Abstract
We report here our systematic studies of the dynamics of four redox states of the flavin cofactor in both photolyases and insect Type 1 cryptochromes. With femtosecond resolution, we observed ultrafast photoreduction of oxidized state (FAD) in subpicosecond and of neutral radical semiquinone (FADH•) in tens of picoseconds through intraprotein electron transfer mainly with a neighboring conserved tryptophan triad. Such ultrafast dynamics make these forms of flavin unlikely to be the functional states of the photolyase/cryptochrome family. In contrast, we find that upon excitation the anionic semiquinone (FAD•-) and hydroquinone (FADH-) have longer lifetimes that are compatible with high-efficiency intermolecular electron transfer reactions. In photolyases, the excited active state (FADH-*) has a long (nanosecond) lifetime optimal for DNA-repair function. In insect Type 1 cryptochromes known to be blue-light photoreceptors the excited active form (FAD•-*) has complex deactivation dynamics on the time scale from a few to hundreds of picoseconds, which is believed to occur through conical intersection(s) with a flexible bending motion to modulate the functional channel. These unique properties of anionic flavins suggest a universal mechanism of electron transfer for the initial functional steps of the photolyase/cryptochrome blue-light photoreceptor family.
Introduction
Insect cryptochromes (CRYs) and photolyases belong to the same family of flavoproteins with high sequence identity but perform totally different biological functions.1-6 Insect CRYs function as either core clock proteins (Type 2 CRY) or as circadian photoreceptors (Type 1 CRY). Photolyases repair either cyclobutane pyrimidine dimer (Pyr<>Pyr>) or (6-4) photoproducts induced in DNA by ultraviolet light. The catalytic cofactor in cryptochrome/photolyase is flavin adenine dinucleotide. It has been established that the catalytic state of flavin in vivo is an anionic hydroquinone form FADH- in photolyases.6,7 The entire repair dynamics of a Pyr<>Pyr by an electron-transfer redox cycle have been recently mapped out by monitoring the evolution of the flavin redox status during catalysis.8 The catalytic function is completed in 560 ps with a radical electron transfer (ET) mechanism.8-10 The anionic active form (FADH-) with one negative charge is essential to donating one electron to damaged DNA for repair function.6-8 In most photolyases, the flavin becomes oxidized to FAD or neutral radical semiquinone (FADH•) when the enzyme is purified under aerobic conditions. These forms of the enzyme are inactive but can be photoreduced to the active form by intraprotein ET from neighboring aromatic residues on a time scale of picoseconds.11-14 However, this intraprotein ET appears to be irrelevant to the enzyme function in vivo because mutations that block the intraprotein ET do not affect catalysis in vivo.15
Two types of CRYs have been found in insects, named Type 1 CRY and Type 2 CRY, respectively.5 Some insects contain Type 1 only, some contain Type 2 only, and some others contain both. It has been shown that Type 1 CRYs function as photoreceptors to set the circadian clock while Type 2 CRYs function as light-independent repressors in the molecular clockwork of insects that possess them.5 Recently, Type 1 CRYs of mosquito (Anopheles gambiae, AgCRY1), Chinese oak silk moth (Antheraea pernyi, ApCRY1), monarch butterfly (Danaus plexippus, DpCRY1), and fruit fly (Drosophila melanogaster, DmCRY) were purified as recombinant proteins expressed in insect cells.5,16-19 These CRYs, purified under aerobic conditions and under yellow light, contain oxidized FAD that is easily reduced to the stable anionic radical semiquinone (FAD•-) with blue light. This photoreduction occurs through intraprotein ET with the conserved Trp triad and moreover, as in photolyases, blocking the intraprotein ET pathway by site-directed mutations does not affect the photoreceptor function of Type 1 CRYs in vivo. Therefore, it was concluded that these photosensory pigments must contain the flavin in FAD•- form in vivo and it was proposed that the excited FAD•- semiquinone rather than the excited FAD initiated CRY photosignaling.16,17 We find it striking that both photolyases and insect Type 1 CRYs use the anionic states of flavin, FADH- and FAD•-, respectively, as their cofactors. This similarity suggests that ET reactions to substrates could be the initial functional steps for Type 1 CRYs, as they are for photolyases, because both anionic forms are ideal electron donors upon blue-light excitation.
In this work, we systematically examine the excited-state dynamics of all redox states of flavin ranging from oxidized FAD, to anionic radical FAD•-, to neutral radical FADH•, and finally to fully reduced anionic FADH- with femtosecond resolution in insect Type 1 CRYs and in photolyases. By comparing the dynamics of photophysics and photochemistry of the flavin cofactor in both classes of proteins, we were able to determine their time scales and redox behaviors and thus gain some insight into the physiologically relevant redox state(s) of flavin in Type 1 CRYs and potential initial steps of signaling by this class of blue-light photoreceptors. Here, we will focus on ultrafast studies of three insect Type 1 CRYs (AgCRY1, ApCRY1 and DpCRY1) and their various mutants as well as Escherichia coli Py<>Pyr photolyase (EcPhr) and Arabidopsis thaliana (6-4) photolyase [At(6-4)].
Results
Steady-state Spectroscopic Properties of Type 1 CRYs
In Figure 1 we show typical absorption and emission spectra of wild-type AgCRY1 and a mutant of this protein with the C→N replacement of the amino acid residue across N5 of the isoalloxazine ring. In Figure 2 we present sequence alignment of flavin binding sites of Type 1 CRYs analyzed in this study with a Pyr<>Pyr photolyase, a (6-4) photolyase, a plant CRY, and a protozoon CRY. In this figure, we also show the local X-ray structure of E. coli photolyase flavin cofactor and its neighboring Trp triad20 which is conserved across the spectrum of all CRYs and photolyases.
