Abstract
The major histocompatibility complex (MHC) is a dense region of immune genes with high levels of polymorphism, which are arranged in haplotype blocks. Traditional models of balancing selection (i.e. overdominance and negative frequency dependence) were developed to study the population genetics of single genes. However, the MHC is a multigene family surrounded by linked (non-neutral) polymorphisms, and not all of its features are well explained by these models. For example, (i) the high levels of polymorphism in small populations, (ii) the unexpectedly large genetic differentiation between populations, (iii) the shape of the allelic genealogy associated with trans-species evolution, and (iv) the close associations between particular MHC (human leucocyte antigen, HLA) haplotypes and the approximately 100 pathologies in humans. Here, I propose a new model of MHC evolution named Associative Balancing Complex evolution that can explain these phenomena. The model proposes that recessive deleterious mutations accumulate as a ‘sheltered load’ nearby MHC genes. These mutations can accumulate because (i) they are rarely expressed as homozygotes given the high MHC gene diversity and (ii) purifying selection is inefficient with low recombination rates (cf. Muller's ratchet). Once fixed, these mutations add to balancing selection and further reinforce linkage through epistatic selection against recombinants.
Keywords: major histocompatibility complex, human leucocyte antigen, balancing selection, linkage disequilibrium, disease associations, plant self-incompatibility loci (S-loci)
1. Introduction
The major histocompatibility complex (MHC) has important biological functions with respect to resistance to infectious diseases, mate choice, kin recognition and reproductive success (Bernatchez & Landry 2003; Piertney & Oliver 2006). Selection by parasites and sexual selection are thought to maintain the unusually high levels of MHC polymorphism, and two types of balancing selection have been proposed (Piertney & Oliver 2006). The overdominance model states that polymorphism is maintained because heterozygous individuals are able to recognize a wider variety of parasites (Doherty & Zinkernagel 1975). The negative frequency-dependent selection model assumes that, because selection favours parasites that can avoid recognition by the most common MHC variants, rare MHC alleles offer better parasite resistance (Clarke & Kirby 1966). However, a number of characteristics of MHC and its evolution are not well explained by these traditional models of balancing selection, and these observations will be briefly discussed next.
Selection pressures by parasites tend to vary across generations (Little 2002). In episodes with little or no parasite selection, the MHC polymorphism is prone to erosion by random genetic drift, particularly in small populations. Traditional models of balancing selection, which assume parasites are the only agent of selection, often require unrealistically large selection coefficients (S≥0.2) to explain the observed MHC polymorphism in small populations (Aguilar et al. 2004; van Oosterhout et al. 2006). This suggests that additional selective forces might be operating on the MHC, which help to maintain this high level of polymorphism.
Genes under balancing selection are expected to display a greater effective migration rate than neutral genes (Muirhead 2001). Consequently, traditional models predict that the MHC should show much less apparent population subdivision than neutral genes (Takahata 1993). Several studies have compared the level of population divergence (FST or GST) based on the MHC and neutral microsatellite loci and found that both type of loci displayed surprisingly similar levels of genetic differentiation (Bernatchez & Landry 2003). To account for this discrepancy between the theory and the data, it has been suggested that (i) microsatellite differentiation is misleadingly low (Hedrick 1999), (ii) selection at the MHC loci studied is very weak (s≪0.01) and the alleles effectively neutral, or (iii) local adaptations increase the population differentiation at the MHC (Richman et al. 2003).
Some MHC alleles are shared between long-diverged species, a phenomenon known as trans-species polymorphism (Klein et al. 1993; Bos & Waldman 2006). Genes that display trans-species polymorphisms are distinct from other (neutrally evolving) genes in two important aspects: the age (or coalescent time) of extant alleles and the shape (or topology) of the allelic genealogy. The traditional models show that the coalescent time of alleles under balancing selection can span many millions of years, which is much older than that of neutral alleles (Takahata 1990; Takahata & Nei 1990). This theoretical prediction is thus consistent with this particular aspect of trans-species polymorphism. Takahata (1990) showed, however, that the topology of an allelic genealogy under balancing selection is similar to that of a neutral gene genealogy but with a different time scale. He derived a constant scaling factor with which the allelic genealogy of an overdominant locus resembles that of a neutral genealogy (Takahata 1990). A similar scaling factor has been derived for negative frequency dependence at a gametophytic S-locus by Vekemans & Slatkin (1994).
Crucially, however, genealogies of MHC genes (Richman et al. 2001; Dorak et al. 2002), gametophytic plant self-incompatibility genes (Uyenoyama 1997) and mating type genes of fungi (May et al. 1999) all show a consistent deviation in their topology in that their terminal branches are significantly too long (Richman 2000). This implies that the alleles are not only relatively old, but also that the extant alleles are too much diverged from each other. The evolutionary process that is responsible for long terminal branches of balanced genealogies has been the subject of much debate (Richman 2000). Wakeland et al. (1990) proposed that the persistence of highly divergent MHC alleles over millions of years could be explained when assuming that the highly differentiated alleles confer an advantage in presenting a broader spectrum of antigens to the immune system (see also She et al. 1990). Uyenoyama (2003, 2005) suggested that the long terminal branches in the S-allele genealogies are consistent with the expression of recessive deleterious mutations at sites closely linked to the S-locus in zygotes that are produced by two closely related S-alleles. Since mutant alleles are incompatible with their parental allele, it reduces the branching process (rate of bifurcation) of S-allele lineages (Uyenoyama 2003, 2005).
Traditional models of MHC evolution based on overdominance and negative frequency-dependent selection were developed to understand the evolution and population genetics of a single immune gene. These models ignore the potential role of linkage and epistatic gene–gene interactions. However, the genomic structure of the human MHC (human leucocyte antigen, HLA) is characterized by islands of high linkage disequilibria (LD) interspersed by recombination hot spots, which predispose the MHC to epistasis and genetic hitch-hiking. Epistasis is evident from the differences in disease phenotype caused by distinct combinations of alleles at multiple loci (Gregersen et al. 2006). Furthermore, the MHC genes are surrounded by linked genetic variation (single nucleotide polymorphisms, SNPs) that is associated with more diseases than any other part of our genome (de Bakker et al. 2006; Shiina et al. 2006). This suggests that this linked peri-MHC region is under strong selection and that it could play a potentially important role in the evolution of this multigene family.
