Abstract
Resolving the relationships among animal phyla is a key biological problem that remains to be solved. Morphology is unable to determine the relationships among most phyla and although molecular data have unveiled a new evolutionary scenario, they have their own limitations. Nuclear ribosomal genes (18S and 28S rDNA) have been used effectively for many years. However, they are considered of limited use for resolving deep divergences such as the origin of the bilaterians, due to certain drawbacks such as the long-branch attraction (LBA) problem. Here, we attempt to overcome these pitfalls by combining several methods suggested in previous studies and routinely used in contemporary standard phylogenetic analyses but that have not yet been applied to any bilaterian phylogeny based on these genes. The methods used include maximum likelihood and Bayesian inference, the application of models with rate heterogeneity across sites, wide taxon sampling and compartmentalized analyses for each problematic clade. The results obtained show that the combination of the above-mentioned methodologies minimizes the LBA effect, and a new Lophotrochozoa phylogeny emerges. Also, the Acoela and Nemertodermatida are confirmed with maximum support as the first branching bilaterians. Ribosomal RNA genes are thus a reliable source for the study of deep divergences in the metazoan tree, provided that the data are treated carefully.
Keywords: Bayesian inference, ecdysozoa, deuterostomia, ribosomal genes, long-branch attraction, maximum likelihood
1. Introduction
Resolving the relationships among animal phyla is a key problem in modern biology, since they are instrumental in understanding the evolution of many biological features including, among others, body plans, embryonic development and gene networks. Unfortunately, morphology is unable to clarify the precise relationships among most phyla. Twenty years ago, interest in this field was intensified by the introduction of the small subunit (SSU) RNA gene (18S rDNA or SSU) into metazoan phylogenies (Field et al. 1988; Lake 1990). However, molecular phylogenies also have their own downsides and raise new problems. The SSU's lack of resolving power on some regions of the metazoan tree (Philippe et al. 1994; Abouheif et al. 1998; Adoutte et al. 2000) and the long-branch attraction effect (LBA, Felsenstein 1978) are the major concerns for phylogeny resolution and credibility (Anderson & Swofford 2004).
Different sources of data have been added to the SSU sequences to overcome these drawbacks and new markers have been developed, the most recent being phylogenomics (Philippe & Telford 2006; Dunn et al. 2008). These approaches have generally supported the SSU division of bilaterians, but have not, however, solved all the questions raised. Moreover, the extra information obtained in these studies is usually counteracted by reduced sampling of animal phyla. The most recent and complete analysis (Dunn et al. 2008) shows better resolution within groups such as the Lophotrochozoa that so far have been poorly resolved. Such improvement, however, comes at the price of removing key taxa (i.e. gastrotrichs, gnathostomulids, rotifers, acoelomorphs, bryozoans or chaetognaths) before the final analyses are performed. Despite these shortcomings, a consensus tree of the Bilateria has been portrayed by various authors (Adoutte et al. 2000; Halanych & Passamaneck 2001; Giribet 2002; Balavoine & Adoutte 2003; Halanych 2004; Telford 2006). In this scheme, the Bilateria are divided into three main groups, Lophotrochozoa, Ecdysozoa and Deuterostomia, the first being the most problematic group, owing to its high number of phyla and poor internal resolution. Furthermore, there are some groups that continue to have unsolved affinities that might hold the key to understand essential transitions in the bilaterian tree, namely the Acoelomorpha, the Chaetognatha, the Gnathifera and the Gastrotricha.
The main aim of this study was to combine different approaches to unravel the status of the three big clades, mainly the internal relationships in the Lophotrochozoa and the position of groups of uncertain affinities. For this endeavour, we have attempted to maximize the metazoan phyla sampling, and at the same time, minimize the LBA effect, these being two factors that have formerly been suggested to cause uncertainties and lack of resolution. We applied a careful analysis involving two steps. First, we used several strategies that have been proposed to avoid LBA based on real and simulated data (Anderson & Swofford 2004; Bergsten 2005) using methods less sensitive to LBA, such as maximum likelihood (ML) or Bayesian inference (BI); employing model modifications such as rate heterogeneity across sites with a discrete γ-distribution parameter; using the shortest branched representatives available for each phyla, and searching for the widest taxon sampling. Second, we compartmentalized the analysis of the still problematic lineages (those with extremely long branches or with dubious status such as polyphyly) by removing all of them from the analysis and re-introducing them again one at a time; this allowed evaluation of their respective position and the support they receive without the interaction with other problematic taxa. Surprisingly, despite the numerous SSU and large subunit (LSU) rDNA sequences present in the databases, an extensive analysis of SSUs and LSUs for all bilaterians, now applying these routine phylogenetic methods, has yet to be performed.
2. Material and methods
(a) Taxon sampling, alignment and dataset assembly
(i) ‘Preliminary’ datasets
The SSU alignment used in Wallberg et al. (2004) was downloaded and new sequences were added from GenBank to complete the taxonomic sampling, namely the Nemertodermatida representatives, resulting in a total of 564 SSU representatives. The added SSU sequences were aligned using the Wallberg et al. alignment as a profile (see that paper for a thorough discussion on the alignment methodology). For LSU, 142 sequences from the former studies were aligned to secondary structure with notation modified from Gillespie et al. (2005). Alignments were performed and checked on Bioedit (v. 7.5, Hall 1999). The length-heterogeneous regions for which nucleotide homology could not be granted and the regions containing indels in the majority of sequences were removed prior to the analyses, using a very conservative criterion of retaining only unambiguously conserved blocks. The final alignments contained 1425 sites (out of 3365 nucleotides) for SSU and 2271 sites (out of 6847 nucleotides) for LSU. For accession numbers see additional table in the electronic supplementary material. A phylogeny was inferred from each of these two alignments and patristic distances to these outgroups (by ML with Treepuzzle) were calculated. These distances were used to select, for each gene, the shortest branched representatives of each phyla and these were reassembled in two alignments (SSU and LSU) both with 104 representatives to be used in the subsequent analyses.
(ii) All taxa dataset (All-set)
The SSU and LSU datasets with 104 representatives (28 bilaterian phyla and the outgroup) resulting from the previous step were merged into a combined dataset. Whenever possible, SSU and LSU sequences came from the same species (additional table in the electronic supplementary material). For those representatives lacking LSU sequences, LSUs were replaced with Ns. In the Chaetognatha, the only LSU representative available was combined with the two chosen SSUs.
(iii) Subset datasets
The phylogenetic analyses with the All-set were used to detect groups showing high rates of substitutions (considered fast evolving when the patristic distance to the outgroup was above 0.3, as calculated by ML with Treepuzzle) or presenting dubious status such as the Gastrotricha polyphyly. These groups (from now on dubbed problematic groups) were selected to produce five different subsamples in order to examine more accurately the position of such groups when the others were not present, hence avoiding interactions among them. These clades were removed from the All-set and five subsets were built adding only one of these groups at a time: ‘Acoelomorpha’ (Acoel-set), Gnathifera (Gnat-set, Gnathostomulida lacks LSU), Bryozoa (Bryo-set), Gastrotricha (Gast-set, Gastrotricha lacks LSU), and Chaetognatha (Chaet-set). With these compartmentalized analyses, we also wanted to test the effect of the problematic groups on the general topology.