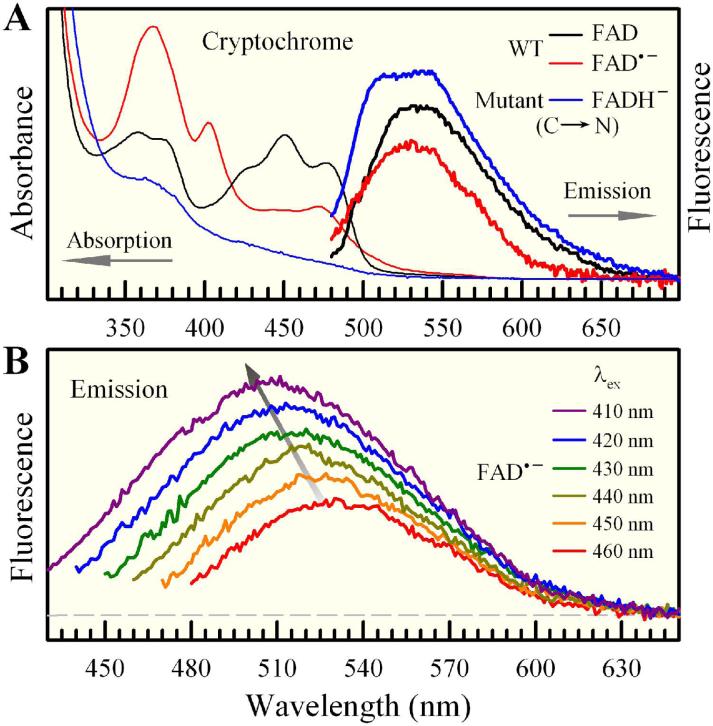
Figure 1.
Steady-state spectra of insect Type 1 CRYs. (A) Absorption and emission spectra of oxidized (FAD), anionic semiquinone (FAD•-) and hydroquinone (FADH-) flavin cofactor in AgCRY1 as a representative of insect Type 1 CRYs. The excitation wavelength for FAD and FADH- was at 400 nm and at 460 nm for FAD•-. The fluorescence of excited FAD contains emission from free FAD in solution. The stable anionic hydroquinone was obtained by mutation of residue C→N near the N5 position of the isoalloxazine ring of flavin. (B) Excitation-wavelength dependence of weak FAD•- emission spectra, indicating deactivation during relaxation. The Raman scattering signals at the blue side of emission peaks were all removed for clarity.
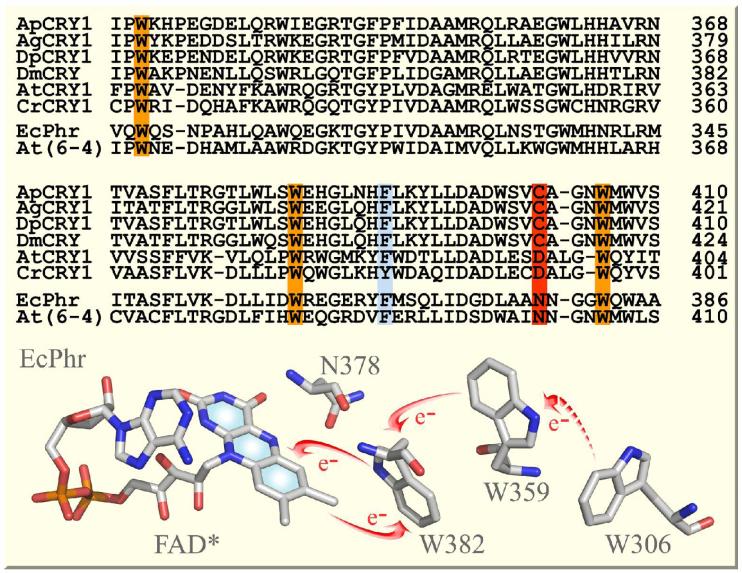
Figure 2.
Sequence alignment of four insect Type 1 CRYs, two plant CRYs, and two photolyases. Ap, Antheraea pernyi (Chinese oak silk moth); Ag, Anopheles gambiae (mosquito); Dp, Danaus plexippus (monarch butterfly); Dm, Drosophila melanogaster (fruit fly); At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Ec, Escherichia coli. Note the conserved tryptophan triad (in orange) for photoreduction through electron transfer across all CRYs and photolyases. Another nearby aromatic residue F or Y (in blue) may also make contributions. Note the critical conserved residue C in insect Type 1 CRYs, N in photolyases and D in plant CRYs (in red) near the N5 position of the isoalloxazine ring, which determines the protonation state of the reduced flavin. Shown at the bottom is the local X-ray structure around the cofactor in E. coli photolyase20 with arrows indicating electron transfer pathways.
When insect Type 1 CRYs are purified, the flavin cofactor is in oxidized state but it is readily reduced to a stable FAD•- with quantum yield (φ) of ~0.2 by blue light,17 considerably higher than that for photoreduction of FADH• in EcPhr (φ=0.05-0.1).21 The photoreduction is accomplished mainly through the conserved Trp triad on picosecond time scales11-17 (see below) although another neighboring aromatic amino acid, phenylalanine (F), may also contribute (Fig. 2).14 Interestingly, the residue near N5 of the isoalloxazine ring is cysteine (C) in all insect Type 1 CRYs and asparagine (N) in all photolyases (Fig. 2).
To examine the roles of the Trp triad and of the amino acid adjacent to N5 of the isoalloxazine ring, we made site-directed mutations in Type 1 CRYs and analyzed them both by steady-state and time-resolved spectroscopic methods. We found that ApCRY1(C402N) and AgCRY1(C413N) are first reduced to FAD•-, then FADH•, and finally to FADH- (see Fig. 1). EcPhr and At(6-4) photolyases containing oxidized FAD go through the same steps of photoreduction: FAD→ FAD•-→ FADH•→FADH-, indicating that photolyases and the mutant CRYs exhibit the same redox behaviors. In all cases, the electron donor is mainly the Trp triad16,17,22 and the proton donor must be related to the neighboring residue Asn and its local environment. Since the pKa of Asn is about 17, clearly it cannot be the proton donor23 and most likely the proton donor is a locally trapped water molecule with a highly acidic proton.20 Interestingly, in both EcPhr and mutant CRYs, during the continuous redox transformation from FAD to FADH- we could observe stable FAD•-, indicating that the proton transfer is slow (at least longer than seconds). Since the electron transfer from the Trp triad occurs ultrafast, the dynamics of proton transfer and electron transfer in this case are decoupled.