Here, I propose a new model of MHC evolution, inspired by theoretical studies of self-incompatibility loci (S-loci) of plants (Uyenoyama 2003, 2005). The model incorporates the impact of selection on the region surrounding the MHC genes. It proposes that (i) recessive disease-causing mutations can accumulate in the MHC in a process similar to Muller's ratchet (Muller 1932) and (ii) natural selection fails to purge these mutations because this so-called ‘sheltered load’ (Stone 2004) only rarely becomes expressed, given the large number of MHC alleles and high gene diversity. Next, I will test predictions made by the model in relation to the disease associations and haplotype block structure of the human MHC (the HLA), the shape of the genealogy associated with trans-species evolution and the population genetics of the MHC in wild populations.
2. Material and methods
(a) Simulation model
Monte Carlo simulations were used to study Associative Balancing Complex (ABC) evolution in diploid sexual populations. The model included one locus under symmetric overdominant selection with selection coefficient (S). de Boer et al. (2004) found that overdominance fails to explain the high degree of MHC polymorphism unless all MHC alleles confer very similar fitness values. However, with identical fitness values for all alleles, symmetric overdominance can maintain a similar level of polymorphism as negative frequency-dependent selection (see also Takahata & Nei 1990). Therefore, in the present study, symmetric overdominance was used to simulate balancing selection.
The overdominant gene mutated with a mutation rate (μ). In the linked genomic region (recombination rate c), recessive deleterious mutations accumulated with selection and dominance coefficient (s and h), and total mutation rate (U) within the entire haplotype block. Fitness was multiplicative across loci.
(b) Model validation
The computer model was validated by comparing the simulation results with theoretically predicted values (Crow & Kimura 1970). The decline in theoretical heterozygosity at generation t was calculated as Het=H0 Π ({1−[1/(2Net+1)]}), where Het is the expected heterozygosity at generation t; H0 is the expected heterozygosity at t=0; and Net is the effective population size at generation t. The decay in LD was calculated as LD=(1−c)t, where c is the recombination rate and t is the generation number. The theoretical predicted fitness values in a mutation–selection balance were calculated as w=e−U for completely recessive mutations (h=0) and w=e−2U for co-dominant mutations (h=0.5), where U is the total mutation rate across the linked region (i.e. haplotype block). The effective number of overdominant alleles (ne) was estimated using ne≈2(NeS)(1/2)/(4.6 log{0.4/[2Neμ/(NeS)(1/2)]})(1/2), when 2Neμ/(NeS)(1/2)<0.1 and ne≈3.7Neμ+(NeS)(1/2), when 2Neμ/(NeS)(1/2)≥0.1, (Crow & Kimura 1970; see figures S5–S8 in the electronic supplementary material).
(c) Epistasis
The effects of positive epistasis on the reinforcement of LD was analysed by simulating haplotypes consisting of two haplotype blocks with given genetic loads of completely recessive (h=0), deleterious mutations (s=0.1–0.4). Both blocks were separated by a recombination hot spot (c=0.01). LD between the haplotype blocks were calculated for every 50 generations over 500 generations (see figure S9 in the electronic supplementary material).
(d) Trans-species polymorphism
The impact of ABC evolution on temporal genetic variation and trans-species polymorphism was analysed by comparing genealogies for genes under symmetric overdominant selection and neutral genes with haplotypes subject to ABC evolution. Populations with size Ne=1000 had a single overdominant (or neutral) gene with mutation rate μ=10−5, and overdominant selection S=0.05 (or S=0). Deleterious mutations (c=0, h=0 s=0.01) accumulated with rate U=10−3. Simulations were stopped when all haplotypes were fixed for approximately one lethal equivalent, i.e. a total mutational load that would be lethal if expressed in homozygotes (Lynch & Walsh 1998). This load was reached after approximately 3×105–1×106 generations.
(e) Population genetics of ABC evolution
I examined whether ABC evolution could explain the high levels of polymorphism detected in small wild populations by simulating the demography of San Nicolas Island foxes (Urocyon littoralis dickeyi; Aguilar et al. 2004) and guppies (Poecilia reticulata; van Oosterhout et al. 2006). Demographic parameters and genetic data reported in these studies were used to establish the selection coefficients required to explain the observed level of MHC polymorphism. Guppy populations were simulated with effective population size Ne=104 and rate of gene flow 2Nm=0.126 (van Oosterhout et al. 2006). Guppies are known to experience strong parasite-mediated selection at the start of wet season rains (van Oosterhout et al. 2007a) and have a generation time of less than six months. Hence, the simulated overdominant selection operated in every alternating generation.
The fox population experienced a two-generation single-pair bottleneck, followed by 16 generations of population growth (with r=0.28) to a final size of n=104 individuals. This bottleneck scenario was necessary to explain monomorphism in 18 microsatellite loci (Aguilar et al. 2004; see text S2 in the electronic supplementary material). Overdominant selection (S=0–0.9) operated during the actual bottleneck generations only (Aguilar et al. 2004). The simulated values of expected heterozygosity and allelic richness were compared with the observed reported values (Aguilar et al. 2004; van Oosterhout et al. 2006).
The effects of ABC evolution and overdominant selection on population differentiation were analysed by simulating a source-sink metapopulation and calculating GST over generations. A sink population with Ne=5000 received unidirectional migration with rate 2Nm=1 from an infinitely large source population. Selection coefficients were S=0.5 for overdominance, and a genetic load of 0.5 lethal equivalents was used to simulate ABC evolution.
