Abstract
Acute apical abscesses and cellulitis are severe endodontic diseases caused by opportunistic bacteria with possible co-infection with latent herpesviruses. The objectives of this study are to identify herpesviruses, including human cytomegalovirus (HCMV), Epstein-Barr virus (EBV), herpes simplex virus-1 (HSV-1) and Varicella zoster virus (VZV), in patients (n=31) presenting with acute apical abscesses and cellulitis of endodontic origin. Primary and nested polymerase chain reaction (PCR) was conducted using virus-specific primers and DNA isolated from cell-free abscess fluid. From patients exhibiting concurrent spontaneous pain (n=28), nine abscesses contained HCMV, two abscesses contained EBV, one abscess contained HSV-1, and no abscesses contained VZV. Control PCR using genomic or recombinant templates demonstrated detection limits to a single genomic copy of HCMV, 100 genomic copies for EBV, and 1-10 copies for HSV-1, with no cross-amplification between herpesviral DNA targets. Nested PCR was required for detection of herpesviral DNA in the abscess specimens, indicating that these viruses were present in low copy number. Filtration of abscess specimens and virus transfer experiments using human fibroblastic MRC-5 cells confirmed the presence of HCMV particles in several abscess specimens. We conclude that herpesviruses are present, but not required for development of acute apical abscesses and cellulitis of endodontic origin.
Keywords: acute apical abscess, cellulitis, apical periodontitis, endodontic infections, herpesviruses, human cytomegalovirus, Epstein-Barr virus, herpes simplex virus, Varicella zoster virus
Introduction
Herpesviruses have recently been implicated in severe oral diseases, including irreversible pulpitis, apical periodontitis, periodontal abscesses, and inflammation of the gingiva and oral mucosa (1-5). Endodontic disease is a polymicrobial, multi-stage inflammatory response initiated by opportunistic microorganisms and exacerbated by an influx of inflammatory cells causing pulpitis and periapical periodontitis (1-5). Acute apical abscess is an inflammatory reaction to pulpal infection and necrosis characterized by rapid onset, spontaneous pain, tenderness of the tooth to pressure, pus formation and associated swelling. Cellulitis is a more advanced staging of acute apical abscess, where the edematous inflammatory response spreads diffusively through connective tissue and fascial planes. Acute endodontic inflammation may be potentially augmented by the influx of inflammatory cells co-infected with latent herpesviruses, with disease progression moderated by potential viral reactivation causing additional impairment of the host immune response (1-6).
Human herpesviruses are currently classified into eight distinct species: herpes simplex virus (HSV) 1 and 2, Varicella zoster virus (VZV), human cytomegalovirus (HCMV), Epstein-Barr virus (EBV) and human herpesvirus 6, human herpesvirus 7, and human herpesvirus 8 or Karposi's sarcoma-associated herpesvirus. EBV is an important human pathogen associated with several diseases, including infectious mononucleosis (7,8), malignant lymphomas and nasopharyngeal carcinoma (9-12), Hodgkin's Disease and lymphoproliferative disorders in immunocompromised individuals (13). EBV can replicate, or may be present in a latent state, in oral epithelial cells and tissues (9-12). Latent EBV infection results in the restricted expression of viral genes and is capable of escaping immune surveillance (7,8). Prolonged periods of latency and periods of reactivation may be partially responsible for the symptomatic and asymptomatic transitioning observed for endodontic disease (14). HCMV and HSV are also present as latent infections in the majority of the human population. HCMV resides in the bone-marrow myeloid progenitor cells during latency, and as active viruses can infect T lymphocytes, macrophages, monocytes, polymorphonuclear leukocytes, endothelial cells and fibroblasts. EBV resides predominantly in B lymphocytes during both primary and latent infections. HSV-1 is most commonly found in the oral cavity as a primary infection and establishes latency within the trigeminal ganglia (15). Herpesviruses, including HCMV, EBV and HSV-1, can be reactivated from a latent infection either spontaneously or concurrent with another infection, or with other stress factors affecting the host immune system (14,16). Shedding of herpesviruses following reactivation can occur in saliva (4), and in the case of HSV-1 appears to occur asymptomatically (17).
In endodontic and periodontal disease, herpesviruses have been proposed to increase the virulence of bacterial pathogens by enhancing adherence and invasiveness into epithelial cells (18-20). HCMV inhibits the expression of macrophage surface receptors, and deters macrophages from destroying invasive gram-negative bacteria (19,20). The EBV latent membrane proteins (LMPs) are known to activate immune signaling pathways to produce several cytokines, and LMP variants have been identified in the oral cavity with varied effects on EBV transmission and persistence (21,22). The profile and abundance of EBV strains in the oral cavity may provide evidence for compartmentalization of specific EBV strains and potential progression of disease (10). EBV type 1 and EBV type 2 can be distinguished by sequence divergence in the latency genes encoding nuclear antigen 2 and 3 (EBNA 2 and EBNA 3) (4,5,9). EBV type 1 appears to retain a greater capacity to induce B cell transformation in vitro (23), potentially due to the stronger ability of type 1 EBNA 2 to induce the EBV transforming protein LMP1 (latent membrane protein 1) gene (23).
While the majority of studies describing the role of herpesviruses in oral disease have focused on periodontitis, there have been no current attempts to examine herpesviruses in acute apical abscesses and cellulitis. In a previous report, we described the association of Epstein-Barr virus with irreversible pulpitis and apical periodontitis (5). In other studies, HCMV and EBV have been identified in periodontal abscesses or localized purulent infections of tissues adjacent to the periodontal pocket (3). The hypothesis for this current study is that herpesviruses, and potentially herpesvirus particles, are present in acute apical abscesses and cellulitis, and that these viruses are associated with radiographic bone destruction or abscess lesion size greater than 5 mm in size. Using purulent specimens collected from patients, we conducted primary and nested polymerase chain reaction (PCR) and virus transfer experiments to determine the presence of herpesviral DNA, or herpesviral particles, in acute apical abscess and cellulitis of endodontic origin.