We also examined the ApCRY1(C402D), ApCRY1(C402A), AgCRY1(C413D), and AgCRY1(C413A) mutants and observed the same photoreduction states as their wild type counterparts, that is, they were only reduced to stable FAD•-, not continuing to FADH• and then FADH-.17 The redox status of FAD photoreduction, the Trp triad and the effects of Cys mutations at the flavin binding site on the flavin photochemistry are listed in Table 1. To put these results in perspective we summarize our recent functional studies in which light-induced proteolysis of Type 1 CRYs in vivo was used as an endpoint for photoreceptor activity:16,17 The C→N mutants showed the same photoinduced proteolysis as the wild type CRYs, excluding the possibility of photoadduction with the cysteine near the N5 position as the initial signaling steps, which has been observed in phototropin,24,25 and suggests that FADH- functions as well as FAD•- in insect Type 1 CRYs. In contrast, the C→A mutants in which flavin is readily photoreduced to FAD•- but reoxidizes within seconds (more than an order of magnitude faster than their wild-type counterparts) showed no photoreceptor activity in vivo,17 indicating that a stable FAD•- but not oxidized FAD must be the active form of flavin in Type 1 CRYs. In support of this conclusion, Trp triad mutations that block FAD→FAD•- photoreduction in vitro do not affect the photoreceptor function of Type 1 CRYs.16,17 Fig. 1B shows the excitation-wavelength dependence of fluorescence spectra of AgCRY1, indicating that the relaxation of excited FAD•- does not follow conventional Kasha's law and its (and other Type 1 CRYs) photophysics is unique, which may have functional implications (see below).
Table 1.
| Wn |
Wm |
Wf |
ResiduesN5 (reduction states) |
|
|---|---|---|---|---|
| ApCRY1 | 406 | 383 | 328 | 402 C (FAD•-) |
| C→D, A (FAD•-) | ||||
| C→N (FAD•-, FADH•, FADH-) | ||||
| AgCRY1 | 417 | 394 | 339 | 413 C (FAD•-) |
| C→D, A (FAD•-) | ||||
| C→N (FAD•-, FADH•, FADH-) | ||||
| DpCRY1 | 406 | 383 | 328 | 402 C (FAD•-) |
| DmCRY | 420 | 397 | 342 | 416 C (FAD•-) |
| AtCRY1 | 400 | 377 | 324 | 396 D (FADH•, FADH-) |
| CrCRY1 | 397 | 374 | 321 | 393 D (FADH•, FADH-) |
| EcCDP | 382 | 359 | 306 | 378 N (FAD•-, FADH•, FADH-) |
| At6-4 |
406 |
383 |
329 |
402 N (FADH•, FADH-) |
Comparative Analysis of Ultrafast Photoreduction Dynamics of Type 1 CRYs and Photolyases
Because the steady-state photophysical properties of Type 1 CRYs suggest that they may function in a manner analogous to photolyases, we carried out a comparative analysis of photoreduction dynamics of Type 1 CRYs and photolyases. Figure 3A shows the fluorescence transients gated at 530-nm emission peak for three Type 1 CRYs: Under these conditions, the ultrafast decay truly reflects the quenching dynamics of excited FAD with negligible solvation contributions because solvation dynamics mostly appear in the transients gated at the blue side of emission peak.26,27 The transients of the three insect Type 1 CRYs were fitted by a dominant component of 0.97 ps for ApCRY1 and 1.3 ps for both AgCRY1 and DpCRY1 with all their amplitudes of larger than 90% (a less than 10% component with a long-time decay was removed for clarity). The ultrafast photoreduction in ~1 ps must result from ET from a nearby W (Wn) to excited FAD to form Wn+ and FAD•-, consistent with all reported ultrafast ET in oxidized flavoproteins.28,29 In general, when there are aromatic amino acids around excited FAD at short distances (3-4 Å), one electron jumps from W or Y (sometimes F or H, depending on the local environment) to excited FAD to quench flavin fluorescence. Such ultrafast processes are highly driven by favorable negative free-energy changes, for example,ΔG≈-1.44 eV for ET from W.30 This general observation explains why in phototropin, which initiates photosignaling by formation of a cysteinyl-flavin adduct, there are no aromatic amino acids within 4 Å radius of the FMN cofactor.31,32
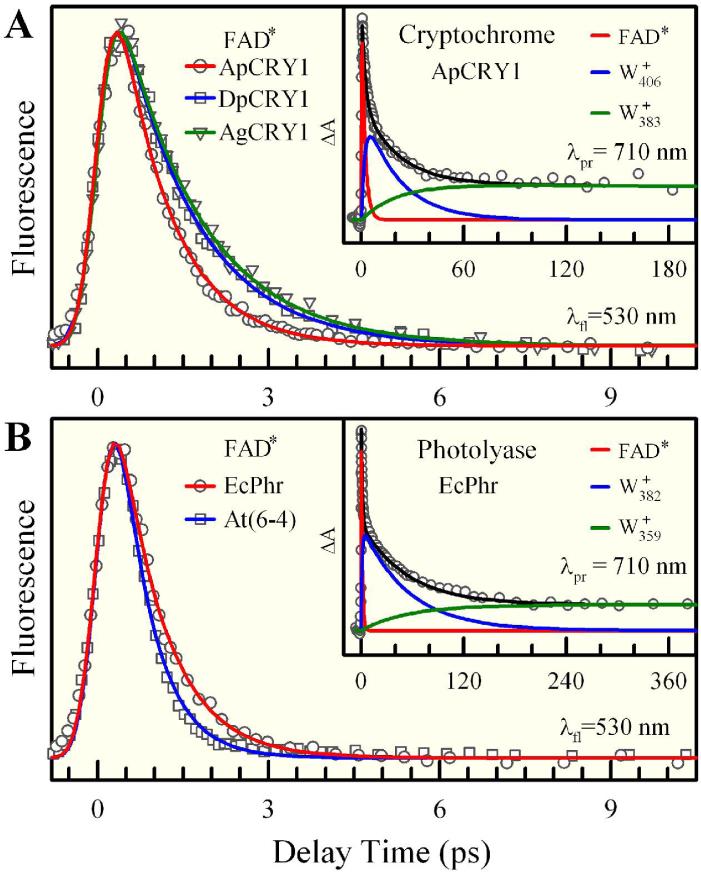
Figure 3.
Femtosecond-resolved oxidized flavin dynamics in insect Type 1 CRYs and photolyases. Normalized fluorescence transients for (A) ApCRY1, DpCRY1 and AgCRY1 and (B) EcPhr and At(6-4), all gated at 530 nm around the emission peaks. Note that for clarity a long-time component with less than 10% of the total amplitude was subtracted from all transients. These signals represent ultrafast ET dynamics with neighboring tryptophans. Shown in the insets are the transient-absorption detection of ET intermediates and charge recombination dynamics; see text for details.
After ultrafast charge separation in the photolyase/CRY family flavoproteins two processes will occur, either charge recombination to close a futile ET cycle or electron hopping from the middle tryptophan (Wm) of the triad (Fig. 2) to finish photoreduction and self-restore the active state FAD•-. In the inset of Fig. 3A, we show the transient-absorption dynamics for ApCRY1 where we have probed for all putative intermediates. At wavelengths longer than 550 nm, FAD•- has no absorption and we only detected excited FAD and tryptophan cation W+. At 710-nm probing, we observed decay signals with three exponential components: 1.8 ps (59%) for excited FAD, 20 ps (31%) for Wn+, and ~ns (10%) for Wm+. The forward ET dynamics detected by transient absorption (2 ps) is usually slower than that by femtosecond-resolved fluorescence measurement (1 ps)33 due to different accessible ranges of nuclear configurations. The W+ signal probed at 710 nm was also observed in our recent studies of ultrafast ET between excited FMN and W in flavodoxin. Thus, knowing the total decay rate of Wn+ and the branching ratio of Wm+, the charge recombination between FAD•- and Wn+ was found to occur in 29 picoseconds and the hopping from Wm to Wn+ in 62 picoseconds. For ApCRY1, Wn+ is W406+ and Wm+ is W383+ (Table 1). For other probe wavelengths from 550 to 710 nm, all the signals of ApCRY1 can be globally fitted with the three decay dynamics of excited FAD (~2 ps), Wn+ (~20 ps) and Wm+ (~ns) with different amplitudes. We observed similar dynamics of charge recombination in 28 and 32 ps for AgCRY1 and DpCRY1, respectively (Fig. 3A). Finally, the dynamics of all C→A, N, and D mutants analyzed revealed similar forward ET in about 1 ps and back ET in tens of picoseconds.
Figure 3B shows the photoreduction dynamics of two photolyases, EcPhr and At(6-4), containing oxidized FAD for comparison with the dynamics of cryptochromes. In general these photolyases exhibit dynamics similar to those of CRYs: Fluorescence transients gated at the 530-nm emission peak show a dominant ultrafast component of 0.8 ps (≥95% of the total amplitude) for EcPhr and 0.5 ps (~100%) for At(6-4). These even faster ET rates in photolyases compared to Type 1 CRYs reflect the different local environments that affect the redox potential of excited flavin or ET distances. We also examined charge recombination using transient-absorption detection. The result for EcPhr using 710-nm probe is shown in the inset of Fig. 3B. Similar to the Type 1 CRYs, the transient can be fitted by three exponential decays: 1.2 ps (65%) for excited FAD, 54 ps (28%) for Wn+ (W382+), and ~ns (7%) for Wm+ (W359+) and the derived charge recombination and electron hopping occur in 71 ps and 220 ps, respectively. The observed slower dynamics of back electron transfer in photolyases than in Type 1 CRYs probably result from different local redox environments and also slow solvation at the flavin binding sites.8 Importantly, Type 1 CRYs and photolyases exhibit similar photophysical properties in that photoreduction of FAD to FAD•- through a neighboring W occurs in ~1 ps and charge recombination takes tens of picoseconds.
The photoreduction of FADH• to FADH- in EcPhr has been extensively studied:11-14 The forward ET dynamics takes about tens of picoseconds, slower than the forward rate of FAD reduction (~1 ps) due to the different driving forces, ΔΔG (FADH•-FAD)≈0.44 eV.34 The dynamics of charge recombination is a matter of some debate13,35 but our recent studies with mutants of the ET pathway reveal that back ET also occurs in tens of picoseconds and not in the subpicosecond region reported previously.35 Of interest, when we examined the reduction of FADH• in Type 1 CRYs using an C→N mutant [AgCRY1(C413N)], we observed a forward ET dynamics in ~20 ps (Table 2). Thus, the photoreduction of semiquinone FADH• both in photolyases and Type 1 CRYs occurs in tens of picoseconds through the conserved Trp triad. Hence, it could be argued that if the photochemistry of excited flavin cofactor is the initial signaling step, neither FAD nor FADH• can be the functional flavin because excited states of these forms are readily quenched on the picosecond time scale by neighboring aromatic residues such as tryptophan or tyrosine.9,10,36 In contrast, the anionic states, FAD•- and FADH-, are ideal for carrying out photochemistry because the excited states of these forms are long-lived and have high reduction potentials which are not conducive for quenching by accepting electrons from neighboring aromatic amino acids but are optimal for donating an electron to substrates.
Table 2.
Time scales of flavin photoredox cylces in a Type 1 cryptochrome and a Pyr<>Pyr photolyasea
All time scales are in picosecond.
Time scales are for insect ApCRY1 and the deactivation dynamics of hydroquinone (hq) is for the mutant C413N of AgCRY1.
Time scales are for E. coli CPD photolyase.
Time scales of deactivation processes are averages of triple exponential decays, <τ> = .
Deactivation Dynamics and Conformational Control in Type 1 CRYs
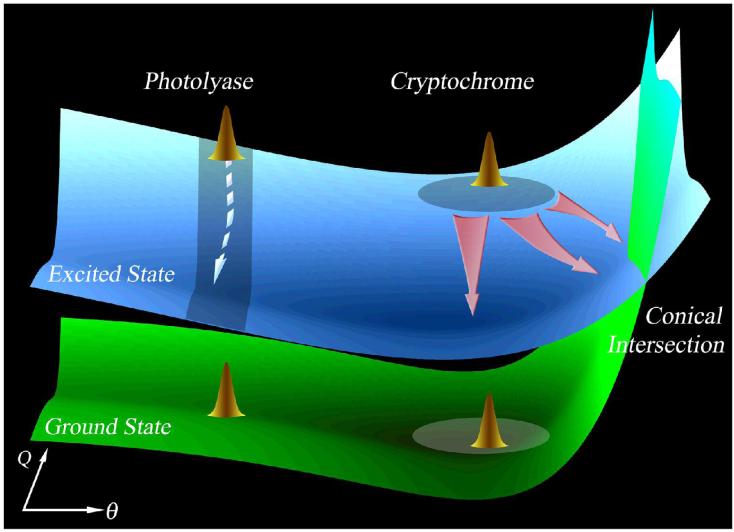
The FAD•- form of Type 1 CRYs is relatively stable16 and in vivo studies suggest that this form represent the active ground state.16,17 Figure 4A shows the fluorescence transient gated at 550 nm for ApCRY1 as a typical representative of Type 1 CRYs. The transient exhibits three exponential decay components: 2.2 ps (41%), 30 ps (35%) and 530 ps (24%). Without a substrate and without the possibility of quenching by ET from neighboring amino acids or intrachromophore ET (the electron transfer from excited FAD•- to the adenine moiety of flavin molecule is not favorable, ΔG≈0 eV),37 the observed multiple decays truly reflect the deactivation dynamics of excited FAD•- in ApCRY1. As shown in Fig. 1B, the peaks of weak emission from excited FAD•- shift to the blue with higher excitation energy, indicating that excited molecules do not completely relax to the lowest excited state and the molecules deactivate during relaxation. Such multiple deactivation processes mostly occur through conical intersection(s) (CIs) often observed in large organic molecules such as DNA bases which have been extensively studied recently.38,39 Figure 5 schematically shows such deactivation processes on a multiple-dimension potential energy landscape, resulting in multiple-exponential decays.
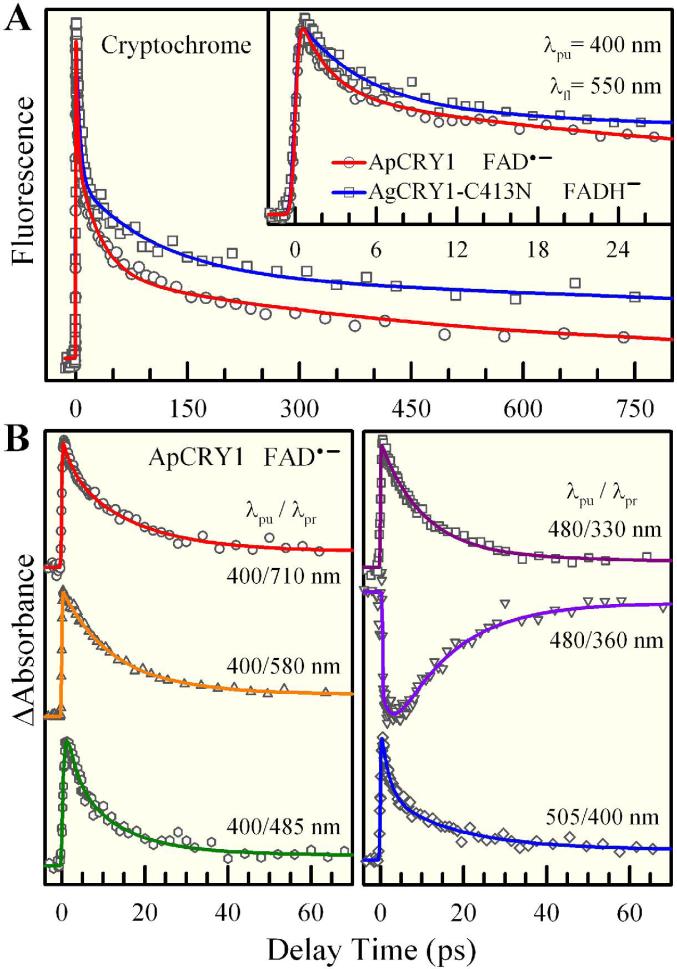
Figure 4.
Femtosecond-resolved anionic flavin dynamics (FAD•- and FADH-) in insect Type 1 CRYs. (A) Normalized fluorescence transients of FAD•- for wild-type ApCRY1 and FADH- for the mutant AgCRY1(C413N) gated at 550-nm emission. (B) Transient-absorption detection of excited FAD•- for various pump/probe schemes, showing similar excited-state dynamics as probed with fluorescence in (A) and the corresponding ground-state formation. All signals are about 10-3-10-4 absorbance unit changes.
Figure 5.
Schematic potential energy landscape of excited-state anionic active states (FAD•- and FADH-) in photolyases and insect Type 1 CRYs with a flexible bending motion in either the ground or excited state to access CI(s) and result in multiple decays. In photolyases, the bending motion is restricted at the local region and the flavin molecule cannot access the CI(s), leading to a long (nanosecond) lifetime. The bending coordinate (θ) could be defined for the “butterfly” motion of the isoalloxazine ring along the N5-N10 axis.