(iv) Basic dataset (Basic-set)
This dataset simultaneously excluded the five problematic groups defined in the previous step. This dataset comprised 88 sequences from SSU and 87 from LSU (that lacks Micrognathozoa representatives) for 22 bilaterian phyla.
(b) Phylogenetic analyses
The Akaike information criterion was used in ModelTest (v. 3.6, Posada & Crandall 1998) to determine the evolutionary model best fitting each dataset. The specified model (GTR+Γ+I) was applied in all the algorithms where it was available. BI trees were inferred with a parallelized version of MrBayes software (v. 3.1, Ronquist & Huelsenbeck 2003). BI analyses were performed with and without partitioning the dataset into the two ribosomal genes (in the former case, unlinking the estimation of Statefreq, revmat, Pinvar and shape parameters for each partition) and with and without the covarion model. In all the cases, 3 000 000 generations were run in two independent analyses with a sample frequency of 1000, allowing the two runs to converge onto the stationary distribution. To obtain the consensus tree and the BI supports, 1 000 000 generations were removed to avoid including trees sampled before likelihood values had reached a plateau. RaxML (Stamatakis 2006), Treefinder (Jobb 2007) and PhyMl (Guindon & Gascuel 2003) were used to infer ML trees, with 1000 bootstrap replicates and the GTR+Γ+I model; in RaxML, a random topology was used as a starting tree and the support values were obtained with the Rapid Bootstrap algorithm. Neighbour joining trees were estimated using Mega with 1000 bootstrap replicates using the Kimura two-parameter model and pairwise-deletion option.
Competing topologies were evaluated for different datasets. Alternative topologies were based on the previous morphological or molecular studies indicated in the footnote) or were variations based on our analyses (table 1). The alternative trees were constructed using Treeview (v. 1.6.6, Page 1996) and PAUP (Swofford 2000) was used to calculate the site likelihoods for all trees and prepare the input dataset for Consel. Consel (v. 0.1i, Shimodaira & Hasegawa 2001) was run to perform the approximately unbiased test (AU, Shimodaira 2002; RELL; 1000 replicates; Shimodaira 2002).
Table 1.
Summary of status of the main clades in different datasets. (Bayesian posterior probabilities and RaxML bootstrap supports are shown. —, not applicable; NR, not recovered. Names in bold indicate the problematic phyla added in the independent datasets.)
| Clade | all taxa dataset (figure 1) | sub-sets (figure 4 in the electronic supplementary material) | basic dataset (figure 4f in the electronic supplementary material) |
|---|---|---|---|
| Acoela split from rest of bilaterians | 1.0–99 | 1.0–99 | — |
| Protostomia | 0.97–64 | 1.0–100 | |
| Lophotrochozoa | 0.62–53 | 1.0–98 | |
| Gnathostomulida as Lophotrochozoa | 0.62–53 | 0.73–53 | — |
| Gastrotricha inside Lophotrochozoa | 0.82–56 | 1.0–81 | — |
| Micrognathozoa+Rotifera | — | 1.0–99 | |
| Rotifera+Acantocephala | 0.98–79 | 0.91–78 | — |
| clade III: (Rotifera+Acantocephala)+Micrognathozoa | 0.83–77 | 1.0–96 | — |
| Cycliophora+Entoprocta | 1.0–92 | 1.0–98 | |
| (Cycliophora+Entoprocta)+Platyhelminthes | — | 0.99–58 | |
| (Cycliophora+Entoprocta)+Bryozoa | 0.61–19 | 0.95–69 | — |
| clade II: ((Cycliophora+Entoprocta)+Bryozoa)+Platyhelminthes | 0.61–15 | 1.0–68 | — |
| clade I (Trochozoa including Phoronida+Brachiopoda) | 1.0–65 | 1.0–63 | |
| ((Annellida+Sipuncula)+Echiura)+((Phoronida+ Brachipoda)+Mollusca) | 0.82–32 | 0.99–49 | |
| Annelida+Sipuncula | 1.0–77 | 1.0–76 | |
| (Annelida+Sipuncula)+Echiura | 0.97–49 | 1.0–56 | |
| (Phoronida+Brachiopoda) | 1.0–72 | 1.0–83 | |
| (Phoronida+Brachiopoda)+Mollusca | 0.63–24 | 0.84–54 | |
| Ecdysozoa | 0.65–44 | 1.0–97 | |
| Panarthropoda (Arthropoda+Tardigrada) | 1.0–85 | 1.0–99 | |
| Nematoida (Nematoda+Nematomorpha) | NR–NR | 0.65–54 | |
| Scalidophora (Priapulida+Kinorhyncha) | 1.0–91 | 1.0–100 | |
| Chaetognatha as Ecdysozoa | 0.65–44 | 1.0–90 | — |
| Deuterostomia | 0.68–42 | 0.59a–25a | |
| Ambulacraria (Hemichordata+Echinodermata) | 1.0–100 | 1.0–100 | |
| Chordata | NR–NR | 0.92a–38a |
Group only recovered when Urochordata are removed.
3. Results
NJ, Treefinder and PhyMl gave similar results, in many cases showing clear differences when compared with BI and RaxML trees. As a general trend, the first (NJ, Treefinder and PhyMl) result in some groupings contradicted by widely accepted clades (see examples and a discussion in figure 3 in the electronic supplementary material), mostly showing a tendency to group long branches together. A distance-based method such as NJ was expected to be prone to artefactual groupings, but to our surprise, similar results were found for many PhyMl and Treefinder topologies. Both programs start the heuristic search with a NJ inferred tree that could trap the search in local minima close to the topology of the NJ algorithm, pointing to inefficiency in the heuristic search. Given the worries about artefacts affecting these methods, we relied on RaxML as well as BI results (from all datasets), together with comparison of topologies, to define which clades were robustly recovered in our analyses.
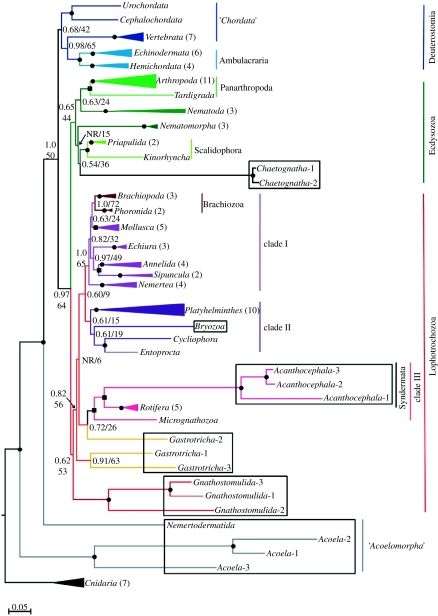
BI and RaxML phylogenies agreed for all the datasets analysed, recovering the same basic topology across all datasets. No differences were seen in the topologies recovered by BI when covarion was used or when BI estimates were unlinked for the SSU and LSU partitions of the matrix. Figure 1 shows the BI and RaxML topology obtained from the All-set; the problematic groups are boxed (see §2a(iii); further analysed in figure 4 in the electronic supplementary material). Figure 4 in the electronic supplementary material (a) to (e) shows the trees obtained when the subsets including only one of the problematic groups were analysed. Finally, another dataset excluding all the problematic taxa (Basic-set, figure 4f in the electronic supplementary material) was used to test the effect on the support values when all the problematic groups were excluded.
