Abstract
To regulate their internal environments, organisms must adapt to varying ion levels in their diet. Adult Drosophila were exposed to dietary salt stress, and their physiological, survival, and gene expression responses monitored. Insects continued to feed on NaCl-elevated diet, although levels >4% wt/vol ultimately proved fatal. Affymetrix microarray analysis of flies fed on diet containing elevated NaCl showed a phased response: the earliest response was widespread upregulation of immune genes, followed by upregulation of carbohydrate metabolism as the immune response was downregulated, then finally a switch to amino acid catabolism and inhibition of genes associated with the reproductive axis. Significantly, the online transcriptomic resource FlyAtlas reports that most of the modulated genes are predominantly expressed in hindgut or Malpighian (renal) tubule, implicating these excretory tissues as the major responders to salt stress. Three genes were selected for further study: the SLC5 symporter CG2196, the GLUT transporter CG6484, and the transcription factor sugarbabe (previously implicated in starvation and stress responses). Expression profiles predicted by microarray were validated by quantitative PCR (qPCR); expression was mapped to the alimentary canal by in situ hybridization. CG2196::eYFP overexpression constructs were localized to the basolateral membrane of the Malpighian (renal) tubules, and RNAi against CG2196 improved survival on high-salt diet, even when driven specifically to just principal cells of the Malpighian tubule, confirming both this tissue and this transporter as major determinants of survival upon salt stress. Accordingly, CG2196 was renamed salty dog (salt).
Keywords: Malpighian tubule, microarray, stress response
salt stress can disrupt internal homeostasis, so most organisms can robustly defend their internal milieu against fluctuations in external salt over a limited range. Work, particularly in yeast (1, 5, 6, 29, 34, 35) and in plants (3, 20, 23, 35, 36, 41), suggests a variety of mechanisms, particularly the synthesis or regulation of osmolytes, such as glycerol; the increase of K+ uptake and Na+ efflux at the plasma membrane; and Na+ accumulation at the vacuole (involving proton pumping ATPases, Na+-efflux pumps, plasma membrane proteins that operate as sensors of turgor loss and signaling cascades that regulate these responses).
The worm, Caenorhabditis elegans, is a simple multicellular organism compatible with FACS-like high-throughput screening. In C. elegans, massive salt loading caused upregulation of glycerol 3-phosphate dehydrogenase (25), so allowing the accumulation of glycerol as a protective osmolyte, exactly as in yeast (24), where the high osmolarity glycerol stress pathway (HOG) is under control of the Hog1 kinase (24). Trehalose is another key osmolyte in bacteria, yeast and plants (17); and in C. elegans, osmotic stress increases expression of the synthetic enzyme trehalose 6-phosphate synthase (tps1 and tps2), with a corresponding increase in trehalose levels (26).
In insects, mechanisms of salt tolerance are less well understood. In the aquatic larvae of the mosquito Aedes aegypti, there is an interplay between salinity, feeding, and hemolymph levels of the diuretic amine 5-hydroxytryptamine (12). Most larval mosquitoes live in fresh or brackish water, and so they are specialized for uptake of ions as a scarce resource. The size of the ion-scavenging anal papillae varies inversely with the ionic content of the water, suggesting that these are key osmoregulatory organs (13, 31, 42). By contrast, some species of insect survive in and around saline lakes and show particular specializations for handling of ions (like magnesium) that are present in massive excess (7). The insect body plan is thus adaptable to a wide range of environmental salt levels.
Although Drosophila does not normally encounter extremes of salinity, its advantages as a genetic model in which gene expression can both be studied and manipulated (15, 16) make it an attractive species to study. Previously, mutation of the inebriated putative neurotransmitter/osmolyte transporter (ine) was shown to confer reduced tolerance to dietary salt (21). Overexpression of either of the two isoforms of ine (ine-P1 or ine-P2) rescued this osmotic stress response defect. The osmotic stress-sensitive phenotype of the ine mutants was suggested to result from an inability to accumulate osmolytes within the Malpighian tubule and hindgut. Osmotic stress can also be induced by dehydration and rehydration. In Drosophila populations selected for enhanced desiccation resistance (2, 19), flies excrete some sodium during desiccation but retain ∼85% of the whole body sodium content, 83% of potassium, and 60% of chloride (19), implying that defense against desiccation is not purely by loss of ionic osmolytes.
The aim of this study was to investigate the response of adult Drosophila to acute salt stress, using Affymetrix microarrays and subsequent molecular, transgenic, and physiological validation of genes implicated in the response.
MATERIALS AND METHODS
Drosophila.
Flies were kept on standard medium in tubes at 25°C, 12:12 h photoperiod, and 55% relative humidity. Wild-type (Oregon R) flies were used for obtaining genomic DNA, cDNA, and for performing fluid secretion assays. Microinjections to produce transgenic lines were performed in a w1118 background. Transgenes under control of yeast upstream activating sequences (UAS) were driven ubiquitously by crossing to the Act5C-GAL4/CyO line (line 4414 from Bloomington Drosophila stock center) and in Malpighian tubule principal cells with the c42-GAL4 line (33).
Generation of transgenic RNA interference flies.
Inserts of ∼500 bp, directed against nonconserved regions of genes of interest, were cloned into the pWIZ vector (27) and germ-line transformed into w1118 embryos, according to standard procedures. This produced transgenic flies, in which hairpin ds-RNA could be expressed in cells of choice under control of appropriate GAL4 driver lines (8).
Dietary salt substitution.
For the preparation of the “salt” food, the required salt (NaCl or KCl) was dissolved in 100 ml of normal food just after its preparation, mixed, and the diet left to set. Where appropriate, indigo carmine dye (at a concentration of 1.66 g/l) was also added immediately after the diet preparation, but before the food set, to provide a visible marker of ingestion. Diet was freshly prepared to avoid any changes in the concentration of the salts due to evaporation. For dissection, we anaesthetized flies by chilling them briefly on ice before dissecting out tubules in Schneider's medium (Invitrogen).
Microarrays.
Seven-day-old flies were transferred either into food with 4% NaCl added or normal food. The flies were left for 4, 8, 16, or 32 h to feed, and after that 100 flies from each treatment were anaesthetized on ice and homogenized in 300 μl of TRIzol. This whole procedure was repeated two more times to produce three biological replicates for each sample. All the samples were stored directly at −80°C and processed according to standard protocols. Samples of 20 μg total RNA were reverse-transcribed, then in vitro transcribed, according to Affymetrix standard protocols. The quality of the complementary RNA (cRNA) was also checked on an Agilent RNA Bioanalyzer, with a sample in which the broad cRNA peak exceeded the height of the low-molecular-weight degradation peak taken to be satisfactory. Samples were then run on the Affymetrix Drosophila genome I array under standard conditions: the experiment thus comprised 24 arrays. Quality control was performed as described in a previous paper from our lab (39).
Bioinformatics.