To confirm such photophysical deactivation processes and search for any possible photochemical reaction intermediates, we examined the dynamics of excited FAD•- using transient-absorption detection with a wide range of probing wavelengths from UV 300 nm to visible 710 nm. The results are nearly similar for the wild type and various mutants of all three insect Type 1 CRYs. Figure 4B shows typical transients with several pump/probe wavelengths for ApCRY1: All transients can be globally fitted with three exponential decay components: 2-2.5 ps, 10-16 ps and a long component (>100 ps). These multiple decay dynamics are similar to excited-state deactivation processes observed by femtosecond-resolved fluorescence detection and described by a complex energy landscape (Fig. 5). In addition to the long-time component, for λpu/λpr=400/710, 400/485, and 505/400 nm, we observed decay dynamics in ~2 ps and ~15 ps, and at λpu/λpr=400/580 and 480/330 nm, we mainly observed a decay component of ~11 ps, and finally for λpu/λpr=480/360 nm, we observed ground-state FAD•- formation in ~15 ps on a time scale similar to that of the decay dynamics. These pump/probe schemes sampled different configurations on the energy landscape and the observed dynamics are robust with multiple decays on similar time scales.
By making the C→N replacement near the N5 position, we can completely reduce FAD to stable FADH- (Table 1), as shown in AgCRY1(C413N) (Fig. 1A). In Fig. 4A we also show the fluorescence transient of excited FADH- in this mutant gated at 550 nm. Similar to the wild type, the transient of the mutant exhibits multiple exponential decays of 5 ps (47%), 105 ps (28%) and 2.3 ns (25%) and reflects the deactivation dynamics of excited FADH- through a similar mechanism of conical intersection(s) as observed for excited FAD•-. Recently, in our studies on excited-state dynamics of free FADH- in buffer, we observed multiple decays similar to those of FAD•- and FADH- in insect Type 1 CRYs, with strong excitation-wavelength dependence of emission. In light of this observation and other relevant evidence,40,41 a flexible “butterfly” bending motion of the isoalloxazine ring is proposed to access CI(s) for multiple deactivation processes, as shown in Fig. 5.
Discussion
We report here our systematic studies of the dynamics of the flavin cofactor in four different redox states and two redox pairs (FAD/FAD•- and FADH•/FADH-) in three insect cryptochromes and two photolyases. In light of our findings we discuss below some special features of flavin photochemistry in photolyase/cryptochrome family and the relevance of the photochemistry to functions of these photoreceptors.
Photoredox Cycles and Anionic States
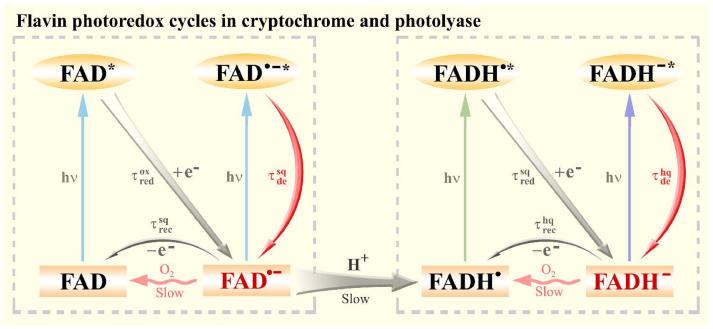
Flavin has rich chemistry with five common redox states: FAD, FAD•-, FADH•, FADH-, and FADH2. Currently available data indicate that the first four states in the forms of two redox pairs FAD/FAD•- and FADH•/FADH- may be relevant to the photoreceptor activities of cryptochromes and photolyases. Figure 6 shows the universal photoredox cycles of both Type 1 CRYs and photolyases in vitro and in the absence of substrates. Blue light reduces FAD to FAD•- in about 1 ps through charge separation with a neighboring tryptophan. The reaction then bifurcates to either charge recombination with the tryptophan in 28-71 ps or electron hopping in 60-220 ps along the trp triad. Under aerobic conditions, the stable FAD•- is slowly reoxidized to FAD in minutes. The fate of FAD•- depends on the amino acid residue in proximity of N5 of the isoalloxazine ring: in Type 1 CRYs which contain a Cys at that position the photoreaction stops at the FAD•- level. In contrast, in photolyases which contain Asn at the position the FAD•- gradually evolves to FADH• through a proton transfer and then is further photoreduced to FADH- in 10-20 ps through the nearby tryptophan. As in the FAD/FAD•- photoredox cycle, the redox pair either reverts to the original form by charge recombination or proceeds to electron hopping along the Trp triad. The time scales of these flavin photoredox cycles are listed in Table 2.
Figure 6.
A universal scheme of four-state flavin photoredox cycle in CRYs and photolyases. The two neutral states (FAD and FADH•) of the flavin cofactor can be photoreduced into two anionic active states (FAD•- and FADH-) through the neighboring tryptophan triad. The two redox pairs (FAD/FAD•- and FADH•/FADH-) can be interconverted by the intermediacy of a key residue N (or D in plant CRYs) near the N5 position of the isoalloxazine ring through proton transfer in seconds. The ground-state conversions from anionic forms to neutral forms with oxygen occur in minutes.