3. Results and discussion
(a) Verbal model
The effective rate of recombination in the MHC is reduced by functional epistasis (Gregersen et al. 2006) and haplotype block (haploblock) structure (Stenzel et al. 2004). This reduces the efficiency of purifying selection to remove recessive deleterious mutations (Haddrill et al. 2007). Some of those mutations become fixed in all copies of a particular haploblock in a process analogous to Muller's ratchet (Muller 1932). Selection against these mutations is further hampered by the fact that the MHC typically has a high gene diversity and large number of alleles (Piertney & Oliver 2006). This means that recessive deleterious mutations only rarely become expressed in a homozygous state, and that they constitute a so-called sheltered load (Stone 2004). However, if expressed, such mutations may cause disease, particularly if they occur in neighbouring functional genes that hitch-hike with the polymorphic MHC gene. Consistent with the model, disease-associated haplotypes linked to the human MHC (the HLA) tend to differ from other haplotypes by multiple SNPs (Shiina et al. 2006), and disease mutations are partially recessive (Gaya et al. 2006). The large number of disease-associated mutations in the MHC (de Bakker et al. 2006) and the linked peri-MHC region (Okamoto et al. 2003; Shiina et al. 2006) thus help to maintain the MHC polymorphism. Assuming that deleterious mutations occur in functional genes, the model predicts that there are more diseases associated to regions with high gene content (such as the HLA-B and HLA-C loci) than to less gene-dense regions (e.g. the HLA-A). This prediction is consistent with the data on disease mapping in humans (Shiina et al. 2006).
The population genetic effects of the accumulated recessive deleterious mutations are twofold (figure 1). First, it increases selection against individuals that are homozygous for the same haploblock. Consequently, if a haplotype becomes very common so that it regularly occurs in a homozygote condition, purifying selection will reduce its frequency. Selection against homozygous mutants can thus maintain a balanced MHC polymorphism, even without strong selection by parasites. Second, it leads to the reinforcement of LD in haploblocks by epistatic selection. Positive epistasis between recessive deleterious mutations in the same haplotype reduces the effective recombination rate (figure 2). The recombinant haplotypes are selected against because they express part of their genetic load when combining with either parental haplotype. Mutations continue to accumulate, which further increases balancing selection and reinforces linkage by epistatic selection against recombinants.
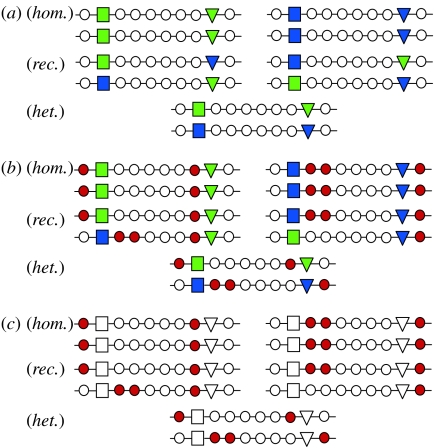
Figure 1.
Schematic of ABC evolution. Positive epistatic selection maintains moderate linkage between combinations of functional immune genes (coloured squares and triangles) that work well together (Gregersen et al. 2006). At (a) t=t0: the homozygotes (hom.) show a low fitness (overdominant selection), the recombinants (rec.) a low fitness (epistatic selection) and the heterozygotes (het.) a high fitness (heterosis). At (b) t=t1: recessive deleterious mutations (red circles) accumulate in haploblocks because purifying selection is inefficient when recombination rates are low (Haddrill et al. 2007). These mutations fix in a process similar to Muller's ratchet (Muller 1932). Once fixed, these mutations increase the efficacy of balancing selection and further reinforce linkage through epistatic selection against recombinants. At t=t1, the homozygotes have a very low fitness (overdominant and purifying selection), the recombinants a very low fitness (epistatic and purifying selection) and heterozygotes a high fitness (heterosis and no purifying selection). At (c) t=t2: even in the absence of balancing selection on the actual immune genes (open squares and triangles), purifying selection operates against the recessive deleterious mutations when expressed in homozygous condition, thus maintaining the polymorphism in the population. Homozygotes and recombinants have a low fitness (purifying selection), and heterozygotes have a high fitness (no purifying selection).
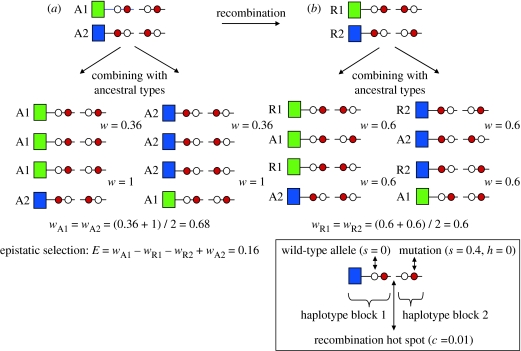
Figure 2.
Epistatic selection (E) against the recombinant haplotypes can prevent the breakdown of LD by recombination between haploblocks. In this example, there are two haploblocks separated by a recombination hot spot with a recombination rate (c=0.01). Each haplotype block contains a genetic load unique to the block (red circles), consisting of a completely recessive deleterious mutation (s=0.4, h=0). The immune genes (squares) have no fitness effect. The fitness effects of mutations are multiplicative across loci. The fitness of individuals homozygous for both blocks equals w=(1−0.4)2=0.36. Completely heterozygous individuals have a fitness of w=1. Recombinant haplotypes have a fitness equal to w=(1−0.4)=0.6, because they always carry one mutant in homozygous condition. This assumes that recombinants are rare and combine with ancestral haplotypes only. The strength of epistatic selection is given by the fitness difference between the (a) ancestral and (b) recombinant haplotypes (E=1−0.6−0.6+0.36=0.16) (see Crow & Kimura 1970).
(b) Epistasis
Once epistatic selection significantly exceeds the recombination rate, it can prevent the breakdown of LD between two haploblocks and extinguish recombination hot spots (see figure S9 in the electronic supplementary material). The accumulation of those linked mutations increases LD and may result in the expansion of haplotype blocks. This could explain the region of recombination suppression found surrounding the S-loci in plants (Uyenoyama 2005; Charlesworth et al. 2006).
