Abstract
Purpose
To create a breast cancer risk estimation model based on the descriptors of National Mammography Database (NMD) format using logistic regression that can aid in decision-making for early detection of breast cancer.
Material and Methods
Institutional Review Board waived this HIPAA-compliant retrospective study from requiring informed consent. We created two logistic regression models based on the mammography features and demographic data for 62,219 consecutive cases of mammography records from 48,744 studies in 18,270 patients reported using the Breast Imaging-Reporting and Data System (BI-RADS) lexicon and NMD format between 4/5/1999 and 2/9/2004. State cancer registry outcomes matched with our data served as the reference standard. The probability of cancer was the outcome in both models. Model-2 was built using all variables in Model-1 plus radiologists’ BI-RADS assessment codes. We used 10-fold cross-validation to train and test the model and calculate the area under the receiver operating characteristic (ROC) curves (Az) to measure the performance. Both models were compared to the radiologists’ BI-RADS assessments.
Results
Radiologists achieved an Az value of 0.939 ± 0.011. The Az was 0.927 ± 0.015 for Model-1 and 0.963 ± 0.009 for Model-2. At 90% specificity, the sensitivity of Model-2 (90%) was significantly better (P<0.001) than that of radiologists (82%) and Model-1 (83%). At 85% sensitivity, the specificity of Model 2 (96%) was significantly better (P<0.001) than that of radiologists (88%) and Model-1 (87%).
Conclusions
Our logistic regression model can effectively discriminate between benign and malignant breast disease and identify the most important features associated with breast cancer.
Introduction
Mammography, accepted as the most effective screening method in the detection of early breast cancer, still has limited accuracy and significant interpretation variability that decreases its effectiveness [1-6]. The use of computer models can help by 1) detecting abnormalities on mammograms [7-10], 2) estimating the risk of breast cancer for improved sensitivity and specificity of diagnosis [11-16], and 3) identifying high risk populations for screening, genetic testing or participation in clinical trials [17-22]. This study focuses on the second goal: the use of a computer aided diagnostic (CADx) model for risk estimation to aid radiologists in breast cancer diagnosis.
CADx models can quantify the risk of cancer using demographic factors and mammography features already identified by a radiologist or a computer aided detection (CAD) model. CADx models estimate the probability (or risk) of disease that can be used for improved decision making by physicians and patients [23-25]. Previous studies on CADx tools use either small subset of data, suspicious mammograms, or mammograms recommended for biopsy [11-15]. Though most of these studies show that CADx tools are efficient in predicting the outcome as benign or malignant disease, none show the effectiveness of CADx models when applied to mammography data collected during the daily clinical practice. In addition, previous studies used biopsy results as the reference standard; whereas, we use a match with our state cancer registry. To the best of our knowledge, our study is the first one to develop and test a logistic regression based CADx model on consecutive mammograms from a breast imaging practice incorporating BI-RADS descriptors.
As the variables that help predict breast cancer increase in number, physicians have to rely on subjective impressions based on their experience to make decisions. Using a quantitative modeling technique such as logistic regression, to predict the risk of breast cancer, may help radiologists manage the large amount of information available, make better decisions, detect more cancers at early stages, and reduce unnecessary biopsies. The purpose of this study was to create a breast cancer risk estimation model based on demographic risk factors and BI-RADS descriptors available in the NMD format using logistic regression that can aid in decision-making for improved early detection of breast cancer.
Methods
The Institutional Review Board determined that this retrospective HIPAA-compliant study was exempt from requiring informed consent. We used variables collected in National Mammography Database (NMD) format [26] to develop a CADx model. NMD is a recommended format for collecting practice level mammography audit data to monitor and standardize performance nationally. The NMD includes Breast Imaging Reporting and Data System (BI-RADS) descriptors [27, 28].
Subjects
We collected data from all screening and diagnostic mammography examinations that were performed at Medical College of Wisonsin, Milwaukee, an academic, tertiary-care medical center, between April 5, 1999 and February 9, 2004. Our database included 48,744 mammography examinations (477 malignant and 48,267 benign) performed on 18,270 patients (Table 1) having the mean population age of 56.8 years (range: 18-99 years). Our dataset consisted of 65,892 records, where each record may represent: (1) a mammography lesion (benign or malignant) when observed on the mammogram; and (2) a single record of demographic factors only, if nothing is observed on the mammogram. The data was entered using PenRad® mammography reporting/tracking data system by technologists and radiologists. There were a total of eight radiologists--four of whom were general radiologists with some mammography background, two radiologists were fellowship trained, and two others had a lengthy experience in breast imaging. Their experience ranged between 1-35 years, and the number of mammograms interpreted by them ranged between 49-22,219. All mammography observations were made by radiologists; all demographic factors were recorded by technologists. This facility used a combination of digital and film mammography (approximately 75% film mammography). No CAD tool was used for lesion detection. Mean glandular dose was not available at the time of study.
Table 1.
Distribution of study population
| Factors | Benign (%) | Malignant (%) | Total |
|---|---|---|---|
| Number of Mammograms | 48,267 (100) | 477 (100) | 48,744 |
| Age Groups | |||
| < 45 | 9,529 (20) | 66 (14) | 9,595 |
| 45-49 | 7,524 (16) | 49 (10) | 7,573 |
| 50-54 | 7,335 (15) | 56 (12) | 7,391 |
| 55-59 | 6,016 (12) | 71 (15) | 6,087 |
| 60-64 | 4,779 (10) | 59 (12) | 4,838 |
| ≥ 65 | 13,084 (27) | 176 (37) | 13,260 |
| Breast Density | |||
| Predominantly fatty | 7,226 (15) | 61 (13) | 7,287 |
| Scattered fibroglandular | 19,624 (41) | 201 (42) | 19,825 |
| Heterogeneously dense | 17,032 (35) | 174 (36) | 17,206 |
| Extremely dense tissue | 4,385 (9) | 41 (9) | 4,426 |
| BI-RADS Code | |||
| 1 | 21,094 (44) | 0 (0) | 21,094 |
| 2 | 10,048 (21) | 13 (3) | 10,061 |
| 3 | 8,520 (18) | 32 (7) | 8,552 |
| 0 | 8,148 (17) | 130 (27) | 8,278 |
| 4 | 364 (1) | 137 (29) | 501 |
| 5 | 93 (0) | 165 (35) | 258 |
The clinical practice we analyzed routinely converts screening examinations to diagnostic mammography examinations when an abnormality is identified; therefore, practice performance parameters were calculated in aggregate because these examinations could not be accurately separated. Specifically, we measured recommended performance parameters (cancer detection rate, early stage cancers detection rate, and abnormal interpretation rate) for all mammograms in our dataset.
In contrast to our practice performance audit based on mammograms, the analysis of the classification accuracy of the logistic regression model and radiologists was conducted at the record level. Since breast cancer classification actually happens at the record level (i.e. each finding on mammography will require a decision to recall or biopsy), we target this level of granularity to help improve radiologists’ performance. We clearly indicate when analyses in this manuscript are based on mammograms versus records.