As well as routine analysis by Affymetrix proprietary software (MAS 5.0), array data were analyzed using FunAlyse, a pipeline based on the Bioconductor package, and using robust multichip average for low-level normalization with subsequent calculation of rank products under random permutations. This method provides reliable estimates of fold change, significance, and false discovery rate (FDR), and a sharply improved performance in experiments with limited numbers of arrays (10). Significant changes were assessed for pairs of samples (salt vs. normal diet) at each time point, and gene ontology terms overrepresented in gene lists identified by iterative group analysis (iGA) (9).
Quantitative PCR.
Real-time quantitative reverse transcription PCR (qPCR) was used for further validation of the microarray results. Seven-day-old adult flies were collected and placed 1) in normal food, 2) in 4% NaCl food, and 3) in vials empty of food. After 8 h, flies were collected and anaesthetized in ice, and the tissue of interest was dissected. Total RNA was extracted (Qiagen RNeasy Kit) and cDNA prepared according to standard protocols (Invitrogen, Superscript II). Experiments were controlled against genomic DNA contamination with a no-reverse-transcriptase sample and were normalized against rp49. The samples were replicated independently six to eight times. Data were then expressed as fold difference of stress-treated samples compared with controls ± SE.
Cell type-specific RNA interference knockdown of gene expression.
For downregulation of CG2196 expression, CG2196-RNA interference (RNAi) flies were crossed to the c42-GAL4 (tubule principal cell specific) or actin-GAL4/CyO (ubiquitous expression) driver lines, and the offspring from the crosses were used to validate the extent of knockdown by qPCR. Six repeats were used for each of the different fly lines, and the experiments ware repeated twice more (three biological repeats) in total. Normalization of the results was performed against rp49 controls.
In situ hybridizations.
An expressed sequence tag (EST) clone of the CG2196 gene was obtained (GH19680 EDM1 133-6918917 Drosophila Gene Collection Clone 1) and used to prepare digoxygenin-labeled DNA probes as described previously (18). In situ hybridization was performed as described previously (18).
Survival of RNAi flies.
RNAi male flies were crossed with c42 virgin female flies in cages and for 10 days, eggs were collected each morning, counted, and transferred to vials with food containing 4% NaCl. The percentage of flies surviving long enough to reach the adult stage was compared with the parental lines. At least 12 vials from each line were collected. Two repeats of the full experiment were performed.
Generation of CG2196::eYFP-overexpressing flies.
Flies overexpressing a construct of the CG2196 gene tagged with enhanced yellow fluorescent protein (eYFP) were generated using the Gateway system. The entry and destination vectors used were obtained from the Drosophila Gateway Vector collection (37). The open reading frame of the gene was obtained from an EST clone and inserted in-frame into the Gateway destination vector pTWV, so encoding fluorescently COOH-terminal tagged CG2196, and flies germ-line transformed as described previously. The construct is under control of the UAS promoter, so allowing cell-specific expression under control of appropriate GAL4 driver lines.
Expression in S2 cells.
Drosophila S2 cells were grown in Schneider's medium according to standard protocols. Genes of interest were cloned the destination vector pAWV (Drosophila Gateway Vector Collection). This vector contains a constitutive ActinC5 promoter. S2 cells were transfected according to the Invitrogen protocol as described previously (32). Approximately 3 × 106 cells were used in each transfection. The cells were harvested in PBS and were allowed to attach to poly-l-lysine-treated glass slides. Cells were counterstained with the nuclear stain DAPI and washed repeatedly before being viewed in a Zeiss 510 Meta confocal microscope.
Statistics.
For fluid secretion experiments, significance of results was determined with an unpaired Student's t-test, taking the critical value of P as 0.05 (two-tailed). For microarray experiments, the FDR for significance was taken as 5%.
RESULTS
Do flies eat salty food?
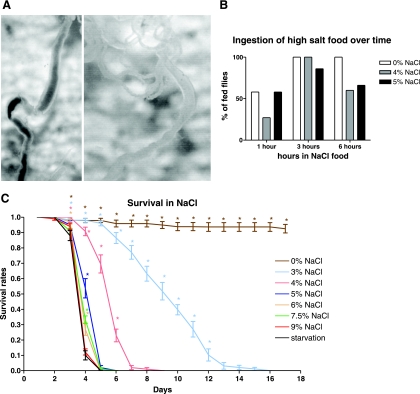
Before studying the effects of dietary salt loading, it is important to establish that the diet remains palatable; otherwise, the effects of salt loading will be obscured by those of starvation or desiccation. Accordingly, flies were exposed to diet labeled with blue dye and containing various loadings of NaCl, for various times and scored for blue color in their guts. The results of this experiment can be seen in Fig. 1B. After 1 h with the food containing no extra NaCl, approximately half of the flies had blue dye in their gut (Fig. 1A). Flies in the food containing 4% or 5% NaCl also appeared with blue dye in their gut, which means that they had also eaten the food. After 3 h, almost all of flies had blue dye in their guts. After 6 h, all the flies in the food without NaCl supplementation, and at least half of those on 4 and 5% NaCl food show a full blue gut (Fig. 1A), confirming that, although food intake is reduced on high-salt diet, unpalatability is not a major issue.
Fig. 1.
Continued ingestion an survival rates on NaCl-supplemented diet. A: example of Indigo carmine in gut after 3 h of feeding with 4% NaCl food (left) compared with 6 h gut that would be scored as clear (right). B: ingestion of high-salt food over time. Flies in 0, 4, and 5% NaCl food, which contained the blue dye Indigo Carmine, were dissected after 1, 3, and 6 h and scored for blue gut contents. C: survival rates in days for flies fed with 0, 3, 4, 5, 6, 7.5, and 9% NaCl compared with starvation. Increasing NaCl significantly reduces survival [Kaplan-Meier analysis with log-rank test for trend, taking P = 0.05 as the critical level (2-tailed)]. *Significantly different from the starvation control.
Survival on high-NaCl diet.
Having established that flies continue to ingest diet containing high levels of NaCl, it is appropriate to examine survival. These results are shown in Fig. 1C. From the graph it is evident that flies fed with 3 or 4% NaCl food survive much better than those that are starved. These results are comparable to those reported previously (21), although our work reports a greater range of salt concentrations and monitors survival over a time course, rather than just at 4 days. Additionally, our wild-type stock survived marginally better on high salt; at 5% NaCl (wt/vol) equivalent to 0.85 M, 50% of flies survived to 4 days compared with 0% reported previously for 0.8 M (21). Although the difference is significant (probably reflecting strain differences), both experiments show a similar pattern of reduction of survival with increasing NaCl concentration. Flies fed with NaCl food up to 5% survive significantly better than the starved flies, but those exposed to higher salt levels die almost as fast as flies deprived of food altogether. This suggests that short-term salt-handling mechanisms can offer partial protection against salt levels elevated by up to 5% for at least a few days. Together with the previous results, a behavioral adaptation to high salt is apparent: insects provided with high-salt diet continue to feed (and thus survive better than insects with no food) but feed less avidly than flies on normal diet. In the subsequent microarray time-course experiment, shorter times (0–8 h) can thus be taken as representative of acute salt exposure, whereas longer time points (16–32 h) might be expected to show additional effects of reduced food intake.
