Abstract
Elite suppressors (ES) are a rare subset of HIV-1–infected individuals who are able to maintain HIV-1 viral loads below the limit of detection by ultra-sensitive clinical assays in the absence of antiretroviral therapy. Mechanism(s) responsible for this elite control are poorly understood but likely involve both host and viral factors. This study assesses ES plasma-derived envelope glycoprotein (env) fitness as a function of entry efficiency as a possible contributor to viral suppression. Fitness of virus entry was first evaluated using a novel inducible cell line with controlled surface expression levels of CD4 (receptor) and CCR5 (co-receptor). In the context of physiologic CCR5 and CD4 surface densities, ES envs exhibited significantly decreased entry efficiency relative to chronically infected viremic progressors. ES envs also demonstrated slow entry kinetics indicating the presence of virus with reduced entry fitness. Overall, ES env clones were less efficient at mediating entry than chronic progressor envs. Interestingly, acute infection envs exhibited an intermediate phenotypic pattern not distinctly different from ES or chronic progressor envs. These results imply that lower env fitness may be established early and may directly contribute to viral suppression in ES individuals.
Author Summary
The majority of HIV-1–infected individuals experience high plasma viral loads and CD4+ T cells loss in the absence of antiretroviral therapy. However, a very rare and important subset of individuals termed elite suppressors is able to maintain HIV-1 plasma viral loads below the limit of viral detection in the absence of treatment. The reasons behind this ability to control the virus are poorly understood, but they likely involve both an effective host immune response against HIV-1 and factors related to the virus itself. Here, we analyze the function of the HIV-1 coat protein or envelope glycoprotein from a group of elite suppressors. HIV-1 envelope mediates entry into the host cell via interaction with the cellular receptors CD4 and CCR5. Envelopes from elite controllers interacted with these receptors inefficiently compared to those from individuals with detectable viral loads. These inefficient interactions by elite suppressor envelopes led to slow rates of entry into host cells. Envelopes from acutely infected individuals were not significantly different from elite suppressors or chronically infected individuals. These findings suggest that the decreased envelope efficiency may contribute to viral control in elite suppressors.
Introduction
A minor subset of HIV-1–infected individuals maintains stable CD4+ T cell counts in the absence of antiretroviral therapy. A small proportion of these long-term nonprogressors (LTNPs), termed elite suppressors (ES), control plasma viral loads to <50 copies/ml [1]. Mechanism(s) responsible for this elite control are poorly understood but likely involve host and viral factors. Studies have explored the contributions of the innate and adaptive immune responses, host genetic polymorphisms, and viral dynamics (reviewed in [2]). For example, the major histocompatibility complex class (MHC) I group B alleles HLA-B27, -B51, and –B57 have been strongly associated with slower rates of HIV-1-associated disease progression [3]–[6]. Although these HLA-B alleles are overrepresented in ES and LTNPs, they are only expressed in a subset of these individuals indicating that the presence of these alleles is not necessary to suppress viremia and that other factors are likely involved [4],[7].
Although much previous work on ES has focused on host factors, less is known about viral fitness in these individuals. The impact of viral attenuation on disease progression was first described in a cohort of LTNPs infected by a common donor with virus containing a deletion in the nef gene [8],[9]. Investigation of other LTNP cohorts has shown both the presence [10],[11] and absence [12],[13] of defective nef genes. In other cohorts, the presence of viruses with reduced replication capacity has been associated with slower disease progression [14]–[19]. This viral attenuation could be the result of divergent evolution as a result of direct selective pressure by the host immune response [16]–[19]. However, recent work has shown that replication-competent viruses can be recovered from ES individuals indicating that ES harbor functional virus [20]. Furthermore, large scale sequencing of ES viruses yielded no identifiable common genetic defects [21]. Investigating the relative fitness of viral quasispecies in ES will help determine whether viral fitness is influencing disease outcome in these individuals.
Low HIV-1 genetic diversity in ES may be indicative of the presence of lower fitness variants [22]. Sequence analysis of functional envelope glycoprotein ES clones showed significantly decreased env diversity compared to individuals with chronic viremia suggesting that viruses in these patients experience minimal viral replication and diversification [23]. Lack of env diversification suggests that ES envs may be closely related in genotype and phenotype to the founder virus establishing infection.
In this study we have performed rigorous phenotypic analysis on subtype B env clones from ES plasma virus to determine whether env fitness may be contributing to viral suppression in ES. A novel cell line was utilized to show that ES env clones exhibit low CD4 receptor and CCR5 co-receptor usage and slow fusion kinetics compared to chronic infection envs. Analysis of control viruses indicated that these characteristics directly correlated to reduced replication capacity in vitro. Acute infections envs were intermediate in their entry efficiency and not significantly different from either chronic or ES envs. This study provides the first direct evidence that decreased env function is a property of ES and that this may contribute to viral suppression.
Materials and Methods
Ethics statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Board of Johns Hopkins School of Medicine and Rockefeller University hospitals. All patients provided written informed consent for the collection of samples and subsequent analysis.
Patient profiles
The elite suppressor and chronic progressor patients have been previously described [23] (Table 1). Patients identified with acute/early HIV-1 infection have been previously described [24]. The estimated duration of infection was calculated 2 weeks prior to the onset of acute retroviral illness unless the patient could identify a precise high risk event. Table 1 contains relevant enrollment data for all acute/early infection patients. Elite suppressors were defined as individuals who maintained viral load to below 50 copies of RNA/ml plasma in the absence of retroviral therapy yet were Western blot positive for infection. Informed consent was obtained prior to phlebotomy. The protocol for ES/CP or acute/early infection was approved by an institutional review board of the Johns Hopkins University School of Medicine and the Aaron Diamond AIDS Research Center, respectively.
Table 1. Patient characteristics of elite suppressors, chronic progressors, and acutely infected individuals.