We used cancer registry matching as the reference standard in this study. All newly diagnosed cancer cases are reported to the Wisconsin Cancer Reporting System. This registry collaborates with several other state agencies to collect a range of data including demographic information, tumor characteristics, treatment and mortality. Data exchange agreements with 17 other state cancer registries yield data for Wisconsin residents receiving care in other states. We sent 65,892 records in the database to the cancer registry and received back 65,904 records after their matching protocol. Additional 12 records were returned to us because of the duplication of records for the patients diagnosed with more than one cancer. We developed an automated process that confirmed whether the cancer matched the assigned abnormality. This process ensured that the record indicated the same side, the same quadrant, and diagnosed at most 12 months after the mammography date. If there was more than one record indicating the same side and quadrant, then the matching was done manually. We used a 12-month follow-up period as the reference standard because it has been recommended as a sufficient interval to identify false negatives in mammography practice audits [27, 28]. We removed 299 records, which belonged to 188 mammograms from 124 women, because they could not be matched due to missing laterality or quadrant information from either the cancer registry (117 records) or the mammography structured report (182 records) (Table 2). Of the unmatched 299 records, 183 records represented a second record identifying a finding in women who already have a cancer matched to the registry. The remaining 116 records consisted of: 38 BI-RADS 1, 24 BI-RADS 2, 22 BI-RADS 3, 21 BI-RADS 0, 4 BI-RADS 4, and 7 BI-RADS 5. We then removed 101 duplicates. Finally, we removed 3,285 records that had BI-RADS assessment categories 0, 3, 4, 5 (indicating a finding) that did not have descriptors recorded in the record. The final sample consisted of 62,219 (510 malignant; 61,709 benign) records.
Table 2.
Data Processing
| Removed | Total | Malignant | |
|---|---|---|---|
| Mammography records reported to ______ Cancer Registry System | 65,892 | ||
| Records from ________ Cancer Registry System | (12)* | 65,904 | |
| Records unmatched with the registry± | 299 | 65,605 | 546 |
| Number of duplicate records | 101 | 65,504 | 532 |
| Records with missing features (but expected) in the structured reports (i.e. BIRADS 3, 0, 4, 5 with no masses and calcifications) | 3,285 | 62,219† | 510† |
Additional records were returned because of duplication of records for the patients diagnosed with more than one cancer
Laterality, quadrant position was not available in the NMD or Registry data
Data used to build logistic regression model
Statistical Analysis
Model Construction
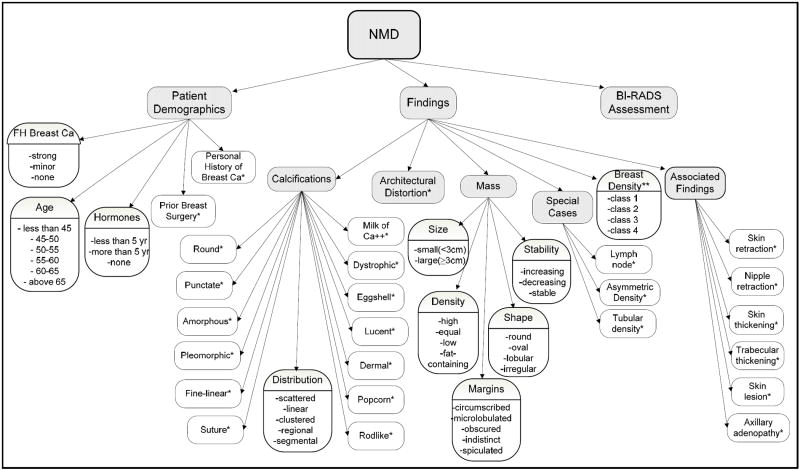
Logistic regression, a statistical approach to predict the presence of a disease based on available variables (symptoms, imaging data, patient history, etc.), has been successfully used for prediction and diagnosis in medicine [29, 30]. To build a breast cancer risk estimation model, we mapped the variables collected by physicians in their daily clinical practice (based on BI-RADS descriptors in the NMD format) to 36 discrete variables. Figure 1 shows the schema of these variables used to build the model. We constructed two risk estimation models. Both models used the presence or absence of breast cancer as the dependent variable, and the 36 variables mentioned above were used as independent variables to build the model. Model-2 included these same variables plus the BI-RADS assessment categories assigned by the radiologists. There are more than 600 possible 2-way interaction effects in each model. We did not include any interaction term in our models.
Fig. 1.
Descriptors of National Mammography Database entered to build a logistic regression model for breast cancer prediction
*Binary variable with categories – “Present” or “Not Present”
**class1: predominantly fatty, class 2: scattered fibroglandular, class 3: heterogeneously dense, and class 4: extremely dense tissue.
Before model construction, we grouped BI-RADS categories 1 and 2 as “BI-RADS 1 or 2” because these cases had a low frequency of malignancy. The logistic regression model was built using the R statistical software [31]. We used forward selection based on the Chi-square test of the change in residual deviance. We used a cutoff of P < 0.001 for adding new terms. This stringent criterion was used to avoid including terms that while statistically significant because of the large sample size are not clinically important. The p-values listed in Tables 3a and 3b are from Chi-square tests of the significance of each term entered last. The importance of each term in predicting breast cancer can be assessed using the odds ratios provided in the tables. The details of logistic regression (including the interpretation of odds ratios) are discussed in Appendix-1.
Table 3a.
