did not affect mutagenesis at G/C bases (apart for a small reduction in C to G mutagenesis), but, surprisingly, A/T mutagenesis was almost completely abolished, thus implying that Polη recruitment by the MSH2–MSH6 complex requires PCNA monoubiquitination (Langerak et al. 2007).
TLS can, however, be dissociated from PCNA ubiquitination, as shown recently in the DT40 cell line (Edmunds et al. 2008). Other DNA clamps have been described as well, which, as suggested by Langerak et al. (2007), could be mobilized instead of PCNA. The 9-1-1 complex, which interacts in yeast with Polζ (Sabbioneda et al. 2005), has recently been shown to mediate an alternative DNA damage response after becoming similarly monoubiquitinated by the Rad6–Rad18 complex (Fu et al. 2008). The precise molecular requirements for the replicative bypass of abasic sites in mammalian B cells thus remain to be determined.
7. Conclusions
Ig gene hypermutation is a unique physiological process, in which many repair pathways are developmentally mobilized in response to an internally induced DNA damage. Some of these pathways, e.g. the recruitment of TLS polymerases as active mutagenic agents, may be uniquely co-opted during the centroblastic programme of gene activation. Other unexpected aspects, such as the behaviour of PCNA for TLS polymerase recruitment, may rather reveal our incomplete knowledge of the in vivo DNA damage tolerance processes.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘DNA deamination in immunity, virology and cancer’.
References
- Aoufouchi S., Faili A., Zober C., D'Orlando O., Weller S., Weill J.C., Reynaud C.A. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. doi:10.1084/jem.20070950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell P.D., Woo C.J., Wei K., Li Z., Martin A., Sack S.Z., Parris T., Edelmann W., Scharff M.D. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 2004;5:224–229. doi: 10.1038/ni1031. doi:10.1038/ni1031 [DOI] [PubMed] [Google Scholar]
- Delbos F., De Smet A., Faili A., Aoufouchi S., Weill J.C., Reynaud C.A. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. doi:10.1084/jem.20050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F., Aoufouchi S., Faili A., Weill J.C., Reynaud C.A. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2007;204:17–23. doi: 10.1084/jem.20062131. doi:10.1084/jem.20062131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., Rada C., Neuberger M.S. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. doi:10.1038/sj.emboj.7600939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstorf C.A., Clark A.B., Lin Q., Kissling G.E., Yuan T., Kucherlapati R., McGregor W.G., Kunkel T.A. Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl Acad. Sci. USA. 2006;103:18 083–18 088. doi: 10.1073/pnas.0605247103. doi:10.1073/pnas.0605247103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds C.E., Simpson L.J., Sale J.E. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. doi:10.1016/j.molcel.2008.03.024 [DOI] [PubMed] [Google Scholar]
- Ehrenstein M.R., Rada C., Jones A.M., Milstein C., Neuberger M.S. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc. Natl Acad. Sci. USA. 2001;98:14 553–14 558. doi: 10.1073/pnas.241525998. doi:10.1073/pnas.241525998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A., Aoufouchi S., Weller S., Vuillier F., Stary A., Sarasin A., Reynaud C.A., Weill J.C. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med. 2004;199:265–270. doi: 10.1084/jem.20031831. doi:10.1084/jem.20031831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili, A., Stary, A., Delbos, F., Weller, S., Aoufouchi, A., Sarasin, A., Weill, J.-C. & Reynaud, C.-A. Submitted. An XPV syndrome without mutations in the coding sequence of DNA polymerase η suggests a dominant role of Pol η in immunoglobulin gene hypermutation.