| Patient | Year of diagnosis | Sampling date | Duration of infectiona | Plasma viral load (RNA copies/ml) | CD4 count (cells/µl) |
| Elite Suppressors | |||||
| ES2 | 1986 | 5/04 | 18 years | <50 | 383 |
| ES3 | 1991 | 3/04 | 13 years | <50 | 677 |
| ES4 | 1996 | 8/04 | 8 years | <50 | 837 |
| ES7 | 1994 | 1/05 | 11 years | <50 | 1,125 |
| ES8 | 2003 | 6/04, 9/04 | 1 year | <50 | 458 |
| ES9 | 1999 | 3/04, 8/04 | 5 years | <50 | 800 |
| ES10 | 2002 | 3/04 | 2 years | <50 | 900 |
| Chronic Progressors | |||||
| C61 | 1999 | 9/04 | 5 years | 19,100 | 1,261 |
| C62 | 1998 | 9/04 | 6 years | 33,300 | 481 |
| C93 | 2001 | 3/05 | 4 years | 47,270 | 402 |
| C94 | 1999 | 2/05 | 6 years | 22,898 | 351 |
| C96 | 2001 | 3/05 | 4 years | 12,500 | 400 |
| C98 | 2004 | 3/05 | 1 year | 17,838 | 426 |
| C109 | 2002 | 5/05 | 3 years | 61,000 | 222 |
| Acute Infection | |||||
| 502 | – | – | 19 days | 31,622,777 | 306 |
| 503 | – | – | 17 days | 588,844 | 531 |
| 504 | – | – | 32 days | 2,691,535 | 152 |
| 506 | – | – | 43 days | 467,735 | 341 |
| 508 | – | – | 26 days | 389,045 | 266 |
| 510 | – | – | 16 days | 1,584,893 | 581 |
| 512 | – | – | 25 days | 3,388,442 | 745 |
| 514 | – | – | 26 days | 676,083 | 291 |
| 516 | – | – | 51 days | 295,121 | 371 |
| 517 | – | – | 38 days | 87,096 | 588 |
| 518 | – | – | 15 days | 21,379,621 | 226 |
| 519 | – | – | 22 days | 1,819,701 | 322 |
| 520 | – | – | 40 days | 257,040 | 512 |
| 522 | – | – | 21 days | 3,235,937 | 583 |
| 523 | – | – | 28 days | 173,780 | 273 |
| 526 | – | – | 26 days | 100,000 | 205 |
| 527 | – | – | 21 days | 134,896 | 538 |
| 528 | – | – | 16 days | 12,022,644 | 438 |
| 529 | – | – | 19 days | 28,840,315 | 537 |
| 530 | – | – | 30 days | 6,456,542 | 469 |
Time of infection expressed in approximate years for ES and CP patients from diagnosis to sampling. Time of infection for acute infection samples estimated by 14 days after the onset of acute seroconversion symptoms or identification of high risk transmission event, and sampling was conducted <4 weeks post-presentation and initiation of HAART.
Generation of pseudotype virus and infection of Affinofile cells
Envelope expression vectors were generated as previously described [23]. Envelope pseudotypes were generated by cotransfection of 293T cells with the 1 µg of the luciferase-encoding pseudotyping vector pNLLuc.AM and 1 µg of envelope expression vector. Cells were washed after 24 h, and pseudoviruses were collected after a subsequent 48 h. Relative particle numbers were determined by limiting dilution reverse transcriptase assay. Viruses were characterized as exclusively CCR5-utilizing by comparison of infectivity of U87-CD4/CCR5 and U87-CD4/CXCR4 cells, as previously described [25]. Affinofile cells were generated by selection of 4 vector stable cells (Johnston et al., submitted). CCR5 expression is controlled by a two vector ecdysone-inducible promoter. pVgRXR encodes the VgEcR fusion protein under control of the CMV promoter, and the RXR open reading frame under control of the RSV 5′ long terminal repeat. pIND-CCR5 encodes CCR5 under control of the minimal heat shock promoter with inducible control provided by five repeats of the glucocorticoid receptor DNA binding domains (5×E/GRE). Addition of the ecdysone derivative ponasterone A (the inducer) results in recruitment of a transcriptional coactivator to the 5×E/GRE element and activation of transcription of the CCR5 ORF. CD4 expression is inducibly regulated by the TREx expression system (Invitrogen). Cells contain pcDNA5-TO-CD4 and transcription of the CD4 ORF is controlled by the addition of the tetracycline analog minocycline. Single cell clones were isolated to generate cell populations with consistent levels of induction upon stimulation of CD4 and CCR5 expression.
Affinofile cells were plated at a density of 10,000 cells per well in a 96-well plate and allowed to adhere for 48 hours. Cells were induced in a matrix pattern to express CD4 and CCR5. Minocycline was added to cells in 2-fold dilutions over 6 separate dilutions (5 ng/ml–0 ng/ml) to induce CD4 expression. Ponasterone A was added in 2-fold dilutions over 6 separate dilutions from a final concentration of 4 µM to 0 µM to induce CCR5 expression. This matrix results in 36 unique CD4 and CCR5 induction surface concentrations. Each drug concentration was induced in triplicate. Cells were induced for 24 hours prior to infection. Cells were then exposed to pseudovirus for 48 h, washed with PBS, and lysed with Glo lysis buffer (Promega, Inc.). Maximal infection was considered luciferase activity generated by infection at the highest CD4 and highest CCR5 concentration. To control for effects caused directly by minocycline and/or ponasterone A on viral infectivity, U87-CD4/CCR5 cells were treated with a similar matrix of both drugs. CCR5 and CD4 expression levels were unchanged by flow cytometry, and no changes in infectivity of Yu-2 and SF162 envelope pseudoviruses were noted, thus variation in infectivity was assumed to be due to variations in receptor expression levels (Johnston et al, submitted).
Kinetic fusion and reverse transcription assays
For the kinetic fusion assay, HIV-1 pseudoviruses bearing either ES or chronic envelopes were spinonculated onto U87-CD4/CCR5 cells. 2.5×106 cells were spin-infected with pseudovirus-containing supernatant for 90 min at 1,200×g at 4°C. The cells were washed twice with cold phosphate buffered saline (PBS) to remove unbound virions. Cells were resuspended in cold medium and split into 96-well plates (50 µl/well). Virus-cell mixes were synchronized for entry by addition of 130 µl of 37°C medium, and then ENF at 10 µM was added to each well in 20 µl of medium at fixed-time intervals after addition of warm medium, which is defined as tim = 0 for synchronization of viral replication. Cells were incubated for 48 h and then treated with lysis buffer and luciferase activity was determined. For the kinetic fusion assay 6 hours was used as 100% or maximal luciferase activity. For the reverse transcription assay, Affinofile cells were induced with 5 ng/ml minocycline 24 hours prior to infection. ES or chronic pseudoviruses were synchronously added to cells. Efavirenz (EFV) was added at a concentration of 1 µM to each well at fixed time intervals after the addition of virus. For reverse transcription assay, 12 hours was used as 100% or maximal luciferase activity.
Entry inhibitor susceptibility
Affinofile cells were induced 24 hours prior to infection with 5 ng/ml minocycline. Cells were incubated with serial 10-fold dilutions of either chemokine (CCL5 [50 nM to 0.1 nM]) or drug (ENF (T-20) [1 µM to 0.1 nM], TAK-779 [1 µM to 0.1 nM]) for 1 h prior to the addition of virus. Cells were incubated for 48 h, washed with PBS, lysed, and luciferase activity determined. Plots of luciferase activity versus drug concentration were used to determine IC50 values for each pseudovirus. Luciferase activity without drug was used as maximal or 100% infection value.