Figure 1.
Bayesian and RaxML topology (GTR+Γ+I) for All-set (104 metazoan representatives, Cnidaria as the outgroup). Posterior probabilities (PP) and ML bootstrap values (BV) are indicated with a bullet (PP=1.0 and BV>90%) or a square (PP=1.0 and 75%<BV≤90%) on the node; lower values are indicated. See table 1 to compare the effects of removing problematic taxa on BVs. NR stands for a node not resolved in the BI consensus tree. Problematic taxa are boxed (see §2). Monophyletic phyla are collapsed (triangle size proportional to number of representatives included) and the monophyly of each phylum has maximum support (except for gastrotrichs). The scale bar indicates the number of changes per site in ML inference. For species names corresponding to each terminal see additional table in the electronic supplementary material data. Circles, Bayesian posterior probability (BPP): 1.0 and ML bootstrap support (BS)>90%; squares, BPP: 1.0 and ML BS between 75% and 90%.
The overall topology of the tree was consistent between the All-set (figure 1), the subsets and Basic-set (figure 4 in the electronic supplementary material). However, most of the nodal supports increased in the subsets and even more so in the Basic-set (except for the Deuterostomia), as shown in table 1. The fact that the supports did not decrease when long branches were removed clearly indicates that high supports in the All-set are not a consequence of a LBA artefact misleading the method. The position of the problematic groups in the All-set tree is consistent with their position in the subset phylogenies (compare figure 1 with figure 4 in the electronic supplementary material), although again the subsets showed notably higher support for these groups (table 1).
(a) Comparison of topologies
For each dataset, the best tree was statistically compared against alternative trees (table 2). Concerning the subsets, all the alternative topologies tested were significantly worse than the original tree for all the sets with two exceptions: (i) the test based on the Gast-set (hypotheses 9–11) rejects the original polyphyletic Gastrotricha in favour of their monophyly, despite the fact that the former is found in BI and ML trees, and (ii) the hypothesis placing chaetognaths as a sister group to ecdysozoans (hypothesis 13) can not be rejected. The All-set allowed the same alternative hypotheses tested in the previous datasets to be studied, as well as new ones. In general, the All-set allowed the rejection of fewer hypotheses than the previous analysis (table 2).
Table 2.
Topology test results. AU (approximately unbiased test) p-values; in bold, the original tree obtained by BI against which alternative hypotheses were tested.
| Ln likelihood | AU | ||
|---|---|---|---|
| independent datasets | |||
| 1. | Acoelomorphadataset original tree (basal bilaterians, figure 4a in the electronic supplementary material) | −64 053.3559 | best |
| 2. | Acoelomorpha sister group to Ambulacraria; Philippe et al. (2007) | −64 104.8488 | 0.001a |
| 3. | Acoelomorpha sister group to Platyhelminthes; Baguñà & Riutort (2004) | −64 136.2360 | 0.001a |
| 4. | Gnathifera dataset original tree (paraphyletic, figure 4b in the electronic supplementary material) | −62 365.0881 | best |
| 5. | Gnathifera monophyletic; Haszprunar (1996) | −62 405.8423 | 0.003a |
| 6. | Bryozoadataset original tree (sister group toEntoprocta+Cycliophora, figure 4c in the electronic supplementary material) | −59 151.7621 | best |
| 7. | Bryozoa moved to be sister group to (Phoronida+Brachiopoda), sister group to Molluscab | −59 175.2591 | 0.008a |
| 8. | Lophophorata=Bryozoa+Entoprocta+(Phoronida+Brachiopoda), sister group to Mollusca; Hyman (1959) | −59 203.2772 | 0.002a |
| 9. | Gastrotrichadataset original tree (polyphyletic, figure 4d in the electronic supplementary material) | −59 152.5900 | 0.006a |
| 10. | Gastrotricha monophyletic and sister group to Lophotrochocozoab | −59 115.3544 | best |
| 11. | Gastrotricha monophyletic and sister group to Ecdysozoa; Schmidt-Rhaesa (2003) | −59 131.5161 | 0.003a |
| 12. | Chaetognathadataset original tree (sister group to Scalidophora, figure 4e in the electronic supplementary material) | −59 610.9699 | best |
| 13. | Chaetognatha sister group to Ecdysozoa; Zrzavy et al. (1998) | −59 617.9625 | 0.058 |
| 14. | Chaetognatha sister group to Lophotrochozoa; Matus et al. (2006) | −59 624.3330 | 0.045a |
| 15. | Chaetognatha sister group to Protostomia; Marletaz et al. (2006) | −59 628.0121 | 0.000a |
| all taxa dataset | |||
| 16. | best tree (figure 1) | −72 590.3401 | best |
| 17. | Nematoida monophyletic, sister group to Panarthropoda; Mallatt & Giribet (2006) | −72 590.6644 | 0.658 |
| 18. | Nematoida+(Scalidophora+Panarthropoda); Glenner et al. (2004, 2005) | −72 595.0884 | 0.342 |
| 19. | (Scalidophora+Panarthropoda) and paraphyletic Nematoidab | −72 601.3860 | 0.032a |
| 20. | ((Scalidophora+Chaetognatha)+Nematoida)+Panarthropodab | −72 592.0407 | 0.474 |
| 21. | ((Scalidophora+Chaetognatha)+Panarthropoda) and paraphyletic Nematoidab | −72 598.6865 | 0.055 |
| 22. | Chaetognatha +((Scalidophora+Nematoida)+Panarthropoda)b | −72 599.7541 | 0.017a |
| 23. | Nematoida +((Scalidophora+Chaetognatha)+Panarthropoda)b | −72 592.3215 | 0.532 |
| 24. | Chaetognatha sister group to Ecdysozoa; Zrzavy et al. (1998) | −72 594.8507 | 0.231 |
| 25. | Chaetognatha sister group to Lophotrochozoa; Matus et al. (2006) | −72 598.2265 | 0.224 |
| 26. | Chaetognatha sister group to Protostomia; Marletaz et al. (2006) | −72 602.4018 | 0.008a |
| 27. | Gastrotricha monophyletic, splitting after Gnathostomulida in the Lophotrochozoab | −72 603.7584 | 0.078 |
| 28. | Gastrotricha monophyletic, sister group to Ecdysozoa; Schmidt-Rhaesa (2003) | −72 619.5605 | 0.003a |
| 29. | Bryozoa sister group to (Phoronida+Brachiopoda), together sister group to Molluscab | −72 614.8445 | 0.012a |
| 30. | Lophophorata=Bryozoa+Entoprocta+(Phoronida+Brachiopoda), sister group to Mollusca; Hyman (1959) | −72 662.9733 | 0.000a |
| 31. | Acoelomorpha sister group to Platyhelminthes; Baguñà & Riutort (2004) | −72 700.9191 | 0.000a |
| 32. | Acoelomorpha sister group to Ambulacraria; Philippe et al. (2007) | −72 693.3459 | 0.000a |
| 33. | Deuteromia monophyletic; Cavalier-Smith (1998) | −72 610.9121 | 0.107 |
| 34. | Gnathifera monophyletic, sister group to the rest of Lophotrochozoab | −74 836.0737 | 0.000a |
| 35. | Platyzoa (without Acoela), sister group to the rest of Lophotrochozoa; Giribet et al. (2000) | −74 363.6413 | 0.000a |
| 36. | Platyzoa (including Acoela), sister group to the rest of Lophotrochozoa; Giribet et al. (2000) | −75 801.0545 | 0.000a |
Hypothesis rejected when p<0.05 for AU test.