| Model-1 (Multivariable model with BI-RADS categories excluded) | |||
|---|---|---|---|
| Risk Factors | Beta | Odds ratio (95 % C.I.1) | p-value |
| Masses Stability | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Increasing | 0.63 | 1.88 (1.37 to 2.60) | |
| Stable | -1.19 | 0.30 (0.18 to 0.50) | |
| Decreasing | -0.74 | 0.48 (0.26 to 0.87) | |
| Masses Shape | 0.0003 | ||
| None | 0.00 | 1 (referent) | |
| Irregular | 0.84 | 2.31 (1.32 to 4.04) | |
| Oval | -0.12 | 0.89 (0.49 to 1.60) | |
| Round | -0.02 | 0.98 (0.51 to 1.89) | |
| Lobular | 0.62 | 1.87 (0.89 to 3.89) | |
| Cannot Discern | -0.70 | 0.50 (0.17 to 1.48) | |
| Masses Margins | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Circumscribed | -0.93 | 0.39 (0.21 to 0.74) | |
| Cannot Discern | 0.27 | 1.32 (0.72 to 2.42) | |
| Ill-defined | 1.41 | 4.10 (2.49 to 6.76) | |
| Spiculated | 2.90 | 18.24 (10.67 to 31.20) | |
| Microlobulated | 0.63 | 1.88 (0.74 to 4.82) | |
| Masses Density | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Cannot Discern | 0.80 | 2.23 (1.25 to 3.97) | |
| Equal | 0.74 | 2.10 (1.13 to 3.88) | |
| Low | 0.63 | 1.88 (0.73 to 4.88) | |
| High | 2.27 | 9.67 (5.59 to 16.71) | |
| Masses Size | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Small | 1.20 | 3.33 (2.32 to 4.77) | |
| Large | 0.90 | 2.46 (1.36 to 4.45) | |
| Skin Retraction | <0.0001 | ||
| Not Present | 0.00 | 1 (referent) | |
| Present | -1.45 | 0.23 (0.11 to 0.49) | |
| Calcification Distribution | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Clustered | 0.64 | 1.89 (1.20 to 2.96) | |
| Regional | 1.10 | 3.01 (1.21 to 7.46) | |
| Scattered | 0.89 | 2.44 (0.31 to 19.22) | |
| Linear | 1.13 | 3.11 (1.15 to 8.44) | |
| Segmental | 3.58 | 35.71 (10.79 to 118.15) | |
| Pleomorphic Calcifications | <0.0001 | ||
| Not Present | 0.00 | 1 (referent) | |
| Present | 2.37 | 10.68 (7.17 to 15.93) | |
| Fine linear Calcifications | <0.0001 | ||
| Not Present | 0.00 | 1 (referent) | |
| Present | 0.89 | 2.44 (1.61 to 3.69) | |
| Age | 0.2216 | ||
| Age < 45 | 0.00 | 1 (referent) | |
| Age 45-50 | -0.02 | 0.98 (0.65 to 1.48) | |
| Age 51-54 | -0.20 | 0.82 (0.51 to 1.32) | |
| Age 55-60 | 0.26 | 1.30 (0.88 to 1.92) | |
| Age 61-64 | 0.18 | 1.20 (0.77 to 1.88) | |
| Age ≥ 65 | 0.22 | 1.25 (0.87 to 1.78) | |
| History of Breast Cancer | <0.0001 | ||
| No History | 0.00 | 1 (referent) | |
| History of DC or LC | 2.90 | 18.16 (14.38 to 22.93) | |
CI = confidence interval
There are a number of possible sources of correlation in these data. Findings from a particular radiologist may be more similar than findings from different radiologists, findings within a patient may be more similar than those from different patients, and findings within the same mammography visit of a patient may be more similar than those during other mammography visits of the same patient. We investigated models where radiologist is included as a random effect and compared it to our models where radiologist is excluded from the models. We found no substantial differences in the coefficients for the other terms in the model due to including radiologist as a random effect. Thus we choose the simpler model without radiologist. We were unable to test random effects for patient or for mammogram within patient because the expected number of cancers for each patient is very small. Random effects models tend to be biased in these circumstances [32]. Instead, we relied on our stringent criterion of P < 0.001 for inclusion in the model to avoid the overly optimistic p-value which occur when the variance of the parameters is reduced by positive correlation induced by clustered data. The parameter estimates themselves are unbiased regardless of the form of the variance.
In order to demonstrate that BI-RADS descriptors substantively contribute to prediction accuracy in Model 2, we also constructed a secondary model (Model-3). Model 3 leaves out these descriptors and includes only patient demographic factors (age, history of breast cancer, family history of breast cancer, history of surgery, breast density, and hormone therapy) and BI-RADS assessment categories as independent variables to test whether performance declines. The details of Model-3 are provided in Appendix-2.
Model Evaluation
We used 10-fold cross validation technique to evaluate the predictive performance of the two models. This methodology avoids the problem of validating the model on the same data used to estimate the parameters by using separate estimation and evaluation subsets of the data. Specifically, we divided the dataset into ten subsets (with approximately one tenth of benign abnormalities and one tenth of malignant abnormalities in each subset or “fold”) such that all abnormalities associated with a single patient were assigned to the same fold. This ensured that all folds are independent of each other. We started with the first nine folds (omitting the 10th fold) to estimate the coefficients of the independent variables (training), and predicted the probability of cancer on the 10th fold (testing). Then we omitted the 9th fold (used as the testing set) and trained the model using the other nine folds. Similarly, we tested on each fold. Finally, we combined all test sets to obtain a full-test set, and evaluated the overall performance of the model using the full-test set. Note that for inclusion of variables in the final model, we used whole dataset (62,219 records), which gave us best possible estimates of the variables from the available data.
Performance Measures
We measured the performance of the two models using the outcome (i.e. the probability of cancer) of the full-test set obtained by 10-fold cross-validation. We plotted and measured area under the receiver operating characteristics (ROC) curve of Model-1 and Model-2 using the probability of cancer. We measured the performance of radiologists using BI-RADS assessment code assigned to each mammography record. We first ordered BI-RADS assessment codes by likelihood of breast cancer (1, 2, 3, 0, 4, 5), generated an ROC curve, and measured its area (Az) using a nonparametric method [33]. We compared the performance of the two models to that of radiologists using Delong’s nonparametric method [34] for comparing two or more areas under ROC curves obtained from the same dataset.
For the purpose of assessing the sensitivity and specificity of radiologists, we classified BI-RADS categories 1, 2, 3 as negative; and BI-RADS categories 0, 4, 5 as positive [28]. We compared the sensitivity of the two models to the radiologists’ sensitivity at 90%-specificity, and specificity of the two models to the radiologists’ specificity at 85%-sensitivity with the corresponding confidence intervals estimated using the efficient score method (corrected to continuity) [35]. Note that the points--sensitivity at 90%-specificity and specificity at 85%-sensitivity on the radiologists’ ROC curve were not observed in practice; they were obtained from the linear interpolation of the two neighboring discrete points. We used these levels of sensitivity and specificity because they represent the minimal performance thresholds for screening mammography [36]. We also estimated the number of true positive and false negative records at 90%-specificity by multiplying the sensitivity (of radiologists, Model-1 and Model-2) with the total number of malignant records. Similarly, we estimated the number of false positive and true negative records at 85%-sensitivity by multiplying the specificity (of radiologists, Model-1 and Model-2) with the total number of benign records. Finally, we identified the most important predictors of the breast cancer using the odds ratio given in the output of the two models when built on the whole dataset (62,219 records).
Results
Practice Performance
We found the following distribution of breast tissue density; predominantly fatty tissue, 14%; scattered fibroglandular tissue, 41%; heterogeneously dense tissue, 36%; and extremely dense tissue 9% (Table 1). At the mammogram level, the cancer detection rate was 9.8 cancers per 1000 mammograms (477 cancers for 48,744 mammograms). The abnormal interpretation rate was 18.5 % (9037 of 48,744 mammograms). Of all cancers detected, 71.9% were early stage (0 or 1) and only 25.9 % had lymph node metastasis. Radiologists demonstrated a sensitivity of 90.5% and a specificity of 82.2% as estimated from BI-RADS assessment categories on the mammogram level.
In Model-1, 10 independent variables (mammographic features and demographic factors) were found to be significant in predicting breast cancer (Table 3a). The most important predictors associated with breast cancer as identified by this model were spiculated mass margins, high mass density, segmental calcification distribution, pleomorphic calcification morphology, and history of invasive carcinoma. Age was not found to be a significant predictor, but it was included in the model because of its clinical relevance. In Model-2, which included BI-RADS assessment categories, 9 independent variables were significant in predicting the risk of breast cancer (Table 3b). The most important predictors associated with breast cancer as identified by this model were BI-RADS assessment codes 0, 4 and 5, segmental calcification distribution, and history of invasive carcinoma. Note that the inclusion of BI-RADS assessment codes in Model-2 removed some of the significant predictors found in Model-1 and added other. We tested for the significance of variables in both the models (as shown in Table 3a and Table 3b) using the whole dataset. Among demographic factors, none of the models found family history of breast cancer and use of hormones as significant predictors of breast cancer. Among imaging descriptors, none of the models found breast density, architectural distortion, and amorphous calcification morphology as significant predictors of breast cancer.