Flow cytometry
Peripheral blood mononuclear cells (PBMCs) were isolated from heparin-treated venous whole blood from HIV–seronegative donors by Ficoll-paque density gradient centrifugation (GE Healthcare, Piscataway, NJ). Isolated PBMCs were washed twice in wash buffer [phosphate-buffered saline (PBS) supplemented with 2% fetal bovine serum (FBS), 0.1% glucose, 12 mM HEPES, and penicillin (100 U/m) and streptomycin (100 µg/ml)] and activated in RPMI 1640 (Mediatech, Inc., Manassas, VA) supplemented with 10% FBS, penicillin/streptomycin, and 2 µg/ml phytohemmaglutinin (PHA, Sigma-Aldrich, St. Louis, MO) and 100 U/ml interleukin 2 (IL-2, Invitrogen, Carlsbad, CA) for 3 days at 37°C and 5% CO2. Total PBMCs were subsequently maintained in RPMI 1640 supplemented with 10% FBS, pen/strep, and 100 U/ml IL-2. For flow cytometry experiments, a total of 10° PBMCs were collected 4 days post-stimulation by centrifugation at 2500 rpm×10 minutes and washed once in FACS staining buffer (PBS with 2% FBS, 0.5% bovine serum albumin, and 0.02 sodium azide). The cells were incubated in either an anti-CD4 antibody [Fluorescein isothiocyanate(FITC)-conjugated anti-CD4, BD Biosciences Pharmingen, San Jose, CA] or FITC-conjugated IgG1 isotype control (BD Biosciences Pharmingen), or an anti-CCR5 antibody [Phycoerythrin(PE)-conjugated anti-CCR5 clone CTC5, R&D Systems, Minneapolis, MN] or PE-conjugated IgG2B isotype control (R&D Systems). All antibodies were incubated at a final concentration of 12.8 µg/ml for 30 minutes at room temperature in FACS staining buffer. Stained PBMCs were washed again in FACS staining buffer and samples were analyzed on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) with Cellquest software. For assessment of Affinofile cell receptor expression levels relative to PBMCs, 5.0×105 cells were added to a 6-well plate and allowed to adhere for 48 hours in Dulbecco's Modified Eagle's Medium (DMEM, Mediatech, Inc.) supplemented with 10% FBS, pen/strep, and 50 µg/ml blasticidin (Sigma). Cells were stimulated with 20 ng/ml minocycline (Sigma) for 24 hours and subsequently recovered from plates with 3 mM EDTA in PBS. Cells were washed, stained, and analyzed by flow cytometry as reported above for PBMCs. Flow cytometry of PBMCs and Affinofile cells was performed in the same experiment. Results were analyzed by Flow-Jo software and receptor expression levels reported as events relative to mean fluorescence intensity.
Pseudovirus incorporation of envelope glycoproteins
Envelope-pseudotyped viruses prepared with ES or CP envelopes by transfection of 293T cells were quantified by limiting dilution reverse transcriptase activity. Equivalent virion numbers were pelleted by centrifugation at 38,000×g for 2 hours at 4°C. Supernatant was removed and virion pellets were lysed in SDS lysis buffer [40 mM Tris-HCl (pH 6.8), 10% glycerol, 10% ß-mercaptoethanol, 1% SDS]. Virus lysates were separated on SDS-10% polyacrylamide gels and transferred to nitrocellulose. Membranes were blocked with gelatin and proteins were detected either with a mouse monoclonal anti-gp120 antibody that recognizes a conserved C2 region linear epitope (B13, courtesy of Dr. Bruce Chesebro, NIAID ) or HIV-Ig (courtesy AIDS Research and Reference Reagent Program). Primary antibodies were detected with horseradish peroxidase-conjugated goat-anti-mouse or goat-anti-human secondary antibodies, respectively (Pierce Biotechnology, Rockford, IL), revealed with the ECL Plus Western Detection kit (Pierce Biotechnology) and exposed to X-ray film.
Statistical analysis
Data were analyzed by the UCLA Statistical/Biomathematical Consulting Clinic using repeated measures ANOVA, simultaneously taking into account effects due to disease group (ES vs CP), CD4 level, and CCR5 level. Within each group, the intrapatient variability in relative infection was small and did not differ significantly between the ES and CP groups. Therefore, results are reported with interpatient variance. For evaluation of surface plots between groups ES and CP, P values are given using the average of clones for a given patient as a single value or using each individual clone as a single value. For groups of 3 or more (ES, CP, and acute) we evaluated independent means by One-way ANOVA using the Kruskal-Wallis test and Dunns post-test for data that did not pass a normalcy test. For drug sensitivity and kinetic analysis statistics were performed using each individual clone as a single value. We considered a P value of <0.05 as statistically significant.
Results
Measuring HIV-1 entry efficiency and fitness by varying CD4 and CCR5 cell surface concentrations
The ability of HIV-1 to infect a cell is largely influenced by surface expression of CD4 and CCR5 [26]–[32]. This study evaluates a previously described cohort of 38 independent full-length plasma env clones derived from 7 ES individuals [23]. The env clones from this cohort expressed similar levels of protein by Western blot (Figure S1) and readily infected the indicator cell line TZM-bl demonstrating their functionality [23]. As described below, observed differences in entry efficiency could not be explained by any minor variations in Env levels on the virus.
As with most cell lines, TZM-bls express CD4 at levels comparable to primary activated CD4+ T cells (approximately 65,000–100,000 molecules/cell), however CCR5 expression is significantly higher than on primary T lymphoctyes (approximately 500 to 7000 molecules/cell) [26], [33]–[37]. CCR5 expression also varies widely from patient to patient not only in absolute number of cells expressing CCR5, but also in CCR5 density/cell [26], [27], [33]–[35]. This study utilizes the Affinofile system, a novel cell line with independent dual-inducible surface expression of CD4 and CCR5 (Figure 1, Figure S1) (Johnston et al., submitted). The ability to modulate receptor and co-receptor expression on the Affinofile cells provides a more physiologic measure of HIV-1 entry efficiency. Since the ability of HIV-1 to infect a cell is largely influenced by cell surface levels of CD4 and CCR5, it is important to consider expression levels when evaluating infectivity [26]–[32].
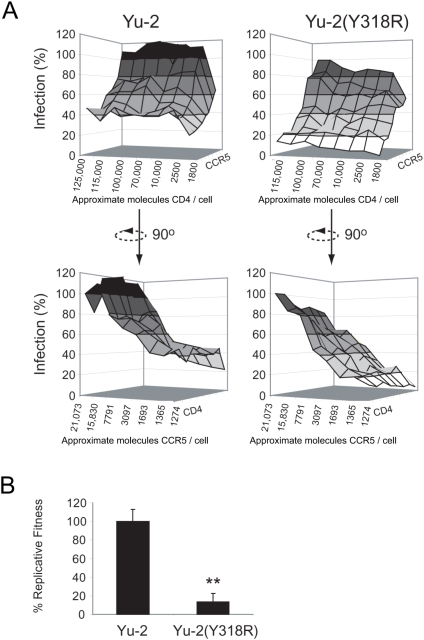
Figure 1. Affinofile cells allow for the analysis of both CD4 and CCR5 usage and are a surrogate marker of fitness.