Microarray of acute NaCl stress.
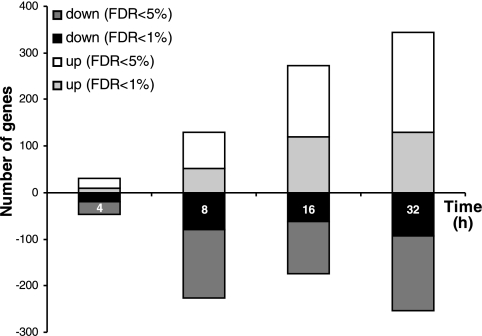
From these results, a microarray experiment was devised comparing matched sets of flies transferred from normal diet to either normal or 4% salt diet and sampled after 4, 8, 16, and 32 h. The numbers of genes both up- and downregulated increased steadily with time, as summarized in Fig. 2. The changed genes are listed in Supplementary Table S1,1 and iGA of all changed gene families is shown in Table 1. The iGA gene list shows some compelling features. When viewed as a time series, the response can be resolved into several phases. Initially, there is a prominent immune response, with multiple genes (metchnikowin; drosocin; diptericin; attacin; immune-induced molecules 1, 10, and 23; PGRP-SB1) upregulated at 4 h and downregulated at 8 and 16 h. At 8 and 16 h, the major response is an upregulation of genes associated with carbohydrate metabolism. Conspicuously, about half of the Drosophila α-glucosidase (maltase) gene family is upregulated, together with an α-amylase and the zinc finger transcription factor sugarbabe (sug), a transcription factor that has previously been implicated in starvation and sugar stress responses (45). Sugarbabe upregulation after feeding is thought to downregulate sugar metabolism (45), so its downregulation here is consistent with upregulation of carbohydrate metabolism.
Fig. 2.
Numbers of genes up- and downregulated at different times from start of salt exposure, at 1 and 5% false discovery rate (FDR).
Table 1.
Iterative Group Analysis of gene families up- and downregulated by dietary salt stress
| Gene Ontology Code | Group |
% Gene Family Changed at Sample Time, h |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 8 | 16 | 32 | |||||||
| A. Upregulated gene families | ||||||||||
| 0008745-N-acetylmuramoyl-l-alanine amidase activity | immune | 25 | ||||||||
| 0006955-immune response | immune | 11 | ||||||||
| 0045087-innate immune response | immune | 14 | ||||||||
| 0042742-defense response to bacterium | immune | 14 | ||||||||
| 0050829-defense response to Gram-negative bacterium | immune | 18 | ||||||||
| 0050830-defense response to Gram-positive bacterium | immune | 9 | ||||||||
| 0019731-antibacterial humoral response | immune | 25 | ||||||||
| 0009253-peptidoglycan catabolic process | immune | 25 | ||||||||
| 0042834-peptidoglycan binding | immune | 25 | ||||||||
| 0030342-1-a,25-dihydroxyvitamin D3 (1,25-(OH)2D3) 24-hydroxylase activity | 100 | |||||||||
| 0004558-alpha-glucosidase activity | sugar | 25 | 42 | 42 | 50 | |||||
| 0043169-cation binding | 10 | 10 | 16 | 14 | ||||||
| 0004613-phosphoenolpyruvate carboxykinase (GTP) activity | 100 | 100 | 100 | 100 | ||||||
| 0004611-phosphoenolpyruvate carboxykinase activity | 100 | 100 | 100 | 100 | ||||||
| 0006094-gluconeogenesis | sugar | 40 | ||||||||
| 0017076-purine nucleotide binding | 40 | |||||||||
| 0004364-glutathione transferase activity | 13 | |||||||||
| 0008643-carbohydrate transport | sugar | 9 | 9 | |||||||
| 0005355-glucose transmembrane transporter activity | sugar | 18 | 24 | |||||||
| 0005351-sugar:hydrogen ion symporter activity | 11 | 14 | ||||||||
| 0006006-glucose metabolic process | sugar | 27 | 27 | 27 | ||||||
| 0005792-microsome | 4 | |||||||||
| 0009063-amino acid catabolic process | AA | 17 | ||||||||
| 0016798-hydrolase activity, acting on glycosyl bonds | sugar | 11 | ||||||||
| 0005996-monosaccharide metabolic process | sugar | 22 | 17 | |||||||
| 0009113-purine base biosynthetic process | 75 | 75 | ||||||||
| 0006144-purine base metabolic process | 9 | 15 | ||||||||
| 0003939-L-iditol 2-dehydrogenase activity | 100 | 100 | ||||||||
| 0006164-purine nucleotide biosynthetic process | 40 | 50 | ||||||||
| 0006189-'de novo' IMP biosynthetic process | 43 | 57 | ||||||||
| 0006012-galactose metabolic process | sugar | 50 | 50 | |||||||
| 0008652-amino acid biosynthetic process | AA | 10 | ||||||||
| 0004477-methenyltetrahydrofolate cyclohydrolase activity | 100 | |||||||||
| 0009072-aromatic amino acid family metabolic process | AA | 38 | ||||||||
| 0004488-methylenetetrahydrofolate dehydrogenase. (NADP+) activity | 100 | |||||||||
| 0006559-L-phenylalanine catabolic process | AA | 50 | ||||||||
| B. Downregulated gene families | ||||||||||
| 0005730-nucleolus | 12 | |||||||||
| 0008143-poly(A) binding | 20 | |||||||||
| 0005616-larval serum protein complex | 33 | |||||||||
| 0005344-oxygen transporter activity | 14 | |||||||||
| 0045735-nutrient reservoir activity | 25 | |||||||||
| 0046870-cadmium ion binding | 67 | 67 | ||||||||
| 0005786-signal recognition particle, endoplasmic reticulum targeting | 27 | |||||||||
| 0004035-alkaline phosphatase activity | 23 | |||||||||
| 0005316-high affinity inorganic phosphate:sodium symporter activity | 22 | |||||||||
| 0005048-signal sequence binding | 75 | |||||||||
| 0006621-protein retention in ER | 100 | |||||||||
| 0050830-defense response to Gram-positive bacterium | immune | 17 | ||||||||
| 0045087-innate immune response | immune | 15 | 10 | |||||||
| 0006955-immune response | immune | 13 | 7 | |||||||
| 0042742-defense response to bacterium | immune | 16 | 11 | |||||||
| 0050829-defense response to Gram-negative bacterium | immune | 27 | 18 | |||||||
| 0019731-antibacterial humoral response | immune | 32 | 21 | |||||||
| 0005784-translocon complex | 43 | 43 | ||||||||
| 0042562-hormone binding | 67 | 67 | ||||||||
| 0007321-sperm displacement | reprod. | 50 | 75 | |||||||
| 0045861-negative regulation of proteolysis | 50 | 63 | ||||||||
| 0007610-behavior | 13 | 23 | ||||||||
| 0004182-carboxypeptidase A activity | 17 | |||||||||
| 0007305-vitelline membrane formation in chorion-containing eggshell | reprod. | 57 | ||||||||
| 0007343-egg activation | reprod. | 33 | ||||||||
| 0008316-structural constituent of vitelline membrane | reprod. | 100 | ||||||||
| 0004181-metallocarboxypeptidase activity | 33 | |||||||||
| 0004263-chymotrypsin activity | 18 | 23 | ||||||||
| 0004806-triacylglycerol lipase activity | 11 | 16 | ||||||||
| 0008970-phospholipase A1 activity | 30 | 20 | ||||||||
| 0016045-detection of bacterium | immune | 33 | ||||||||
| 0004867-serine-type endopeptidase inhibitor activity | 12 | |||||||||
| 0016298-lipase activity | 16 | |||||||||
| 0046692-sperm competition | reprod. | 50 | ||||||||
| 0042600-chorion | reprod. | 38 | ||||||||
| 0005213-structural constituent of chorion | reprod. | 78 | ||||||||
| 0007306-eggshell chorion formation | reprod. | 36 | ||||||||
Gene families that are significantly (by iterative group analysis, Ref. 9) overrepresented in either the up- or downregulated groups are ordered by time of onset and are highlit with the following names: immune, immune/defence response; reprod, reproduction; sugar, carbohydrate metabolism; AA, amino acid metabolism. Note that, because of the limitations and hierarchical nature of gene ontology annotation, there is substantial overlap between the genes reported under related headings. Nonetheless, the trends should be clear.
Sugar metabolism as a general response to salt loading.
The marked changes in genes involved in carbohydrate metabolism are evocative of studies of high-salt diet in other species. In both yeast and C. elegans, salt loading causes the accumulation of glycerol as a protective osmolyte (25) (24). In yeast (24) the HOG stress pathway is under control of the Hog1 kinase (24). Although the Drosophila Hog1 homolog mpk2 does not change significantly in our experiment (Supplementary Table S1), this does not preclude a posttranscriptional modulation of glycerol accumulation, so we cannot absolutely exclude it as a mechanism in insects.
Trehalose is another key osmolyte in bacteria, yeast, and plants (17); and in C. elegans, osmotic stress increases expression of the synthetic enzyme trehalose 6-phosphate synthase (tps1 and tps2), with a corresponding increase in trehalose levels. Interestingly, although no early changes in trehalose metabolism are found in the array, there are two significant changes at 16 h: the synthetic enzyme trehalose 6-phosphate synthase 1 (tps1) is downregulated, whereas the catabolic α-trehalase (CG16965) is upregulated. Accordingly, trehalose does not appear to be a key osmolyte in response to acute salt stress in this insect. Rapid conversion of glucose to trehalose on passage through the insect midgut has classically been seen as a way of maintaining a favorable concentration gradient for glucose absorption from the gut (38), so it is possible that, in insects, trehalose has lost its primitive role as an osmolyte.
Another key change in osmotically stressed worms is the upregulation of chaperones of the heat shock protein (hsp) family, presumably to maintain protein folding under changing ionic strength. However, hsps are underrepresented in the change list in Drosophila (Supplementary Table S1): only hsp68 is found to change significantly and then only at a single time point. The impression is thus gained that Drosophila, with its relatively sophisticated osmoregulatory system, can defend its internal environment effectively against dietary salt loading for an extended period (of several days), before death ensues.
Ion transport and salt loading.
Ion transport genes, perhaps the most obvious candidates for change, are relatively modestly represented; one putative sodium/halide cotransporter is upregulated, while another does not change hugely. However, the major insect epithelial transport genes [Na+, K+ ATPase, V-ATPase, Na+/H+ exchangers, Na+/K+/2Cl− cotransporters (40)] do not feature in the list.
Which tissues are critical in salt response?.
These microarray experiments were performed on whole flies; although this design reduces the sensitivity of the experiment to tissue-specific changes of expression (11), it also avoids preconceptions as to the major tissues involved in salt response. However, since the work was performed, the FlyAtlas online resource has become available, allowing basal expression levels to be ascertained for any Drosophila gene across 14 tissues and life stages (11). So, although our original data were obtained from whole flies, it is possible to see from expression in FlyAtlas whether the 19 most significantly changed genes (as called by MAS 5.0) are housekeeping (generally expressed), or whether they are tissue specific, and whether any particular tissues emerge as common candidates for salt response. The results (Table 2) are remarkable; not only are both up- and downregulated genes all highly tissue-specifically expressed, but they almost all show highest expression in either the hindgut or Malpighian (renal) tubules, the two key osmoregulatory tissues of the excretory cycle (4), with some others specific to the midgut. Our model for response to high levels of salt, therefore, is a massive upregulation of carbohydrate metabolism (perhaps reflecting sharply increased energy demands) in the hindgut and Malpighian tubules, perhaps under control of sugarbabe (a transcription factor previously implicated in dietary stress response, and which is predominantly expressed in the midgut, tubules, hindgut, and fat body), while other tissues are relatively unaffected.
Table 2.
The genes modulated by dietary salt are predominantly expressed in tubule and hindgut
| Treatment |
Number of Genes With Maximal Expression in Tissue |
Total | χ2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | Head | Crop | Midgut | Tubule | Hindgut | Ovary | Testis | Acc | l_fat | tag | car | Fly | |||
| All FlyAtlas | 1,783 | 463 | 1,134 | 1,324 | 791 | 571 | 2,563 | 2,655 | 2,386 | 1,518 | 900 | 1,856 | 144 | 18,088 | |
| 4 h up | 0 | 0 | 0 | 2 | 25 | 22 | 0 | 0 | 1 | 0 | 5 | 7 | 2 | 64 | 426 |
| 8 h up | 0 | 0 | 0 | 2 | 25 | 22 | 0 | 0 | 1 | 0 | 4 | 7 | 2 | 63 | 432 |
| 16 h up | 0 | 2 | 1 | 10 | 33 | 40 | 5 | 0 | 2 | 0 | 16 | 26 | 0 | 135 | 467 |
| 32 h up | 1 | 7 | 1 | 14 | 37 | 54 | 6 | 0 | 4 | 0 | 26 | 43 | 0 | 193 | 534 |
| 4 h down | 1 | 1 | 1 | 0 | 0 | 6 | 2 | 0 | 0 | 0 | 1 | 4 | 0 | 16 | 52 |
| 8 h down | 1 | 12 | 2 | 3 | 36 | 23 | 15 | 8 | 4 | 32 | 24 | 52 | 129 | 341 | 3,273 |
| 16 h down | 1 | 7 | 1 | 1 | 48 | 12 | 3 | 8 | 0 | 10 | 1 | 8 | 4 | 104 | 638 |
| 32 h down | 0 | 6 | 0 | 0 | 49 | 10 | 3 | 33 | 0 | 19 | 1 | 6 | 5 | 132 | 545 |
At each time point, each probe set with significantly changed (false discovery rate < 5%) expression was screened against the Flyatlas.org expression dataset, and the tissue with maximum expression of that probe set was identified. The total scores for each tissue were then compared with the profile obtained from all genes in FlyAtlas, and χ2 was calculated. For 12 d.f., the critical value of χ2 is 21, so the genes up- and downregulated at all time points are significantly nonrandomly drawn from the different tissues: specifically, excretory system (tubule and hindgut, shown in bold) genes are massively overrepresented. Abbreviations for tissues: tag, thoracicoabdominal ganglion; acc, accessory gland; car, carcass; l_fat, larval fat body; fly, whole fly. Details of the FlyAtlas dataset have been published previously (11).