During ABC evolution, epistasis purges recombinants because they are selected against when unifying with either parental haplotype (figure 2; see figure S9 in the electronic supplementary material). The recombinants have a low fitness because they share deleterious mutations with both parental haplotypes (these mutations are indicated by the red circles in figure 2). This results in negative epistasis and reduces the effective recombination rates. The model thus explains why evolutionarily successful recombination events between deeply diverged haplotypes in the human MHC class II region have been relatively rare (Raymond et al. 2005). ABC evolution furthermore appears to be consistent with the genomic architecture of the HLA in that only ‘hot’ recombination spots will be maintained, while epistatic selection extinguishes rare recombination events. This can create polymorphic frozen blocks in islands of high LD that hinder the identification of the actual disease-causing mutations (Dawkins et al. 1999).
(c) Trans-species polymorphism
Computer simulations were run to analyse the allelic genealogy of haplotypes subjected to ABC evolution, overdominance and neutral evolution (figure 3a–c). For ABC evolution, the selection coefficients of the recessive deleterious mutations were s=0.01. Overdominance and neutral evolution were simulated with S=0.05 and S=0, respectively. In figure 3a–c, the overall divergence among extant alleles can be calculated by summing the mutations accumulated since coalescence (on the x-axis). The coalescence time of alleles can be found by observing the age of the last common ancestor (on the y-axis). In the ABC genealogy, the new mutant and parental haplotypes coexisted for very short periods (dotted lines in figure 3a), a finding that is consistent with the results of the model for self-incompatibility in the S-locus of flowering plants (Uyenoyama 2003, 2005). Some alleles diverged by only two mutations from the ancestral allele and persisted for t>100Ne generations in the population (e.g. H1 in figure 3a). The ABC genealogy is characterized by long terminal branches. This is reminiscent of trans-species polymorphism (Klein et al. 1993; Bos & Waldman 2006) and the topology is similar to the genealogies of S-loci of plants (Uyenoyama 1997), mating type genes of fungi (May et al. 1999) and MHC genes in vertebrates (Richman et al. 2001; Dorak et al. 2002). At generation 3×105, the extant alleles of the ABC genealogy differed from each other by a combined total of 30 mutations. This compares with a divergence of only six mutations among the extant alleles in the overdominant genealogy (figure 3a,b; see table S1 in the electronic supplementary material).
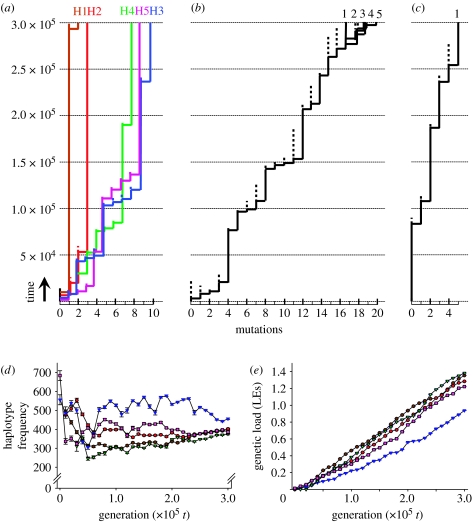
Figure 3.
Genealogies, haplotype frequencies and genetic loads of simulated populations over 3×105 generations. Each step in the genealogies (a–c) represents a mutation in the MHC gene. Total number of accumulated MHC mutations and time are shown on the x- and y-axes, respectively. Dotted vertical lines represent the ancestral MHC allele that is replaced by its derived mutant (solid vertical line). Only ancestral alleles with extant descendants are shown. (a) ABC evolution results in long terminal branches and large differentiation between extant alleles. (b) Overdominant selection results in a rapid turnover rate of alleles and little allelic differentiation. (c) Neutral evolution results in low level of polymorphism in the population (see table S1 and text S1 in the electronic supplementary material). (d) Marked unevenness in haplotype frequencies in the population under ABC evolution is consistent with the empirical data on the MHC (Richman 2000). (e) Deleterious mutations continue to accumulate in each haplotype, which might drive the birth and death process of multigene evolution (Nei & Rooney 2005). Brown circles, H1; red circles, H2; blue triangles, H3; green triangles, H4; pink squares, H5.
What causes the marked difference in the shape of the allelic genealogy of genes evolving under balancing selection and ABC evolution? During overdominant selection, alleles can be replaced by any new MHC mutant, and this increases the allelic turnover rate and reduces the maximum persistence time of alleles. All extant alleles have an approximately similar probability to go extinct once a mutant MHC allele has invaded. By contrast, with ABC evolution, the invasion of a new mutant MHC allele does not reduce the persistence time of other extant haplotypes in the population. Rather, novel MHC mutants tend to go extinct, or they replace their own parental haplotypes with which they share the same deleterious mutations. The evolution of each haplotype lineage (e.g. H1–H5 in figure 3a) is virtually independent from the population genetic processes (i.e. the mutation–selection–drift balance) taking place in the other haplotype lineages. Consequently, ABC evolution results in longer terminal branches and more allelic differentiation than overdominance.
Allele frequency distributions are markedly uneven in populations when simulating ABC evolution (figure 3d). Although there are several explanations for the unevenness of allele frequency distributions, such as amplification bias of certain MHC alleles, asymmetric overdominance and selective inequality of alleles, it is also predicted by the ABC model. According to this model, the unevenness is caused by differences between haplotypes in their genetic load (figure 3e). Haplotypes with a relatively low genetic load showed a high frequency (e.g. H3 in figure 3d,e) because purifying selection was not as stringent for individuals that were homozygous for such haplotypes. The simulated data are thus consistent with the observation that while many MHC alleles tend to be maintained at appreciable frequencies, their distributions are significantly uneven in populations (Richman 2000).
ABC evolution predicts that deleterious mutations continue to accumulate in each haplotype (figure 3e). It might be responsible for the large number of MHC pseudogenes found in the genomes of many vertebrates and drive the birth and death process of multigene evolution (Nei & Rooney 2005). Possibly, gene conversion could reduce genetic loads if a short track of sequence is converted by a donor sequence without mutations, or if interallelic gene conversion occurs between paralogous genes (Ohta 1989; Chen et al. 2007). This generates new variation in genetic load within a haplotype lineage, which can be used by natural selection to remove the haploblock copies with the heaviest load. Gene conversion can thus potentially reverse Muller's ratchet.
(d) MHC polymorphism
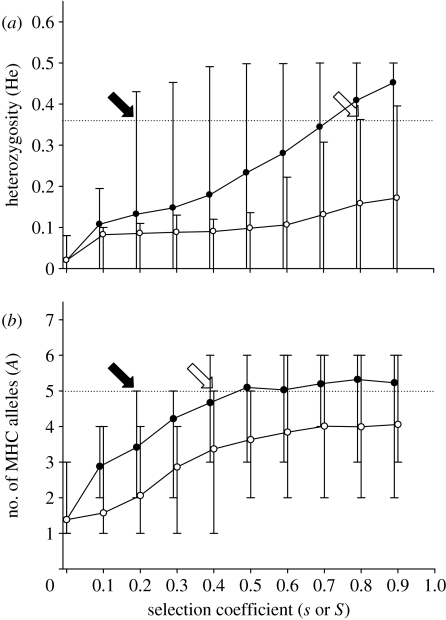
ABC evolution is particularly efficient in maintaining MHC polymorphism because purifying selection acts in all generations, independent of temporal fluctuating selection pressures by parasites. Simulation of ABC evolution of the MHC in foxes showed that genetic polymorphism observed by Aguilar et al. (2004) can be maintained with moderate selection coefficients (purifying selection s≥0.2 across all linked loci; figure 4a). However, near-lethal selection against homozygotes (overdominant selection S≥0.8) acting in the bottleneck generations was required to explain the observed level of MHC polymorphism (figure 4a; see also Aguilar et al. 2004).
Figure 4.
Genetic diversity with ABC evolution (filled circles) and overdominant selection (open circles) in simulated fox and guppy populations across a range of selection coefficients. Selection coefficients necessary to explain the observed MHC diversity are considerably lower under continual selection with ABC evolution than under temporary overdominant selection. (a) Mean (5–95% CL) expected heterozygosity (He) at the MHC in simulated populations of the fox (U. l. dickeyi) after a two-generation single-pair bottleneck. The heterozygosity after the bottleneck (He=0.36, Aguilar et al. 2004) is indicated by the dotted line. Genetic polymorphism in simulated fox populations can be maintained with selection coefficients (s≥0.2) with ABC evolution (black arrow). However, considerably stronger selection (S≥0.8) is necessary to explain the observed gene diversity (open arrow), assuming that overdominant selection operates during the bottleneck generations only (Aguilar et al. 2004). (b) The mean (5–95% CL) number of simulated MHC alleles in samples of 21 guppies (P. reticulata) in populations under ABC evolution (purifying selection every generation) and overdominant selection (operating in alternating generations) (van Oosterhout et al. 2007a). The dotted line at A=5 shows the number of observed alleles at each locus (van Oosterhout et al. 2006). Selection coefficients s≥0.2 (ABC evolution, black arrow) and S≥0.4 (overdominance, open arrow) are required to explain the observed MHC polymorphism.
Also, without the selection pressure by parasites (S=0), simulated populations can maintain a high level of MHC polymorphism with ABC evolution. For example, the simulated MHC polymorphism is similar to that observed in wild guppies (see van Oosterhout et al. 2006) when purifying selection s=0.2 acts on the linked mutations (figure 4b). This selection intensity translates in a genetic load of 0.2 lethal equivalents, which is only a fraction of the genetic load observed in most vertebrates (Lynch & Walsh 1998), and falls well within the load estimated for this guppy population (van Oosterhout et al. 2007b). Assuming that intense parasite selection operates only during heavy rains at the start of the wet season (van Oosterhout et al. 2007a), strong overdominant selection can only act every alternate generation. Consequently, a much higher overdominant selection coefficient (S≥0.4) is required to maintain the observed variation at the MHC (figure 4b). Combinations of (moderate) overdominance (S=0.2) and purifying selection (s=0.15) with ABC evolution are also consistent with the observed MHC polymorphism.
(e) Temporal fluctuations in MHC allele frequencies
Balancing selection can thus be partitioned into overdominant selection (operating on the immune genes) and purifying selection (on the linked recessive deleterious mutations). This implies that ABC evolution can moderate allele frequency fluctuations over time. The increase in frequency of a ‘high-resistance’ allele by positive selection is counteracted by purifying selection against this haplotype when common, because it then often expresses its recessive deleterious mutations in homozygote condition. This evolutionary ‘cost of resistance’ might explain why only relatively few studies detect genotype frequency changes in response to parasite selection (Little 2002; but see Westerdahl et al. 2004), and why temporal patterns of host–parasite coevolution are difficult to demonstrate (Woolhouse et al. 2002). Indeed, the coevolutionary arms race between hosts and parasites rarely results in fixation of a single genetic variant that confers superior resistance, but rather, in a dynamic ‘Red Queen’ process (Bell 1982). ABC evolution can operate alongside other evolutionary mechanisms and contribute towards the dynamics of host–parasite coevolution.
(f) Population differentiation of the MHC
According to the traditional models of balancing selection, genes experiencing overdominance or negative frequency-dependent selection are expected to show little population differentiation (Takahata 1993). However, this prediction is discordant with many empirical MHC studies (Muirhead 2001; Bernatchez & Landry 2003; Richman et al. 2003; see §1). As predicted by theory, simulations show that allele frequencies of genes under overdominant selection quickly become homogenized in large populations (Ne=5000) that receive migrants (see figure S10 in the electronic supplementary material). However, these populations remain genetically differentiated over considerable time during ABC evolution (GST≫0). The reason is that although the MHC alleles may have diverged, the linked haploblock of MHC alleles with common ancestry will continue to share their ancestral deleterious mutations. A novel MHC allele carried by a migrant will be amidst diverged MHC alleles with a largely similar sheltered load. Consequently, a novel immigrant allele will be selected against when unifying with a native allele of common ancestry, as many of its recessive deleterious mutations will become expressed. The sharing of a similar sheltered load retards the homogenization of gene pools, which means that both populations can remain genetically differentiated for their MHC despite ongoing gene flow. It has been suggested that local adaptation to different parasite communities may explain the unexpectedly large differentiation of the MHC (e.g. Bernatchez & Landry 2003). ABC evolution offers an alternative (complementary) explanation for this finding.
(g) Applicability of ABC evolution
The relative importance of ABC evolution is likely to vary between taxa. Recent comparative genomic studies of complex diseases show that the MHC is also associated with genetic disorders in non-human vertebrates (Kennedy et al. 2006, 2008; Mangalam et al. 2008). The population genetics of the immune genes of mammals other than humans may thus be affected by the ABC evolution as well.
In teleosts, many MHC genes (or MH genes) have been scattered throughout the genome by translocation events (Kuroda et al. 2002). Nevertheless, certain segments containing teleost MH show a similarly high gene density as the human MHC, and the genes involved in antigen presentation are linked in the so-called ‘core’ regions (Palti et al. 2007). Comparative studies suggest that the clustering of these MH genes is the result of selection pressure requiring coevolution and coregulation of the MHC-related genes (Kelley et al. 2005), which theoretically makes this genomic organization also predisposed to the ABC evolution.
Noteworthy is the polymorphic B-locus in the MHC of chicken, which is simple and compact, with 19 genes found in 92 kilobases of DNA (Kaufman et al. 1999). This compact structure is hypothesized to lead to very tight coevolution (Kaufman et al. 1999). Alternatively, natural selection might favour a compact MHC with few genes as this reduces the target of mutation and, hence, the mutational load. Interestingly, compared with the mammalian MHC, the chicken MHC is known to lead to stronger associations with resistance and susceptibility to infectious pathogens (Kaufman et al. 1999). Perhaps in chickens, the signal of parasite-mediated selection is not obscured by the ABC evolution and selection acting on the deleterious mutations linked to the MHC.
The ‘minimum essential’ MHC of chickens (and other birds) suggests that the ABC evolution is not a prerequisite for polymorphism, and that traditional models of balancing selection may offer a better explanation for high gene diversity in such cases. Furthermore, I propose that the ABC evolution can act alongside balancing selection on the actual immune genes, and that compared with the existing models, it can offer a plausible explanation for several remarkable aspects of the MHC.
(h) Future research
The model of ABC evolution makes predictions that can be tested using various approaches. For example, reduced parasite pressure and the absence of mate choice in ‘parasite-free’ captive populations are not predicted to result in a neutral decline in genetic variation, as selection continues to act on the mutations that are linked to the MHC. Zygotes produced in crosses between parents with identical MHC alleles are predicted to deviate from Mendelian ratios also in the absence of parasites and mate choice. These deviations can be used experimentally to estimate the haplotype-specific genetic load or sheltered load (Stone 2004). Pre- and postcopulatory sexual selection, including the abortion of MHC homozygous embryos, may be an important proximate mechanism that facilitates ABC evolution. ABC evolution predicts that the level of population differentiation (FST) depends on the demography and history of both populations. Furthermore, different MHC (or HLA) alleles with a common phylogenetic history may express shared deleterious mutations in homozygous condition, which, according to the model, could result in genetic disorders or pathologies similar to those observed in homozygous individuals. ABC evolution might also be applicable to other gene families with haplotype block structure and/or genes under balancing selection, such as the disease resistance (R) genes, self-incompatibility genes in plants (Uyenoyama 1997) and mating-type genes of fungi (May et al. 1999).
Acknowledgments
The author would like to thank Deborah Charlesworth, David Weetman, seven anonymous reviewers and members of the Molecular Ecology laboratory of the University of Hull for their valuable comments and discussions.