(A) Affinofile cells were infected with Luciferase-encoding envelope psudotyped viruses bearing the envelope of the neurotropic lab strain Yu-2 or a V3-crown mutant [Yu-2(Y318R)] with reduced CCR5 usage. Surface plots were constructed using infectivity data derived from luciferase activity. Percent infection (y-axis) was derived from the luciferase activity at each receptor expression combination divided by the luciferase activity at maximal CD4/CCR5 expression levels. The x-axis depicts approximate CD4 expression level described as approximate molecules CD4/cell as assessed by quantitative flow cytometry (top 2 panels), and the z-axis depicts approximate CCR5 expression levels (bottow 2 panels). (B) Relative replicative fitness of full length HIV-1 viruses in multiple replication cycle in primary PHA/IL-2–activated peripheral blood mononuclear cells. The replicative fitness of full length Yu-2 and Yu-2(Y318R) was determined by pairwise infection and quantification of relative outgrowth. The y-axis represents percent replicative fitness of Yu-2(Y318R) relative to Yu-2.
Receptor usage as measured by the Affinofile system was validated as a surrogate marker of entry fitness. Yu-2 and a V3 crown mutant of Yu-2 [Yu-2(Y318R)] known to affect CCR5 usage were evaluated for infectivity using Affinofile cells induced at each pairwise combination of [Minocycline] and [Ponasterone A] (42 unique combinations) (Figure S1). Three-dimensional surface plots were generated from luciferase activity expressed as a function of virus infectivity at each combination of CCR5 and CD4, which was confirmed by flow cytometry (Figure 1A). CD4 and CCR5 surface levels at each drug combination are given as an average level calculated from a pool of cells expressing a range of CD4 and CCR5 molecules (Figure S4). Reduced infectivity of the Yu-2(Y318R) variant over the wild type was observed over a range of CCR5 and CD4 (Figure 1A). This decreased ability of Yu-2(Y318R) to infect cells expressing low CCR5 is consistent with a 90% reduction in replicative fitness measured by competitive replication assays in peripheral blood mononuclear cells (PBMCs) (Figure 1B, p<0.01, unpaired student's t-test). The direct relationship between entry efficiency using the Affinofile system and replicative fitness in human PBMCs has been validated for multiple primary HIV-1 isolates. Generally, viruses of increased replicative fitness display increased infectivity of cells expressing low CCR5, CD4, or both CCR5 and CD4 in the Affinofile system (Johnston et al, submitted).
To assess relative infectivity of chronic and ES env clones, pseudotyped viruses carrying a non-LTR driven luciferase were generated for each clone. Pseudotyped viruses generated from 38 independent plasma virus env glycoprotein clones from 7 ES and 32 independent plasma virus clones from 7 chronic progressors (CP) were evaluated for infectivity at each pairwise drug combination described above and surface plots were generated (Figure S2 and Figure S3). The percent infection defines the infection at each surface CCR5/CD4 level relative to a 100% infection at the highest CD4 and CCR5 surface density. This method permits the direct comparison of CD4 and CCR5 usage by each env clone and provides a rapid and efficient way to measure viral env replicative fitness.
ES are less efficient than CP env clones in utilizing both CCR5 and CD4 for entry
Relative infectivity of 38 independent ES env clones and 32 independent CP env clones was ascertained at multiple combinations of CCR5 and CD4 density (Figure 2A–2F and Figure 3A–3F). Values for each of the env clones tested are shown as well as for Yu-2 and SF162. Varying CD4 levels with constant CCR5 (Figure 2A–2F) or varying CCR5 levels with constant CD4 (Figure 3A–3F) consistently demonstrated that ES env clones supported lower levels of infection than the CP clones.
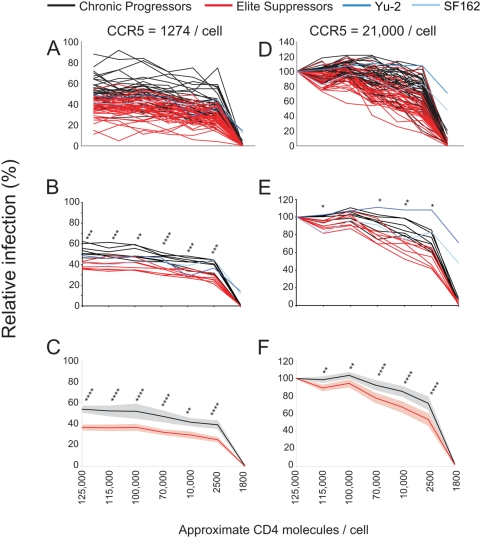
Figure 2. ES clones show reduced entry efficiency at fixed CCR5 surface concentrations.
(A–C,D–F) Affinofile cells were induced to express approximately 1274 CCR5 molecules/cell (A–C) or approximately 21,073 CCR5 molecules/cell (D–F) and CD4 molecules ranging from approximately 1,800 to 125,000 molecules/cell prior to infection. Induced Affinofile cells were infected with equivalent amounts of pseudotyped viruses. Relative infection of all CP (black) and ES (red) clones are shown with SF162 and YU-2 included as positive controls (blue). Relative infection is expressed as a proportion of maximal infection at the highest CD4 (approximately 125,000 molecules/cell) and CCR5 (approximately 21,073 molecules/cell) level. (B,E) Infectivity of clones from individuals were averaged and plotted as a single line for chronic and ES individuals. P values are given for patient averages. (n = 7) (C,F) Average of all individual chronic or elite clones at minimal (C) and maximal (F) CCR5 concentrations with CD4 concentrations ranging from approximately 1,800 to 125,000 molecules/cell. Affinofile cells were induced as described above for (A,B). P values as calculated by unpaired student's t test are shown above each CD4 expression condition. P values are represented as follows: P<0.05 (*), P<0.01 (**), P<0.001 (***), P<0.0001 (****). 95% confidence intervals are shown for chronic (grey) and ES (pink).
Figure 3. ES clones show reduced entry efficiency at fixed CD4 surface concentrations.
(A–C,D–F) Affinofile cells were induced to express threshold (approximately 2,500) CD4 molecules/cell (A–C) or maximal (approximately 125,000) CD4 molecules/cell (D–F) and CCR5 molecules ranging from approximately 1274 to 21,073 molecules/cell prior to infection. Infection is performed as described in Figure 2. (B,E) Infectivity of clones from individuals were averaged and plotted as a single line for chronic and ES individuals. P values are given for patient averages. (n = 7) (C,F) Average of all individual chronic or elite env clones at threshold (C) and maximal (F) CD4 concentrations with CCR5 concentrations ranging from approximately 1274 to 21,073 molecules/cell. P values are represented as follows: P<.05 (*), P<.01 (**), P<.001 (***), P<.0001 (***). 95% confidence intervals are shown for chronic (grey) and ES (pink).