Validation of selected genes.
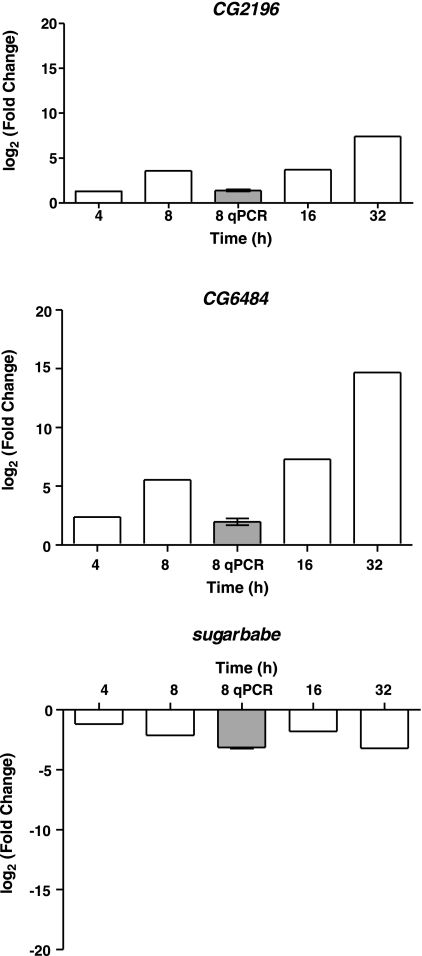
To encompass a range of the responses induced by salt loading, three genes were selected for further study: the SLC5 cotransporter CG2196 (annotated in FlyBase as a sodium/halide symporter), the GLUT4/8-like sugar transporter CG6484, and the transcription factor sugarbabe. According to FlyAtlas (Table 3A), basal expression of CG2196 is virtually tubule-specific, whereas CG6484 is massively upregulated in midgut, and sugarbabe is rather more widely expressed. The expression patterns of CG2196 and CG6486 were validated further by qPCR, confirming the massive expression of these genes in the tubules and midgut respectively (Table 3B). Whole fly quantitative PCRs for the three genes were performed for the 8 h time point and compared with the time-course microarray data (Fig. 3). In all cases, there was reasonable agreement. CG2196 and CG6484 were consistently and increasingly upregulated over time, whereas sugarbabe was consistently downregulated. Thus, based on this limited sample the array, FlyAtlas and qPCR data are concordant.
Table 3.
Genes selected for further study
|
A. FlyAtlas expression levels across multiple tissues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Brain | Head | Tag | Crop | Midgut | Tubule | Hindgut | Ovary | Testis | Acc | Car | L_fat | Fly |
| CG2196 | 7 | 4 | 5 | 7 | 8 | 7,248 | 70 | 4 | 6 | 11 | 21 | 5 | 275 |
| CG6484 | 6 | 22 | 4 | 7 | 2,738 | 12 | 32 | 4 | 8 | 523 | 161 | 6 | 178 |
| sugarbabe | 10 | 197 | 10 | 8 | 584 | 455 | 95 | 5 | 6 | 8 | 371 | 550 | 137 |
|
B. qPCR validation of CG2196 and CG6484 expression in Malpighian tubules, guts, and heads | |||
|---|---|---|---|
|
Relative Expression Level (w.r.t. rp49) |
|||
| Gene | Head | Midgut | Malpighian tubules |
| CG2196 | 4.8×10−5±1.2×10−5 | 3.6×10−5±4.5×10−6 | 0.129±0.008 |
| CG6484 | 2.38×10−7±1.8×10−8 | 8.7×10−5±1.5×10−5 | 3.3×10−8±4.2×10−9 |
In A, the FlyAtlas expression profile across multiple tissues is shown. The maximal signal for each gene is shown in bold. Details of the FlyAtlas data set have been published previously (11). In B, data are shown as means ± SE (n = 3 independent biological replicates).
Fig. 3.
Time course of expression of selected genes and validation at 8 h. For each gene of interest (CG2196, sugarless, and CG6844), mean fold change (salt relative to normal diet) were plotted at 4, 8, 16, and 32 h. Bars with heavy lines are significantly different from no-change hypothesis at FDR <5%. Array fold changes are also compared with qPCR measurements (shaded boxes) at 8 h (means ± SE, n = 3; where error bars are not visible, they are too small to plot).
Role of CG2196/salty dog.
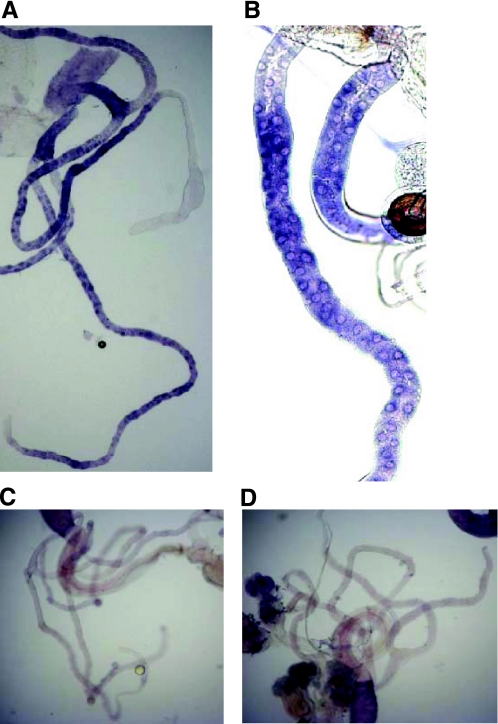
CG2196 was selected for further study, as it was predicted to represent a sodium/halide symporter by the computer annotation of the Drosophila genome, and because it was highly specifically expressed in the Malpighian tubules, one of the key tissues for response to salt (Table 2). According to Drosophila tradition for whimsical but informative names, the gene was named salty dog (salt). Firstly, the expression of salt within the tubule was established by in situ hybridization (Fig. 4). Expression was seen in the principal cells (not the stellate cells) along the length of the tubule but particularly in the main segment. The initial segment was not stained. This localization is reasonable, because the principal cells are the major active ion-transporting cell type in this tissue (15). No staining was seen in other tissues or with the sense control.
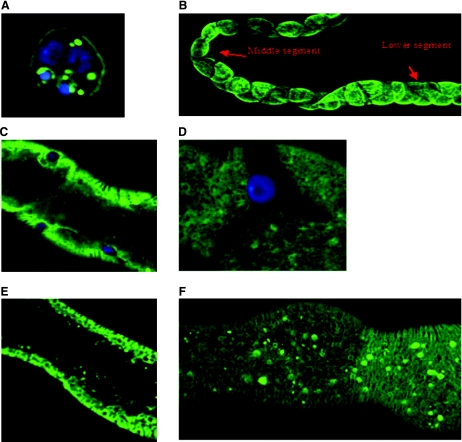
Fig. 4.