Radiologists achieved an Az of 0.939 ± 0.011 as measured by the BI-RADS assessment code assigned to each record. Model-1 achieved Az of 0.927 ± 0.015, which was not significantly different (P=.104) than the radiologists’ Az. Model-2 performed significantly better (P<.001) than radiologists, with Az of0.963 ± 0.009 and Model-1 (P<.001) (Figure 2).
Fig. 2.
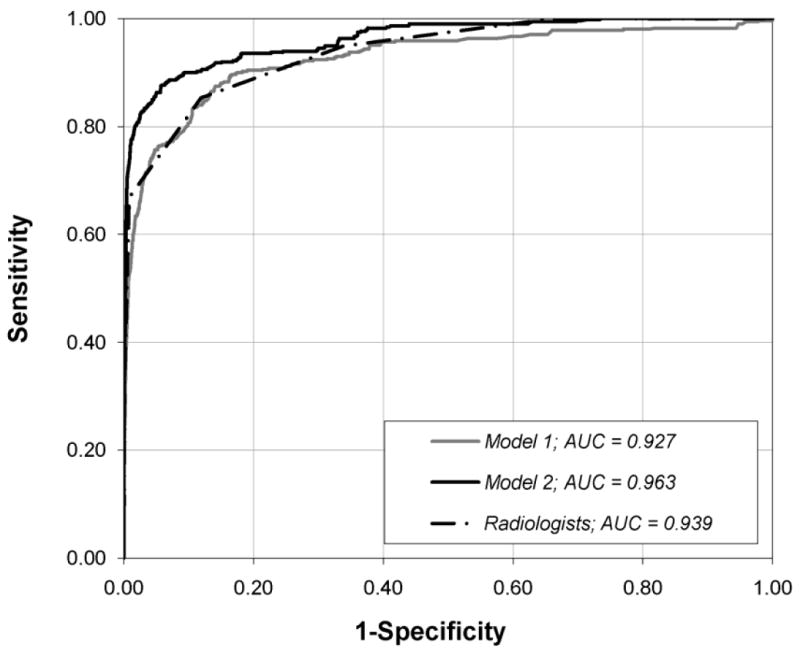
Graph shows ROC curves constructed from the output probabilities of Model-1 and Model-2, and Radiologist’s BI-RADS assessment categories. AUC = Area under the curve.
At the abnormality level, we found that at 90% specificity, the sensitivity of Model-2 was 90.2% (95% CI: 87.2%-92.6%) that was significantly better (P < .001) than that of the radiologists at 82.2% (95% CI: 78.5%-85.3%) and Model-1 at 80.7% (95% CI: 77.0%-84.1%). Table 4a illustrates that Model-2 identified 41 more cancers than the radiologists at this level of specificity. At a fixed sensitivity of 85%, the specificity of Model-2 at 95.6% (95% CI: 95.4%-95.8%) was also significantly better (P < .001) than the radiologists at 88.2% (95% CI: 87.9%-88.5%) and Model-1 at 87.0% (95% CI: 86.7%-87.3%) respectively. Table 4b illustrates that Model-2 decreased the number of false positives by 4,567 when compared to radiologists’ performance.
Table 4.
| a: Performance at 90% specificity | ||
|---|---|---|
| True Positive (95% CI) | False Negative (95% CI) | |
| Radiologist | 419 (400 to 435) | 91 (75 to 110) |
| Model-1 | 412 (393 to 429) | 98 (81 to 117) |
| Model-2 | 460 (445 to 472) | 50 (38 to 65) |
|
| ||
| b: Performance at 85% sensitivity | ||
|
| ||
| False Positive | True Negative | |
|
| ||
| Radiologist | 7,282 (7,126 to 7,441) | 54,427 (54,268 to 54,583) |
| Model-1 | 8,002 (7,837 to 8,207) | 53,687 (53,502 to 53,872) |
| Model-2 | 2,715 (2,592 to 2,839) | 59,994 (58,870 to 59,117) |
We now illustrate the use of the logistic regression models to estimate the probability of cancer using three cases:
Case 1
A 45 years old woman presented with a circumscribed oval mass of equal density on her baseline mammogram. She was assigned BI-RADS 4 assessment code by the radiologist on this abnormality. Model-1 and Model-2 estimated her probability of cancer equal to 0.05% (95% CI = 0.01% to 0.23%). and 1.79% (95% CI = 0.27% to 11.11%), respectively. Biopsy of this case turned out to be benign. This is a classic example of a probably benign finding with an estimate of breast cancer of less than 2%.
Case 2
A 52 year old woman with a history of breast cancer had a mammogram which demonstrated an ill-defined, oval-shaped mass (< 3 cm), which was increasing in size and had equal density. Radiologist assigned BI-RADS 3 assessment code. The probability of malignancy for this finding using Model-1 was 30.6% (95% CI = 8.2% to 68.6%) and for Model-2 was 3.6% (95% CI = 0.7% to 17.4%). Biopsy revealed malignancy. This case illustrates superior predictive ability for Model-1 because the BI-RADS code was not correct and misled Model-2.
Case 3
A 60 year old woman with a family history of breast cancer had a mammogram which demonstrated a mass with a spiculated margin and irregular shape. Model-1 estimated her probability of cancer equal to 51.2% (95% CI = 24.4% to 78.3%). This abnormality was assigned BI-RADS 5 assessment code. Model-2 estimated her probability of cancer equal to 69.7% (95% CI = 33.5% to 91.2%). Biopsy outcome of this case was malignant. This case represents a straightforward case of malignancy where a correct BI-RADS code increases the probability of malignancy using Model 2.
Discussion
We constructed two breast cancer risk estimation models based on the NMD format descriptors to aid radiologists in breast cancer diagnosis. Our results show that the combination of a logistic regression model and radiologists’ assessment performs better than either alone in discriminating between benign and malignant lesions. The ROC curve of Model-1, which only includes demographic factors and mammography observations, overlaps and intersects with the radiologists at certain points in the curve, showing that one is not always better than the other. On the other hand, Model-2, which also includes radiologists’ impression, clearly dominates the other two ROC curves indicating better sensitivity and specificity at all threshold levels. Adding radiologists’ overall impression (BI-RADS category) in Model-2, we could identify more malignant lesions and avoid false positive cases as compared to the performance of Model-1 and radiologists alone.