- Franklin A., Blanden R.V. The strand bias paradox of somatic hypermutation at immunoglobulin loci. Trends Immunol. 2008;29:167–172. doi: 10.1016/j.it.2008.01.008. doi:10.1016/j.it.2008.01.008 [DOI] [PubMed] [Google Scholar]
- Frey S., Bertocci B., Delbos F., Quint L., Weill J.C., Reynaud C.A. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. doi:10.1016/S1074-7613(00)80594-4 [DOI] [PubMed] [Google Scholar]
- Fu Y., Zhu Y., Zhang K., Yeung M., Durocher D., Xiao W. Rad6–Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133:601–611. doi: 10.1016/j.cell.2008.02.050. doi:10.1016/j.cell.2008.02.050 [DOI] [PubMed] [Google Scholar]
- Gan G.N., Wittschieben J.P., Wittschieben B.O., Wood R.D. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. doi:10.1038/cr.2007.117 [DOI] [PubMed] [Google Scholar]
- Goodman M.F., Creighton S., Bloom L.B., Petruska J. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. doi:10.3109/10409239309086792 [DOI] [PubMed] [Google Scholar]
- Gueranger, Q., Stary, A., Aoufouchi, S., Faili, A., Sarasin, A., Reynaud, C.-A. & Weill, J.-C. 2008 Role of DNA polymerases eta, iota and zeta in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair (Amst.)7, 1551–1562. (doi:10.1016/j.dnarep.2008.05.012) [DOI] [PubMed]
- Hagen L., et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. doi:10.1038/sj.emboj.7601958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J.G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. doi:10.1084/jem.20052227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P.L., Wing J., Lehmann A.R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. doi:10.1016/S1097-2765(04)00259-X [DOI] [PubMed] [Google Scholar]
- Langerak P., Nygren A.O., Krijger P.H., van den Berk P.C., Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J. Exp. Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. doi:10.1084/jem.20070902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., Schatz D.G. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. doi:10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Gearhart P.J. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 2004;200:61–68. doi: 10.1084/jem.20040691. doi:10.1084/jem.20040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Wersto R.P., Ohkumo T., Kondo Y., Yokoi M., Masutani C., Hanaoka F., Gearhart P.J. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl Acad. Sci. USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. doi:10.1073/pnas.0501852102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Saribasak H., Yokoi M., Hanaoka F., Gearhart P.J. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst.) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. doi:10.1016/j.dnarep.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. doi:10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Ohashi E., Bebenek K., Matsuda T., Feaver W.J., Gerlach V.L., Friedberg E.C., Ohmori H., Kunkel T.A. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39 678–39 684. doi: 10.1074/jbc.M005309200. doi:10.1074/jbc.M005309200 [DOI] [PubMed] [Google Scholar]
- Pavlov Y.I., Rogozin I.B., Galkin A.P., Aksenova A.Y., Hanaoka F., Rada C., Kunkel T.A. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase eta during copying of a mouse immunoglobulin kappa light chain transgene. Proc. Natl Acad. Sci. USA. 2002;99:9954–9959. doi: 10.1073/pnas.152126799. doi:10.1073/pnas.152126799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. doi:10.1038/nature00862 [DOI] [PubMed] [Google Scholar]
- Phung Q.H., Winter D.B., Cranston A., Tarone R.E., Bohr V.A., Fishel R., Gearhart P.J. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. doi:10.1084/jem.187.11.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung Q.H., Winter D.B., Alrefai R., Gearhart P.J. Hypermutation in Ig V genes from mice deficient in the MLH1 mismatch repair protein. J. Immunol. 1999;162:3121–3124. [PubMed] [Google Scholar]
- Rada C., Ehrenstein M.R., Neuberger M.S., Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. doi:10.1016/S1074-7613(00)80595-6 [DOI] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. doi:10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., Neuberger M.S. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. doi:10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Reynaud C.A., Bertocci B., Frey S., Delbos F., Quint L., Weill J.C. Mismatch repair and immunoglobulin gene hypermutation: did we learn something? Immunol. Today. 1999;20:522–527. doi: 10.1016/s0167-5699(99)01540-6. doi:10.1016/S0167-5699(99)01540-6 [DOI] [PubMed] [Google Scholar]
- Sabbioneda S., Minesinger B.K., Giannattasio M., Plevani P., Muzi-Falconi M., Jinks-Robertson S. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:38 657–38 665. doi: 10.1074/jbc.M507638200. doi:10.1074/jbc.M507638200 [DOI] [PubMed] [Google Scholar]
- Unniraman S., Schatz D.G. Strand-biased spreading of mutations during somatic hypermutation. Science. 2007;317:1227–1230. doi: 10.1126/science.1145065. doi:10.1126/science.1145065 [DOI] [PubMed] [Google Scholar]
- Weill J.C., Reynaud C.A. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. 2008;8:302–312. doi: 10.1038/nri2281. doi:10.1038/nri2281 [DOI] [PubMed] [Google Scholar]
- Wiesendanger M., Kneitz B., Edelmann W., Scharff M.D. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2–MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. doi:10.1084/jem.191.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K., Rada C., Neuberger M.S. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. doi:10.1084/jem.20061067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Winter D.B., Kasmer C., Kraemer K.H., Lehmann A.R., Gearhart P.J. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2001;2:537–541. doi: 10.1038/88740. doi:10.1038/88740 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yuan F., Wu X., Wang M., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. doi:10.1093/nar/28.21.4138 [DOI] [PMC free article] [PubMed] [Google Scholar]