At the highest CD4 and lowest CCR5 expression level, ES clones averaged 36.7% while CP clones were reliably higher averaging 53.3% (Figure 2A–2C). Infectivity differences were significant for each CD4 concentration (P values ranged from 0.01 to <0.0001, repeated measures ANOVA) at a fixed high or low CCR5 level regardless if individual env clones were evaluated (Figure 2C and 2F) or if the env clones were averaged for a given individual and compared as patient averages (Figure 2B and 2E). Additionally, consistent with previous data, the neurotropic envs SF162 andYU-2 readily infected cells expressing sub-threshold levels of CD4 while the primary isolates could not (Figure 3A–3F) [38]. Taken together, these results reveal that ES clones inefficiently infect cells expressing low CCR5 in the presence of threshold or higher levels of CD4 compared to CP clones. Additionally, the discrepancy in infectivity between ES and chronic clones at fixed, high CCR5 levels indicates that ES clones also require higher levels of CD4 to achieve similar infection as chronic clones.
To further evaluate CCR5 usage independent of CD4 expression, infection was determined at minimal and maximal CD4 levels as CCR5 expression was varied. At each concentration of CCR5 below maximal examined, ES clones infected significantly less efficiently than chronic clones (Figure 3D–3F). Therefore, even in the presence of optimal CD4 concentrations, ES clones inefficiently utilize CCR5 for entry.
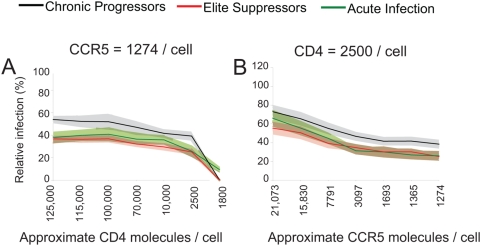
ES env clones do not differ significantly from acute infection env clones in CD4 and CCR5 utilization
Differences in receptor and co-receptor utilization of ES and chronic progressor envs were most significant when infection was performed in the context of (1) low CCR5 and varying CD4 levels or (2) low CD4 and varying CCR5 levels. Thus, these conditions were repeated to examine entry efficiency of 23 pseudotyped env plasma clones from 20 acutely infected individuals (Figure 4A and 4B). At low CCR5 expression, acute envs averaged an intermediate pattern of infectivity compared to ES and chronic envs, but these differences were not significant (Figure 4A). Similar results were obtained when infections were performed at minimal surface CD4 levels (Figure 4B). Consistent with previous reports using similar systems, these results indicate that acute envs show a broad pattern of infectivity which is not significantly different from chronic or ES envs [39].
Figure 4. Acute env clones show no significant receptor or co-receptor utilization difference from chronic or elite envs.
(A) Affinofile cells were induced to express minimal CCR5 (approximately 1274 molecules/cell) and CD4 molecules ranging from approximately 1,800 to 125,000 molecules/cell or (B) threshold (approximately 2,500) CD4 molecules/cell and CCR5 molecules ranging from approximately 1,274 to 21,073 molecules/cell prior to infection. Induced Affinofile cells were infected with equivalent amounts of pseudotyped viruses. Average of all acute (green), chronic (black), and elite (red) clones are shown. 95% confidence intervals are given for acute (light green), chronic (grey) and ES (pink).
Susceptibility of env clones to CCL5, TAK-779, or ENF
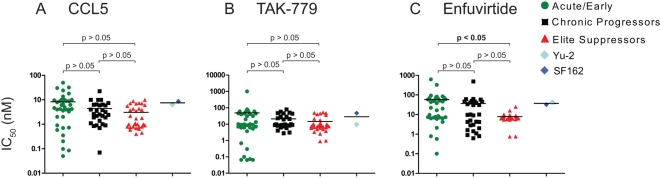
Previous studies suggest that major differences in entry efficiency may impact on susceptibility to various entry inhibitors [25], [40]–[42]. Thus ES, CP, and acute clones were tested for their susceptibility to the natural CCR5 ligand CCL5 (RANTES), the small molecule CCR5 antagonist TAK-779, and the fusion inhibitor enfuvirtide (ENF) in Affinofile cells induced to express CD4 and CCR5 to levels that closely mimic primary CD4+ T cells (approximately 125,000 molecules CD4/cell and 1274 molecules CCR5/cell) (Figure S4). Susceptibility to CCL5 did not differ significantly between chronic and ES env clones (Figure 5A). Interestingly, acute clone IC50 values ranged from 0.05 to 50 nM (1,000-fold) with an average of 8.25 nM. The range of IC50 values for acute clones was much larger relative to ES clones whose values ranged from 0.4 to 10 nM (25-fold), with an average of 3.05 nM. Although similar trends were observed with the small molecule CCR5 antagonist TAK-779, clones were not significantly different with average IC50 values of 48.8 nM for acute, 21.0 nM for chronic, and 14.7 nM for ES (Figure 5B). These results show that ES, CP, and acute envs have no remarkable differential susceptibility to CCL5 or TAK-779 however the broad range of IC50 values for acute envs highlights the variability in env phenotypes associated with acute infection.
Figure 5. ES, CP, and acute clones show no differential susceptibility to CCL5, TAK-779, or ENF.
(A–C) ES, acute, and chronic clones were used to infect Affinofile cells induced to express approximately 125,000 molecules CD4 and approximately 1,274 molecules CCR5/cell. (A) CCL5 was added in concentrations ranging from 0.1–50 nM, incubated for 1 h, and infected. Plots represent percent replication relative to no drug. 50 percent inhibitory concentrations of drug (IC50 values) were computed from the curves and plotted for acute, chronic, ES, and Yu-2 or SF162 envelopes. (B) TAK-779 was added to the induced Affinofile cells at concentrations ranging from 0.1 nM–1 µM. Plots were used to determine IC50 values for each envelope. (C) Enfuvirtide was added to cells at concentrations ranging from 0.1 nM–1 µM.
Finally, susceptibility of clones to ENF was evaluated (Figure 5C). IC50 values for acute clones again showed a broad range from 0.10 to 623 nM (>5000 fold range) with an average of 58.13 nM. Again, ES (33-fold range) clones exhibited a significantly narrower range of IC50 values compared to acute clones. The average IC50 value for acute envs was significantly greater than for ES (P<0.05, ANOVA, Kruskal-Wallis test). Overall, ES clones showed trends towards increased susceptibility to entry inhibitors consistent with decreased entry efficiency. These susceptibility profiles suggest that ES clones have a high degree of phenotypic similarity indicated by the narrow range of susceptibility to entry inhibitors. Conversely, chronic and especially acute envs showed broad ranges of susceptibility indicative of their diverse entry phenotypes.