Our computer model is different in various ways when compared to the existing mammography computer models. The existing models in literature can be categorized in the following ways: (1) for detecting abnormalities present on the mammograms, (2) for estimating the risk of breast cancer based on the mammographic observations and patient demographic information, and (3) for predicting the risk of breast cancer to identify high risk population. The first category of models is used to identify abnormalities on the mammograms, whereas, our model provides the interpretation of mammography observations after they are identified. The models in the second category, in which we classify our model, have used (a) suspicious findings recommended for biopsy for training and evaluation and/or (b) biopsy results as the reference standard. For example, one study constructed a Bayesian Network using 38 BI-RADS descriptors and by training the model on 111 biopsies performed on suspicious calcifications, they found Az = 0.919 [37]. Another study developed linear discriminant analysis and artificial neural network models using a combination of mammographic and sonographic features, and found Az = 0.92 [16]. In contrast, our computer model was trained and evaluated on consecutive mammography examinations and used registry match as the reference standard. The third category of models (risk prediction models) have been built using consecutive cases, but they only included demographic factors and breast density in their model [19, 21, 22]; and cannot be directly compared to our model. In addition, our model differs from these risk prediction models by estimating the risk of cancer at a single time point (i.e. at the time of mammography) instead of risk prediction over an interval in the future (e.g. over the next 5 years). In contrast to their findings, our model did not find breast density as a significant predictor of breast cancer. This could be due to the fact that the risk of breast cancer is explained by more informative mammographic descriptors in our logistic regression model. Our model reinforces previously known mammography predictors of breast cancer – irregular mass shape, ill-defined and spiculated mass margins, fine linear calcifications, and clustered, linear and segmental calcification distributions [38]. In addition, we found increasing mass size and high mass density as significant predictors, which have not been demonstrated in the literature to our knowledge. Of note, our results reflect a single practice and must be viewed with some caution with respect to their generalizability as significant variability has been observed in interpretive performance of screening and diagnostic mammography [5, 6].
We developed two risk estimation models by excluding (Model-1) and including (Model-2) BI-RADS assessment codes. Though Model-2 performed significantly better than Model-1 in discriminating between benign and malignant lesions, Model-2 may have weaknesses as a stand-alone risk estimation tool if the assessed BI-RADS category is incorrect. If the BI-RADS assessment category does not agree with the findings, Model-1 and Model-2 used jointly will show a high level of disagreement in the prediction of breast cancer (as in example case 2) and potentially indicate this error. When the radiologist’s BI-RADS code is correct (i.e. when there is an agreement between the prediction of Model-1 and Model-2), Model-2 would be a better model for breast cancer prediction. In future work, we plan to estimate the level of disagreement between the two models and investigate the possible use of these models as complimentary tools.
Our secondary model (Model-3) showed that the exclusion of the BI-RADS descriptors significantly impairs the performance of the logistic regression model, underscoring the need for the collection of these variables at a clinical practice.
It is common for clinical data sets to contain a substantial number of “missing” data. While complete data is ideally better, it is rarely encountered in the real world. There is no perfect way to handle missing data but there are two possibilities: (1) impute the missing descriptor depending on the fraction of various possible values of the descriptor or (2) assume that the missing descriptor was not observed by radiologists and mark it as “not present”. While building the model, we made the decision to label all of the missing data as “not present”; therefore, while testing/applying the model on a new case the missing descriptors should be treated as “not present”. Our approach to handle missing data is appropriate for mammography data where radiologists often leave the descriptors blank if nothing is observed on the mammogram.
To the best of our knowledge, no prior studies discuss a logistic regression based CADx model incorporating mammography descriptors from consecutive mammograms from a breast imaging practice. The use of logistic regression model has some attractive features when compared with artificial intelligence prediction tools (e.g. artificial neural networks, Bayesian networks, support vector machines). Logistic regression can identify important predictors of breast cancer using odds ratios and generate confidence intervals which provide additional information for decision-making.
Our models’ performance depends on the ability of the radiologists to accurately identify findings on mammograms. Therefore, based on literature, the performance may be higher in facilities where the majority of the mammograms are read by mammography-subspecialists as compared to general radiologists[39]. However, with appropriate training [40], general radiologists in combination with the model may approach the accuracy of subspecialty trained mammographers. Decreasing variability in mammography interpretation, one of the underlying motivations of this research, can only be realized with further development of tools such as our model and research to validate accuracy, effectiveness, and generalizability. We consider this work only a first step toward this goal.
We could not compare practice parameters directly with the literature because screening and diagnostic examinations could not be separated for this database. Our prediction Model-2 shows a significant improvement over radiologists’ assessment in classifying abnormalities when built on a mix of screening and diagnostic data. The model’s performance may differ when built separately on screening and diagnostic mammograms. For screening mammograms, the incidence is low and descriptors are less exact due to general imaging protocols, hence may result in less accurate model parameters. In contrast, for diagnostic mammograms, the model parameters may be more accurate since more descriptors can be observed because of additional specialized views. In addition, the performance our existing model may differ when tested on screening and diagnostic mammograms separately. The model may perform better when tested on the diagnostic exams, but worse when tested on the screening exams.
Our risk estimation models are designed to aid radiologists, not act as a substitute. The improvement in the model’s performance by adding BIRADS assessments in this manuscript indeed suggests that the radiologist integration of the imaging findings summarized by the BIRADS assessment categories does augment predictions based on the observed mammographic features. However, the LR model contributes an additional measure of accuracy over and above that provided by the BI-RADS assessment categories as evidenced by improved performance as compared to the radiologists alone.
The objective of our model is to aid decision-making by generating a risk prediction for a single point in time (at mammography). As we were designing the study, we did not want to increase the probability of breast cancer based on future events but only on variables identified at the time of mammography. For this reason, we excluded unmatched BI-RADS 1 cases from our analyses, which represented either undetected cancer (present on the mammogram but not seen) or an interval cancer (not detectable on the mammogram). The inclusion of these cases may have erroneously increased the probability of malignancy by considering future risks rather than making a prediction at a single time point based on mammography features alone. However, the exclusion of these cases may have erroneously decreased the estimated probability of malignancy, given that at least some of the false negative cancers were likely present at the time of the mammogram - especially those in women with dense breasts; which is a limitation of our model.
Our models provide the probability of cancer as the outcome that can be used by radiologists for making appropriate patient management decisions. The use of such models has a potential to reduce the mammography interpretive variability across practices and radiologists. Our models also facilitate shared decision-making by providing probability of cancer, which can be better understood by patients than BI-RADS categories. In the future, we will test our models’ performance on other mammography practices to evaluate their generalizability. We will also include potentially important interaction effects that deserve particular attention. Note that including interaction effects will further improve the performance of our models.
In conclusion, we found that our logistic regression models (Model-1 and Model-2) can effectively discriminate between benign and malignant lesions. Furthermore, we have found that the radiologist alone or the logistic regression model incorporating only mammographic and demographic features (Model-1) are inferior to Model-2 which incorporates the model, the features, and the radiologist’s impression as captured by the BI-RADS assessment categories. Our study supports that further research is needed to define how radiologists and computational models can collaborate, each adding valuable predictive features, experience and training to improve overall performance.