Hypotheses partially based on bibliography but modified to accommodate the topology obtained from our dataset.
4. Discussion
(a) Methodological approach
The analyses shown here represent, to our knowledge, the largest animal dataset of SSU and LSU sequences analysed to date using probabilistic methods. Overall, our results confirm that a combination of wide taxon sampling, the use of short-branched representatives and analyses that take into account the flaws of ribosomal genes still allows them to furnish new answers. ML and BI algorithms place long branches deep inside the ingroup, as clearly shown in the All-set (figure 1) and the subset analyses (figure 4 in the electronic supplementary material). If the LBA effect were active, the long branches would appear near the outgroup or close to one another. In our view, this suggests that LBA generally does not affect our results obtained with BI and ML.
Regarding the compartmentalized and the basic-set analyses, the trees show topologies that are consistent with the All-set, although with higher support (table 1). Moreover, the topology comparison test (table 2) for subsets generally rejects the alternative hypotheses, while in some cases; the same hypothesis cannot be rejected for the All-set. This could merely be an effect of the reduced taxon sampling or it could stem from the simultaneous presence of fast-evolving sequences in the All-set leading to homoplasy. Homoplasy, while not misleading the inference method, would reduce the proportion of sites supporting a node and would make the differences among alternative topologies non-significant. Taken together, these findings suggest that compartmentalized analyses are an adequate strategy to deal with simultaneous problematic groups in a phylogeny.
(b) Bilaterian phylogeny
BI and ML results from all datasets, together with the comparison of topologies, were used to define which clades were robustly recovered in our analyses. These groups are summarized in the tree depicted in figure 2. For the first time, to our knowledge, in such a comprehensive SSU+LSU analysis, the monophyly of protostomia is recovered, and remarkably Lophotrochozoa and Ecdysozoa also appear with high support on most of the subsets (figure 4 in the electronic supplementary material). The most noticeable improvement when compared with previous studies is the increase in resolution obtained within the Lophotrochozoa, mainly in the subset trees (with the exception of the Gnathifera dataset).
Figure 2.
Summary tree from our results. See text for discussion.
Although, beyond the scope of the present paper, some of the relationships observed outside the lophotrochozoans are worthy of comment, such as the problematic behaviour of Ciona sequences that results in a non-monophyletic Deuterostomes in some analyses, and especially the position of the Chaetognatha and the Acoelomorpha. To avoid an extremely long discussion, these commentaries have been placed in the electronic supplementary material.
(i) Lophotrochozoa
The lophotrochozoans are of special interest because they include the greatest body plan diversity of the three main bilaterian superclades. In fact, in its original node-based definition, this group included the last common ancestor of annelids, molluscs and lophophorates and all their descendants (Halanych et al. 1995). In some posterior comprehensive analyses involving many phyla, Lophotrochozoa refers to an extended group including many other phyla (such as Gnathostomulida, Platyhelminthes, Rotifera, etc.), owing to the very basal Bryozoa position in the resulting phylogenies. However, since most of the phyla included do not fit the original definition of having either trochophora larvae (Trochozoa) or a lophophore (Lophophorata), and the composition of the Lophotrochozoa is in a state of flux due to the unsettled situation regarding the Bryozoa, some authors propose avoiding this name. Until a better name is agreed, we prefer to avoid introducing more confusion by using new names. Hence, we have used Lophotrochozoa sensu stricto to name the group resulting from applying the original node-based definition to our final tree (clade I and II), and to avoid using new names we will refer to the extended assemblage (including Gnathostomulida, Gastrotricha and clade III) as Lophotrochozoa sensu lato.
Clade I receives high support in the analyses of all the datasets. It is an assemblage constituted by phyla with spiral cleavage (nemertines, annelids, molluscs, echiurans and sipunculans) and two lophophorate phyla with radial cleavage (brachiopods and phoronids). Although, affinities among these groups have already been hinted at in previous studies (Zrzavy et al. 1998; Giribet et al. 2000; Peterson & Eernisse 2001; Winchell et al. 2002; Passamaneck & Halanych 2006), the internal phylogeny shown here was not recovered in any of them. The recent phylogenomic study based on 150 genes (Dunn et al. 2008) shows the same group with similar relationships and also points to the inclusion of brachiopods and phoronids. The most basal group in clade I is Nemertea that are known to bear a coelomic cavity (Turbeville et al. 1992) and the hox signatures of lophotrochozoans (de Rosa et al. 1999; Balavoine et al. 2002). Next to the nemerteans, we find a highly supported (Echiura+(Annelida+Sipunculida)) group. A close relationship between echiurans and annelids has been proposed both on morphological grounds (Nielsen 1995; Hessling & Westheide, 2002) and molecular data (McHugh 1997; Giribet et al. 2000; Peterson & Eernisse 2001; Mallatt & Winchell 2002; Bourlat et al. 2008). In turn, sipunculans also have developmental affinities to annelids (Clark 1969; Rice 1985), a relationship that is supported by recent mtDNA and multigenic studies (Boore & Staton 2002; Struck et al. 2007; Bourlat et al. 2008) but that has never been proposed in previous SSU and LSU studies (Mallatt & Winchell 2002; Passamaneck & Halanych 2006). The sister group to the annelids assemblage is a clade made up by Mollusca and Phoronida+Brachiopoda. The brachiopod–phoronid affinity has already been shown on the basis of SSU data (Cohen et al. 1998; Cohen 2000; Peterson & Eernisse 2001) and mitochondrial gene data (Stechmann & Schlegel 1999; Helfenbein & Boore 2004; ). Former SSU+LSU analysis suggested the close relationship of brachiopods and phoronids to molluscs (Mallatt & Winchell 2002), a placement corroborated in our trees and suggested by paleontological evidence (Vinther & Nielsen 2005), while multigenic analyses have related brachiopods and phoronids to nemertea (Bourlat et al. 2008; Dunn et al. 2008) or the phoronida to a non-monophyletic mollusc group and brachiopods to nemertines (Bourlat et al. 2008).