Delayed fusion and reverse transcription kinetics exhibited by ES–derived env clones
Infection data of cells expressing sub-maximal concentrations of CD4 and CCR5 indicates that ES-derived env clones require higher levels of receptor and co-receptor for efficient entry. This requirement for higher receptor levels could suggest that these envs also exhibit differences in the rates of HIV-1 entry into host cells [42],[43].
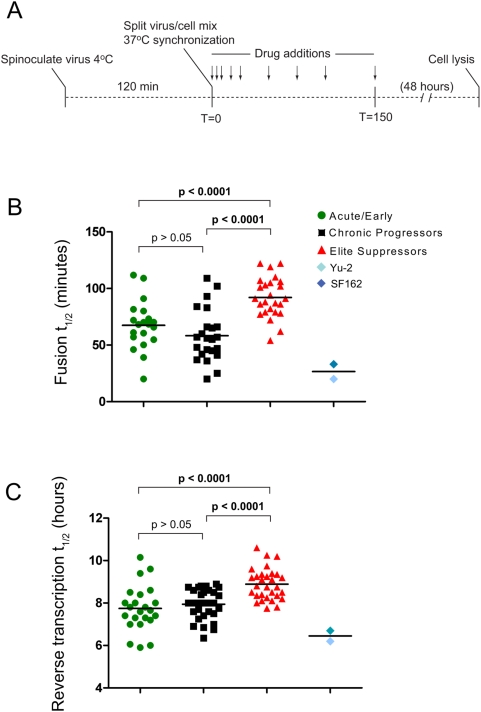
To assess host cell entry kinetics, U87-CD4/CCR5 cells were first spinoculated with virus at a temperature non-permissive for viral fusion with the host cell. Enfuviritide (ENF) was added once to each well at a concentration of 10 µM at various times after the cells were shifted to temperatures permissive for viral fusion (Figure 6A). Viruses that have completed the final step in HIV-1 entry (six helix bundle formation) are ENF insensitive and will continue the viral replication cycle regardless of the addition of ENF. Thus, this assay permits determination of the entry kinetics of each env clone.
Figure 6. ES clones show slower fusion and reverse transcription kinetics compared to chronic and acute clones.
(A) U87-CD4/CCR5 cells were spinoculated with equivalent amounts of pseudotyped virus at a temperature non-permissive for entry. Infections were synchronized by simultaneous addition of warm medium. ENF (10 µM) was added once to wells at given times post-infection. All infections are given as a percent of maximal infection. (B) Average T1/2 fusion values in minutes for acute (green), chronic (black), and ES (red). (C) Equivalent amounts of pseudotyped virus were simultaneously added to Affinofile cells induced to express approximately 125,000 molecules CD4 and 1274 molecules CCR5/cell. EFV (1 µM) was added at various times post-infection. Average T1/2 reverse transcription values in hours are shown. YU-2 and SF162 are included as controls (B,C).
Fusion kinetics were measured for each ES, CP, and acute clone. ES clones fused with an average T1/2 of 92.1 minutes while acute clones averaged a T1/2 of 67.5 minutes and chronic clones a T1/2 of 58.3 minutes (Figure 6B). This delay in ES fusion kinetics was significant when compared with acute and CP clones (P<.0001 and P<.0001 respectively, One-way ANOVA). Therefore, even in the presence of saturating levels of CD4 and CCR5, ES clones do not complete entry processes as efficiently and exhibit slower kinetics than both CP and acute env clones.
This delay in entry kinetics was maintained during subsequent steps of the replication cycle as indicated by a kinetic reverse transcription assay. For these analyses, Efavirenz (a non-nucleoside reverse transcriptase inhibitor) was added at various times post-infection to arrest infection events which have not completed reverse transcription. As expected, ES derived env clones completed reverse transcription slower (mean T1/2 of 8.89 hours) than acute and CP clones [mean T1/2 values of 7.74 (P<.0001) and 7.95 (P<.0001) respectively with One-way ANOVA] (Figure 6C). Due to the isogenic background of the pseudotyping virus these results suggest that delays in reverse transcription are the result of delays in entry processes. ES env clones exhibit a kinetic lag in entry processes which are maintained during downstream events in the viral life cycle.
Discussion
This study represents an evaluation of intrinsic phenotypic characteristics of full-length functional subtype B env quasispecies derived from ES plasma. Envelope glycoproteins from ES clearly exhibited reduced capacity to support HIV-1 entry into host cells compared to CP envs. Given the wide range in entry efficiencies observed with envs derived from acute infections it is possible that relatively lower fitness env variants are selected early in infection in ES. The impact of this observed entry deficiency with ES clones is still not fully understood but decreased replicative fitness and lack of diversification in ES viruses suggests these individuals may have contracted a less fit HIV-1 variant or these low fitness variants are selected for early in infection[7],[44],[45].
To date, phenotypic studies of ES viruses have been difficult to perform due to the low amount of virus in these individuals. Analysis of minor differences in env function has been confounded by the use of cell lines expressing non-physiologic amounts of co-receptor (CCR5). Given the high degree of variability in expression of CCR5 among patients it is important to evaluate env function over a wide range of CCR5 levels [26],[27],[34],[35]. Detailed analyses of env function in the presence of physiologic levels of CD4 and CCR5 was possible in this study through the use of the novel Affinofile system. Although ES and chronic individuals each harbored quasispecies with different receptor utilization phenotypes, ES clones from each individual showed an average decreased entry efficiency compared to chronic clones over almost all CD4 and CCR5 expression levels. These differences between chronic and ES clones were most dramatic at low CCR5 surface levels. Thus, this low fitness phenotype could be further accentuated in vivo in an individual expressing low CCR5 levels or possibly higher levels of CCR5 ligands which have been associated with viral control [46]–[48].
Several reports have suggested a correlation between susceptibility to entry inhibitors and relative env fitness [25],[40]. ES, CP, and acute clones exhibited a diverse range of IC50 values consistent with previous data [40],[41],[43],[49],[50]. However, acute clones showed consistently the most variation in susceptibility indicating the diverse phenotypes associated with early infection [40],[43],[49]. Conversely, the low range of IC50 values for ES clones underscores their phenotypic homogeneity. Due to reported differences between primary cells and cell lines entry inhibitor susceptibility assays were performed in the Affinofile cells induced to express CD4 levels and CCR5 levels that closely mimic primary CD4+ T cells (approximately 125,000 molecules CD4/cell and approximately 1274 molecules CCR5/cell) [51]. Additionally, it would be expected that variations in CD4 utilization would result in variations in susceptibility to soluble CD4 (sCD4) [52]. However, consistent with previous reports showing the relative resistance of primary isolate viruses to sCD4 [53], meaningful inhibition of ES, CP, and acute infection envelopes was not observed at maximal achievable concentrations of sCD4 (25 µg/ml).
Previous studies have also highlighted an association between receptor utilization profiles and susceptibility to neutralizing antibodies. It is possible that ES envs display altered susceptibility to broadly neutralizing antibodies given their observed entry phenotype. Previous studies suggest that antibody binding and neutralization have a kinetic component [54]. It may potentially be generalized that slow fusing viruses, independent of the mechanism, may be more susceptible to neutralizing antibodies that act with a kinetic dependence. However, it has been shown that ES individuals generate low titers of neutralizing antibodies against autologous virus and thus the role of neutralizing antibodies in maintenance of low level viremia in ES is nominal [23].
The host entry process is thought to be a rate limiting step in HIV-1 replication. It was thus important to determine if poor entry efficiency by ES clones leads to a reduced rate of entry kinetics. ES env clones were found to fuse on average over 1.5 times slower than chronic or acute clones in the presence of saturating levels of both CD4 and CCR5. As result of poor entry efficiency, this kinetic delay was maintained during subsequent steps of the retroviral lifecycle. Compounded effects of inefficient CD4 and CCR5 usage by ES clones likely contributes to kinetic delays in entry processes and thus overall decreased replicative fitness. The delayed entry kinetics may be even more important in vivo if there is a limited time frame over which entry can occur due to competing inhibitory processes such as the binding of neutralizing antibodies or the presence of CCR5 ligands.
Despite genotypic differences in both the virus and the ES host, viral quasispecies in different ES individuals are remarkably similar phenotypically. This implies that poor envelope function is a common feature in ES individuals. This result is in sharp contrast to data from acute infection envs where clones exhibited much phenotypic diversity. Potentially, HIV-1 infection in ES may be established by lower fitness env(s) which are present in a subset of acutely infected individuals. Alternatively, HIV-1 infection in ES may be established by phenotypically diverse envs and early pressure from the immune results in the outgrowth of lower fitness escape variants. At present, no data exists on the natural history of acute infection of ES. It remains unclear whether these individuals experience typical high level viremia that is subsequently reduced to an undetectable setpoint or control their viral load from the onset of infection. It would be of great interest to be able to address this significant gap in our understanding of viral dynamics in elite control of viremia. Lower env fitness is likely not sufficient to mediate absolute viral suppression. Viral control could be achieved in those individuals who are also able to mount a potent immune response and/or are genetically predisposed to better control HIV-1 viremia. In these individuals, viral replication and diversification of early infection viruses required to achieve efficient receptor utilization by env quasispecies may never be attained. Lower fitness of other viral factors may also be contributing to reduced replication and lower viral load. Full understanding of the in vivo impact of lower env fitness in ES will require further study however this data underscores the important contribution of viral factors in elite HIV-1 suppression.
Supporting Information
ES and chronic envs show no difference in Env virion incorporation.
(7.47 MB EPS)
Affinofile cells allow for the analysis of both CD4 and CCR5.
(3.03 MB EPS)
Full surface plots for chronic progressor (CP) and elite suppressor (ES) envelopes. (A–O) Surface plots are arranged by patient, including Yu-2 and SF-162 control strains, indicated in blue. CP envs are indicated in grayscale, and ES envs are indicated in red.
(1.45 MB PDF)
Affinofile cells induced to express physiologic levels of CD4 and CCR5 express similar surface levels as CD4+ PBMCs.
(0.80 MB EPS)
Acknowledgments
Virus work at Case Western Reserve University was performed in the biosafety level 2 and 3 facilities of the Case/UH Centers for AIDS Research (AI25879). We would like to thank Joel N. Blankson for helpful discussions and advice on the manuscript. Additionally, the authors wish to thank E. M. Landaw and D. Markovic at the UCLA statistics center for help in statistical analysis.
Footnotes
The authors have declared that no competing interests exist.
KGL was supported by a postdoctoral fellowship from the Francis Goelet Foundation, the Mathilde Krim fellowship (106999-43-RFRL) from amfAR, the Foundation for AIDS Research, and Infectious Diseases Training Grant T32 AI07024. MAL was supported by the Case Western Reserve University Medical Scientist Training Program Grant T32 GM07250. BL was supported by UCLA AIDS Institute, the UCLA Center for AIDS Research (AI 28697), and NIH grant R01 AI52021. SJ was supported in part by a NIH Virology & Gene Training grant (T32 AI 060567). RFS was supported by the Doris Duke Charitable Foundation and the Howard Hughes Medical Institute. MM was supported by National Institutes of Health grants R01AI047033, AI067854, and U01AI041534, and Rockefeller University CCTS, Grant Number UL1 RR024143. EJA was supported by NIAID, NIH grants AI49170, AI57005, and AI058894. The funders had no role in the preparation, review, or approval of the manuscript.
References
- 1.Hubert JB, Burgard M, Dussaix E, Tamalet C, Deveau C, et al. Natural history of serum HIV-1 RNA levels in 330 patients with a known date of infection. The SEROCO Study Group. Aids. 2000;14:123–131. doi: 10.1097/00002030-200001280-00007. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Kaslow RA, Carrington M, Apple R, Park L, Munoz A, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 4.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. Aids. 2003;17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 6.Brettle RP, McNeil AJ, Burns S, Gore SM, Bird AG, et al. Progression of HIV: follow-up of Edinburgh injecting drug users with narrow seroconversion intervals in 1983–1985. Aids. 1996;10:419–430. [PubMed] [Google Scholar]
- 7.Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203:1357–1369. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 9.Churchill MJ, Rhodes DI, Learmont JC, Sullivan JS, Wesselingh SL, et al. Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J Virol. 2006;80:1047–1052. doi: 10.1128/JVI.80.2.1047-1052.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 11.Salvi R, Garbuglia AR, Di Caro A, Pulciani S, Montella F, et al. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J Virol. 1998;72:3646–3657. doi: 10.1128/jvi.72.5.3646-3657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Zhang L, Ho DD. Biological characterization of nef in long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1995;69:8142–8146. doi: 10.1128/jvi.69.12.8142-8146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Zhang L, Ho DD. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1995;69:93–100. doi: 10.1128/jvi.69.1.93-100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tersmette M, Gruters RA, de Wolf F, de Goede RE, Lange JM, et al. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tersmette M, Lange JM, de Goede RE, de Wolf F, Eeftink-Schattenkerk JK, et al. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;1:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 16.Allen TM, O'Connor DH, Jing P, Dzuris JL, Mothe BR, et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 17.Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, et al. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Gladden AD, Altfeld M, Kaldor JM, Cooper DA, et al. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J Virol. 2007;81:193–201. doi: 10.1128/JVI.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81:2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miura T, Brockman MA, Brumme CJ, Brumme ZL, Carlson JM, et al. Genetic Characterization of Human Immunodeficiency Virus type 1 in Elite Controllers: Lack of gross genetic defects or common amino acid changes. J Virol. 2008 doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joos B, Trkola A, Fischer M, Kuster H, Rusert P, et al. Low human immunodeficiency virus envelope diversity correlates with low in vitro replication capacity and predicts spontaneous control of plasma viremia after treatment interruptions. J Virol. 2005;79:9026–9037. doi: 10.1128/JVI.79.14.9026-9037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey JR, Lassen KG, Yang HC, Quinn TC, Ray SC, et al. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J Virol. 2006;80:4758–4770. doi: 10.1128/JVI.80.10.4758-4770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehandru S, Poles MA, Tenner-Racz K, Manuelli V, Jean-Pierre P, et al. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J Virol. 2007;81:599–612. doi: 10.1128/JVI.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobritz MA, Marozsan AJ, Troyer RM, Arts EJ. Natural variation in the V3 crown of human immunodeficiency virus type 1 affects replicative fitness and entry inhibitor sensitivity. J Virol. 2007;81:8258–8269. doi: 10.1128/JVI.02739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anton PA, Elliott J, Poles MA, McGowan IM, Matud J, et al. Enhanced levels of functional HIV-1 co-receptors on human mucosal T cells demonstrated using intestinal biopsy tissue. Aids. 2000;14:1761–1765. doi: 10.1097/00002030-200008180-00011. [DOI] [PubMed] [Google Scholar]
- 27.Nokta MA, Li XD, Nichols J, Mallen M, Pou A, et al. Chemokine/CD4 receptor density ratios correlate with HIV replication in lymph node and peripheral blood of HIV-infected individuals. Aids. 2001;15:161–169. doi: 10.1097/00002030-200101260-00004. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore JP. Coreceptors: implications for HIV pathogenesis and therapy. Science. 1997;276:51–52. doi: 10.1126/science.276.5309.51. [DOI] [PubMed] [Google Scholar]
- 30.Kabat D, Kozak SL, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, et al. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J Virol. 2005;79:4828–4837. doi: 10.1128/JVI.79.8.4828-4837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng G, Sellers MT, Mosteller-Barnum M, Rogers TS, Shaw GM, et al. Lamina propria lymphocytes, not macrophages, express CCR5 and CXCR4 and are the likely target cell for human immunodeficiency virus type 1 in the intestinal mucosa. J Infect Dis. 2000;182:785–791. doi: 10.1086/315790. [DOI] [PubMed] [Google Scholar]
- 35.Olsson J, Poles M, Spetz AL, Elliott J, Hultin L, et al. Human immunodeficiency virus type 1 infection is associated with significant mucosal inflammation characterized by increased expression of CCR5, CXCR4, and beta-chemokines. J Infect Dis. 2000;182:1625–1635. doi: 10.1086/317625. [DOI] [PubMed] [Google Scholar]
- 36.Choudhry V, Zhang MY, Harris I, Sidorov IA, Vu B, et al. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochem Biophys Res Commun. 2006;348:1107–1115. doi: 10.1016/j.bbrc.2006.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heredia A, Gilliam B, DeVico A, Le N, Bamba D, et al. CCR5 density levels on primary CD4 T cells impact the replication and Enfuvirtide susceptibility of R5 HIV-1. Aids. 2007;21:1317–1322. doi: 10.1097/QAD.0b013e32815278ea. [DOI] [PubMed] [Google Scholar]
- 38.Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, et al. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology. 2007;360:105–119. doi: 10.1016/j.virol.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagar M, Kirkegaard E, Lavreys L, Overbaugh J. Diversity in HIV-1 envelope V1–V3 sequences early in infection reflects sequence diversity throughout the HIV-1 genome but does not predict the extent of sequence diversity during chronic infection. AIDS Res Hum Retroviruses. 2006;22:430–437. doi: 10.1089/aid.2006.22.430. [DOI] [PubMed] [Google Scholar]
- 40.Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, et al. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci U S A. 2002;99:16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves JD, Miamidian JL, Biscone MJ, Lee FH, Ahmad N, et al. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J Virol. 2004;78:5476–5485. doi: 10.1128/JVI.78.10.5476-5485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platt EJ, Durnin JP, Kabat D. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J Virol. 2005;79:4347–4356. doi: 10.1128/JVI.79.7.4347-4356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safarian D, Carnec X, Tsamis F, Kajumo F, Dragic T. An anti-CCR5 monoclonal antibody and small molecule CCR5 antagonists synergize by inhibiting different stages of human immunodeficiency virus type 1 entry. Virology. 2006;352:477–484. doi: 10.1016/j.virol.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Bailey JR, Zhang H, Wegweiser BW, Yang HC, Herrera L, et al. Evolution of HIV-1 in an HLA-B*57-positive patient during virologic escape. J Infect Dis. 2007;196:50–55. doi: 10.1086/518515. [DOI] [PubMed] [Google Scholar]
- 45.Bailey JR, O'Connell K, Yang HC, Han Y, Xu J, et al. Transmission of human immunodeficiency virus type 1 from a patient who developed AIDS to an elite suppressor. J Virol. 2008;82:7395–7410. doi: 10.1128/JVI.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 47.Cooke GS, Tosh K, Ramaley PA, Kaleebu P, Zhuang J, et al. A polymorphism that reduces RANTES expression is associated with protection from death in HIV-seropositive Ugandans with advanced disease. J Infect Dis. 2006;194:666–669. doi: 10.1086/505875. [DOI] [PubMed] [Google Scholar]
- 48.Dolan MJ, Kulkarni H, Camargo JF, He W, Smith A, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 49.Labrosse B, Labernardiere JL, Dam E, Trouplin V, Skrabal K, et al. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J Virol. 2003;77:1610–1613. doi: 10.1128/JVI.77.2.1610-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rusert P, Kuster H, Joos B, Misselwitz B, Gujer C, et al. Virus isolates during acute and chronic human immunodeficiency virus type 1 infection show distinct patterns of sensitivity to entry inhibitors. J Virol. 2005;79:8454–8469. doi: 10.1128/JVI.79.13.8454-8469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ketas TJ, Kuhmann SE, Palmer A, Zurita J, He W, et al. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364:281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, et al. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology. 2004;321:8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Daar ES, Li XL, Moudgil T, Ho DD. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci U S A. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laakso MM, Lee FH, Haggarty B, Agrawal C, Nolan KM, et al. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 2007;3:e117. doi: 10.1371/journal.ppat.0030117. doi:10.1371/journal.ppat.0030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ES and chronic envs show no difference in Env virion incorporation.
(7.47 MB EPS)
Affinofile cells allow for the analysis of both CD4 and CCR5.
(3.03 MB EPS)
Full surface plots for chronic progressor (CP) and elite suppressor (ES) envelopes. (A–O) Surface plots are arranged by patient, including Yu-2 and SF-162 control strains, indicated in blue. CP envs are indicated in grayscale, and ES envs are indicated in red.
(1.45 MB PDF)
Affinofile cells induced to express physiologic levels of CD4 and CCR5 express similar surface levels as CD4+ PBMCs.
(0.80 MB EPS)