Table 3b.
| Model-2 (Multivariable model with BI-RADS categories included) | |||
|---|---|---|---|
| Risk Factors | Beta | Odds ratio (95 % C.I.1) | p-value |
| Masses Stability | 0.0002 | ||
| None | 0.00 | 1 (referent) | |
| Increasing | 0.54 | 1.71 (1.21 to 2.42) | |
| Stable | -0.04 | 0.96 (0.55 to 1.68) | |
| Decreasing | -0.96 | 0.38 (0.19 to 0.78) | |
| Masses Margins | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Circumscribed | -0.41 | 0.66 (0.38 to 1.14 | |
| Cannot Discern | 0.41 | 1.51 (0.89 to 2.55) | |
| Ill-defined | 0.76 | 2.13 (1.38 to 3.29) | |
| Spiculated | 0.77 | 2.16 (1.27 to 3.69) | |
| Microlobulated | 0.10 | 1.11 (0.41 to 2.95) | |
| Masses Size | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Small | 1.13 | 3.10 (2.15 to 4.48) | |
| Large | 0.42 | 1.51 (0.78 to 2.95) | |
| Sp-Lymph Node | <0.0001 | ||
| Not Present | 0.00 | 1 (referent) | |
| Present | -1.73 | 0.18 (0.07 to 0.45) | |
| Sp-Asymmetric Density | 0.0002 | ||
| Not Present | 0.00 | 1 (referent) | |
| Present | 0.78 | 2.18 (1.54 to 3.08) | |
| Calcification Distribution | <0.0001 | ||
| None | 0.00 | 1 (referent) | |
| Clustered | 1.09 | 2.98 (2.00 to 4.43) | |
| Regional | 0.92 | 2.51 (0.95 to 6.62) | |
| Scattered | 0.60 | 1.82 (0.14 to 23.48) | |
| Linear | 0.40 | 1.49 (0.49 to 4.54) | |
| Segmental | 2.82 | 16.73 (3.76 to 74.48) | |
| Age | <0.0001 | ||
| Age < 45 | 0.00 | 1 (referent) | |
| Age 45-50 | 0.03 | 1.04 (0.66 to 1.64) | |
| Age 51-54 | 0.02 | 1.02 (0.61 to 1.72) | |
| Age 55-60 | 0.77 | 2.16 (1.39 to 3.36) | |
| Age 61-64 | 0.66 | 1.93 (1.29 to 2.87) | |
| Age ≥ 65 | 0.70 | 2.01 (1.21 to 3.35) | |
| History of Breast Cancer | <0.0001 | ||
| No History | 0.00 | 1 (referent) | |
| History of DC or LC | 2.40 | 11.05 (8.56 to 14.27) | |
| BI-RADS2 | <0.0001 | ||
| BI-RADS 1 or 2 | 0.00 | 1 (referent) | |
| BI-RADS 3 | 1.08 | 2.94 (1.80 to 4.82) | |
| BI-RADS 0 | 3.14 | 23.00 (14.40 to 36.73) | |
| BI-RADS 4 | 5.21 | 183.62 (113.36 to 297.45) | |
| BI-RADS 5 | 6.26 | 522.10 (296.73 to 918.63) | |
CI = confidence interval
BI-RADS = Breast Imaging Reporting and Data Systems
Appendix-1
Binomial (or binary) logistic regression is a form of regression which is used when the dependent variable is dichotomous (e.g. present or absent) and the independent variables are of any type (discrete or continuous). The independent (observed) variables, Xi are related to the dependent (outcome) variable, Y by the following equation:-
| (1) |
Where p = Pr {Y=1} and and p can be calculated by taking the inverse of the Logit (p) as shown in the following equation:
| (2) |
where p represents the probability of the presence of disease (e.g. probability of cancer) when the findings X1, X2,… Xn, (e.g. calcification types, breast density, and age) are identified. βi is the coefficient of the independent variable Xi that is estimated using the available data (training set). Only significant variables (p-value ≤ α) are included in the model. Variables can be added by stepwise, forward, or backward selection methods. Odds ratio is commonly used to interpret the effect of independent variables on the dependent variable, which is estimated by exp (βi). For example, if β1 is the coefficient of variable “prior history of breast surgery”, then exp (β1) is the odds ratio corresponding to the prior history of surgery, i.e. the odds that the patient has malignant lesion increases by the factor of exp (β1), if the patient ever had breast surgery, and all other independent variables remain fixed. More details of logistic regression and its application to medical field can be found in [29, 41, 42].
Appendix-2
In order to assess the contribution of mammography descriptors in our model, we constructed Model-3 which excluded these variables. We included the remaining variables: patient demographic factors (age, history of breast cancer, family history of breast cancer, history of surgery, breast density, and hormone therapy) and BI-RADS assessment categories. Only three variables were found significant in predicting the risk of cancer (Appendix 2 Table 1) with BI-RADS assessment codes as the most important predictor.
Appendix 2 Table 1.
Model-3 (Multivariable model with patient demographic factors and BI-RADS categories only)
| Risk Factors | Beta | Odds ratio (95 % C.I.1) | p-value |
|---|---|---|---|
| BI-RADS2 | <0.0001 | ||
| BI-RADS 1 or 2 | 0.00 | 1 (referent) | |
| BI-RADS 3 | 1.62 | 5.07 (3.15 to 8.18) | |
| BI-RADS 0 | 3.02 | 20.43 (13.20 to 31.63) | |
| BI-RADS 4 | 5.97 | 389.18 (250.95 to 603.55) | |
| BI-RADS 5 | 7.01 | 1112.24 (691.79 to 1788.22) | |
| Age | <0.0001 | ||
| Age < 45 | 0.00 | 1 (referent) | |
| Age 45-50 | -0.03 | 0.97 (0.62 to 1.52) | |
| Age 51-54 | -0.03 | 0.97 (0.59 to 1.60) | |
| Age 55-60 | 0.65 | 1.92 (1.25 to 2.95) | |
| Age 61-64 | 0.49 | 1.63 (0.99 to 2.68) | |
| Age ≥ 65 | 0.54 | 1.71 (1.16 to 2.51) | |
| History of Breast Cancer | <0.0001 | ||
| No History | 0.00 | 1 (referent) | |
| History of DC or LC | 2.27 | 9.64 (7.60 to 12.23) |
CI = confidence interval
BI-RADS = Breast Imaging Reporting and Data Systems
We measured the performance of our model using ROC curves and precision-recall (PR) curves (Appendix 2 Figures 1 and 2). We used PR curves in addition to ROC curves to gain more insights on the performance of our model as PR curves have higher discriminative power than ROC curves in case of skewed data [43, 44]. The precision measures the positive predictive value (PPV) and recall measures the sensitivity of a test. We plotted and measured the area under the PR curve (APR) of the three models (Model-1, Model-2 and Model-3) and radiologists using the probability of cancer and BI-RADS assessment codes, respectively [43].
Appendix 2 Fig. 1.
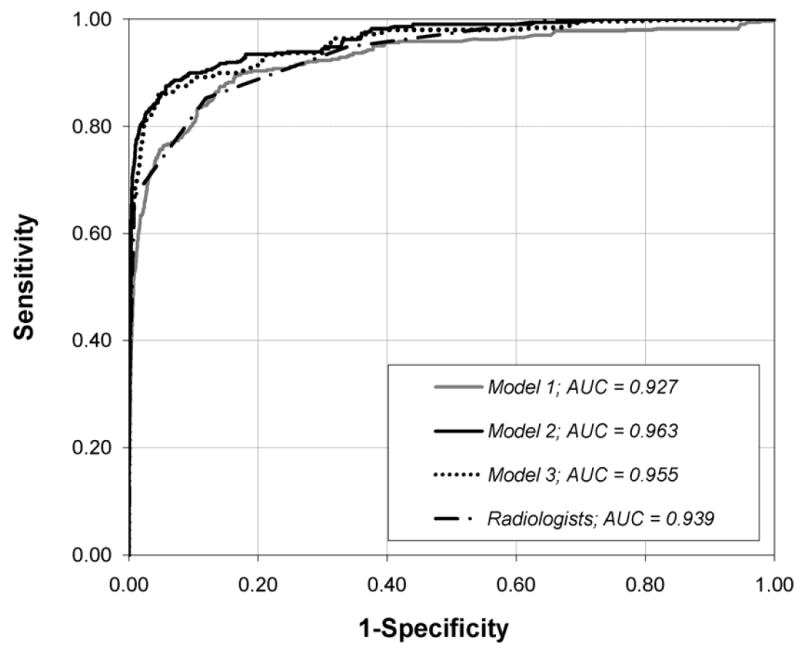
Graph shows ROC curves constructed from the output probabilities of Model-1, Model-2 and Model-3, and Radiologist’s BI-RADS assessment categories.