Materials And Methods
Collection and Categorization of Endodontic Specimens
Endodontic abscess specimens were collected from patients seeking dental care at the OHSU School of Dentistry. The protocol for collection of endodontic abscess specimens was reviewed and approved by the OHSU Institutional Review Board (IRB). All patients admitted for treatment with swelling associated with an acute apical abscess or cellulitis were included in this study. Specimen collections were obtained using informed consent and patients were anonymously coded for identification. The diagnostic terminology was based on the guidelines from the American Board of Endodontics (2007) Pulpal & Periapical Diagnostic Terminology and referenced in Ingle's Endodontics, 6th Edition (24). Prior to the needle aspiration of the abscess or cellulitis, the overlying surface was disinfected using Povidone-Iodine U.S.P. Pads™ (Professional Disposables, Inc., Orangeburg, NY). The clinical samples were then aseptically aspirated with a sterile needle and immediately transported to the lab for freezing. The abscess material is composed predominantly of fluid, potentially containing virus particles, in addition to polymorphonuclear neutrophilic leukocytes, mononuclear cells, bacterial by-products and cellular debris. Patients were categorized according to presence of spontaneous pain, radiographic lesion size, abscess lesion size and presence of cellulitis.
DNA Extraction and PCR Conditions
Abscess specimens were pre-clarified by centrifugation to remove debris, prior to DNA extraction using the EasyXpress Viral Nucleic Acid Release kit from Express Biotech International (Thurmont, MD). Using control recombinant or viral genomic DNA templates (see below), we optimized PCR conditions and selected Platinum Taq DNA High Fidelity (Invitrogen) for HCMV and HSV-1 amplifications, the Pfx50 DNA polymerase (Invitrogen) for EBV amplifications and Go Taq Green Master Mix (Promega, Madison, WI) for VZV amplifications. PCR was conducted with an initial denaturation step at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 seconds, annealing at 57°C for 30 seconds and extension at 72°C for 30 seconds, with a final extension at 72°C for 5 minutes followed by a 4°C soak. Nested PCR, following the same cycling parameters defined in primary PCR, was used to amplify low herpesviral DNA copy number found in the abscess specimens and to increase the sensitivity of PCR fragment detection.
Primers and DNA Controls
Primers used in primary and nested PCR for amplification of herpesviral sequences are displayed in Table 1. Primers described in Table 1 were described in part in our earlier study (5). Primers were designed to amplify sequences from HCMV pp65 lower matrix phosphoprotein (UL 83 gene; accession number NC 001347), EBV BLR F2 gene product (EBV structural protein; EBV type 1, accession number NC 007605; EBV type 2, NC 009334), HSV-1 RL2 immediate early gene product (latency; accession number NC 001806), and VZV ORF 29 gene product (gp31; accession number NC 001348). DNA controls include 1) recombinant plasmids containing HCMV pp65 (7.12 kb), (provided by Mark Stinski, University of Iowa, Iowa City, IA; Richard Longnecker, Northwestern University, Chicago, IL), 2) genomic DNA from the EBV-containing Namalwa cell line (contains 2 EBV copies per cell; provided by Astrid Meerbach, Institute of Virology and Antiviral Therapy, Friedrich-Schiller University Jena, Germany), 3) HSV-1 nucleocapsid DNA (152,261 bp) and VZV genomic DNA (provided by Randall Cohrs, University of Colorado Health Science Center, Denver, CO) and 4) genomic DNA from HCMV-transformed lymphoid cells and culture medium containing HCMV laboratory strain AD169 (provided by Sunwen Chou, Department of Veterans Affairs Medical Center, Portland, OR).
Table 1. Primer sequences of HCMV, HSV, EBV, VZV and Actin.
| Target Molecule1 | Accession Number | Primer Description | Primer Location2 | Primer Sequence | Size of Amplicon |
|---|---|---|---|---|---|
| HCMV3 | Primary upstream | 121314→121333 | 5′ TCACCTGCATCTTGGTTGCG 3′ | ||
| NC_001347 | Primary downstream | 121622→121603 | 5′ TGCCGCTCAAGATGCTGAAC 3′ | 309 bp | |
| Nested upstream | 121403→121422 | 5′ GGAAACACGAACGCTGACGT 3′ | |||
| Nested downstreamA | 121582→121563 | 5′ TCAACGTGCACCACTACCGC 3′ | 180 bp | ||
| Nested downstreamB | 121622→121603 | 5′ TGCCGCTCAAGATGCTGAAC 3′ | 220 bp | ||
| EBV type 14 | Primary upstream | 76641→76661 | 5′ CAGCTCCACGCAAAGTCAGAT 3′ | ||
| NC_007605 | Primary downstream | 77122→77101 | 5′ ATCAGAAATTTGCACTTTCTTT 3′ | 482 bp | |
| Nested upstream | 76680→76699 | 5′ TTGACATGAGCATGGAAGAC 3′ | |||
| Nested downstream | 77042→77022 | 5′ CTCGTGGTCGTGTTCCCTCAC 3′ | 363 bp | ||
| EBV type 24 | Primary upstream | 76775→76795 | 5′ CAGCTCCACGCAAAGTCAGAT 3′ | ||
| NC_009334 | Primary downstream | 77256→77235 | 5′ ATCAGAAATTTGCACTTTCTTT 3′ | 482 bp | |
| Nested upstream | 76814→76833 | 5′ TTGACATGAGCATGGAAGAC 3′ | |||
| Nested downstream | 77176→77156 | 5′ CTCGTGGTCGTGTTCCCTCAC 3′ | 363 bp | ||
| HSV-1 | Primary upstream | 120665→120674 | 5′ CCAACACAGACAGGGAAAAG 3′ | ||
| NC_001806 | Primary downstream | 121000→120981 | 5′ GGAACATGCTGTTCGACCAG 3′ | 336 bp | |
| Nested upstream | 120703→120723 | 5′ AGACAGCAAAAATCCCCTGAG 3′ | |||
| Nested downstream | 120898→120880 | 5′ ACGAGGGAAAACAATAAGG 3′ | 196 bp | ||
| VZV | Primary upstream | 51067→51089 | 5′ ACGGGTCTTGCCGGAGCTGGTAT 3′ | ||
| NC_001348 | Primary downstream | 51338→51315 | 5′AATGCCGTGACCACCAAGTATAAT 3′ | 272 bp | |
| Nested upstream | 51099→51118 | 5′ACTCACTACCAGTCATTTCT 3′ | |||
| Nested downstream | 51306→51286 | 5′TTCTGGCTCTAATCCAAGGCG 3′ | 208 bp | ||
| Human β -actin | Upstream | 1141→1162 | 5′ CAGCAGATGTGGATCAGCAAGC 3′ | ||
| NM_00101 | Downstream | 1506→1485 | 5′ AGGATGGCAAGGGACTTCCTGT 3′ | 366 bp |
HCMV: human cytomegalovirus, EBV: Epstein-Barr virus, HSV: herpes simplex virus, VZV: Varicella-zoster virus
Primer locations are relative to the positions in the complete viral genomic sequence referenced in individual accession numbers. Herpesviral genomic sequences and human beta actin sequences were available for download from the US National Library of Medicine (www.pubmed.gov).
In selected cases for nested PCR, nested downstreamB primer was utilized instead of nested downstreamA.
Primary and nested primers for EBV type 1 and type 2 are conserved for both EBV types. Thus, our EBV PCR primers simultaneously recognize and amplify fragments for both EBV type 1 and type 2.
Other Recombinant DNA Methods and Statistical Analysis
PCR products were electrophoresed in 1.2% agarose gels containing ethidium bromide (0.5 μg/ml) and were sequenced using an ABI 3130xl DNA analyzer (Molecular Microbiology and Immunology Research Core Facility, OHSU School of Medicine). t-tests (Microsoft Excel software) were conducted to assess statistical significance between categorical data.
Virus Particle Detection and Virus Infection Procedures
For detection of herpesvirus particles, the abscess specimens were subjected to filtration (0.45 μm) to exclude cells potentially containing dormant herpesviruses, followed by DNase I treatment (10 U, 37°C, 30 minutes) to degrade any extracellular DNA, prior to use in primary PCR and nested PCR. The human lung fibroblast MRC-5 cell line (American Type Culture Collection, Bethesda, MD) was grown in Eagle's Minimum Essential Medium (Mediatech, Inc., Manassas, VA) in the presence of 10% fetal bovine serum (Hyclone, Thermo Scientific, Logan, Utah) and antibiotics/antimycotics (Hyclone; 100 U/ml penicillin G, 100 U/ml streptomycin, and 0.25 μg/ml amphotericin B; all at final concentrations in medium). MRC-5 cells were plated in 12-well trays and were grown overnight to 40% confluency. Just prior to virus adsorption, the abscess specimens (50 μl), previously determined to contain HCMV DNA by PCR analyses, were mixed with an equivalent volume of serum-free medium. The composite solutions were filtered (0.22 μm) to exclude mammalian cells that could contain dormant herpesviruses and also any potential bacterial contaminants that would affect cell culture. Cultures were then inoculated with the filtered abscess specimens (0.1 ml volume), and adsorption of potential herpesviruses were allowed to occur for 6 hours, followed by replacement with serum-containing medium for a 48-hour infection period. MRC-5 cells were also infected with pre-clarified medium containing the HCMV laboratory strain AD169 to serve as a positive control, or subjected to mock infection with saline to serve as a negative control. Genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA), which is based on digestion and extraction using proteinase K and subsequent elution of nucleic acids on spin columns using chaotropic salts. Primary and nested PCR were conducted using HCMV primers.
Results
Presence of Herpesviruses in Acute Apical Abscesses and Cellulitis
A total of 31 specimens were collected from patients (age range 14-81 years; average age 47.5 years; 22 males and 9 females). Out of these 31 patients, 12 individuals demonstrated presence of cellultis. Table 2 illustrates the incidence of herpesviruses in acute apical abscesses and cellulitis. HCMV DNA was present in 29% of the patients with acute abscesses (9/31), compared to similar incidence in healthy pulp controls (8/19 or 42.1%; data obtained from reference 5). EBV DNA was present in two out of 31 abscess specimens (6.5%) compared to zero incidence in healthy pulp controls (0/19; data obtained from reference 5). HSV-1 DNA was found in one of 31 abscess specimens (3.22%) compared to similar incidence in healthy pulp controls (1/19 or 5.26%; data obtained from reference 5). No abscess specimens contained VZV. Out of the 12 patients presenting with cellulitis, HCMV, EBV, HSV-1 and VZV were found in 16.7%, 8.3%, 0%, and 0%, respectively, of the specimens examined (Table 2). In all positive identifications confirming the presence of HCMV, EBV, and HSV-1 in the abscess specimens, the electrophoretic sizes of the PCR products matched closely with the predicted sizes based on sequence, and matched the sizes of the PCR products generated by the recombinant or genomic viral DNA controls. PCR fragments were also directly verified by sequence analysis and were aligned with herpesviral genomic DNA sequences obtained from the National Library of Medicine / National Institutes of Health (www.pubmed.gov; see Table 1). The EBV primers were designed to recognize identical target sequences between EBV types 1 and 2, and generated PCR fragments of identical size; in addition, the 363 bp fragments amplified during nested PCR were nearly identical in sequence to the two EBV strains. However, the sequence for the EBV PCR fragments had one mismatch compared to the corresponding EBV type 1 sequence (362 / 363 matches or 99.7% identity) versus four mismatches compared to the corresponding EBV type 2 sequence (359 / 363 matches or 99.2% identity). Even though the sequence of the amplified region obtained from EBV type 1 and EBV type 2 are too similar to definitively distinguish the two EBV strains, it appears more likely that the abscess specimens may have contained EBV type 1.
Table 2. Incidence of Herpesviruses in Endodontic Abscess and Healthy Pulp1.
| Endodontic Specimen | N | HCMV | EBV | HSV | VZV |
|---|---|---|---|---|---|
| HEALTHY PULP2 | |||||
| 19 | 8 (42.1) | 0 | 1 (5.26) | 0 | |
| ACUTE APICAL ABSCESSES | |||||
| 31 | 9 (29.0) | 2 (6.5) | 1 (3.2) | 0 |
Values are given as n (% incidence).
Values for healthy pulp were obtained from reference 5
Herpesvirus DNA is Present in Low Copy Number in Abscess Specimens
Using defined herpesviral DNA in primary PCR, we observed no cross amplification of fragments when using herpesvirus primers not matched to its specific herpesvirus template. Using recombinant or genomic DNA of defined size or copy number, we also conducted 10-fold limiting dilutions of HCMV, EBV or HSV-1 DNAs from 100 (or 1) copy to 105 copies for use as templates in primary PCR. The defined DNA molecules included the HCMV pp65 recombinant plasmid (7.12 kb), the Namalwa human genomic DNA containing two EBV genomes per cell, and HSV-1 nucleocapid DNA (genomic size = 152261 bp). Using primary PCR, we detected PCR product in all dilutions at 103 – 104 copies or greater (Figure 1. Panels A, C and E). When aliquots of the primary PCR reaction were used as templates for nested PCR, we detected the smaller nested PCR fragments from dilutions originally containing only a single genomic copy for HCMV, 100 genomic copies of EBV, and 10 - 100 genomic copies for HSV-1 (Figure 1, Panels B, D and F). All abscess specimens required nested PCR for detection of amplified DNA, indicating that all positive samples contained less than 10 – 100 copies of herpesviral genomic DNA per five microliter of abscess specimen used as template in the primary PCR. Thus our limit of detection using nested PCR is 0.2 copies per μl [or 1 copy / 5 microliters] for HCMV DNA, 20 genomic copies per μl [100 copies / 5 microliters] for EBV DNA, and 2 genomiccopies per μl [10 copies / 5 microliters] for HSV-1 DNA.
Figure 1. Detection Limits for Primary and Nested PCR.
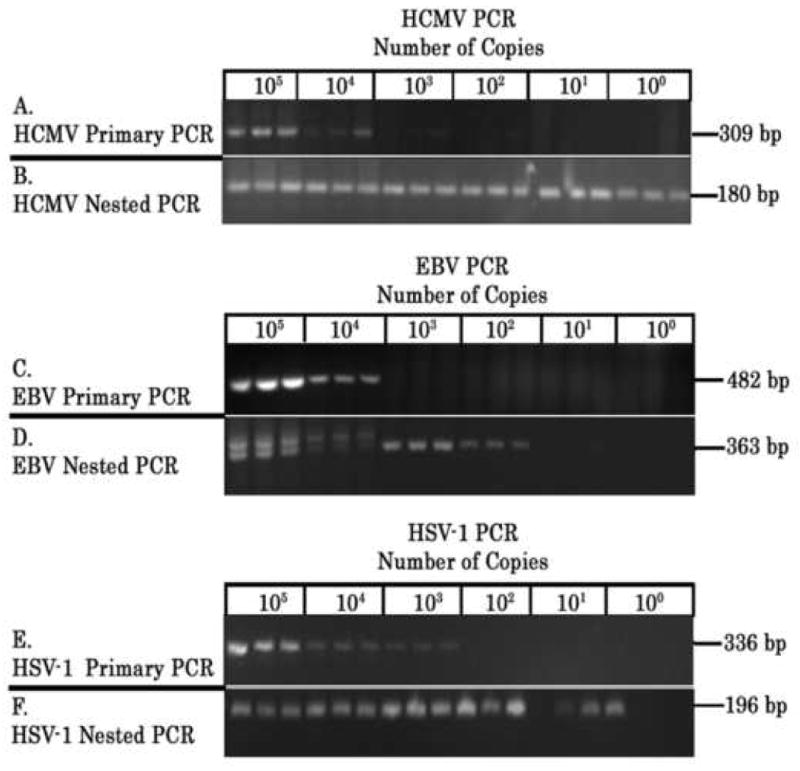
Recombinant and genomic DNA were diluted to contain 100 – 105 copies of HCMV, EBV, or HSV-1 DNA and subjected to primary PCR using specific herpesviral primers in three replicate reactions. The primary PCR reactions were then used as templates to conduct nested PCR. DNA controls include 1) recombinant plasmids containing HCMV pp65 (7.12 kb), 2) genomic DNA from the EBV-containing Namalwa cell line (contains 2 EBV copies per cell), and 3) HSV-1 nucleocapsid DNA (152,261 bp). PCR products were electrophoresed in 1.2% agarose gels containing ethidium bromide (0.5 μg/ml) and visualized by UV transillumination. HCMV primary and nested PCR (Panels A and B, respectively), EBV primary and nested PCR (Panels C and D, respectively), and HSV-1 primary and nested PCR (Panels E and F, respectively) are displayed. PCR fragment size (in bp) is displayed in the right margin of each panel. In some cases, primary PCR product used as template for nested PCR are visualized upon electrophoresis of aliquots of the nested PCR (see HCMV Nested PCR and EBV Nested PCR in Panels B and D, respectively).
Herpesviruses Detected as Intact Virus Particles
The initial screening for presence of herpesviral DNA was conducted using abscess specimens pre-clarified of cell debris by centrifugation. We subsequently subjected the abscess specimens to filtration to definitively exclude any cells potentially containing dormant herpesviruses, followed by DNase I treatment to degrade any extracellular DNA released from lysed cells, prior to use in primary PCR and nested PCR. Intact herpesvirus particles contain virion DNA encased within an icosahedral protein capsid and further wrapped within a lipid envelope, making the virion DNA resistant to direct digestion with DNase I. Using this procedure, we detected HCMV PCR fragments in eight abscess specimens and HSV-1 PCR fragment in one abscess specimen, supporting our contention that abscess fluids contained intact herpesvirus particles (Figure 2, Panel A). Using this procedure, HCMV was not detected in one abscess specimen (specimen 26) previously determined to be HCMV positive using non-filtered abscess material and nested PCR.
Figure 2.
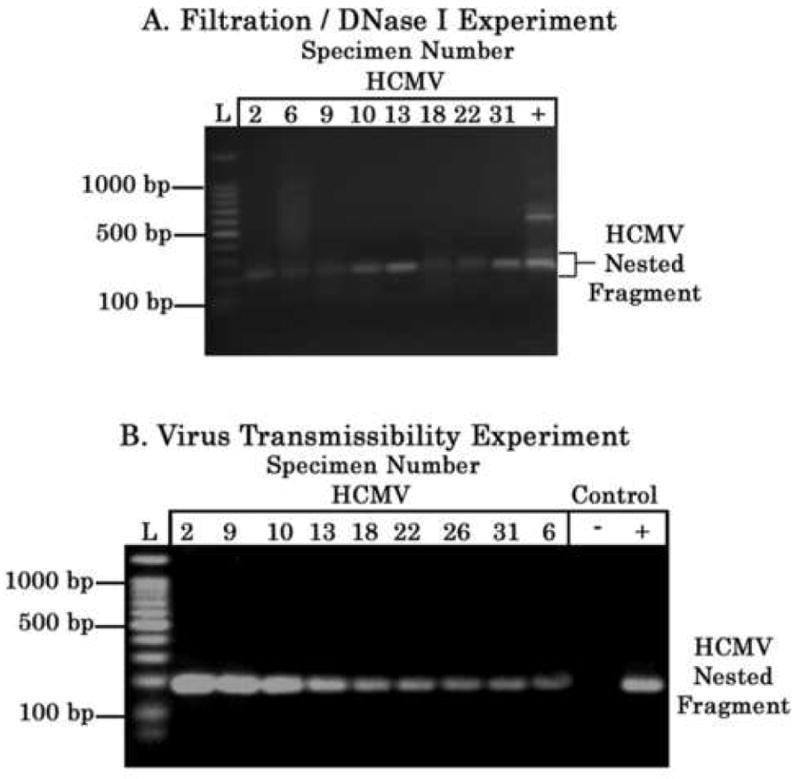
A. Filtration and DNase I Experiment for Detection of HCMV DNA. Abscess specimens were subjected to filtration (0.45 μm) to exclude cells potentially containing dormant herpesviruses, followed by DNase I treatment (10 U, 37°C, 30 minutes) to degrade any extracellular DNA released from lysed cells, prior to use in primary and nested PCR using HCMV primers. PCR products were electrophoresed in 1.2% agarose gels containing ethidium bromide (0.5 μg/ml) and visualized by UV transillumination. Nested PCR products are displayed. 100 bp ladder (L; Promega, Madison, WI) is displayed in the extreme left lane of the panel; selected ladder fragment sizes are denoted. The positive control (denoted by + symbol) consists of nested PCR using genomic DNA from HCMV-transformed lymphoid cells as template. Note that the lower molecular weight band in the + control lane is the HCMV nested PCR fragment. B. Virus Transmissibility Experiment. Abscess fluids were pre-filtered (0.22 μm to exclude cells and bacteria) and subjected to virus adsorption (6 hours) and infection of MRC-5 cells for 48 hours, prior to conducting PCR analyses. Each of the MRC-5 cultures was subjected to genomic DNA extraction, and primary and nested PCR using the HCMV primers. PCR products were electrophoresed in 1.2% agarose gels containing ethidium bromide (0.5 μg/ml) and visualized by UV transillumination. Nested PCR products are displayed. All nine cultures yielded PCR fragments following nested PCR, indicating that the abscess specimens contained low levels of HCMV particles. MRC-5 cells were infected with pre-clarified culture medium containing HCMV laboratory strain AD169 to serve as a positive control (denoted as + symbol), and also subjected to mock infection with saline to serve as a negative control (denoted as – symbol); genomic DNA extracted from control cultures, when used for nested PCR using HCMV primers, yielded HCMV-specific fragments in the positive control and no fragments in the negative control (see + and - control lanes, respectively). 100 bp ladder (L; Promega, Madison, WI) is displayed in the left margin of the panel; selected ladder fragment sizes are denoted.
In order to determine virus transmissibility, nine abscess specimens, previously determined in the initial screening to contain HCMV DNA by PCR analysis, were filtered and inoculated onto MRC-5 cell cultures. All nine cultures yielded PCR fragments following nested PCR, indicating that all nine abscess specimens contained low but detectable levels of intact HCMV particles (Figure 2. Panel B). As expected, genomic DNA extracted from the positive control MRC-5 culture infected with pre-clarified medium containing the HCMV laboratory strain AD169, when used in nested PCR, amplified HCMV-specific fragments (Figure 2, Panel B). Genomic DNA extracted from the mock-infected culture, when used in nested PCR, did not amplify any HCMV-specific fragments.
Herpesviruses in Acute Apical Abscesses Subdivided According to Presence of Spontaneous Pain, Sizes of Radiographic Bone Destruction and Abscess Lesion, and Presence of Cellulitis
The patients were grouped according to the presence of spontaneous pain, sizes of radiographic bone destruction and abscess lesion (≥ 5 mm or < 5 mm) and presence of cellulitis (Table 3). Twenty-eight (28) patients experienced spontaneous pain, including the 9 patients with abscesses containing HCMV and the 2 patients with abscesses containing EBV. Twenty-three (23) patients had radiographic bone destruction of ≥ 5 mm and 8 patients had radiographic bone destruction of < 5 mm. From the patients with radiographic bone destruction ≥ 5 mm, 26.1% of the patients contained HCMV DNA, 4.3% of the patients contained EBV DNA or HSV-1 DNA, and none of the patients contained VZV DNA. From the patients with radiographic bone destruction < 5 mm, 37.5% of the patients contained HCMV DNA, 2.5% of the patients contained EBV or HSV-1 DNA, and none of the patients contained VZV DNA. Thirty (30) patients had abscess lesion sizes ≥ 5 mm and one patient had abscess lesion sizes of < 5 mm. Twelve (12) patients contained cellulitis of endodontic origin.
Table 3. Clinical Observations and Incidence of Herpesviruses in Acute Apical Abscesses1.
| Clinical Observations | Lesion Size | N | HCMV | EBV | HSV | VZV |
|---|---|---|---|---|---|---|
| Presence of Spontaneous Pain2 | 28 | 9 (32.1) | 2 (7.1) | NA2 | 0 | |
| Radiographic Lesion Size | ||||||
| ≥ 5 mm | 23 | 6 (26.1) | 1 (4.3) | 1 (4.3) | 0 | |
| < 5 mm | 8 | 3 (37.5) | 1 (12.5) | 0 | 0 | |
| Abscess Lesion Size | ||||||
| ≥ 5 mm | 30 | 8 (26.7) | 2 (6.7) | 1 (3.3) | 0 | |
| < 5 mm | 1 | 1 (100.0) | 0 | 0 | 0 | |
| Presence of Cellulitis | 12 | 2 (16.7) | 1 (8.3) | 0 | 0 |
Values are given as n (% incidence).
28 patients experienced spontaneous pain, including 9 patients with abscesses containing HCMV and 2 patients with abscesses containing EBV. Data describing presence or absence of spontaneous pain for the patient containing HSV-1 are unavailable (NA = not available).
Discussion
Herpesviruses in Endodontic Disease
Herpesviruses have been implicated in the pathoses of symptomatic and asymptomatic apical periodontitis lesions (1-3). Acute apical abscesses represent one subcategory of symptomatic apical periodontitis. Sabeti et al. (1) identified herpesvirus in periapical granulomatous tissues, and detected herpesviruses in large symptomatic periapical lesions at higher incidence compared to small asymptomatic periapical lesions. Yildirim et al. (18) demonstrated the presence of herpesviruses and bone resorption-inducing cytokines in periapical lesions of deciduous teeth. Saboia-Dantas et al. (14) found HCMV and EBV in apical periodontitis lesions, with higher prevalence in human immunodeficiency virus-positive patients. Andric et al. (25) detected HCMV in the cystic wall, and identified HCMV in both inflammatory and non-inflammatory odontogenic cysts. In this current study, we analyzed clinical specimens from 31 patients exhibiting acute apical abscesses and cellulitis, and conducted primary and nested PCR to determine the presence and association of herpesviruses with spontaneous pain, sizes of radiographic bone destruction and abscess lesion, and cellulitis. HCMV, EBV, HSV-1, and VZV were found in 29.0%, 6.5%, 3.22% and 0% of the abscess specimens, respectively. The prevalence of herpesviruses in cellulitis, which represents a more advanced staging of endodontic disease because of the diffuse nature of the abscess, does not appear to be different than the virus prevalence in more focalized abscesses. Out of the 28 patients experiencing spontaneous pain, 9 patients contained HCMV and 2 patients contained EBV. While there were some apparent differences in the incidence of HCMV, EBV, HSV-1 or VZV between groups containing small radiographic lesions (< 5 mm) versus large radiographic lesions (≥ 5 mm), none of these differences were statistically significant based on t-test analyses (Table 3). This is interesting in contrast to the findings of Sabeti et al. (1), who have demonstrated that HCMV and EBV occur at higher frequency in periapical lesions with large radiographic bone destruction (5 mm × 7 mm) compared to smaller sizes. In addition, the statistical comparison examining differences in incidence of HCMV, EBV, HSV-1 or VZV between groups containing small abscess lesion sizes (< 5 mm) versus large abscess lesion sizes (≥ 5 mm) was not possible because of the small size of the group (n=1) for the abscess lesion sizes of < 5 mm.
Low Prevalence of Herpesviruses in Selected Acute Apical Abscesses
Sabeti et al. (1) identified HCMV in almost all patients examined that were previously treated with apical periodontitis. Yildirim et al. (18) found HCMV and EBV DNA in 58% and 67% of the periapical specimens, respectively, from deciduous teeth. In our prior study (5) and contrary to Sabeti et al. (1), we found the incidence frequency of HCMV in apical periodontitis specimens to be lower (4 out of 30 specimens or 13.3% with HCMV) and also statistically indistinguishable from the incidence observed in healthy pulp specimens (8 out of 19 pulp specimens or 42.1% with HCMV DNA). For our current study and in contrast to the findings of Sabeti et al. (1), we find that herpesviruses and herpesvirus particles are present but not required for the development of acute abscesses and cellulitis of endodontic origin. Our results were more consistent with the findings of Saboia-Dantas et al. (14) and Sunde et al. (4). Saboia-Dantas et al. (14) identified HCMV and EBV in 23% and 31% of patients with apical periodontitis, respectively. Sunde et al. (4) found EBV to be the predominant herpesvirus in apical periodontitis (50% incidence) and were not able to detect HCMV in their specimens. Sunde et al. (4) proposed that the high incidence of HCMV and EBV found in some studies were influenced by the surgical technique and potential contamination from the marginal area. In addition, Sunde et al. (4) found that the incidence of EBV in apical periodontitis specimens decreased significantly when using submarginal incisions to avoid marginal contamination. In our acute apical abscesses, we also found a higher incidence of HCMV (29%) compared to our apical periodontitis specimens (13.3%), but lower to what we have previously identified in healthy pulp (42.1%; see reference 5).
Limit of Detection of Herpesviral DNA Using Nested PCR
Our limit of detection using nested PCR is 0.2 copies per μl for HCMV DNA, 20 copies per μl for EBV DNA, and 2 copies per μl for HSV-1 DNA. Botero et al. (26) recently determined that nested PCR was more sensitive than real-time PCR for detection of HCMV DNA, and was able to detect 0.71 copies / μl HCMV DNA using nested PCR, consistent with our detection limit of 0.2 copies / μl HCMV DNA. In addition, because of our inability to detect less than 0.2 copies / μl HCMV DNA, 20 copies / μl EBV DNA or 2 copies / per μl HSV-1 DNA using nested PCR, we cannot preclude the possibility that some abscess specimens may contain trace amounts of HCMV, EBV or HSV-1 genomic DNA.
Viral Load and Endodontic Disease
HCMV was detected in nine out of 31 abscess specimens, with viral numbers being quite low, probably on the order of < 2000 particles per μl (based on primary PCR detection limit of 104 copies / 5 μl = 2000 copies; see Figure 1, Panel A). Botero et al. (18) demonstrated that active HCMV infection occurred in only a single patient out of 44 individuals with chronic or aggressive periodontitis, indicating that active HCMV infection does not occur frequently in periodontal pockets. Botero et al. (26) stated that HCMV reactivation may be limited to only a few sites within periodontitis patients. In contrast, Contreras and Slots (27) demonstrated active HCMV infection in 4 out of 9 periodontitis patients using reverse transcription - PCR for detection of mRNA encoding HCMV late major capsid protein. HCMV and EBV load in blood has been quantified to be variable in several herpesviral-associated diseases (4,28-30), and even low levels of virus have been found to be clinically relevant. Our virus transmissibility and filtration / DNase I treatment experiments support our contention that intact HCMV particles are present in abscess fluid. However, we cannot formally rule out the possibility that HCMV particles adsorbed to virus receptors on the external surface of MRC-5 cell membranes, but did not undergo penetration and replication in the infected cells, or if residual HCMV DNA in the abscess fluid underwent DNA-mediated transfection into MRC-5 cells; either of these scenarios, the latter being very unlikely, may lead to amplification of HCMV-specific fragments upon nested PCR. In addition, the detection of HCMV-specific PCR fragments for specimen 26 in the virus transmissibility experiment, but not in the experiment using DNA extracted directly from filtered abscess fluids, may have been the result of amplification of HCMV copy number during the extended infection in MRC-5 cells, allowing subsequent detection of HCMV by nested PCR.
High virus titers are not necessarily considered to be causative factors for disease; for example, in the case of circoviruses, small DNA viruses normally found in pigs and birds, high viral titers were previously implicated in hepatitis, but more recently are believed to simply be the result of generalized viremia and impaired immune status (31). We found that the incidence of HCMV in acute apical abscess (29.0%) was higher than the incidence of HCMV in apical periodontitis (13.3%; stated in reference 5). Saygun et al. (3) proposes that herpesvirus reactivation may preferentially occur at periodontal abscesses. We believe that this may also apply to acute apical abscesses of endodontic origin where we observe higher incidence of HCMV in endodontic abscesses compared to incidence of HCMV in apical periodontitis. Periodontal abscesses not only serve as potential sites of herpesvirus reactivation, but appear to develop at sites with high bacterial pathogen load and low levels of antiviral T lymphocytes (3,32). Although herpesviruses may not be the primary determinant in the formation of acute apical abscesses, herpesvirus reactivation may play a contributory role in the polymicrobial infection process leading to immune impairment, inflammation, and acute endodontic disease. Thus, herpesviruses and herpesvirus particles may appear in selected cases of acute apical abscesses and cellulitis, but are not required for the development of this form of endodontic pathosis. The exact role of herpesviruses in the inflammatory process leading to endodontic disease will be need to be explored by further experimentation using in vitro or animal model systems.
Acknowledgments
This work was supported by research funds from the American Association of Endodontists Foundation (fellowship award to HL) and the Oregon Clinical and Translational Research Institute [OCTRI; grant number UL1 RR024140 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research]. VC was a 2007 OCTRI Student Research Fellow and is a 2008 OSLER Student Research Fellow. YC is a 2008 OCTRI Summer Research Fellow. KK, JCB and CAM are supported with funds provided by the OHSU School of Dentistry. The authors thank Drs. Tom Shearer and Jack Clinton for their encouragement and support. The authors thank Rebecca Sauerwein, Ying Bai, Tyler Finlayson, and Jennifer Kimmell for their technical assistance during various phases of this project, and for their encouragement and generous provision of time and ideas.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sabeti M, Valles Y, Nowzari H, Simon JH, Kermani-Arab V, Slots J. Cytomegalovirus and Epstein-Barr virus DNA transcription in endodontic symptomatic lesions. Oral Microbiol Immunol. 2003;18(2):104–108. doi: 10.1034/j.1399-302x.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- 2.Slots J, Nowzari H, Sabeti M. Cytomegalovirus infection in symptomatic periapical pathosis. Int Endod J. 2004;37(8):519–24. doi: 10.1111/j.1365-2591.2004.00816.x. [DOI] [PubMed] [Google Scholar]
- 3.Saygun I, Yapar M, Ozdemir A, Kubar A, Slots J. Human Cytomegalovirus and Epstein-Barr virus type 1 in periodontal abscesses. Oral Microbiol Immunol. 2004;19(2):83–7. doi: 10.1046/j.0902-0055.2002.00118.x. [DOI] [PubMed] [Google Scholar]
- 4.Sunde PT, Olsen I, Enersen M, Beiske K, Grinde B. Human cytomegalovirus and Epstein-Barr virus in apical and marginal periodontitis: a role in pathology? J Med Virol. 2008;80(6):1007–1011. doi: 10.1002/jmv.21180. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Chen V, Chen Y, Baumgartner JC, Machida CA. Herpesviruses in endodontic pathoses: Association of Epstein-Barr Virus with irreversible pulpitis and apical periodontitis. J Endod. 2008 doi: 10.1016/j.joen.2008.09.017. manuscript in press. [DOI] [PubMed] [Google Scholar]
- 6.Siqueira JF, Rocas IN, Souto R, de Uzeda M, Colombo AP. Microbiological evaluation of acute periradicular abscesses by DNA-DNA hybridization. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;92(4):451–457. doi: 10.1067/moe.2001.118620. [DOI] [PubMed] [Google Scholar]
- 7.Chen T, Hudnall SD. Anatomical mapping of human herpesvirus reservoirs of infection. Mod Pathol. 2006;19(5):726–737. doi: 10.1038/modpathol.3800584. [DOI] [PubMed] [Google Scholar]
- 8.Puchhammer-Stockl E, Gorzer I. Cytomegalovirus and Epstein-Barr virus subtypes – The search for clinical significance. J Clin Virol. 2006;36(4):239–48. doi: 10.1016/j.jcv.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Sitki-Green D, Edwards RH, Webster-Cyriaque J, Raab-Traub N. Identification of Epstein-Barr virus strain variants in hairy leukoplakia and peripheral blood by use of a heteroduplex tracking assay. J Virol. 2002;76(19):9645–56. doi: 10.1128/JVI.76.19.9645-9656.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sitki-Green D, Covington M, Raab-Traub N. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J Virol. 2003;77(3):1840–1847. doi: 10.1128/JVI.77.3.1840-1847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walling DM, Etienne W, Ray AJ, Flaitz CM, Nichols CM. Persistence and transition of Epstein-Barr virus genotypes in the pathogenesis of oral hairy leukoplakia. J Infect Dis. 2004;190(2):387–95. doi: 10.1086/421708. [DOI] [PubMed] [Google Scholar]
- 12.Slots J, Saygun I, Sabeti M, Kubar A. Epstein-Barr virus in oral diseases. J Periodontal Res. 2006;41(4):235–244. doi: 10.1111/j.1600-0765.2006.00865.x. [DOI] [PubMed] [Google Scholar]
- 13.Shibata Y, Hoshino Y, Hara S, Yagasaki H, Kojima S, Nishiyama Y, Morishima T, Kimura H. Clonality analysis by sequence variation of the latent membrane protein 1 gene in patients with chronic active Epstein-Barr virus infection. J Med Virol. 2006;78(6):770–779. doi: 10.1002/jmv.20622. [DOI] [PubMed] [Google Scholar]
- 14.Saboia-Dantas CJ, Coutrin de Toledo LF, Sampaio-Filho HR, Siqueira JF., Jr Herpesviruses in asymptomatic apical periodontitis lesions: an immunohistochemical approach. Oral Microbiol Immunol. 2007;22(5):320–325. doi: 10.1111/j.1399-302X.2007.00362.x. [DOI] [PubMed] [Google Scholar]
- 15.Heling I, Morag-Hezroni M, Marva E, Hockman N, Zakay-Rones Z, Morag A. Is herpes simplex virus associated with pulp/periapical inflammation? Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;91(3):359–61. doi: 10.1067/moe.2001.113162. [DOI] [PubMed] [Google Scholar]
- 16.Slots J, Contreras A. Herpesviruses: a unifying causative factor in periodontitis? Oral Microbiol Immunol. 2000;15(5):277–280. doi: 10.1034/j.1399-302x.2000.150501.x. [DOI] [PubMed] [Google Scholar]
- 17.Miller CS, Danaher RJ. Asymptomatic shedding of herpes simplex virus (HSV) in the oral cavity. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2008;105(1):43–50. doi: 10.1016/j.tripleo.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Yildirim S, Yapar M, Kubar A, Slots J. Human cytomegalovirus, Epstein-Barr virus and bone resorption-inducing cytokines in periapical lesions of deciduous teeth. Oral Microbiol Immunol. 2006;21(2):107–11. doi: 10.1111/j.1399-302X.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- 19.Sabeti M, Slots J. Herpesviral-bacterial coinfection in periapical pathosis. J Endod. 2004;30(2):69–72. doi: 10.1097/00004770-200402000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Stashenko P, Teles R, D'Souza R. Periapical inflammatory responses and their modulation. Crit Rev Oral Biol Med. 1998;9(4):498–521. doi: 10.1177/10454411980090040701. [DOI] [PubMed] [Google Scholar]
- 21.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, Kieff E. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64(9):4084–92. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fafi-Kremer S, Morand P, Germi R, Ballout M, Brion JP, Genoulaz O, Nicod S, Stahl JP, Ruigrok RW, Seigneurin JM. A prospective follow-up of Epstein-Barr virus LMP1 genotypes in saliva and blood during infectious mononucleosis. J Infect Dis. 2005;192(12):2108–11. doi: 10.1086/498215. [DOI] [PubMed] [Google Scholar]
- 23.Lucchesi W, Brady G, Dittrich-Breiholz O, Kracht M, Russ R, Farrell PJ. Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J Virol. 2008;82(15):7456–7466. doi: 10.1128/JVI.00223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handysides RA, Jaramillo DE, Ingle JI. Examination, evaluation, diagnosis and treatment planning. In: Ingle J, Bakland L, Baumgartner J, editors. Ingle's Endodontics. 6th. Hamiliton, CA: BC Decker, Inc.; 2008. p. 526. [Google Scholar]
- 25.Andric M, Milasin J, Jovanovic T, Todorovic L. Human cytomegalovirus is present in odontogenic cysts. