Abstract
Polystomatid flatworms are parasites of high host specificity, which mainly infect amphibian hosts. Only one polystome species has so far been recorded from Madagascar despite the high species richness and endemicity of amphibians on this island. Out of the 86 screened Malagasy frog species, we recovered polystomes from 25 in the families Ptychadenidae and Mantellidae. Molecular phylogenetic analysis uncovered an unexpected diversity of polystome species belonging to two separate clades: one forming a lineage within the genus Metapolystoma, with one species in Ptychadena and several species in the mantellid host genera Aglyptodactylus and Boophis; and the second corresponding to an undescribed genus that was found in the species of the subfamily Mantellinae in the family Mantellidae. The phylogenetic position of the undescribed genus along with molecular dating suggests that it may have colonized Madagascar in the Late Mesozoic or Early Cainozoic. By contrast, the more recent origin of Metapolystoma in Madagascar at ca 14–2 Myr ago strongly suggests that the ancestors of Ptychadena mascareniensis colonized Madagascar naturally by overseas dispersal, carrying their Metapolystoma parasites. Our findings provide a striking example of how parasite data can supply novel insights into the biogeographic history of their hosts.
Keywords: Madagascar, Amphibia, Mantellidae, Ptychadenidae, Polystomatidae, biogeography
1. Introduction
Ever since Myers et al. (2000) first identified 25 geographical areas worldwide which qualify as ‘hot spots’ for biodiversity conservation, Madagascar has repeatedly been ranked high in terms of species diversity, endemism and threats to its fauna and flora. This island has been separated from Africa in the west for ca 165–158 Myr (Rabinowitz et al. 1983; Briggs 2003) and for ca 96–84 Myr from India in the east (Storey et al. 1995; Briggs 2003), and its biota has thus evolved under isolation for an extended period of time (Goodman & Benstead 2003; Glaw & Vences 2007).
In the last decade, Madagascar and surrounding landmasses are one of the regions that have inspired a major paradigm shift in historical biogeography: the predominance of vicariance explanations of disjunct distributions on different landmasses has shifted to a resurrection of trans-oceanic dispersal as an alternative scenario of equal standing (De Queiroz 2005; McGlone 2005). This shift was mainly triggered by the possibility of reconstructing phylogenetic relationships of organisms, and estimating their ages or divergence, using molecular methods.
In Madagascar, a Cainozoic dispersal from Africa is now considered as the most probable origin for a variety of endemic lineages, whereas others may have reached the island from South America via Antarctica in the Late Cretaceous (Krause et al. 1997; Case 2002; Vences et al. 2003; Poux et al. 2005; Noonan & Chippindale 2006; Yoder & Nowak 2006). Novel data, resulting from the constant refinement of methods that calibrate and calculate molecular clocks, also indicate that some lineages, such as cichlid fishes, may indeed represent old Gondwanan elements (Azuma et al. 2008).
Despite a wealth of phylogenetic data published on the amphibians of Madagascar (Bossuyt & Milinkovitch 2001; Vences et al. 2003; Andreone et al. 2004; Van der Meijden et al. 2004, 2005, 2007; Glaw et al. 2006; Wollenberg et al. 2007, 2008; Kurabayashi et al. 2008), their biogeographic origins are still incompletely known. Madagascar harbours no salamanders or caecilians, whereas six lineages of neobatrachian ranoid frogs are present. These lineages differ in species diversity and altogether comprise approximately 240 described species, all but one native and endemic to Madagascar. Molecular age estimates are unambiguous in suggesting that the hyperoliid genus Heterixalus originated by overseas dispersal from Africa 30–19 Myr ago (Vences et al. 2003), and it is almost certain that the dicroglossid Hoplobatrachus tigerinus was introduced by humans from India (Guibé 1953; Kosuch et al. 2001). On the contrary, it remains disputed which odysseys account for the arrival of (i) the ancestors of microhylids and mantellids and (ii) the ptychadenid Ptychadena mascareniensis on Madagascar. Molecular data estimate ages (including confidence intervals) of stem splits for the two lineages of microhylids and for mantellids, ca 112–49 Myr ago (Vences et al. 2003; Bossuyt et al. 2006; Van Bocxlaer et al. 2006; Van der Meijden et al. 2007). It has been hypothesized that they reached Madagascar in a mass-concerted dispersal event at the end of the Cretaceous or in the Palaeocene (Van Bocxlaer et al. 2006), at least in the case of mantellids that originated from Asia owing to their nested position in a largely Asian clade of frogs (Van der Meijden et al. 2005). On the other hand, P. mascareniensis was considered an introduced species in Madagascar until molecular data indicated large genetic variability among Malagasy populations, and strong divergence from African populations (Vences et al. 2004; Measey et al. 2007). Although their origin is considered to stem from recent transoceanic dispersal, the available data are not fully conclusive owing to incomplete sampling from Africa.
Coevolution patterns in close parasite–host associations are one example of independent datasets that can most convincingly contribute to biogeographic reconstructions. In amphibians, a particularly close association exists within a group of parasitic platyhelminths of the class Monogenea, the polystomes. In addition to amphibians, these worms infect hosts such as the Australian lungfish, freshwater turtles and the African hippopotamus. They are globally distributed and probably arose during the ecological transition between actinopterygians and sarcopterygians, ca 425 Myr ago (Verneau et al. 2002). Although a number of host switches need to be assumed in their evolution, polystomes usually are very host specific. In fact, a molecular phylogeny as well as molecular estimates of divergence times has been found to be in accordance with the sequence of break-up and fragmentation of Gondwanaland (Badets et al. submitted). Ancestral polystome lineages have been successively isolated in the Australian plate, Indian subcontinent and finally in African and South American continents. Badets et al. (submitted) concluded that the presence of polystome parasites in specific anuran host clades, and in discrete geographical areas, may track the occurrence and radiation of amphibians over ancient and recent geological periods.
To date only a single polystome species, Metapolystoma brygoonis, has been found from an amphibian in Madagascar, i.e. P. mascareniensis (Euzet & Combes 1964). Here we report a spectacular and previously unknown diversity of Malagasy polystomes based on new samples obtained from over 20 mantellid frog species. The molecular phylogenetic relationships of these parasites indicate their dual colonization of Madagascar. We argue that the host association and relationships of these parasites favour a natural origin of P. mascareniensis in Madagascar and may hold the key to fully understand the biogeographic origins of the Mantellidae.