AUC = Area under the curve.
Appendix 2 Fig. 2.
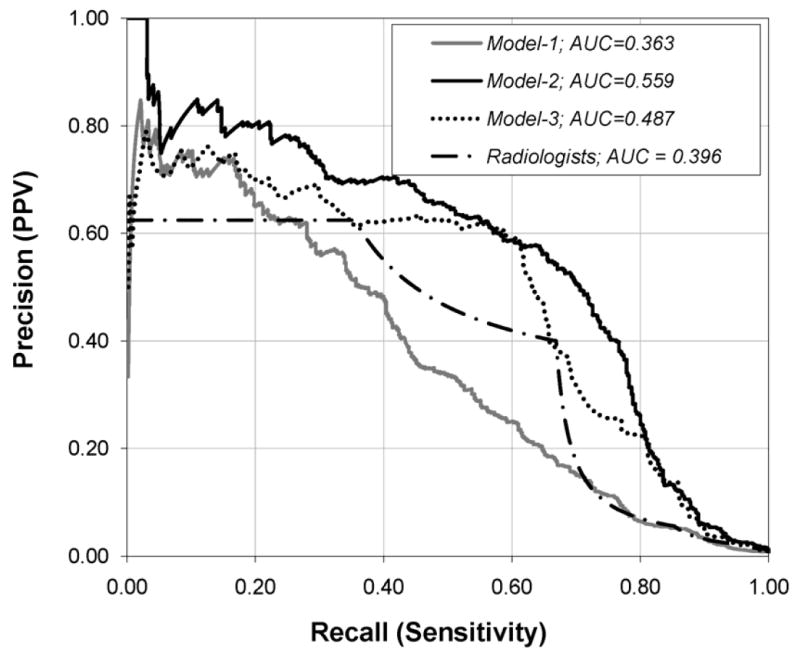
Graph shows PR curves constructed from the output probabilities of Model-1, Model-2 and Model-3, and Radiologist’s BI-RADS assessment categories.
AUC = Area under the curve.
Model-3 achieved an Az, and APR which were significantly higher than that of Model-1 and radiologists (all P < 0.001). More importantly, Model 3—excluding descriptors—performed significantly worse (P<0.001) than that of Model-2—including descriptors—in terms of Az and APR (Appendix 2 Table 2). Thus, the inclusion of mammographic descriptors significantly contributes to the superior performance of Model 2.
Appendix 2 Table 2.
Comparison of the area under the ROC and PR curves
| AROC! | APR!! | |
|---|---|---|
| Radiologists | 0.939 ± 0.011 | 0.396 ± 0.027 |
| Model-1 (demographics + descriptors) | 0.927 ± 0.015 | 0.363 ± 0.030 |
| Model-2 (demographics + descriptors + assessments) | 0.963 ± 0.009 | 0.559 ± 0.026 |
| Model-3 (demographics + assessments) | 0.955 ± 0.011 | 0.487 ± 0.028 |
Az = Area under the receiver operating characteristic curve
APR = Area under the precision-recall curve
References
- 1.Kopans DB. The positive predictive value of mammography. American Journal of Roentgenology. 1992;158:521–526. doi: 10.2214/ajr.158.3.1310825. [DOI] [PubMed] [Google Scholar]
- 2.Barlow WE, Chi C, Carney PA, et al. Accuracy of screening mammography interpretation by characteristics of radiologists. JNCI Journal of the National Cancer Institute. 2004;96:1840–1850. doi: 10.1093/jnci/djh333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerlikowske K, Grady D, Barclay J, et al. Variability and accuracy in mammographic interpretation using the American College of Radiology Breast Imaging Reporting and Data Systems. Journal of the National Cancer Institute. 1998;90:1801–1809. doi: 10.1093/jnci/90.23.1801. [DOI] [PubMed] [Google Scholar]
- 4.Elmore JG, Miglioretti DL, Reisch LM, et al. Screening Mammograms by Community Radiologists: Variability in False-Positive Rates. JNCI Cancer Spectrum. 2002;94:1373–1380. doi: 10.1093/jnci/94.18.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miglioretti DL, Smith-Bindman R, Abraham L, et al. Radiologist Characteristics Associated With Interpretive Performance of Diagnostic Mammography. JNCI Journal of the National Cancer Institute. 2007;99:1854. doi: 10.1093/jnci/djm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taplin S, Abraham L, Barlow WE, et al. Mammography Facility Characteristics Associated With Interpretive Accuracy of Screening Mammography. JNCI Journal of the National Cancer Institute. 2008;100:876. doi: 10.1093/jnci/djn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freer TW, Ulissey MJ. Screening mammography with computer-aided detection: prospective study of 12,860 patients in a community breast center. Radiology. 2001;215:781–786. doi: 10.1148/radiol.2203001282. [DOI] [PubMed] [Google Scholar]
- 8.Dean JC, Ilvento CC. Improved Cancer Detection Using Computer-Aided Detection with Diagnostic and Screening Mammography: Prospective Study of 104 Cancers. American Journal of Roentgenology. 2006;187:20–28. doi: 10.2214/AJR.05.0111. [DOI] [PubMed] [Google Scholar]
- 9.Cupples TE, Cunningham JE, Reynolds JC. Impact of Computer-Aided Detection in a Regional Screening Mammography Program. Am Roentgen Ray Soc. 2005:944–950. doi: 10.2214/AJR.04.1300. [DOI] [PubMed] [Google Scholar]
- 10.Birdwell RL, Bandodkar P, Ikeda DM. Computer-aided Detection with Screening Mammography in a University Hospital Setting 1. Radiology. 2005;236:451–457. doi: 10.1148/radiol.2362040864. [DOI] [PubMed] [Google Scholar]
- 11.Baker JA, Kornguth PJ, Lo JY, Williford ME, Floyd CE., Jr Breast cancer: prediction with artificial neural network based on BI-RADS standardized lexicon. Radiology. 1995;196:817–822. doi: 10.1148/radiology.196.3.7644649. [DOI] [PubMed] [Google Scholar]
- 12.Bilska-Wolak AO, Floyd CE., Jr Development and evaluation of a case-based reasoning classifier for prediction of breast biopsy outcome with BI-RADS™ lexicon. Medical Physics. 2002;29:2090. doi: 10.1118/1.1501140. [DOI] [PubMed] [Google Scholar]
- 13.Burnside ES, Rubin DL, Shachter RD. Using a Bayesian network to predict the probability and type of breast cancer represented by microcalcifications on mammography. Medinfo. 2004;11:13–17. [PubMed] [Google Scholar]
- 14.Fischer EA, Lo JY, Markey MK. Bayesian Networks of BI-RADS Descriptors for Breast Lesion Classification. IEEE EMBS. 2004 doi: 10.1109/IEMBS.2004.1403858. [DOI] [PubMed] [Google Scholar]
- 15.Markey MK, Lo JY, Floyd CE. Differences between Computer-aided Diagnosis of Breast Masses and That of Calcifications 1. RSNA. 2002:489–493. doi: 10.1148/radiol.2232011257. [DOI] [PubMed] [Google Scholar]
- 16.Jesneck JL, Lo JY, Baker JA. Breast mass lesions: computer-aided diagnosis models with mammographic and sonographic descriptors. Radiology. 2007;244:390–398. doi: 10.1148/radiol.2442060712. [DOI] [PubMed] [Google Scholar]
- 17.Claus EB, Risch N, Thompson WD. Autosomal dominant inheritance of early-onset breast cancer. Implications for risk prediction. Cancer. 1994;73:643–651. doi: 10.1002/1097-0142(19940201)73:3<643::aid-cncr2820730323>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Colditz GA, Rosner B. Cumulative risk of breast cancer to age 70 years according to risk factor status: data from the Nurses’ Health Study. Am J Epidemiol. 2000;152:950–964. doi: 10.1093/aje/152.10.950. [DOI] [PubMed] [Google Scholar]
- 19.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 20.Taplin SH, Thompson RS, Schnitzer F, Anderman C, Immanuel V. Revisions in the risk-based Breast Cancer Screening Program at Group Health Cooperative. Cancer. 1990;66:812–818. doi: 10.1002/1097-0142(19900815)66:4<812::aid-cncr2820660436>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 21.Barlow WE, White E, Ballard-Barbash R, et al. Prospective Breast Cancer Risk Prediction Model for Women Undergoing Screening Mammography. JNCI Journal of the National Cancer Institute. 2006;98:1204–1214. doi: 10.1093/jnci/djj331. [DOI] [PubMed] [Google Scholar]
- 22.Tice JA, Cummings SR, Smith-Bindman R, Ichikawa L, Barlow WE, Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Ann Intern Med. 2008;148:337–347. doi: 10.7326/0003-4819-148-5-200803040-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vyborny CJ, Giger ML, Nishikawa RM. Computer-aided detection and diagnosis of breast cancer. Radiol Clin North Am. 2000;38:725–740. doi: 10.1016/s0033-8389(05)70197-4. [DOI] [PubMed] [Google Scholar]
- 24.Doi K, Macmahon H, Katsuragawa S, Nishikawa RM, Jiang Y. Computer-aided diagnosis in radiology: potential and pitfalls. European journal of radiology. 1999;31:97–109. doi: 10.1016/s0720-048x(99)00016-9. [DOI] [PubMed] [Google Scholar]
- 25.Freedman AN, Seminara D, Gail MH, et al. Cancer risk prediction models: a workshop on development, evaluation, and application. J Natl Cancer Inst. 2005;97:715–723. doi: 10.1093/jnci/dji128. [DOI] [PubMed] [Google Scholar]
- 26.Osuch JR, Anthony M, Bassett LW, et al. A proposal for a national mammography database: content, purpose, and value. American Journal of Roentgenology. 1995;164:1329–1334. doi: 10.2214/ajr.164.6.7754870. [DOI] [PubMed] [Google Scholar]
- 27.Breast Imaging Reporting And Data System (BI-RADS) 3. Reston VA: American College of Radiology; 1998. [Google Scholar]
- 28.Breast Imaging Reporting And Data System (BI-RADS) 4. Reston VA: American College of Radiology; 2004. [Google Scholar]
- 29.Bagley SC, White H, Golomb BA. Logistic regression in the medical literature: Standards for use and reporting, with particular attention to one medical domain. Journal of Clinical Epidemiology. 2001;54:979–985. doi: 10.1016/s0895-4356(01)00372-9. [DOI] [PubMed] [Google Scholar]
- 30.Gareen IF, Gatsonis C. Primer on Multiple Regression Models for Diagnostic Imaging Research. Radiology. 2003;229:305–310. doi: 10.1148/radiol.2292030324. [DOI] [PubMed] [Google Scholar]
- 31.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2005 [Google Scholar]
- 32.Moineddin R, Matheson FI, Glazier RH. A simulation study of sample size for multilevel logistic regression models. BMC Medical Research Methodology. 2007;7:34. doi: 10.1186/1471-2288-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 34.DeLong E, DeLong D, Clarke-Pearson D. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 35.Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Statistics in Medicine. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 36.Bassett LW, Hendrick RE, Bassford TL. Public Health Service, US Department of Health and Human Services. Rockville, Md: Agency for Health Care Policy and Research; 1994. Quality determinants of mammography. Clinical practice guideline. No. 13. [Google Scholar]
- 37.Burnside ES, Rubin DL, Fine JP, Shachter RD, Sisney GA, Leung WK. Bayesian Network to Predict Breast Cancer Risk of Mammographic Microcalcifications and Reduce Number of Benign Biopsy Results: Initial Experience. Radiology. 2006;240:666. doi: 10.1148/radiol.2403051096. [DOI] [PubMed] [Google Scholar]
- 38.Liberman L, Abramson AF, Squires FB, Glassman JR, Morris EA, Dershaw DD. The breast imaging reporting and data system: positive predictive value of mammographic features and final assessment categories. AJR Am J Roentgenol. 1998;171:35–40. doi: 10.2214/ajr.171.1.9648759. [DOI] [PubMed] [Google Scholar]
- 39.Sickles E, Wolverton D, Dee K. Performance Parameters for Screening and Diagnostic Mammography: Specialist and General Radiologists. Radiology. 2002;224:861–869. doi: 10.1148/radiol.2243011482. [DOI] [PubMed] [Google Scholar]
- 40.Berg WA, D’Orsi CJ, Jackson VP, et al. Does Training in the Breast Imaging Reporting and Data System (BI-RADS) Improve Biopsy Recommendations or Feature Analysis Agreement with Experienced Breast Imagers at Mammography? Radiology. 2002 doi: 10.1148/radiol.2243011626. 224301162. [DOI] [PubMed] [Google Scholar]
- 41.Kleinbaum DG. Logistic regression: a self-learning text. Springer New York: 1994. [Google Scholar]
- 42.Hosmer D, Lemeshow S. Applied Logistic Regression. New York: John Wiley & Sons, Inc; 1989. [Google Scholar]
- 43.Davis J, Goadrich M. The relationship between Precision-Recall and ROC curves. Proceedings of the 23rd international conference on Machine learning. 2006:233–240. [Google Scholar]
- 44.Chhatwal J, Burnside ES, Alagoz O. Receiver Operating Characteristic (ROC) Curves versus Precision-Recall (PR) Curves In Models Evaluated With Unbalanced Data. Proceedings of the 29th Annual Meeting of the Society for Medical Decision Making. 2007 [Google Scholar]