In clade II, the Bryozoa cluster with Entoprocta+Cycliophora with maximum support and Platyhelminthes are the most basal phyla. Former SSU studies have already shown that Bryozoa are not closely related to lophophorates (Littlewood et al. 1998; Cohen 2000) and recent multigenic analyses linked them to Entoprocta (Hausdorf et al. 2007) or to nemertines and brachiopods (Bourlat et al. 2008). Cycliophora have also been related to entoprocts in morphological analyses (Funch & Kristensen 1995; Zrzavy et al. 1998; Sørensen et al. 2000) and in the most recent SSU+LSU study (Passamaneck & Halanych 2006), but SSU studies left their position open (Winnepenninckx et al. 1998; Giribet et al. 2000; Peterson & Eernisse 2001; Giribet et al. 2004). Therefore, independent molecular studies have suggested the relationship of bryozoans and cycliophorans with entoprocts, this study being the first proposing a strong supported clade that groups them all together. Regarding Platyhelminthes (Catenulida+Rhabditophora), molecular phylogenies have shown them as basal lophotrochozoans (Ruiz-Trillo et al. 1999; Peterson & Eernisse 2001) or situated within the Platyzoa (Giribet et al. 2000; Passamaneck & Halanych 2006). In our tree, flatworms appear in an unprecedented new position as a sister group to the Bryozoa+(Cycliophora+Entoprocta) clade, with high support in the Basic-set and Bryo-set (table 2). Albeit platyhelminths and cycliophorans share a negative trait, the acoelomate condition, this does not hold true for the pseudocelomate bryozoans and entoprocts. No obvious morphological synapomorphies exist for clade II.
(ii) Lophotrochozoa sensu lato
Gnathostomulida are the first branching phylum of the Lophotrochozoa s.l. in the BI and ML trees for All-set and Gnat-set (figure 4b in the electronic supplementary material). Unfortunately, this relationship never shows significant support, probably due to the absence of LSU sequences for this phylum. Previous molecular studies related them to ecdysozoa (Littlewood et al. 1998; Zrzavy et al. 1998) or situated them within Platyzoa (close to rotifers, acanthocephalans and cycliophorans, Giribet et al. 2000), while morphology placed them close to rotifers and acanthocephalans forming the Gnathifera (Rieger & Tyler 1995; Ahlrichs 1997). Regarding the Gastrotricha, they appear to be polyphyletic, but their monophyly can not be rejected by the comparison of topologies test. Gastrotrich SSU sequences have presented conflicting results in previous studies, showing them to be either a polyphyletic group within lophotrochozoans (Giribet et al. 2004; Manylov et al. 2004) or monophyletic in the most recent study (but with low support, Todaro et al. 2006). Their problematic nature, together with the lack of gastrotrich LSU in our dataset, may explain our failure to recover their monophyly. Despite their polyphyly, their clustering with the rest of the lophotrochozoans (excluding gnathostomulids) has maximum support (figure 4c and additional table in the electronic supplementary material) and any relationship to ecdysozoa is rejected by the comparison of topologies (table 2).
Clade III includes Rotifera, Acantocephala and Micrognathozoa. The relationship among Rotifera and Acanthocephala is solidly recovered in our trees despite the very long branches of acanthocephalans, a clade suggested by morphology (see examples in Schmidt-Rhaesa 2003) and SSU (Syndermata, Garey et al. 1996; Garey & Schmidt-Rhaesa 1998; Zrzavy et al. 1998). With regards to Micrognathozoa, morphology related them to gnathostomulids and rotifers (clade Gnathifera, based on homologous jaw elements, Kristensen & Funch 2000; Sørensen 2003), although recent molecular data are more ambiguous (Giribet et al. 2004). Our analyses clearly recover the clade (Micrognathozoa+(Rotifera+Acanthocephala)) with maximum support in the Gnat-set (figure 4b in the electronic supplementary material).
Overall, our phylogeny of the Protostomia and the comparison of topologies tests do not recover proposals such as Gnathifera (Gnathostomulida+Micrognathozoa (Rotifera+Acanthocephala); Ahlrichs 1997; Sørensen et al. 2000; Nielsen 2001), Cycloneuralia (sensu lato, Gastrotricha+Nematoida+Scalidophora; Schmidt-Rhaesa et al. 1998; Sørensen et al. 2000; Nielsen 2001; Peterson & Eernisse 2001; Zrzavy 2003), Neotrichozoa (Gastrotricha+Gnathostomulida; Zrzavy et al. 1998) and Platyzoa (Cavalier-Smith 1998; Giribet et al. 2000; Garey 2001; Passamaneck & Halanych 2006). However, if acoels are excluded from the Platyzoa definition (Giribet et al. 2000), the platyzoan representatives could be seen as a paraphyletic assemblage at the base of the Lophotrochozoa made up by Gnathostomulida (Gastrotricha+((Micrognathozoa (Rotifera+Acanthocephala))+Lophotrochozoa ss). Hence, the characters that have been proposed as synapomorphies for the Platyzoa may be reconsidered as plesiomorphic states for the Lophotrochozoa s.l.
(c) Evolutionary implications
The results presented here (figure 2) have some interesting evolutionary implications. First, the paraphyletic branching of the acoels and nemertodermatids at the base of the bilaterians suggests that the last common ancestor of all bilaterians, however, different from present-day acoels and nemertodermatids, was a small, benthonic, acoelomate worm with an anterior concentration of nerve cells (primitive brain), a blind gut, mesoderm that forms the musculature and mesenchymal cells and direct development. Second, the early branching of gnathostomulids within the lophotrochozoans agrees with their acoelomate nature and its presumed lack of a permanent anus that may be plesiomorphies shared with acoelomorphs and diploblasts. Next to gnathostomulids branch the gastrotrichs that are also acoelomate worm-like animals, but with a through-gut with anus. According to this scenario, gnathostomulids and gastrotrichs may be an intermediate state between an acoel-like ancestor and the more complex lophotrochozoans, as suggested by a recent study of mouth and anus evolution (Hejnol & Martindale 2008).
Finally, this phylogenetic scheme clearly demonstrates that some morphological features, such as the presence and type of coelomic cavities or the type of cleavage (for example, the multiple apparition of radial cleavage in bilaterians, including the case of the radial Brachiozoa within a clade of spiralian animals) that are classically considered as good phylogenetic characters for the metazoa, have appeared independently more than once.
5. Conclusions
To summarize, this study demonstrates that the combination of broad taxon sampling, short-branched sequences and the application of adequate methodologies to avoid LBA, together with careful compartmentalized analyses of problematic taxa allows a phylogenetic hypothesis of the bilaterian animals to be inferred with better resolution than previous similar studies. Furthermore, the vast taxonomic sampling available for ribosomal genes allowed us to test the position of some key clades that have been poorly examined for the new genetic markers. Altogether, these observations point to the fact that ribosomal RNA genes are still a reliable source for the study of deep divergences in the metazoan tree.