2. Material and methods
(a) Sampling of hosts and parasites
Frogs were collected at night using flashlights. Field collections were carried out in February–March 2005, 2006 and 2007, to coincide with peak anuran activity periods during the rainy season. A variety of species were collected in diverse localities, including Ranomafana and Isalo National Parks, Ambohitantely Special Reserve, the Ankaratra Massif and the surroundings of Andasibe and An'Ala. Host classification follows recent revisions (Glaw & Vences 2006, 2007; Glaw et al. 2006).
Frogs were anaesthetized using ethyl-4-aminobenzoate (MS222), and dissected using a field dissecting microscope in order to check for the presence of polystome flatworms in the urinary bladder. Specimens earmarked for genetic analyses were preserved in 95 per cent ethanol. We altogether recovered parasites from 25 out of the total 86 species of Malagasy frogs screened for polystomes, and they all belonged to the Ptychadenidae and Mantellidae. Thus far we have found no polystomes from Malagasy hyperoliids or microhylid hosts. Based on their morphological features, the parasites were assigned to two different genera, Metapolystoma and an undescribed new genus herein referred to as ‘Madapolystoma’. A formal description of this genus, including adequate morphological data, is going to be published separately; by setting the name ‘Madapolystoma’ in quotation marks and not in italics we use it as a conditional name in the sense of the International Code of Zoological Nomenclature and thus do not coin a nomenclaturally available new name herein.
About 14 polystome specimens from 12 Malagasy host species were included in the molecular analysis. The dataset was complemented with 23 species of the genera Eupolystoma, Metapolystoma, Polystoma, Wetapolystoma, Diplorchis and Parapolystoma (see the electronic supplementary material, appendix A), most of them from previous molecular studies and representative of global polystome diversity in neobatrachian hosts (Bentz et al. 2006; Badets et al. submitted). Finally, in order to explore the phylogenetic relationships within Madagascan Metapolystoma in more detail, we analysed five individuals of four Metapolystoma species and three individuals of two African Polystoma species from partial mitochondrial cytochrome c oxidase subunit I (COI) sequences.
(b) Molecular methods
Specimens were dried and incubated for 1.5 hours at 55°C in 100 μl of Chelex 10 per cent and Proteinase K (final concentration 1 mg ml−1). Extraction reactions were stopped at 100°C for 15 min and DNA preserved at −20°C until use. We amplified the complete 18S rRNA gene in two overlapping fragments of approximately 1 kb each, with the forward F18: 5′-ACCTGGTTGATCCTGCCAGTAG-3′ and reverse 18RG, 5′-CTCTCTTAACCATTACTTCGG-3′ primers for the 5′ terminal end and the forward 18F3: 5′-GGACGGCATGTTTACTTTGA-3′ and reverse IR5: 5′-TACGGAAACCTTGTTACGAC-3′ primers for the 3′ terminal end; and a portion of the 28S rRNA gene in two overlapping fragments of approximately 1 kb and 500 bp, respectively, with the forward LSU5′: 5′-TAGGTCGACCCGCTGAAYTTAAGCA-3′ and reverse IR14: 5′-CATGTTAAACTCCTTGGTCCG-3′ primers for the 5′ terminal end and the forward IF15: 5′-GTCTGTGGCGTAGTGGTAGAC-3′ and reverse LSU3′: 5′-TAGAAGCTTCCTGAGGGAAACTTCGG-3′ primers for the 3′ terminal end. COI was amplified with the forward L-CO1p, 5′-TTTTTTGGGCATCCTGAGGTTTAT-3′ and reverse H-Cox1p2, 5′-TAAAGAAAGAACATAATGAAAATG-3′ primers (Littlewood et al. 1997), yielding a PCR product of approximately 400 bp. PCRs for all 18S, 28S and COI fragments were conducted following the same amplification procedure: one initial step of 5′ at 95°C for long denaturation; 35 cycles of 1′ at 95°C for denaturation, 2′ at 48°C for annealing and 2′ at 72°C for elongation; one final step of 10′ at 72°C for terminal elongation. Each PCR reaction was repeated three times in a final volume of 25 μl with 3 μl of genomic DNA and PCR reagents of the kit GoTaq FlexiDNA Polymerase from Promega (Charbonnières-les-Bains, France) according to the supplier recommendations. PCR products were subsequently purified with the kit Wizard SV Gel and PCR Clean-Up System of Promega and sent to Macrogen Inc. (Seoul, Korea) for sequencing. The F18-18RG PCR portion was sequenced with the reverse 18RC: 5′-TACGAGCTTTTTAACTGCAG-3′ and 18RG primers while the 18F3-IR5 portion was sequenced with the Forward 18F3 and S1: 5′-ATTCCGATAACGAACGAGACT-3′ primers. The LSU5′-IR14 PCR fragment was sequenced with the reverse IR13: 5′-GTCGTGGCTTACACCCTGAGG-3′ and IR14 primers while the IF15-LSU3′ fragment was sequenced with the forward IF15 primer. COI was sequenced with both PCR primers. Newly determined sequences were deposited in GenBank under accession numbers FM897262 to FM897301.
(c) Sequence analysis
Sequences were first corrected for reading errors with the software Sequencher 4.5 of Gene Codes Corporation and then individually aligned with related sequences using the command ED of the MUST package (Philippe 1993) and each observed substitution was verified. Finally, the 18S and 28S sequences were aligned manually with the program ED with all existing sequences selected for phylogenetic analyses. 18S and 28S alignments were visually inspected and gap penalties were introduced when necessary to optimize sequence similarities. Blocks of indels were preferred to multiple single events in the most variable regions considering that one gap of several deletions was a priori more likely than several gaps of one deletion each. Gaps and ambiguous aligned regions were excluded when estimating molecular divergences within the 28S rRNA gene, as well as for the phylogenetic analyses.
As the polystome species from Madagascar have not yet been taxonomically evaluated, we calculated uncorrected pairwise divergences (p-distances) based on the 28S rDNA sequences, using PAUP* v. 4.0b9 (Swofford 2002) as an estimate of their genetic and species diversity in the light of the proposed molecular level of polystome species delineation by Du Preez et al. (2007).
Phylogenetic reconstructions were performed with PAUP* v. 4.0b9 (Swofford 2002) from a concatenated set of the DNA sequences of the two genes (18S and partial 28S). We performed a maximum-parsimony (MP) heuristic search with random addition of taxa (10 replicates) and tree bisection reconnection (TBR) on all 420 equally weighted informative characters. The robustness of nodes was evaluated through a non-parametric bootstrap analysis with 1000 pseudo-replicates. The most appropriate model of sequence evolution for maximum-likelihood (ML) analysis was selected from the hierarchical likelihood ratio tests implemented in the program Modeltest v. 3.06 (Posada & Crandall 1998): GTR+I+Γ, with empirically determined substitution rates ([A,C]=0.8843; [A,G]=6.8568; [A,T]=1.6346; [C,G]=0.7436; [C,T]=4.5578; [G,T]=1.0000) and nucleotide frequencies (Pi [A]=0.2546; Pi [C]=0.1981; Pi [G]=0.2673; Pi [T]=0.2800), a proportion of invariable sites of 0.6543 and a shape parameter of α=0.8063. The ML analysis and ML parametric bootstrapping was performed on 3402 characters in PAUP*, following a heuristic procedure under the TBR and nearest-neighbour interchange branch swapping options, respectively. Bayesian inference (BI) was conducted using the software MrBayes v. 3.04b (Huelsenbeck & Ronquist 2001), with four chains running for a million generations, sampling each 100 cycles. The first 1000 trees were removed as the burn-in phase upon empirical evaluation. The 50 per cent majority rule consensus tree was computed on the last 9000 trees to obtain the Bayesian posterior probability (BPP) for each association. For MP, ML and BI analyses, sequences of Diplorchis ranae and Parapolystoma bulliense were used as outgroups according to the phylogenetic analyses of Badets et al. (submitted).
Molecular relationships within Metapolystoma were inferred from neighbour-joining (NJ) analysis (p-distance) on the 358 characters sequenced for COI.
(d) Molecular estimates of divergence times
The Multidistribute package (Thorne et al. 1998; Thorne & Kishino 2002) was used to infer divergence times among polystomes following Rutschmann (2005). The topology used as support for molecular calibrations corresponded to the phylogenetic tree inferred from the ML analysis, except for Polystoma gallieni that was placed basal to the African Polystoma and Metapolystoma lineage according to Bentz et al. (2001, 2006), and M. brygoonis, i.e. the polystome of P. mascareniensis, which we arbitrarily placed basal within the Madagascan Metapolystoma group in order to break polytomies. ML parameters were estimated under the F84+Γ model with Baseml (Yang 1997) and ML branch lengths and their variance–covariance matrices were calculated with Estbranches. Rates of molecular evolution and divergence times with their 95% confidence intervals were estimated using Multidivtime using the Markov chain Monte Carlo approach.
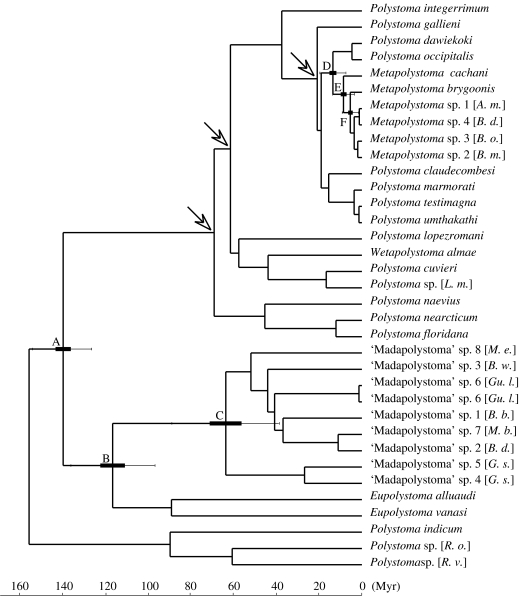
Owing to the lack of fossil calibrations for polystomes, two calibration points were deduced from the biogeographic scenario suggested by Bentz et al. (2001, 2006) and Badets et al. (submitted). Firstly, the root prior was set at 160 Myr ago (s.d.±5 Myr), corresponding to an initial divergence separating Asian and Australian polystomes from all other neobatrachian polystomes (figure 1), hypothetically corresponding to a separation of the western and eastern components of Gondwanaland. Secondly, the divergence of the lineage associating Eurasian and African Polystoma and Metapolystoma from their closest New World relatives was constrained between 65 and 56 Myr ago, reflecting vertebrate exchanges between the two Americas in the Palaeocene (Gayet et al. 1992) and possible dispersal to Eurasia via Beringia. Thirdly, the divergence between P. gallieni and African Polystoma and Metapolystoma was constrained between 25 and 5 Myr ago, reflecting the hypothesized ages of dispersal routes between Eurasia and Africa (Rage 1988; Bentz et al. 2001; Badets et al. submitted).
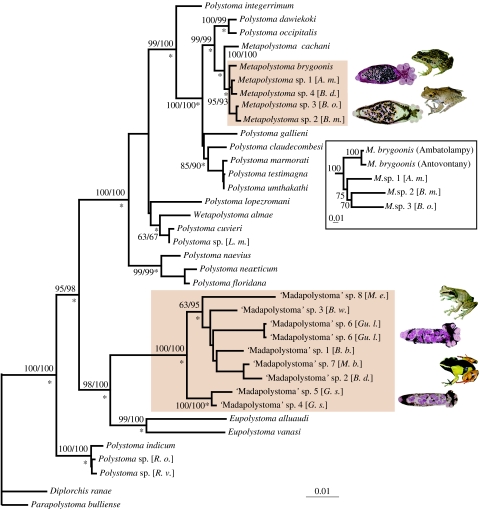
Figure 1.
Best maximum likelihood (ML) tree inferred from the analysis of complete 18S and partial 28S sequences. Abbreviations in brackets refer to host species, from top to bottom: B. m., Boophis madagascariensis; B. o., Boophis occidentalis; A. m., Aglyptodactylus madagascariensis; B. d., Boophis doulioti; L. m., Leptodactylus mystaceus; M. e., Mantella expectata; B. w., Blommersia wittei; Gu. l., Guibemantis liber; B. b., Blommersia blommersae; M. b., Mantella baroni; B. d., Blommersia domerguei; G. s., Gephyromantis sculpturatus; R. o., Rhacophorus omeimontis; R. v., Rhacophorus viridis. Values above or below branches indicate, respectively, maximum parsimony (MP) and ML bootstrap proportions (BPs) after 1000 replicates and asterisks refer to Bayesian posterior probabilities (BPPs) superior to 0.95. The two Malagasy polystome radiations are in brown boxes. Inset photos show M. brygoonis and M. sp. 2 with their host species, P. mascareniensis and Boophis madagascariensis; and two undescribed species of ‘Madapolystoma’ with their host species, Mantella madagascariensis (not included in this analysis, but one of the few host species that yielded a mature ‘Madapolystoma’) and G. liber. The inset tree shows relationships among some Malagasy Metapolystoma based on partial COI sequences, indicating the distinct genetic differentiation and putative basal position of M. brygoonis from the host species P. mascareniensis relative to the specimens recovered from mantellid hosts.
3. Results
(a) Uncovering unknown species diversity in Malagasy polystomes
The encountered parasites belonged to two morphologically clearly distinct groups, one corresponding to the genus Metapolystoma and the second to an undescribed genus (‘Madapolystoma’). p-distances between polystomes from different hosts from 1384 unambiguously aligned positions of 28S revealed, within the Malagasy Metapolystoma group, genetic distances ranging from 0.15 per cent (between the parasites of Aglyptodactylus madagascariensis and Boophis doulioti) to 1.0 per cent (between the parasites of Boophis madagascariensis and P. mascareniensis). Within ‘Madapolystoma’, divergences ranged from 1.5 per cent (between parasites from Mantella baroni and Blommersia domerguei) to 6.0 per cent (between parasites from B. domerguei and Mantella expectata). Two specimens recovered from the same host (G. liber) had identical sequences, and two other specimens recovered from Gephyromantis sculpturatus diverged by 1.3 per cent.
(b) Double phylogenetic origin of Malagasy polystomes
The MP analysis yielded 28 equal most parsimonious trees with a length of 978 steps and a consistency index of 0.557. Differences between these were restricted to the relationships among the Eurafrican Polystoma–Metapolystoma lineage and the two American polystome groups on one hand and the relationships within ‘Madapolystoma’ on the other. The topology of the best ML tree (tree score=11763.5752) was very similar to the MP and Bayesian consensus trees. Here we therefore depict only the phylogenetic relationships of polystomes as inferred from the ML analysis with bootstrap proportions (BP) assessed from MP and ML, and BPPs (figure 1).
Phylogenetic relationships within polystomes indicate with high support values the basal position of P. indicum (BPP=1.00 and BP=95% and 98% in MP and ML, respectively). Two further clades receiving strong support associate (i) Eupolystoma and ‘Madapolystoma’ (1.00, 98%, 100%) and (ii) the American, Eurasian and African Polystoma, Metapolystoma and Wetapolystoma (BPP=1.00; BP=100% in MP and ML). Metapolystoma is monophyletic and nested within Eurafrican Polystoma (1.00, 100%, 100%). Thus, Malagasy polystomes clustered into two unrelated and robust monophyletic groups, i.e. ‘Madapolystoma’ and Metapolystoma, each related to different non-Malagasy taxa. ‘Madapolystoma’ is the sister group of Eupolystoma and the Malagasy Metapolystoma are monophyletic (1.00, 95%, 93%) and sister to the African Metapolystoma cachani (1.00, 100%, 100%). The NJ analysis that was performed from COI sequences indicates with moderate bootstrap values (75%) that M. brygoonis is basal within the Malagasy Metapolystoma, but an MP analysis with the same dataset did not provide relevant support for this topology.
(c) Radically different ages for Malagasy polystome radiations
Divergence time estimates were calculated for six major nodes that are relevant for understanding the origins of Malagasy polystomes (figure 2). According to our analysis, Eupolystoma and the Malagasy endemic ‘Madapolystoma’ diverged ca 116 Myr ago (node B) and the first crown divergence in ‘Madapolystoma’ (node C) took place ca 63 Myr ago. Metapolystoma originated at ca 14 Myr ago (node D), started diversifying in Africa ca 9 Myr ago (node E), and the Malagasy lineage diverged at ca 5 Myr ago (node F). The two groups of Malagasy polystomes thus have radically different ages—Cretaceous versus Pliocene.
Figure 2.
Ultrametric tree inferred from Multidivtime. Arrows indicate the calibration points used for molecular dating (§2). Means of node estimates are given with their standard deviation and 95% confidence intervals. Node A, 139.4±6.9 (152.2–124.9); node B, 116.2±10.0 (134.6–95.6); node C, 63.3±13.0 (89.8–39.9); node D, 13.5±3.0 (19.8–8.0); node E, 8.6±2.5 (14.2–4.3); node F, 5.4±1.9 (9.8–2.3). Abbreviations in brackets are as given in the legend of figure 1.
4. Discussion
(a) Taxonomy of Malagasy polystomes
Our molecular data provide compelling evidence for the presence of two separate lineages of polystomes in Madagascar. The first of these, Metapolystoma has only been reported from the Afrotropical realm and so far was only known to infect grass frogs belonging to the genus Ptychadena in the family Ptychadenidae. The known species includes M. brygoonis from P. mascareniensis of Madagascar, and M. cachani and M. porosissimae from African hosts Ptychadena longirostris and P. porosissima, respectively. The identification of parasites retrieved from species of Ptychadena and from mantellids of the genera Boophis and Aglyptodactylus (subfamilies Boophinae and Laliostominae) was morphologically unambiguous based on the extended uterus, midbody position of the ovary and the presence of hamuli, and was strongly supported by the molecular phylogeny.
On the other hand, polystome specimens recovered from mantellids of the subfamily Mantellinae differed significantly from all known polystome genera through a combination of characters. Based on the posteriorly positioned ovary, extended uterus and intrauterine development of eggs, it closely resembles the African and Indian polystome genus Eupolystoma, but unlike Eupolystoma it possesses hamuli. The assignment of these taxa to a new, undescribed genus is supported by the molecular data that place them as an ancient monophyletic group that is highly distinct from its closest relatives in the genus Eupolystoma.
Du Preez et al. (2007) suggested that a threshold of 0.07 per cent uncorrected pairwise divergence in the 28S gene is usually indicative of well-differentiated species in the genus Polystoma. If this threshold applies also to Malagasy polystomes, the molecular divergences we observed within Metapolystoma and ‘Madapolystoma’ suggest that most or all of the parasites recovered from different hosts are distinct species. This also supports a high host specificity of these parasites and with Madagascar's rich anuran diversity suggests the possible existence of many undiscovered polystome species from Madagascar.
(b) Origins and evolution of Madagascan polystomes
Our phylogenetic analysis provides unambiguous evidence for a double colonization of Madagascar by polystomes over geological time.
The ‘Madapolystoma’ odyssey: our data support the existence of an endemic Malagasy genus, ‘Madapolystoma’, that is the sister group of Eupolystoma. Using the measure of colonization time windows as proposed by Poux et al. (2005), i.e. using 95% confidence intervals of both stem and crown splits, the origin of ‘Madapolystoma’ in Madagascar is estimated at ca 135–40 Myr ago (Nodes B and C; figure 2).
A straightforward explanation for the origin of ‘Madapolystoma’ is not available, considering the convoluted host–parasite association in related genera. The ‘Madapolystoma’ hosts are all in the endemic Malagasy family Mantellidae, and restricted to the subfamily Mantellinae, which is characterized by derived reproductive modes with non-aquatic egg deposition. The closest relatives of mantellids are the rhacophorids (Bossuyt & Milinkovitch 2001; Van der Meijden et al. 2005; Roelants et al. 2007), found mostly in Asia, and probably the mantellid ancestor colonized Madagascar from there (Van der Meijden et al. 2007). However, polystomes recovered from rhacophorids do not belong to Eupolystoma but were basal Polystoma species such as P. indicum, without close affinities to ‘Madapolystoma’ (figure 1).
Eupolystoma includes only five species that have been reported from various host species of Africa and India (Du Preez et al. 2003). Some of them have been recorded from frogs in the family Bufonidae (genera Amietophrynus and Schismaderma), which belong to the hyloid lineage of neobatrachian frogs, phylogenetically distant from mantellids that are deeply nested in the ranoid lineage. Only two Eupolystoma, E. alluaudi and E. rajai, have been reported from ranoid hosts, namely Pyxicephalus adspersus (Pyxicephalidae) and Rana sp. (of uncertain taxonomic affinities, probably belonging to the present-day families Ranidae and Dicroglossidae). Neither of these anuran families (bufonids, pyxicephalids, ranids, nor dicroglossids) occur naturally in Madagascar and the dicroglossid species H. tigerinus was only recently introduced to this island (Guibé 1953). Considering this intricate situation, two alternative scenarios can be outlined that may account for the origin of ‘Madapolystoma’.
As a first possibility, the ‘Madapolystoma’ ancestor may have been carried to Madagascar by hyloid frogs and switched to the mantellids in situ. Evans et al. (2008) recently published a spectacular giant anuran fossil, Beelzebufo, from the latest Cretaceous of Madagascar which, unlike all recent Malagasy frogs, appears to belong to the hyloids. This demonstrates that representatives of this major lineage have been present in the Mesozoic of Madagascar. However, the hypothesis of ancestry of ‘Madapolystoma’ in these early Malagasy hyloids is hampered by the hypothesized affinities of Beelzebufo with South American ceratophryids and not with bufonids. A historical presence and extinction of bufonids in Madagascar is very unlikely considering that these frogs are known to be very adaptive and strong competitors in relation to other anurans.
Alternatively, mantellid ancestors may have carried the ancestor of ‘Madapolystoma’ when colonizing Madagascar. In this case, the presence of related parasites in rhacophorid frogs is to be expected. Although Eupolystoma is not known from recent rhacophorids, E. rajai and E. alluaudi demonstrate that these parasites can infect ranoid frogs. The split between ‘Madapolystoma’ and its closest Eupolystoma relatives would then correspond to the split between rhacophorids and mantellids at ca 86–56 Myr ago (Vences et al. 2003; Bossuyt et al. 2006; Van Bocxlaer et al. 2006; Van der Meijden et al. 2007), and these parasites probably went extinct in the lineages leading to laliostomine and boophine mantellids, and to recent rhacophorids.
Other hypotheses, such as an origin in microhylid hosts that colonized Madagascar simultaneously with mantellids (Van Bocxlaer et al. 2006) are not impossible, but unlikely, because no polystomes have so far been found in microhylids, neither in our screenings of Malagasy species nor in any other species of this cosmopolitan group. Even more unlikely is a colonization of Madagascar by ‘Madapolystoma’ via Heterixalus, an endemic genus that entered the island much more recently than mantellids (Vences et al. 2003). A crucial factor is the limited knowledge on polystomes from Asia, the Seychelles islands and especially India. A wider screening of the deep endemic frog lineages of India is badly needed. Particular emphasis should be placed on uncovering polystomes of rhacophorids. A hypothetical presence of Eupolystoma in South Asian rhacophorids would provide immediate and unambiguous support for the second scenario outlined above. The prediction is that these Eupolystoma would be phylogenetically sister to ‘Madapolystoma’, rendering Eupolystoma paraphyletic, which could provide an elegant explanation for an odyssey by the ancestors of ‘Madapolystoma’ and of mantellid frogs that lead to the colonization of Madagascar.
The Metapolystoma odyssey: Madagascan Metapolystoma include polystome species that were recovered from host species of two unrelated frog families (Ptychadenidae and Mantellidae). These two frog lineages are not closely related and their divergence dates back far into the Cretaceous or even Jurassic (Van der Meijden et al. 2005; Frost et al. 2006; Roelants et al. 2007). The time window (Poux et al. 2005) for the colonization of Madagascar by Metapolystoma is ca 14.2–2.3 Myr ago. Considering this apparently young age of Metapolystoma, a host switch is required in order to explain its presence in these two disparate frog groups. In this context, it is relevant that the mantellids found to be infected by Metapolystoma (in the subfamilies Boophinae and Laliostominae) are those that conserve a plesiomorphic reproductive mode, i.e. a mating amplexus with deposition of eggs directly into water, while species in the subfamily Mantellinae (hosting ‘Madapolystoma’; see above) have a derived reproduction with non-aquatic egg deposition (Glaw & Vences 2006). Furthermore, at least two of the Metapolystoma host species identified (A. madagascariensis and Boophis tephraeomystax) breed in lentic water bodies and are often syntopic with P. mascareniensis which would favour a host switch.
The young age of Malagasy Metapolystoma also indicates that they diverged from their African relatives much more recently than mantellids diverged from their non-Malagasy relatives in the Late Cretaceous or Palaeocene (Vences et al. 2003; Van der Meijden et al. 2005; Bossuyt et al. 2006; Van Bocxlaer et al. 2006). Thus, it is not probable that Metapolystoma was present in the mantellid ancestor colonizing Madagascar. The time frame of Metapolystoma origin leaves two host groups as candidates to have carried Metapolystoma to Madagascar: hyperoliids and ptychadenids. The ancestor giving rise to the Malagasy hyperoliid genus Heterixalus and the Seychellean genus Tachycnemis diverged from its African relatives at ca 30–19 Myr ago, which is older than the estimated Metapolystoma origin in Madagascar but at least falls within the 95% confidence interval of the origin of these parasites in Africa (figure 2). Thus far our screening of 99 individuals of six Heterixalus species yielded no evidence for infection. On the other hand, five polystome species are known from African hyperoliids. As polystome infections are known to be overdispersed, it cannot be excluded that polystomes will be discovered in Malagasy hyperoliids. Nevertheless, it remains true that hyperoliid hosts so far were found to contain only Polystoma and no Metapolystoma parasites.
On the contrary, ptychadenids are well-established Metapolystoma hosts (see above and electronic supplementary material, appendix A), and in the phylogeny M. cachani, from an African Ptychadena species, stands basal to the Malagasy Metapolystoma (figure 1). Our phylogeny was ambiguous regarding the placement of M. brygoonis relative to the Metapolystoma from mantellid hosts (figure 1). Although NJ analysis inferred from COI sequences favoured a basal position of M. brygoonis within Malagasy Metapolystoma, the MP analyses were not conclusive. More extensive molecular work with inclusion of additional genes is necessary to confirm whether M. brygoonis is indeed the most basal Malagasy Metapolystoma. In such a case, parsimony arguments would be unambiguous in indicating an origin of Metapolystoma from ptychadenid hosts. Nevertheless, such an origin is already now convincingly supported by (i) the evidence for a Metapolystoma divergence too recent to fit with other anuran colonization events of Madagascar and (ii) a parsimony attempt to minimize the number of host switches between genera. Our data thus support the hypothesis that Ptychadena carried Metapolystoma to Madagascar, with a secondary host switch to mantellid frogs that had already diversified on the island. Du Preez & Kok (1992) reported the susceptibility of the host genus Ptychadena as a host for polystome parasites. No less than 52 per cent of known African polystomes have been described from Ptychadena spp. and 26 per cent of known Ptychadena species harbour polystome parasites. Conversely, because Metapolystoma in various mantellid species represent different species with substantial genetic differentiation, this host switch must have occurred significantly prior to the colonization of Madagascar by humans, ca 2300 years ago. The polystome data thus provide additional support for a natural transoceanic colonization of Madagascar by the ancestor of P. mascareniensis (Vences et al. 2004; Measey et al. 2007).
Acknowledgments
Research was carried out at the BETM Laboratory at the University of Perpignan. Permit no. A66040 was obtained from Ministère del'Agriculture et de la Pêche and French Ministère de l'Education Nationale de la Recherch et de la Technologie. Permits for fieldwork in Madagascar were obtained by M.V.
We are grateful to numerous colleagues, students and assistants for their help in the field, in particular to Parfait Bora, James and Carol Patton, Roger-Daniel Randrianiaina, David R. Vieites and Che Weldon. We also thank Martins Aisien for providing specimens of M. cachani from Nigeria. The research was carried out in the framework of collaboration between the authors' institutions and the Département de Biologie Animale of the Université d'Antananarivo, and the Association Nationale pour la Gestion des Aires Protégées, ANGAP. We are grateful to the Malagasy authorities for providing research and export permits. Funding was provided through the CNRS (to O.V.), the Deutsche Forschungsgemeinschaft (to M.V.) and the Volkswagen Foundation (to M.V., L.H. D.P., L.R. and F.G.).
Supplementary Material
Parasite species investigated, their host species, geographical origin, authority and GenBank accession numbers
References
- Andreone F., Vences M., Vieites D.R., Glaw F., Meyer A. Recurrent ecological adaptations revealed through a molecular analysis of the secretive cophyline frogs of Madagascar. Mol. Phylogenet. Evol. 2004;34:315–322. doi: 10.1016/j.ympev.2004.10.013. doi:10.1016/j.ympev.2004.10.013 [DOI] [PubMed] [Google Scholar]
- Azuma Y., Kumazawa Y., Miya M., Mabuchi K., Nishida M. Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. BMC Evol. Biol. 2008;8:e215. doi: 10.1186/1471-2148-8-215. doi:10.1186/1471-2148-8-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badets, M. et al Submitted. Correlating vicariance of parasitic platyhelminths during Gondwanaland break-up to the early evolution of neobatrachian frogs.