Flavin Anions as the Functional Forms in the Photolyase/Cryptochrome Family
As a general rule chromophores must have a long-lived excited state to perform photochemical catalysis with a reasonable quantum yield in biological systems. In line with this notion the lifetime of excited FADH- in E. coli photolyase is ~1 ns8 and, not surprisingly, this enzyme repairs T<>T with a quantum yield of nearly unity.6,8 In contrast, excited FAD•- in Type 1 CRYs exhibits multi-exponential decay with lifetimes in the range of ~2 ps (~40%), 30 ps (~35%), and ~500 ps (~25%) and interestingly even though converting FAD•- to FADH- by site-specific mutagenesis lengthens the lifetimes by a factor of 2-4, it does not change the multi-exponential form of deactivation nor the fraction (75%) of predominantly short-lifetime species [5 ps (47%) and 105 ps (28%)]. These properties of Type 1 CRYs raise two questions: Why is the decay of excited FAD•- (or FADH-) multi-exponential, and secondly, with the predominance of the fast decay routes how can Type 1 CRYs carry out efficient photochemistry necessary for their photoreceptor function? The answer to the first question probably lies in the fact that Type 1 CRYs have higher plasticity42 and the isoalloxazine ring in CRYs is not as rigidly held in the active site as in photolyases.43,44 The ring undergoes “butterfly” bending motion in the ground or excited state that leads to multiple deactivation processes through conical intersection(s) in which the rapid decay pathways predominate. With respect to the efficiency of catalysis by Type 1 CRYs, only some general comments can be made at this point because currently the primary photochemical reaction catalyzed by these photoreceptors is not known. Assuming that the primary event, as in photolyase, is electron transfer, only the ~500 ps component of excited FAD•- (25%) would be expected to carry out catalysis with appreciable quantum yield. This would be in the range of 10-20% of the photolyase quantum yield if one considers the lifetimes and fractions of the molecules that decay with that lifetime in the two photoreceptors. Whether this is the case awaits the identification of the primary photochemical reaction in Type 1 CRYs. However, we do have a quantum yield for an enzymatic reaction that is initiated by photoexcited Type 1 CRYs, the proteolytic degradation of the CRY itself.17 The quantum yield for this process is in the range of φ~10-3, which is clearly within the range of theoretical maximum of about φ~0.2, assuming conventional photochemical reactions. In fact, proteolysis is at least one and perhaps more than one step removed from the primary photochemical event and hence we expect that the actual quantum yield of electron transfer (if that is the mechanism) of Type 1 CRYs may approach the theoretical limit.
Universal Model for Cryptochromes
Recently cryptochromes from plants including Arabidopsis thaliana (AtCRY1 and AtCRY2) and Chlamydomonas reinhardtii (CrCRY) have been purified and analyzed in some detail.45-48 All these CRYs contain flavin in the form of oxidized FAD. Blue-light exposure leads to the sequential reduction FAD→FAD•-→FADH•→FADH-. It has been proposed that, in contrast to photolyases and animal Type 1 CRYs in which FADH- (FAD•-) represents the dark state, in plant CRYs the FAD form represents the dark state and that the FADH• form produced following blue-light exposure is the signaling state.46,47 In support of this proposal it was reported that, in contrast to insect Type 1 CRYs, disrupting the intraprotein electron transfer by mutating the Trp triad residues abolished the photoreceptor function.45 This raises the issue of whether the mechanisms of photoreception/phototransduction are different between animal and plant cryptochromes. While at present we cannot answer this question with any certainty we note that the AtCRY1 Trp triad mutants used to support the proposed plant CRY signaling mechanism exhibited some indication of being misfolded,4 casting some doubt about the interpretation of the negative in vivo data. Moreover, the highly conserved very rapid quenching (~1 ps) of excited state FAD in CRYs we demonstrate in this work raises some theoretical and conceptual problems for different signaling mechanisms between plant and animal CRYs. Clearly, experiments such as the ones reported here for insect Type 1 CRYs must be carried out with plant CRYs to resolve the issue of whether there is a unified mode of action for the entire photolyase/CRY family, including Pyr<>Pyr photolyase, (6-4) photolyase, animal Type 1 CRYs and plant CRYs, or if CRYs in general and plant CRYs in particular employ a signaling mechanism that does not involve electron transfer from enzyme to substrate.
Experimental Section
Protein Preparation
The purifications of all three insect CRYs have been described in detail elsewhere.16,17 Final protein concentrations of 20-50 μM in 50% (v/v) glycerol in a buffer of 50 mM sodium phosphate, 100 mM NaCl, pH 7.4 or of 50 mM Tris-HCl, 100 mM NaCl, 1mM EDTA, 5 mM dithiothreitol, pH 7.5 were used in femtosecond-resolved studies. The purifications of EcPhr and At(6-4) have been described in detail previously with some modifications.49,50 We used the concentrations of 80 μM for EcPhr and 200 μM for At(6-4) in 50% (v/v) glycerol with a buffer of 50 mM sodium phosphate, 100 mM NaCl, pH 7.4 for ultrafast measurements.
Femtosecond Spectroscopy
All the femtosecond-resolved measurements were carried out using the fluorescence up-conversion and transient absorption methods. The experimental layout has been detailed elsewhere.13 Briefly, for femtosecond-resolved fluorescence detection, the pump wavelengths of 400 and 480 nm were generated by doubling of 800 nm or mixing of 800 nm with the signal beam (1200 nm) from an optical parametric amplifier (OPA-800C, Spectra-Physics) in a 0.2 mm thick β-barium borate crystal (BBO, type I). The resulting fluorescence from the sample was gated by another 800-nm beam in a 0.2 mm BBO crystal to obtain fluorescence transients at desired wavelengths. For transient absorption measurements, two OPAs were used to generate all desired signal or idler wavelengths, which were then mixed with an 800-nm beam in a 0.2 mm BBO crystal to produce various pump/probe wavelengths for different detection schemes. The instrument response time is between 400 and 500 fs for fluorescence detection and less than 300 fs for transient-absorption measurements. All experiments were done at the magic angle (54.7°).
Acknowledgment
We thank Prof. Steven M. Reppert for providing the plasmid carrying the cDNA of the Type 1 CRYs used in our study. The work was supported in part by the Packard Foundation Fellowship (DZ) and the National Institute of Health (AS and DZ).