Acknowledgments
This research was supported by DGI-MEC grant BOS2002-02097 and CIRIT grant 2005SGR00578 to J.B. We are grateful to Iñaki Ruiz-Trillo for all the lively discussions and stimulating insights on the subject and to Sara Rojas for the animal drawings included in the final tree. We also wish to thank David Vicente (Barcelona Supercomputing Centre) and Alfred Gil (CESCA) for their kind help with installing programs and resolving problems during execution. M.R. and J.P. designed the study and performed the analyses. M.R., J.P. and J.B. prepared the manuscript. All the three authors read and approved the final manuscript.
Supplementary Material
Sequences analyzed in this study and their GenBank Accesión Numbers
Results obtained with NJ, Treefinder and PhyML
Bayesian and RaxML topologies for the subsets
Discussion on the results for the non-lophotrochozoan clades
References
- Abouheif E., Zardoya R., Meyer A. Limitations of metazoan 18S rRNA sequence data: implications for reconstructing a phylogeny of the animal kingdom and inferring the reality of the Cambrian explosion. J. Mol. Evol. 1998;47:394–405. doi: 10.1007/pl00006397. doi:10.1007/PL00006397 [DOI] [PubMed] [Google Scholar]
- Adoutte A., Balavoine G., Lartillot N., Lespinet O., Prud'homme B., de Rosa R. The new animal phylogeny: reliability and implications. Proc. Natl Acad. Sci. USA. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. doi:10.1073/pnas.97.9.4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlrichs W.H. Epidermal ultrastructure of Seison nebaliae and Seison annulatus, and a comparison of epidermal structures within the Gnathifera. Zoomorphology. 1997;117:41–48. doi:10.1007/s004350050028 [Google Scholar]
- Anderson F.E., Swofford D.L. Should we be worried about long-branch attraction in real data sets? Investigations using metazoan 18S rDNA. Mol. Phylogenet. Evol. 2004;33:440–451. doi: 10.1016/j.ympev.2004.06.015. doi:10.1016/j.ympev.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Baguñà J., Riutort M. The dawn of bilaterian animals: the case of acoelomorph flatworms. BioEssays. 2004;26:1046–1057. doi: 10.1002/bies.20113. doi:10.1002/bies.20113 [DOI] [PubMed] [Google Scholar]
- Balavoine G., Adoutte A. The segmented Urbilateria: a testable scenario. Integr. Comp. Biol. 2003;43:137–147. doi: 10.1093/icb/43.1.137. doi:10.1093/icb/43.1.137 [DOI] [PubMed] [Google Scholar]
- Balavoine G., de Rosa R., Adoutte A. Hox clusters and bilaterian phylogeny. Mol. Phylogenet. Evol. 2002;24:366–373. doi: 10.1016/s1055-7903(02)00237-3. doi:10.1016/S1055-7903(02)00237-3 [DOI] [PubMed] [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. doi:10.1111/j.1096-0031.2005.00059.x [DOI] [PubMed] [Google Scholar]
- Boore J.L., Staton J.L. The mitochondrial genome of the Sipunculid Phascolopsis gouldii supports its association with Annelida rather than Mollusca. Mol. Biol. Evol. 2002;19:127–137. doi: 10.1093/oxfordjournals.molbev.a004065. [DOI] [PubMed] [Google Scholar]
- Bourlat S.J., Nielsen C., Economou A.D., Telford M.J. Testing the new animal phylogeny: a phylum level molecular analysis of the animal kingdom. Mol. Phylogenet. Evol. 2008;49:23–31. doi: 10.1016/j.ympev.2008.07.008. doi:10.1016/j.ympev.2008.07.008 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. A revised six-kingdom system of life. Biol. Rev. 1998;73:203. doi: 10.1017/s0006323198005167. doi:10.1017/S0006323198005167 [DOI] [PubMed] [Google Scholar]
- Clark R.B. Chemical zoology. Academic Press; New York, NY; London, UK: 1969. Systematics and phylogeny: Annelida, Echiura, Sipuncula. [Google Scholar]
- Cohen B.L. Monophyly of brachiopods and phoronids: reconciliation of molecular evidence with Linnaean classification (the subphylum Phoroniformea nov.) Proc. R. Soc. B. 2000;267:225–231. doi: 10.1098/rspb.2000.0991. doi:10.1098/rspb.2000.0991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B.L., Gawthrop A., Cavalier Smith T. Molecular phylogeny of brachiopods and phoronids based on nuclear-encoded small subunit ribosomal RNA gene sequences. Phil. Trans. R. Soc. B. 1998;353:2039–2061. doi:10.1098/rstb.1998.0351 [Google Scholar]
- de Rosa R., Grenier J.K., Andreeva T., Cook C.E., Adoutte A. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772. doi: 10.1038/21631. doi:10.1038/21631 [DOI] [PubMed] [Google Scholar]
- Dunn C.W., et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. doi:10.1038/nature06614 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Biol. 1978;27:401–410. doi:10.2307/2412923 [Google Scholar]
- Field K.G., Olsen G.J., Lane D.J., Giovannoni S.J., Ghiselin M.T., Raff E.C., Pace N.R., Raff R.A. Molecular phylogeny of the animal kingdom. Science. 1988;239:748–753. doi: 10.1126/science.3277277. doi:10.1126/science.3277277 [DOI] [PubMed] [Google Scholar]
- Funch P., Kristensen R.M. Cycliophora is a new phylum with affinities to Entoprocta and Ectoprocta. Nature. 1995;378:711–714. doi:10.1038/378711a0 [Google Scholar]
- Garey J.R. Ecdysozoa: the relationship between Cycloneuralia and Panarthropoda. Zool. Anz. 2001;240:321. doi:10.1078/0044-5231-00039 [Google Scholar]
- Garey J.R., Schmidt-Rhaesa A. The essential role of ‘minor’ phyla in molecular studies of animal evolution. Am. Zool. 1998;38:907–917. doi:10.1093/icb/38.6.907 [Google Scholar]
- Garey J., Near T., Nonnemacher M., Nadler S. Molecular evidence for Acanthocephala as a subtaxon of Rotifera. J. Mol. Evol. 1996;43:287–292. doi: 10.1007/BF02338837. doi:10.1007/BF02338837 [DOI] [PubMed] [Google Scholar]
- Gillespie J.J., Yoder M.J., Wharton R.A. Predicted secondary structure for 28S and 18S rRNA from Ichneumonoidea (Insecta: Hymenoptera: Apocrita): impact on sequence alignment and phylogeny estimation. J. Mol. Evol. 2005;61:114–137. doi: 10.1007/s00239-004-0246-x. doi:10.1007/s00239-004-0246-x [DOI] [PubMed] [Google Scholar]
- Giribet G. Current advances in the phylogenetic reconstruction of metazoan evolution. A new paradigm for the Cambrian explosion? Mol. Phylogenet. Evol. 2002;24:345–357. doi: 10.1016/s1055-7903(02)00206-3. doi:10.1016/S1055-7903(02)00206-3 [DOI] [PubMed] [Google Scholar]
- Giribet G., Distel D.L., Polz M., Sterrer W., Wheeler W.C. Triploblastic relationships with emphasis on the acoelomates and the position of Gnathostomulida, Cycliophora, Plathelminthes, and Chaetognatha: a combined approach of 18S rDNA sequences and morphology. Syst. Biol. 2000;49:539–562. doi: 10.1080/10635159950127385. doi:10.1080/10635159950127385 [DOI] [PubMed] [Google Scholar]
- Giribet G., Sørensen M.V., Funch P., Kristensen R.M., Sterrer W. Investigations into the phylogenetic position of Micrognathozoa using four molecular loci. Cladistics. 2004;20:1–13. doi: 10.1111/j.1096-0031.2004.00004.x. doi:10.1111/j.1096-0031.2004.00004.x [DOI] [PubMed] [Google Scholar]
- Glenner H., Hansen A.J., Sørensen M.V., Ronquist F., Huelsenbeck J.P., Willerslev E. Bayesian inference of the metazoan phylogeny: a combined molecular and morphological approach. Curr. Biol. 2004;14:1644–1649. doi: 10.1016/j.cub.2004.09.027. doi:10.1016/j.cub.2004.09.027 [DOI] [PubMed] [Google Scholar]
- Glenner H., Hansen A.J., Sørensen M.V., Ronquist F., Huelsenbeck J.P., Willerslev E. Erratum: Bayesian inference of the metazoan phylogeny: a combined molecular and morphological approach. Curr. Biol. 2005;15:392–393. doi: 10.1016/j.cub.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Halanych K.M. The new view of animal phylogeny. Annu. Rev. Ecol. Evol. Syst. 2004;35:229–256. doi:10.1146/annurev.ecolsys.35.112202.130124 [Google Scholar]
- Halanych K.M., Passamaneck Y. A brief review of metazoan phylogeny and future prospects in Hox-research. Am. Zool. 2001;41:629–639. doi:10.1668/0003-1569(2001)041[0629:ABROMP]2.0.CO;2 [Google Scholar]
- Halanych K.M., Bacheller J.D., Aguinaldo A.M., Liva S.M., Hillis D.M., Lake J.A. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science. 1995;267:1641–1643. doi: 10.1126/science.7886451. doi:10.1126/science.7886451 [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Haszprunar G. Platyhelminthes and Platihelminthomorpha: paraphyletic taxa. J. Zool. Syst. Evol. Res. 1996;34:41–48. [Google Scholar]
- Hausdorf B., Helmkampf M., Meyer A., Witek A., Herlyn H., Bruchhaus I., Hankeln T., Struck T.H., Lieb B. Spiralian phylogenomics supports the resurrection of Bryozoa comprising Ectoprocta and Entoprocta. Mol. Biol. Evol. 2007;24:2723–2729. doi: 10.1093/molbev/msm214. doi:10.1093/molbev/msm214 [DOI] [PubMed] [Google Scholar]
- Hejnol A., Martindale M.Q. Acoel development indicates the independent evolution of the bilaterian mouth and anus. Nature. 2008;456:382–386. doi: 10.1038/nature07309. doi:10.1038/nature07309 [DOI] [PubMed] [Google Scholar]
- Helfenbein K.G., Boore J.L. The mitochondrial genome of Phoronis architecta: comparisons demonstrate that phoronids are lophotrochozoan protostomes. Mol. Biol. Evol. 2004;21:153–157. doi: 10.1093/molbev/msh011. doi:10.1093/molbev/msh011 [DOI] [PubMed] [Google Scholar]
- Hessling R., Westheide W. Are Echiura derived from a segmented ancestor? Immunohistochemical analysis of the nervous system in developmental stages of Bonellia viridis. J. Morphol. 2002;252:100–113. doi: 10.1002/jmor.1093. doi:10.1002/jmor.1093 [DOI] [PubMed] [Google Scholar]
- Hyman L.H. The coelomate bilateria. vol. 5. McGraw-Hill Book; New York, NY: 1959. The invertebrates: smaller coelomate. Groups Chaetognatha, Hemicordata, Pogonophora, Phoronida, Ectoprocta, Brachiopoda, Sipunculida. [Google Scholar]
- Jobb, G. 2007 Treefinder version of June 2007. Munich, Germany. Distributed by the author at www.treefinder.de
- Kristensen R.M., Funch P. Micrognathozoa: a new class with complicated jaws like those of Rotifera and Gnathostomulida. J. Morphol. 2000;246:1–49. doi: 10.1002/1097-4687(200010)246:1<1::AID-JMOR1>3.0.CO;2-D. doi:10.1002/1097-4687(200010)246:1<1::AID-JMOR1>3.0.CO;2-D [DOI] [PubMed] [Google Scholar]
- Lake J.A. Origin of the metazoa. Proc. Natl Acad. Sci. USA. 1990;87:763–766. doi: 10.1073/pnas.87.2.763. doi:10.1073/pnas.87.2.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood D.T.J., Telford M.J., Clough K.A., Rohde K. Gnathostomulida: an enigmatic metazoan phylum from both morphological and molecular perspectives. Mol. Phylogenet. Evol. 1998;9:72–79. doi: 10.1006/mpev.1997.0448. doi:10.1006/mpev.1997.0448 [DOI] [PubMed] [Google Scholar]
- Mallatt J., Giribet G. Further use of nearly complete 28S and 18S rRNA genes to classify ecdysozoa: 37 more arthropods and a kinorhynch. Mol. Phylogenet. Evol. 2006;40:772–794. doi: 10.1016/j.ympev.2006.04.021. doi:10.1016/j.ympev.2006.04.021 [DOI] [PubMed] [Google Scholar]
- Mallatt J., Winchell C.J. Testing the new animal phylogeny: first use of combined large-subunit and small-subunit rRNA gene sequences to classify the protostomes. Mol. Biol. Evol. 2002;19:289–301. doi: 10.1093/oxfordjournals.molbev.a004082. [DOI] [PubMed] [Google Scholar]
- Manylov O.G., Vladychenskaya N.S., Milyutina I.A., Kedrova O.S., Korokhov N.P., Dvoryanchikov G.A., Aleshin V.V., Petrov N.B. Analysis of 18S rRNA gene sequences suggests significant molecular differences between Macrodasyida and Chaetonotida (Gastrotricha) Mol. Phylogenet. Evol. 2004;30:850–854. doi: 10.1016/S1055-7903(03)00251-3. doi:10.1016/S1055-7903(03)00251-3 [DOI] [PubMed] [Google Scholar]
- Marletaz F., et al. Chaetognath phylogenomics: a protostome with deuterostome-like development. Curr. Biol. 2006;16:R577–R578. doi: 10.1016/j.cub.2006.07.016. doi:10.1016/j.cub.2006.07.016 [DOI] [PubMed] [Google Scholar]
- Matus D.Q., Copley R.R., Dunn C.W., Hejnol A., Eccleston H., Halanych K.M., Martindale M.Q., Telford M.J. Broad taxon and gene sampling indicate that chaetognaths are protostomes. Curr. Biol. 2006;16:R575–R576. doi: 10.1016/j.cub.2006.07.017. doi:10.1016/j.cub.2006.07.017 [DOI] [PubMed] [Google Scholar]
- McHugh D. Molecular evidence that echiurans and pogonophorans are derived annelids. Proc. Natl Acad. Sci. USA. 1997;94:8006–8009. doi: 10.1073/pnas.94.15.8006. doi:10.1073/pnas.94.15.8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. Oxford University Press; Oxford, UK: 1995. Animal evolution: interrelationships of the living phyla. [Google Scholar]
- Nielsen C. 2nd edn. Oxford University Press; Oxford, UK: 2001. Animal evolution: interrelationships of the living phyla. [Google Scholar]
- Page R.D.M. Treeview: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Passamaneck Y., Halanych K.M. Lophotrochozoan phylogeny assessed with LSU and SSU data: evidence of lophophorate polyphyly. Mol. Phylogenet. Evol. 2006;40:20–28. doi: 10.1016/j.ympev.2006.02.001. doi:10.1016/j.ympev.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Peterson K.J., Eernisse D.J. Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 2001;3:170–205. doi: 10.1046/j.1525-142x.2001.003003170.x. doi:10.1046/j.1525-142x.2001.003003170.x [DOI] [PubMed] [Google Scholar]
- Philippe H., Telford M.J. Large-scale sequencing and the new animal phylogeny. Trends Ecol. Evol. 2006;21:614–620. doi: 10.1016/j.tree.2006.08.004. doi:10.1016/j.tree.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Philippe H., Chenuil A., Adoutte A. Can the Cambrian explosion be inferred through molecular phylogeny? Development. 1994;((Suppl. 1994)):15–25. [Google Scholar]
- Philippe H., Brinkmann H., Martinez P., Riutort M., Baguñà J. Acoel flatworms are not Platyhelminthes: evidence from phylogenomics. PLoS One. 2007;8:e717. doi: 10.1371/journal.pone.0000717. doi:10.1371/journal.pone.0000717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. ModelTest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rice M.E. The origins and relationships of lower invertebrates. Oxford University Press; Oxford, UK: 1985. Sipuncula: developmental evidence for phylogenetic inference. [Google Scholar]
- Rieger R.M., Tyler S. Sister–group relationship of Gnathostomulida and Rotifera–Acanthocephala. Invertebr. Biol. 1995;114:186–188. doi:10.2307/3226891 [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I., Riutort M., Littlewood D.T., Herniou E.A., Baguna J. Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. doi:10.1126/science.283.5409.1919 [DOI] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A. Old trees, new trees: is there any progress? Zoology. 2003;106:291–301. doi: 10.1078/0944-2006-00128. doi:10.1078/0944-2006-00128 [DOI] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A., Bartolomaeus T., Lemburg C., Ehlers U., Garey J.R. The position of the Arthropoda in the phylogenetic system. J. Morphol. 1998;238:263–285. doi: 10.1002/(SICI)1097-4687(199812)238:3<263::AID-JMOR1>3.0.CO;2-L. doi:10.1002/(SICI)1097-4687(199812)238:3<263::AID-JMOR1>3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. doi:10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. Consel: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. doi:10.1093/bioinformatics/17.12.1246 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. doi:10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stechmann A., Schlegel M. Analysis of the complete mitochondrial DNA sequence of the brachiopod Terebratulina retusa places Brachiopoda within the protostomes. Proc. R. Soc. B. 1999;266:2043. doi: 10.1098/rspb.1999.0885. doi:10.1098/rspb.1999.0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck T., Schult N., Kusen T., Hickman E., Bleidorn C., McHugh D., Halanych K. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 2007;7:57. doi: 10.1186/1471-2148-7-57. doi:10.1186/1471-2148-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M.V. Further structures in the jaw apparatus of Limnognathia maerski (Micrognathozoa), with notes on the phylogeny of the Gnathifera. J. Morphol. 2003;255:131–145. doi: 10.1002/jmor.10038. doi:10.1002/jmor.10038 [DOI] [PubMed] [Google Scholar]
- Sørensen M.V., Funch P., Willerslev E., Hansen A.J., Olesen J. On the phylogeny of the Metazoa in light of Cycliophora and Micrognathozoa. Zool. Anz. 2000;239:297. [Google Scholar]
- Swofford, D. L. 2000 PAUP*. Phylogenetic analysis using parsimony (*and other methods) v. 4. Sunderland, MA: Sinear Associates.
- Telford M.J. Animal phylogeny. Curr. Biol. 2006;16:R981–R985. doi: 10.1016/j.cub.2006.10.048. doi:10.1016/j.cub.2006.10.048 [DOI] [PubMed] [Google Scholar]
- Todaro M.A., Telford M.J., Lockyer A.E., Littlewood D.T.J. Interrelationships of the Gastrotricha and their place among the Metazoa inferred from 18S rRNA genes. Zool. Scr. 2006;35:251–259. doi:10.1111/j.1463-6409.2006.00228.x [Google Scholar]
- Turbeville J.M., Field K.G., Raff R.A. Phylogenetic position of phylum Nemertini, inferred from 18S rRNA sequences: molecular data as a test of morphological character homology. Mol. Biol. Evol. 1992;9:235–249. doi: 10.1093/oxfordjournals.molbev.a040716. [DOI] [PubMed] [Google Scholar]
- Vinther J., Nielsen C. The early Cambrian Halkieria is a mollusc. Zool. Scr. 2005;34:81–89. doi:10.1111/j.1463-6409.2005.00177.x [Google Scholar]
- Wallberg A., Thollesson M., Farris J.S., Jondelius U. The phylogenetic position of the comb jellies (Ctenophora) and the importance of taxonomic sampling. Cladistics. 2004;20:558–578. doi: 10.1111/j.1096-0031.2004.00041.x. doi:10.1111/j.1096-0031.2004.00041.x [DOI] [PubMed] [Google Scholar]
- Winchell C.J., Sullivan J., Cameron C.B., Swalla B.J., Mallatt J. Evaluating hypotheses of deuterostome phylogeny and chordate evolution with new LSU and SSU ribosomal DNA data. Mol. Biol. Evol. 2002;19:762–776. doi: 10.1093/oxfordjournals.molbev.a004134. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx B.M.H., Backeljau T., Kristensen R.M. Relations of the new phylum Cycliophora. Nature. 1998;393:636–638. doi:10.1038/31377 [Google Scholar]
- Zrzavy J. Gastrotricha and metazoan phylogeny. Zool. Scr. 2003;32:61–81. doi:10.1046/j.1463-6409.2003.00104.x [Google Scholar]
- Zrzavy J., Mihulka S., Kepka P., Bezdek A., Tietz D. Phylogeny of the Metazoa based on morphological and 18S ribosomal DNA evidence. Cladistics. 1998;14:249–285. doi: 10.1111/j.1096-0031.1998.tb00338.x. doi:10.1006/clad.1998.0070 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences analyzed in this study and their GenBank Accesión Numbers
Results obtained with NJ, Treefinder and PhyML
Bayesian and RaxML topologies for the subsets
Discussion on the results for the non-lophotrochozoan clades