- Bentz S., Leroy S., Du Preez L., Mariaux J., Vaucher C., Verneau O. Origin and evolution of African Polystoma (Monogenea: Polystomatidae) assessed by molecular methods. Int. J. Parasitol. 2001;31:697–705. doi: 10.1016/s0020-7519(01)00179-5. doi:10.1016/S0020-7519(01)00179-5 [DOI] [PubMed] [Google Scholar]
- Bentz S., Sinnappah-Kang N.D., Lim L.H.S., Lebedev B., Combes C., Verneau O. Historical biogeography of amphibian parasites, genus Polystoma (Monogenea: Polystomatidae) J. Biogeogr. 2006;33:742–749. doi:10.1111/j.1365-2699.2005.01402.x [Google Scholar]
- Bossuyt F., Milinkovitch M.C. Amphibians as indicators of early tertiary ‘Out-of-India’ dispersal of vertebrates. Science. 2001;292:93–95. doi: 10.1126/science.1058875. doi:10.1126/science.1058875 [DOI] [PubMed] [Google Scholar]
- Bossuyt F., Brown R.M., Hillis D.M., Cannatella D.C., Milinkovitch M.C. Phylogeny and biogeography of a cosmopolitan frog radiation: late Cretaceous diversification resulted in continent-scale endemism in the family Ranidae. Syst. Biol. 2006;55:579–594. doi: 10.1080/10635150600812551. doi:10.1080/10635150600812551 [DOI] [PubMed] [Google Scholar]
- Briggs J.C. The biogeographic and tectonic history of India. J. Biogeogr. 2003;30:381–388. doi:10.1046/j.1365-2699.2003.00809.x [Google Scholar]
- Case J.A. A new biogeographical model for dispersal of Late Cretaceous vertebrates into Madagascar and India. J. Vertebr. Paleont. 2002;22(Suppl. 3):42A. [Google Scholar]
- De Queiroz A. The resurrection of oceanic dispersal in historical biogeography. Trends Ecol. Evol. 2005;20:68–73. doi: 10.1016/j.tree.2004.11.006. doi:10.1016/j.tree.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Du Preez L.H., Kok D.J. The frog genus Ptychadena as host for polystomatid (Monogenea) parasites in Africa. J. Herpetol. Assoc. Afr. 1992;40:47–49. [Google Scholar]
- Du Preez L.H., Tinsley R.C., De Sa R. Polystomatidae (Monogenea) of Southern African Anura: Eupolystoma vanasi n. sp. parasitic in Schismaderma carens (Smith, 1848) Syst. Parasitol. 2003;54:71–79. doi: 10.1023/a:1022169106845. doi:10.1023/A:1022169106845 [DOI] [PubMed] [Google Scholar]
- Du Preez L.H., Verneau O., Gross T.S. Polystoma floridana n. sp. (Monogenea: Polystomatidae), a parasite in the green tree frog, Hyla cinerea (Schneider), of North America. Zootaxa. 2007;1663:33–45. [Google Scholar]
- Euzet L., Combes C. Sur un Polystomatidae (Monogenea) récolté à Madagascar chez Rana mascareniensis Duméril et Bibon. Bull. Soc. Geol. Fr. 1964;89:392–401. [Google Scholar]
- Evans S.E., Jones M.E.H., Krause D.W. A giant frog with South American affinities from the late Cretaceous of Madagascar. Proc. Natl Acad. Sci. USA. 2008;105:2951–2956. doi: 10.1073/pnas.0707599105. doi:10.1073/pnas.0707599105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost D.R., et al. The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 2006;28:1–370. doi:10.1206/0003-0090(2006)297[0001:TATOL]2.0.CO;2 [Google Scholar]
- Gayet M., Rage J.C., Sempere T., Gagnier P.Y. Modalités des échanges de vertébrés continentaux entre l'Amérique du nord et l'Amérique du sud au Crétacé supérieur et au Paléocène. Bull. Soc. Geol. Fr. 1992;163:781–791. [Google Scholar]
- Glaw F., Vences M. Phylogeny and genus-level classification of mantellid frogs (Amphibia Anura) Org. Divers. Evol. 2006;6:236–253. doi:10.1016/j.ode.2005.12.001 (Electr.. Suppl. 11, part 1: 1–3) [Google Scholar]
- Glaw F., Vences M. 3rd edn. Frosch Verlag; Cologne, Germany: 2007. A field guide to the amphibians and reptiles of Madagascar. [Google Scholar]
- Glaw F., Hoegg S., Vences M. Discovery of a new basal relict lineage of Madagascan frogs and its implications for mantellid evolution. Zootaxa. 2006;1334:27–43. [Google Scholar]
- Goodman S.M., Benstead J.P. The University of Chicago Press; Chicago, IL: 2003. The natural history of Madagascar. [Google Scholar]
- Guibé J. Au sujet de l'introduction de Rana tigerina Daudin à Madagascar. Nat. Malgache. 1953;5:241–242. [Google Scholar]
- Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Kosuch J., Vences M., Dubois A., Ohler A., Böhme W. Out of Asia: mitochondrial DNA evidence for an oriental origin of tiger frogs, genus Hoplobatrachus. Mol. Phylogenet. Evol. 2001;21:398–407. doi: 10.1006/mpev.2001.1034. doi:10.1006/mpev.2001.1034 [DOI] [PubMed] [Google Scholar]
- Krause D.W., Prasad G.V.R., von Koenigswald W., Sahni A., Grine F.E. Cosmopolitanism among Gondwanan Late Cretaceous mammals. Nature. 1997;390:504–507. doi:10.1038/37343 [Google Scholar]
- Kurabayashi A., Sumida M., Yonekawa H., Glaw F., Vences M., Hasegawa M. Phylogeny, recombination, and mechanisms of stepwise mitochondrial genome reorganization in mantellid frogs from Madagascar. Mol. Biol. Evol. 2008;25:874–891. doi: 10.1093/molbev/msn031. doi:10.1093/molbev/msn031 [DOI] [PubMed] [Google Scholar]
- Littlewood D.T.J., Rohde K., Clough K.A. Parasite speciation within or between host species? Phylogenetic evidence from site-specific polystome monogeneans. Int. J. Parasitol. 1997;27:1289–1297. doi: 10.1016/s0020-7519(97)00086-6. doi:10.1016/S0020-7519(97)00086-6 [DOI] [PubMed] [Google Scholar]
- McGlone M.S. Goodbye Gondwana. J. Biogeogr. 2005;32:739–740. doi:10.1111/j.1365-2699.2005.01278.x [Google Scholar]
- Measey G.J., Vences M., Drewes R.C., Chiari Y., Melo M., Bourles B. Freshwater paths across the ocean: molecular phylogeny of the frog Ptychadena newtoni gives insights into amphibian colonization of oceanic islands. J. Biogeogr. 2007;34:7–20. doi:10/1111/j.1365-2699.2006.01589.x [Google Scholar]
- Myers N., Mittermeier R.A., Mittermeier C.G., da Fonseca G.A.B., Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. doi:10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- Noonan B.P., Chippindale P.T. Vicariant origin of Malagasy reptiles supports Late Cretaceous Antarctic land bridge. Am. Nat. 2006;168:730–741. doi: 10.1086/509052. doi:10.1086/509052 [DOI] [PubMed] [Google Scholar]
- Philippe H. MUST, a computer package for management utilitarians for sequences and trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. doi:10.1093/nar/21.22.5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Poux C., Madsen O., Marquard E., Vieites D.R., De Jong W.W., Vences M. Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst. Biol. 2005;54:719–730. doi: 10.1080/10635150500234534. doi:10.1080/10635150500234534 [DOI] [PubMed] [Google Scholar]
- Rabinowitz P.D., Coffin M.F., Falvey D. The separation of Madagascar and Africa. Science. 1983;220:67–69. doi: 10.1126/science.220.4592.67. doi:10.1126/science.220.4592.67 [DOI] [PubMed] [Google Scholar]
- Rage J.C. Gondwana Tethys, and terrestrial vertebrates during the Mesozoic and Cainozoic. In: Audley-Charles M.G., Hallam A., editors. Gondwana and Tethys. Geological Society Special Publication; London, UK: 1988. pp. 255–273. [Google Scholar]
- Roelants K., Gower D.J., Wilkinson M., Loader S.P., Biju S.D., Guillaume K., Moriau L., Bossuyt F. Global patterns of diversification in the history of modern amphibians. Proc. Natl Acad. Sci. USA. 2007;104:887–892. doi: 10.1073/pnas.0608378104. doi:10.1073/pnas.0608378104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann F. University of Zurich; Zurich, Switzerland: 2005. Bayesian molecular dating using PAML/multidivtime. A step-by-step manual.http://www.plant.ch [Google Scholar]
- Storey M., Mahoney J.J., Saunders A.D., Duncan R.A., Kelley S.P., Coffin M.F. Timing of hot spot-related volcanism and the breakup of Madagascar and India. Science. 1995;267:852–855. doi: 10.1126/science.267.5199.852. doi:10.1126/science.267.5199.852 [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0b9. Sunderland, MA: Sinauer Associates.
- Thorne J.L., Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. doi:10.1080/10635150290102456 [DOI] [PubMed] [Google Scholar]
- Thorne J.L., Kishino H., Painter I.S. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Van Bocxlaer I., Roelants K., Biju S.D., Nagaraju J., Bossuyt F. Late Cretaceous vicariance in Gondwanan amphibians. PLoS ONE. 2006;1:1–6. doi: 10.1371/journal.pone.0000074. doi:10.1371/journal.pone.0000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meijden A., Vences M., Meyer A. Novel phylogenetic relationships of the enigmatic brevicipitine and scaphiophrynine toads as revealed by sequences from the nuclear Rag-1 gene. Proc. Biol. Sci. 2004;271:S378–S381. doi: 10.1098/rsbl.2004.0196. doi:10.1098/rsbl.2004.0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meijden A., Vences M., Hoegg S., Meyer A. A previously unrecognized radiation of ranid frogs in southern Africa revealed by nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2005;37:674–685. doi: 10.1016/j.ympev.2005.05.001. doi:10.1016/j.ympev.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Van der Meijden A., Vences M., Hoegg S., Boistel R., Channing A., Meyer A. Nuclear gene phylogeny of narrow-mouthed toads (family: Microhylidae) and a discussion of competing hypotheses concerning their biogeographical origins. Mol. Phylogenet. Evol. 2007;44:1017–1030. doi: 10.1016/j.ympev.2007.02.008. doi:10.1016/j.ympev.2007.02.008 [DOI] [PubMed] [Google Scholar]
- Vences M., Vieites D.R., Glaw F., Brinkmann H., Kosuch J., Veith M., Meyer A. Multiple overseas dispersal in amphibians. Proc. R. Soc. Lond. B. 2003;270:2435–2442. doi: 10.1098/rspb.2003.2516. doi:10.1098/rspb.2003.2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vences M., Kosuch J., Rödel M.O., Lötters S., Channing A., Glaw F., Böhme W. Phylogeography of Ptychadena mascareniensis suggests transoceanic dispersal in a widespread African-Malagasy frog lineage. J. Biogeogr. 2004;31:593–601. doi:10.1046/j.1365-2699.2003.01031.x [Google Scholar]
- Verneau O., Bentz S., Sinnappah N.D., Du Preez L., Whittington I., Combes C. A view of early vertebrate evolution inferred from the phylogeny of polystome parasites (Monogenea: Polystomatidae) Proc. R. Soc. B. 2002;269:535–543. doi: 10.1098/rspb.2001.1899. doi:10.1098/rspb.2001.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg K.C., Glaw F., Meyer A., Vences M. Molecular phylogeny of Malagasy reed frogs Heterixalus, and the relative performance of bioacoustics and color-patterns for resolving their systematics. Mol. Phylogenet. Evol. 2007;45:14–22. doi: 10.1016/j.ympev.2007.06.024. doi:10.1016/j.ympev.2007.06.024 [DOI] [PubMed] [Google Scholar]
- Wollenberg K.C., Vieites D.R., Van der Meijden A., Glaw F., Cannatella D.C., Vences M. Patterns of endemism and species richness in Malagasy cophyline frogs support a key role of mountainous areas for speciation. Evolution. 2008;62:1890–1907. doi: 10.1111/j.1558-5646.2008.00420.x. doi:10.1111/j.1558-5646.2008.00420.x [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. http://abacus.gene.ucl.ac.uk/software/paml.html [DOI] [PubMed] [Google Scholar]
- Yoder A.D., Nowak M.D. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu. Rev. Ecol. Evol. Syst. 2006;37:405–431. doi:10.1146/annurev.ecolsys.37.091305.110239 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Parasite species investigated, their host species, geographical origin, authority and GenBank accession numbers