Supplementary Material
Model validation, and the analysis of epistasis and genealogies of trans-species evolution
References
- Aguilar A., Roemer G., Debenham S., Binns M., Garcelon D., Wayne R.K. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl Acad. Sci. USA. 2004;101:3490–3494. doi: 10.1073/pnas.0306582101. doi:10.1073/pnas.0306582101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. University of California Press; Berkeley, CA: 1982. The masterpiece of nature: the evolution and genetics of sexuality. [Google Scholar]
- Bernatchez L., Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. doi:10.1046/j.1420-9101.2003.00531.x [DOI] [PubMed] [Google Scholar]
- Bos D.H., Waldman B. Evolution by recombination and transspecies polymorphism in the MHC class I gene of Xenopus laevis. Mol. Biol. Evol. 2006;23:137–143. doi: 10.1093/molbev/msj016. doi:10.1093/molbev/msj016 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Kamau E., Hagenblad J., Tang C.L. Trans-specificity at loci near the self-incompatibility loci in Arabidopsis. Genetics. 2006;172:2699–2704. doi: 10.1534/genetics.105.051938. doi:10.1534/genetics.105.051938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-M., Cooper D.N., Chuzhanova N., Férec C., Patrinos G.P. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 2007;8:762–775. doi: 10.1038/nrg2193. doi:10.1038/nrg2193 [DOI] [PubMed] [Google Scholar]
- Clarke B.C., Kirby D.R.S. Maintenance of histocompatibility polymorphism. Nature. 1966;211:999–1000. doi: 10.1038/211999a0. doi:10.1038/211999a0 [DOI] [PubMed] [Google Scholar]
- Crow J.F., Kimura M. Harper & Row Publishers; New York, NY: 1970. An introduction to population genetics theory. [Google Scholar]
- Dawkins R., Leelayuwat C., Gaudieri S., Tay G., Hui J., Cattley S., Martinez P., Kulski J. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol. Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. doi:10.1111/j.1600-065X.1999.tb01399.x [DOI] [PubMed] [Google Scholar]
- de Bakker P.I.W., et al. A high resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. doi:10.1038/ng1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer R.J., Borghans J.A.M., Van Boven M., Keşmir C., Weissing F.J. Heterozygote advantage fails to explain the high degree of polymorphism of the MHC. Immunogenetics. 2004;55:725–731. doi: 10.1007/s00251-003-0629-y. doi:10.1007/s00251-003-0629-y [DOI] [PubMed] [Google Scholar]
- Doherty P.C., Zinkernagel R.M. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. doi:10.1038/256050a0 [DOI] [PubMed] [Google Scholar]
- Dorak M.T., Lawson T., Machulla K.G., Mills K.I., Burnett A.K. Increased heterozygosity for MHC class II lineages in newborn males. Genes Immun. 2002;3:263–269. doi: 10.1038/sj.gene.6363862. doi:10.1038/sj.gene.6363862 [DOI] [PubMed] [Google Scholar]
- Gaya D.R., Russell R.K., Nimmo E.R., Satsangi J. New genes in inflammatory bowel disease: lessons from complex diseases? Lancet. 2006;367:1271–1284. doi: 10.1016/S0140-6736(06)68345-1. doi:10.1016/S0140-6736(06)68345-1 [DOI] [PubMed] [Google Scholar]
- Gregersen J.W., Kranc K.R., Ke X., Svendsen P., Madsen L.S., Thomsen A.R., Cardon L.R., Bell J.I., Fugger L. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature. 2006;443:574–577. doi: 10.1038/nature05133. doi:10.1038/nature05133 [DOI] [PubMed] [Google Scholar]
- Haddrill P.R., Halligan D.L., Tomaras D., Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. doi:10.1186/gb-2007-8-2-r18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P.W. Highly variable loci and their interpretations in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. doi:10.2307/2640768 [DOI] [PubMed] [Google Scholar]
- Kaufman J., Milne S., Gobel T.W.F., Walker B.A., Jacob J.P., Auffray C., Zoorob R., Beck S. The chicken B locus is a minimal essential major histocompatibility complex. Nature. 1999;401:923–925. doi: 10.1038/44856. doi:10.1038/44856 [DOI] [PubMed] [Google Scholar]
- Klein J., Satta Y., Takahata N., O'hUigin C. Trans-species MHC polymorphisms and the origin of species in primates. J. Med. Primatol. 1993;22:57–64. [PubMed] [Google Scholar]
- Little J.T. The evolutionary significance of parasitism: do parasite-driven genetic dynamics occur ex silico? J. Evol. Biol. 2002;15:1–9. doi:10.0000/014461999371097 [Google Scholar]
- Kelley J., Walter L., Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–695. doi: 10.1007/s00251-004-0717-7. doi:10.1007/s00251-004-0717-7 [DOI] [PubMed] [Google Scholar]
- Kennedy L.J., Davison L.J., Barnes A., Short A.D., Fretwell N., Jones C.A., Lee A.C., Ollier W.E., Catchpole B. Identification of susceptibility and protective major histocompatibility complex haplotypes in canine diabetes mellitus. Tissue Antig. 2006;68:467–476. doi: 10.1111/j.1399-0039.2006.00716.x. doi:10.1111/j.1399-0039.2006.00716.x [DOI] [PubMed] [Google Scholar]
- Kennedy L.J., et al. Risk of anal furunculosis in German Shepherd dogs is associated with the major histocompatibility complex. Tissue Antig. 2008;71:51–56. doi: 10.1111/j.1399-0039.2007.00964.x. doi:10.1111/j.1399-0039.2007.00964.x [DOI] [PubMed] [Google Scholar]
- Kuroda N., Figueroa F., O'hUigin C., Klein J. Evidence that the separation of MHC class II from class I loci in the zebrafish, Danio rerio, occurred by translocation. Immunogenetics. 2002;54:418–430. doi: 10.1007/s00251-002-0473-5. doi:10.1007/s00251-002-0473-5 [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B. Sinauer; Sunderland, MA: 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- Mangalam A.K., Rajagopalan G., Taneja V., David C.S. HLA class II transgenic mice mimic human inflammatory diseases. Adv. Immunol. 2008;97:65–147. doi: 10.1016/S0065-2776(08)00002-3. doi:10.1016/S0065-2776(08)00002-3 [DOI] [PubMed] [Google Scholar]
- May G., Shaw F., Badrane H., Vekemans X. The signature of balancing selection: fungal mating compatibility gene evolution. Proc. Natl Acad. Sci. USA. 1999;96:9172–9177. doi: 10.1073/pnas.96.16.9172. doi:10.1073/pnas.96.16.9172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muirhead C.A. Consequences of population structure on genes under balancing selection. Evolution. 2001;55:1532–1541. doi: 10.1111/j.0014-3820.2001.tb00673.x. doi:10.1554/0014-3820(2001)055[1532:COPSOG]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Muller H.J. Some genetic aspects of sex. Am. Nat. 1932;66:118–138. doi:10.1086/280418 [Google Scholar]