References
- 1.Cashmore AR. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- 2.Lin C, Shalitin D. Annu. Rev. Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 3.Partch CL, Sancar A. Photochem. Photobiol. 2005;81:1291–1304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 4.Öztürk N, Song S-H, Özgür S, Selby CP, Morrison L, Partch C, Zhong D, Sancar A. In: Cold Spring Harbor Symposium on Quantitative Biology, Vol. LXXII. Stillman B, Stewart D, editors. Cold Spring Harbor Lab Press; New York: 2007. pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 5.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Mol. Biol. Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 6.Sancar A. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 7.Kim S-T, Sancar A, Essenmacher C, Babcock GT. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8023–8027. doi: 10.1073/pnas.90.17.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kao Y-T, Saxena C, Wang L, Sancar A, Zhong D. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong D. Curr. Opin. Chem. Biol. 2007;11:174–181. doi: 10.1016/j.cbpa.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 10.Kao Y-T, Saxena C, Wang L, Sancar A, Zhong D. Cell Biochem. Biophys. 2007;48:32–44. doi: 10.1007/s12013-007-0034-5. [DOI] [PubMed] [Google Scholar]
- 11.Aubert C, Vos MH, Mathis P, Eker APM, Brettel K. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 12.Byrdin M, Villette S, Eker APM, Brettel K. Biochemistry. 2007;46:10072–10077. doi: 10.1021/bi700891f. [DOI] [PubMed] [Google Scholar]
- 13.Saxena C, Sancar A, Zhong D. J. Phys. Chem. B. 2004;108:18026–18033. [Google Scholar]
- 14.Wang H, Saxena C, Quan D, Sancar A, Zhong D. J. Phys. Chem. B. 2005;109:1329–1333. doi: 10.1021/jp044652b. [DOI] [PubMed] [Google Scholar]
- 15.Kavakli IH, Sancar A. Biochemistry. 2004;43:15103–15110. doi: 10.1021/bi0478796. [DOI] [PubMed] [Google Scholar]
- 16.Song S-H, Öztürk N, Denaro TR, Arat NÖ, Kao Y-T, Zhu H, Zhong D, Reppert SM, Sancar A. J. Biol. Chem. 2007;282:17608–17612. doi: 10.1074/jbc.M702874200. [DOI] [PubMed] [Google Scholar]
- 17.Öztürk N, Song S-H, Selby CP, Sancar A. J. Biol. Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 18.Van Vickle-Chavez SJ, Van Gelder RN. J. Biol. Chem. 2007;282:10561–10566. doi: 10.1074/jbc.M609314200. [DOI] [PubMed] [Google Scholar]
- 19.Berndt A, Kottke T, Breitkreuz H, Dvorsky R, Hennig S, Alexander M, Wolf E. J. Biol. Chem. 2007;282:13011–13021. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 20.Park H-W, Kim S-T, Sancar A, Deisenhofer J. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 21.Heelis PF, Sancar A. Biochemistry. 1986;25:8163–8166. doi: 10.1021/bi00373a006. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Heelis PF, Sancar A. Biochemistry. 1991;30:6322–6329. doi: 10.1021/bi00239a034. [DOI] [PubMed] [Google Scholar]
- 23.Olkhova E, Helms V, Michel H. Biophys. J. 2005;89:2324–2331. doi: 10.1529/biophysj.105.062091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christie JM. Annu. Rev. Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 25.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. J. Biol. Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 26.Lu W, Kim J, Qiu W, Zhong D. Chem. Phys. Lett. 2004;388:120–126. [Google Scholar]
- 27.Zhang L, Wang L, Kao Y-T, Qiu W, Yang Y, Okobiah O, Zhong D. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18461–18466. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong D, Zewail AH. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11867–11872. doi: 10.1073/pnas.211440398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mataga N, Chosrowjan H, Shibata Y, Tanaka F, Nishina Y, Shiga K. J. Phys. Chem. B. 2000;104:10667–10677. [Google Scholar]
- 30.Swenson RP, Krey GD. Biochemistry. 1994;33:8505–8514. doi: 10.1021/bi00194a015. [DOI] [PubMed] [Google Scholar]
- 31.Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Biophys. J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crosson S, Moffat K. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan C, Xia T, Becker H-C, Zewail AH. Chem. Phys. Lett. 2005;412:158–163. [Google Scholar]
- 34.Gindt YM, Schelvis JPM, Thoren KL, Huang TH. J. Am. Chem. Soc. 2005;127:10472–10473. doi: 10.1021/ja051441r. [DOI] [PubMed] [Google Scholar]
- 35.Pan J, Byrdin M, Aubert C, Eker APM, Brettel K, Vos MH. J. Phys. Chem. B. 2004;108:10160–10167. [Google Scholar]
- 36.Henzler-Wildman K, Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 37.Seidel CAM, Schulz A, Sauer MHM. J. Phys. Chem. 1996;100:5541–5553. [Google Scholar]
- 38.Sobolewski AL, Domcke W. Euro. Phys. J. D. 2002;20:369–374. [Google Scholar]
- 39.Levine BG, Martínez TJ. Annu. Rev. Phys. Chem. 2007;58:613–634. doi: 10.1146/annurev.physchem.57.032905.104612. [DOI] [PubMed] [Google Scholar]
- 40.Lennon BW, Williams CH, Jr., Ludwig ML. Protein Sci. 1999;8:2366–2379. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Y-J, Ornstein RL. J. Am. Chem. Soc. 1996;118:9402–9408. [Google Scholar]
- 42.Partch CL, Clarkson MW, Özgür S, Lee AL, Sancar A. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- 43.Mees A, Klar T, Gnau P, Hennecke U, Eker APM, Carell T, Essen L-O. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 44.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeugner A, Byrdin M, Bouly J-P, Bakrim N, Giovani B, Brettel K, Ahmad M. J. Biol. Chem. 2005;280:19437–19440. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 46.Bouly J-P, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Galland P, Bittl R, Ahmad M. J. Biol. Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]
- 47.Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, Bittl R, Batschauer A. J. Biol. Chem. 2007;282:14916–14922. doi: 10.1074/jbc.M700616200. [DOI] [PubMed] [Google Scholar]
- 48.Immeln D, Schlesinger R, Heberle J, Kottke T. J. Biol. Chem. 2007;282:21720–21728. doi: 10.1074/jbc.M700849200. [DOI] [PubMed] [Google Scholar]
- 49.Sancar A, Smith FW, Sancar GB. J. Biol. Chem. 1984;259:6028–6032. [PubMed] [Google Scholar]
- 50.Li J, Uchida T, Todo T, Kitagawa T. J. Biol. Chem. 2006;281:25551–25559. doi: 10.1074/jbc.M604483200. [DOI] [PubMed] [Google Scholar]