In situ hybridizations for CG2196 in Malpighian tubules of adult Oregon R flies. Staining with anti-sense probe. A and B: clear staining appears in the ureter and in the principal cells of the main and the lower segment of both the posterior and anterior Malpighian tubules. In most cases, the staining in the main segment of the tubule was stronger compared with that of the lower segment. The initial segment was not stained in both pairs of tubules. C and D: sense controls. In all photographs, the diameter of the tubule can be taken as 35 μm.
Salt localizes to the basolateral plasma membrane.
Drosophila S2 cells were transiently transfected with a salt::eYFP construct using the calcium phosphate transfection method. The S2 cells present a clear staining around the plasma membrane (Fig. 5A). Also, there seems to be staining in vesicles or conglomerations of the protein inside the cell, as frequently observed when proteins are overexpressed. So at least in the S2 cells, the CG2196-eYFP protein is a membrane protein that appears mainly in the plasma membrane.
Fig. 5.
Expression pattern of CG2196-eYFP in vitro and in vivo. A: Drosophila S2 cells were transfected with the CG2196::eYFP construct under control of an actin 5C promoter. There is clear staining in the plasma membrane and in vesicles or conglomerations of the protein inside the cell. B–G: the CG2196::eYFP construct in flies. B: overview of expression driven in tubule principal cells by the c42 GAL4 line. C: the construct under the c42 promoter appears in the principal cells in the infoldings of the basolateral plasma membrane. Nuclei were counterstained blue with DAPI in A–C. D–F: the CG2196::eYFP construct under the Act5C-GAL4 line, which drives ubiquitous expression. Stellate cells of the middle segment do not express this construct (D). In E and F the focus on the cells differs; E was focused midway through the tubular epithelium, whereas F was focused at the top of the tubule. In all photographs, the diameter of the tubule can be taken as 35 μm.
Homozygous CG2196-eYFP flies (insertion in the first chromosome) were crossed with c42-GAL4 or Act5C-GAL4 flies, and expression of the chimeric protein was assessed (Fig. 5). These GAL4 lines drive expression of UAS constructs in either tubule principal cells [where salt is normally expressed (Fig. 4)] or ubiquitously, respectively. When CG2196-eYFP is driven in tubule principal cells of the main and lower segment by c42-GAL4, it is directed to basolateral membrane infoldings (Fig. 5, B and C). Since this promoter does not drive expression in stellate cells or in the initial segment of the tubule, no staining is seen in these.
With the Act5C-Gal4 promoter, the staining in the principal cells is exactly the same as for the c42 promoter. The progeny of the cross (Act5C-GAL4 flies) × (CG2196-eYFP flies) expresses the CG2196-eYFP construct in the principal cells of the main and lower segment again in the foldings of the basolateral membrane and perhaps in a part of the endoplasmic reticulum, while in the stellate cells of the middle segment no staining is observed (Fig. 5D). Thus salt is expressed in the ion-transporting cells of the Malpighian tubule, on the basolateral membrane, consistent with its presumed function as an uptake symporter.
Modulation of salt expression in tubules impacts on organismal survival after salt challenge.
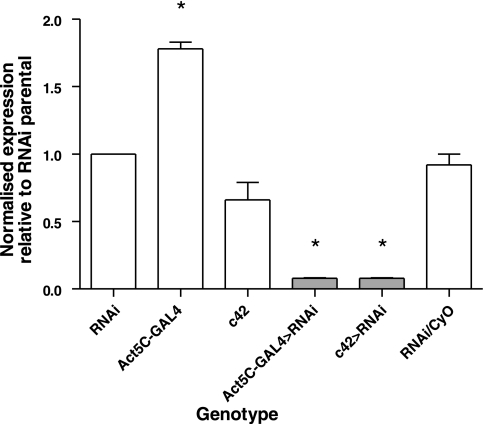
Flies transgenic for a UAS-salt-RNAi construct were generated, and extent of knockdown was assessed by qPCR. UAS-salt-RNAi flies were crossed with the Act5C-GAL4 and c42-GAL4 driver lines, and expression levels of salt in Malpighian tubules compared with parental controls. The results of this experiment are shown in Fig. 6. The construct produces a significant (Student's t-test, taking P < 0.05 as significant) knockdown in salt expression, in both Act5C>salt-RNAi (18.8 times lower than parental RNAi flies), and c42>salt-RNAi (17.7 times less than the salt expression in RNAi flies and 10.7 times less than the salt expression in c42 flies).
Fig. 6.
Quantification of CG2196-RNA interference (RNAi) induced knockdown of CG2196 mRNA levels. UAS-CG2196-RNAi flies were crossed with a ubiquitous (Act5C-GAL4) and Malpighian tubule principal cell-specific (c42) GAL4 driver and tubule mRNA levels measured by qPCR relative to the rp49 reference gene for both parentals and progeny. Although Act5C-GAL4 flies have expression levels rather higher, and c42 slightly lower, than the RNAi parents, both drive RNAi highly efficiently: CG2196 expression in Act5C-GAL4>progeny is 19× less, and in c42-GAL4>RNAi progeny is 18× less, than in RNAi flies. RNAi/CyO control progeny express CG2196 at levels almost exactly the same as the RNAi flies. Data are expressed relatively to the CG2196 expression of the CG2196-RNAi parental line and normalized against the rp49 reference gene. *Significant change from control (the CG2196-RNAi parental line) (Student's t-test, taking P < 0.05 as significant).
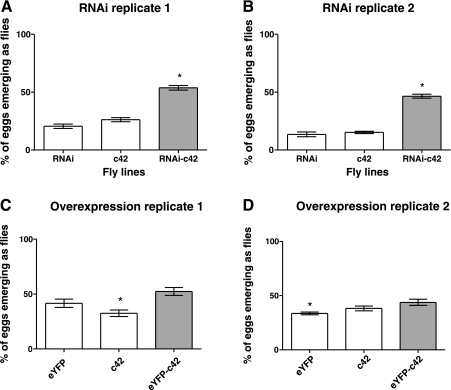
Salt is upregulated in response to salt challenge, so does RNAi-mediated salt knockdown in just the tubule reduce survival of the whole organism when challenged with dietary salt? The progeny of both overexpression and knockdown crosses to the principal cell-specific driver c42 was raised on food containing 4% NaCl, and survival compared with parental controls. The results are shown in Fig. 7. As can be seen from these results, in both repeats overexpression of salt had no consistent effect on survival. Surprisingly, c42>salt-RNAi flies survive better in food containing 4% NaCl than either parental line (RNAi or c42). The percentage of RNAi-c42 flies surviving long enough is more than double of that of any of the parents, and this result is statistically significant (Student's t-test, two-tailed). So, surprisingly, the upregulation of salt on salt challenge observed by microarray and qPCR appears to be counteradaptive.
Fig. 7.
Effect of modulation of tubule CG2196 expression on whole fly survival after salt challenge. Flies were allowed to lay eggs on 4% NaCl diet, then the numbers of progeny reaching adulthood were counted and compared with survival of RNAi, eYFP, or c42 parental lines. The c42 line drives expression selectively in tubule principal cells. For each experimental condition, 2 independent replicates are shown. A and B: tubule-specific RNAi against CG2196 enhances survival on 4% salt. C and D: tubule-specific overexpression of eYFP-tagged CG2196 has no significant impact on expression relative to parental controls. *Significant change from control (Student's t-test, taking P < 0.05 as significant).
DISCUSSION
These results provide the most comprehensive view on salt stress in an insect to date. Previous studies associated salt stress (NaCl and KCl) with the inebriated neurotransmitter/osmolyte transporter (ine) (21), and the studies of dehydration and rehydration (2, 19) demonstrated that Drosophila melanogaster is a very strong osmoregulator. Here, dietary salt stress induced a clear and reproducible sequence of changes in gene expression involving immune/stress response, carbohydrate metabolism, and ion transport pathways. In the longer term (>16 h), effects analogous to starvation were seen, as the reproductive axis was shut down in both males and females. It is interesting to see sugarbabe, a transcription factor associated with dietary sugar loading, also changing in this study; it suggests that sug may play a general role in response to dietary stress.
Salt is a highly tubule-specific gene, expressed on the basolateral plasma membrane of the tubule, and which is strongly up regulated in response to dietary salt. However, this response is inappropriate, because c42>salt-RNAi flies actually survive better on high salt. The most probable reason for this behavior is that the overexpression of salt under salt diet is a miscalculated response of the organism. Such inappropriate responses are commonly encountered in human health and disease: for example, the cytokine storm elicited in healthy humans exposed to influenza can be more dangerous than the original infection. We speculate that flies might sense increasing Na+ levels associated with dietary salt stress and increase Na+ excretion by the tubule by upregulating basolateral Na+ uptake. However, it is possible that the osmolyte cotransported with Na+ by salt is not well handled by the tubule, and so RNAi against salt actually improves the situation.
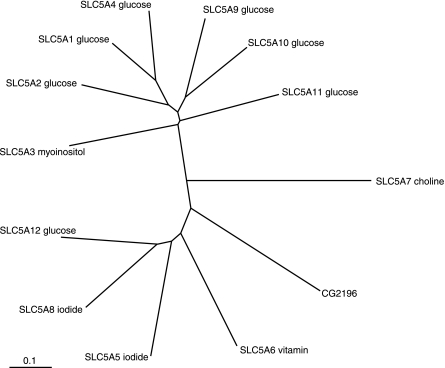
What does salt/salty dog actually encode? Although it is evidently a member of the well-characterized SLC5 (sodium/solute symporter) gene family (43, 44), the answer is surprisingly vague. The Drosophila genome project (flybase.bio.indiana.edu) annotates it as a sodium/iodide symporter. An NCBI BLASTP search against human sequences produces a closest match (E value 3e-87) to SLC5A12, and a second extremely close match (E value 2e-82) to SLC5A8. The former cotransports short chain fatty acids, lactate, and nicotinate with sodium and is found in kidney, small intestine, and skeletal muscle; whereas the latter cotransports short chain fatty acids, lactate, nicotinate, and iodide with sodium and is found in thyroid, kidney, and intestine (43, 44). However, CLUSTALX alignment of the protein sequences with the human SLC5 members (Fig. 8) places salt between SLC5A6, a sodium/vitamin cotransporter, and SLC5A7, a sodium/choline cotransporter. Even this list is not exhaustive, as SLC5 members are also thought to facilitate the transfer of water and organic solutes such as urea. So, based on structure, salt can be considered to be a Na+/solute cotransport but cannot be ascribed a particular transport substrate. Clearly, then, the exact function of salty dog will require extensive further work, as functional workup of SLC5 family members is known to be problematic (43, 44), However, as SLC5 members are uptake transporters, and salty dog is localized to the basolateral membrane of the Malpighian tubule, its upregulation in response to dietary salt would clearly increase sodium clearance from the blood. Although this overexpression does not appear sufficient to confer enhanced survival on the organism on salt challenge (Fig. 7), the observation that knockdown of the gene is actually beneficial (Fig. 7) is intriguing.
Fig. 8.
Which human SLC5 sodium/solute symporter is most similar to CG2196? The reference protein sequence for each of the 12 human SLC5 members was downloaded from NCBI, together with that for CG2196. They were aligned by CLUSTALX with default parameters, and a PHYLIP tree generated with 1,000 bootstrapping. The tree was viewed in Treeview (30).
Perhaps the most compelling aspect of the work is the clear phasing of different classes of response over the experimental period (Table 2), particularly the early induction, then repression, of an immune response. The tubule, although classically considered to be fluid-transporting and osmoregulating tissue (14), actually plays multiple roles in the organism (16). Recently, it has been shown to mount a robust, autonomous immune response to pathogens (28), mediated by the canonical Toll/imd immune pathways (22). So the machinery of the immune response is already in place in the tubules; it is intriguing that it is invoked so early in the salt stress response. We speculate that this finding implies a generality in stress signaling, at least in this tissue; irrespective of the precise nature of the stress, a set of key adaptive genes is quickly upregulated, then selectively turned off as the nature of the stress becomes apparent.
Of course, in nature Drosophila does not face quite the problems of excess salt as, for example, do mosquito larvae that live in hypersaline lakes. But, importantly, the results provide a baseline against which to assess the specialized responses of insects whose evolutionary history has equipped them with the ability to osmoregulate and thrive under continuous osmotic stress.
GRANTS
This work was supported by general funds of the University of Glasgow. The microarray experiment was funded by the UK Biotechnology and Biological Sciences Research Council's Investigating Gene Function initiative.
Supplementary Material
Address for reprint requests and other correspondence: J. A. T. Dow, Integrative & Systems Biology, Faculty of Biomedical and Life Sciences, Univ. of Glasgow, Glasgow G12 8QQ, UK (e-mail: j.a.t.dow@bio.gla.ac.uk).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Adler L, Blomberg A, Nilsson A. Glycerol metabolism and osmoregulation in the salt-tolerant yeast Debaryomyces hansenii. J Bacteriol 162: 300–306, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albers MA, Bradley TJ. Osmotic regulation in adult Drosophila melanogaster during dehydration and rehydration. J Exp Biol 207: 2313–2321, 2004. [DOI] [PubMed] [Google Scholar]
- 3.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258, 1999. [DOI] [PubMed] [Google Scholar]
- 4.Berridge MJ A structural analysis of intestinal absorption. Symp Rent Soc Lond 5: 135–150, 1970. [Google Scholar]
- 5.Blomberg A Metabolic surprises in Saccharomyces cerevisiae during adaptation to saline conditions: questions, some answers and a model. FEMS Microbiol Lett 182: 1–8, 2000. [DOI] [PubMed] [Google Scholar]
- 6.Blomberg A, Adler L. Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J Bacteriol 171: 1087–1092, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley TJ, Philips JE. Regulation of rectal secretion in saline-water mosquito larvae living in waters of diverse ionic composition. J Exp Biol 66: 83–96, 1977. [DOI] [PubMed] [Google Scholar]
- 8.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415., 1993. [DOI] [PubMed] [Google Scholar]
- 9.Breitling R, Amtmann A, Herzyk P. Iterative Group Analysis (iGA): a simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinformatics 5: 34, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Chintapalli VR, Wang J, Davies SA, Dow JAT. Using FlyAtlas to identify better Drosophila models of human disease. Nat Genet 39: 715–720, 2007. [DOI] [PubMed] [Google Scholar]
- 12.Clark TM, Bradley TJ. Malpighian tubules of larval Aedes aegypti are hormonally stimulated by 5-hydroxytryptamine in response to increased salinity. Arch Insect Biochem Physiol 34: 123–141, 1997. [Google Scholar]
- 13.Donini A, Gaidhu MP, Strasberg DR, O'Donnell MJ. Changing salinity induces alterations in hemolymph ion concentrations and Na+ and Cl− transport kinetics of the anal papillae in the larval mosquito, Aedes aegypti. J Exp Biol 210: 983–992, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Dow JAT, Davies SA. The Drosophila melanogaster Malpighian tubule. Adv Insect Physiol 28: 1–83, 2001. [Google Scholar]
- 15.Dow JAT, Davies SA. Integrative physiology and functional genomics of epithelial function in a genetic model organism. Physiol Rev 83: 687–729, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Dow JAT, Davies SA. The Malpighian tubule: rapid insights from post-genomic biology. J Insect Physiol 52: 365–378, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology 13: 17R–27R, 2003. [DOI] [PubMed] [Google Scholar]
- 18.Evans JM, Allan AK, Davies SA, Dow JA. Sulphonylurea sensitivity and enriched expression implicate inward rectifier K+ channels in Drosophila melanogaster renal function. J Exp Biol 208: 3771–3783, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Folk DG, Bradley TJ. Evolved patterns and rates of water loss and ion regulation in laboratory-selected populations of Drosophila melanogaster. J Exp Biol 206: 2779–2786, 2003. [DOI] [PubMed] [Google Scholar]
- 20.Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499, 2000. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Huang Y, Chinnappan R, Bocchini C, Gustin MC, Stern M. The Drosophila inebriated-encoded neurotransmitter/osmolyte transporter: dual roles in the control of neuronal excitability and the osmotic stress response. Genetics 160: 561–569, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silverman N. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol 7: 715–723, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Kizis D, Lumbreras V, Pages M. Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett 498: 187–189, 2001. [DOI] [PubMed] [Google Scholar]
- 24.Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S. Integrative model of the response of yeast to osmotic shock. Nat Biotechnol 23: 975–982, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Lamitina ST, Morrison R, Moeckel GW, Strange K. Adaptation of the nematode Caenorhabditis elegans to extreme osmotic stress. Am J Physiol Cell Physiol 286: C785–C791, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Lamitina ST, Strange K. Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol 288: C467–C474, 2005. [DOI] [PubMed] [Google Scholar]
- 27.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30: 322–329, 2003. [DOI] [PubMed] [Google Scholar]
- 28.McGettigan J, McLennan RK, Broderick KE, Kean L, Allan AK, Cabrero P, Regulski MR, Pollock VP, Gould GW, Davies SA, Dow JA. Insect renal tubules constitute a cell-autonomous immune system that protects the organism against bacterial infection. Insect Biochem Mol Biol 35: 741–754, 2005. [DOI] [PubMed] [Google Scholar]
- 29.Norbeck J, Blomberg A. Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 M NaCl: evidence for osmotic induction of glycerol dissimilation via the dihydroxyacetone pathway. J Biol Chem 272: 5544–5554, 1997. [DOI] [PubMed] [Google Scholar]
- 30.Page RDM TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358, 1996. [DOI] [PubMed] [Google Scholar]
- 31.Patrick ML, Aimanova K, Sanders HR, Gill SS. P-type Na+/K+-ATPase and V-type H+-ATPase expression patterns in the osmoregulatory organs of larval and adult mosquito Aedes aegypti. J Exp Biol 209: 4638–4651, 2006. [DOI] [PubMed] [Google Scholar]
- 32.Radford JC, Davies SA, Dow JAT. Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J Biol Chem 277: 38810–38817, 2002. [DOI] [PubMed] [Google Scholar]
- 33.Rosay P, Davies SA, Yu Y, Sozen A, Kaiser K, Dow JA. Cell-type specific calcium signalling in a Drosophila epithelium. J Cell Sci 110: 1683–1692, 1997. [DOI] [PubMed] [Google Scholar]
- 34.Serrano R, Mulet J, Rios G, Marquez J, de Larrinoa I, Leube M, Mendizabal I, Pascual-Ahuir A, Proft M, Ros R, Montesinos C. A glimpse of the mechanisms of ion homeostasis during salt stress. J Exp Botany 50: 1023–1036, 1999. [Google Scholar]
- 35.Serrano R, Rodriguez-Navarro A. Ion homeostasis during salt stress in plants. Curr Opin Cell Biol 13: 399–404, 2001. [DOI] [PubMed] [Google Scholar]
- 36.Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson CI, Hinrichs T, Johnson LA, Zhao Y, Barolo S. A directional recombination cloning system for restriction- and ligation-free construction of GFP, DsRed, and lacZ transgenic Drosophila reporters. Gene 408: 180–186, 2008. [DOI] [PubMed] [Google Scholar]
- 38.Treherne JE The absorption of glucose from the alimentary canal of the locust, Schistocerca gregaria (Forsk.). J Exp Biol 35: 297–306, 1958. [Google Scholar]
- 39.Wang J, Kean L, Yang J, Allan AK, Davies SA, Herzyk P, Dow JA. Function-informed transcriptome analysis of Drosophila renal tubule. Genome Biol 5: R69, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Kean L, Yang J, Allan AK, Davies SA, Herzyk P, Dow JAT. Function-informed transcriptome analysis of Drosophila renal tubule. Genome Biol 5: R69, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White PJ The molecular mechanism of sodium influx to root cells. Trends Plant Sci 4: 245–246, 1999. [DOI] [PubMed] [Google Scholar]
- 42.Wigglesworth VB The principles of insect physiology. London: Chapman & Hall, 1972.
- 43.Wright EM, Loo DD, Hirayama BA, Turk E. Surprising versatility of Na+-glucose cotransporters: SLC5. Physiology (Bethesda) 19: 370–376, 2004. [DOI] [PubMed] [Google Scholar]
- 44.Wright EM, Turk E. The sodium/glucose cotransport family SLC5. Pflügers Arch 447: 510–518, 2004. [DOI] [PubMed] [Google Scholar]
- 45.Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J 21: 6162–6173, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.