- Nei M., Rooney A.P. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. doi:10.1146/annurev.genet.39.073003.112240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. The mutational load of a multigene family with uniform members. Genet. Res. 1989;53:141–145. doi: 10.1017/s0016672300028020. [DOI] [PubMed] [Google Scholar]
- Okamoto K., et al. Identification of I kappa BL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am. J. Hum. Genet. 2003;72:303–312. doi: 10.1086/346067. doi:10.1086/346067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palti Y., Rodriguez M.F., Gahr S.A., Hansen J.D. Evolutionary history of the ABCB2 genomic region in teleosts. Dev. Comp. Immunol. 2007;31:483–498. doi: 10.1016/j.dci.2006.07.010. doi:10.1016/j.dci.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Piertney S.B., Oliver M.K. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. doi:10.1038/sj.hdy.6800724 [DOI] [PubMed] [Google Scholar]
- Raymond C.K., et al. Ancient haplotypes of the HLA class II region. Genome Res. 2005;15:1250–1257. doi: 10.1101/gr.3554305. doi:10.1101/gr.3554305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman A. Evolution of balanced genetic polymorphism. Mol. Ecol. 2000;9:1953–1963. doi: 10.1046/j.1365-294x.2000.01125.x. doi:10.1046/j.1365-294X.2000.01125.x [DOI] [PubMed] [Google Scholar]
- Richman A.D., Herrera L.G., Nash D. MHC class II beta sequence diversity in the deer mouse (Peromyscus maniculatus): implications for models of balancing selection. Mol. Ecol. 2001;10:2765–2773. doi: 10.1046/j.0962-1083.2001.01402.x. doi:10.1046/j.0962-1083.2001.01402.x [DOI] [PubMed] [Google Scholar]
- Richman A.D., Herrera L.G., Nash D., Schierup M.H. Relative roles of mutation and recombination in generating allelic polymorphism at an MHC class II locus in Peromyscus maniculatus. Genet. Res. Camb. 2003;82:89–99. doi: 10.1017/s0016672303006347. doi:10.1017/S0016672303006347 [DOI] [PubMed] [Google Scholar]
- She J.X., Wakeland E.K., Boehme S. The generation and maintenance of MHC class II gene polymorphism in rodents. Immunol. Rev. 1990;113:207–226. doi: 10.1111/j.1600-065x.1990.tb00042.x. doi:10.1111/j.1600-065X.1990.tb00042.x [DOI] [PubMed] [Google Scholar]
- Shiina T., et al. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics. 2006;173:1555–1570. doi: 10.1534/genetics.106.057034. doi:10.1534/genetics.106.057034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel A., Lu T., Koch W.A., Hampe J., Guenther S.M., De La Vega F.M., Krawczak M., Schreiber S. Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum. Genet. 2004;114:377–385. doi: 10.1007/s00439-003-1075-5. doi:10.1007/s00439-003-1075-5 [DOI] [PubMed] [Google Scholar]
- Stone J.L. Sheltered load associated with S-alleles in Solanum carolinense. Heredity. 2004;92:335–342. doi: 10.1038/sj.hdy.6800425. doi:10.1038/sj.hdy.6800425 [DOI] [PubMed] [Google Scholar]
- Takahata N. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl Acad. Sci. USA. 1990;87:2419–2423. doi: 10.1073/pnas.87.7.2419. doi:10.1073/pnas.87.7.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. Allelic genealogy and human evolution. Mol. Biol. Evol. 1993;10:2–22. doi: 10.1093/oxfordjournals.molbev.a039995. [DOI] [PubMed] [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama M.K. Genealogical structure among alleles regulating self-incompatibility in natural populations of flowering plants. Genetics. 1997;147:1389–1400. doi: 10.1093/genetics/147.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama M.K. Genealogy-dependent variation in viability among self-incompatibility genotypes. Theor. Popul. Biol. 2003;63:281–293. doi: 10.1016/s0040-5809(03)00020-0. doi:10.1016/S0040-5809(03)00020-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama M.K. Evolution under tight linkage to mating type. New Phytol. 2005;165:63–70. doi: 10.1111/j.1469-8137.2004.01246.x. doi:10.1111/j.1469-8137.2004.01246.x [DOI] [PubMed] [Google Scholar]
- van Oosterhout C., Joyce D.A., Cummings S.M., Blais J., Barson N.J., Cable J. Balancing selection, random genetic drift and genetic variation at the major histocompatibility complex (MHC) in two wild populations of guppies (Poecilia reticulata) Evolution. 2006;60:2562–2574. doi:10.1016/j.ijpara.2007.09.01 [PubMed] [Google Scholar]
- van Oosterhout C., Mohammed R.S., Hansen H., Archard G.A., McMullan M., Weese D.J., Cable J. Selection by parasites in spate conditions in wild Trinidadian guppies (Poecilia reticulata) Int. J. Parasitol. 2007a;37:805–812. doi: 10.1016/j.ijpara.2006.12.016. doi:10.1016/j.ijpara.2006.12.016 [DOI] [PubMed] [Google Scholar]
- van Oosterhout C., Smith A.M., Hänfling B., Ramnarine I.W., Mohammed R.S., Cable J. The guppy as a conservation model: implications of parasitism and inbreeding for reintroduction success. Conserv. Biol. 2007b;21:1573–1583. doi: 10.1111/j.1523-1739.2007.00809.x. doi:10.1111/j.1523-1739.2007.00809.x [DOI] [PubMed] [Google Scholar]
- Vekemans X., Slatkin M. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics. 1994;137:1157–1165. doi: 10.1093/genetics/137.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeland E.K., Boehme S., She J.X., Lu C.C., McIndoe R.A., Cheng I., Ye Y., Potts W.K. Ancestral polymorphisms of MHC class II genes: divergent allele advantage. Immunol. Res. 1990;9:115–122. doi: 10.1007/BF02918202. doi:10.1007/BF02918202 [DOI] [PubMed] [Google Scholar]
- Westerdahl H., Hansson B., Bensch S., Hasselquist D. Between-year variation of MHC allele frequencies in great reed warblers: selection or drift? J. Evol. Biol. 2004;17:485–492. doi: 10.1111/j.1420-9101.2004.00711.x. doi:10.1111/j.1420-9101.2004.00711.x [DOI] [PubMed] [Google Scholar]
- Woolhouse M.E.J., Webster J.P., Domingo E., Charlesworth B., Levin B.R. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. doi:10.1038/ng1202-569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Model validation, and the analysis of epistasis and genealogies of trans-species evolution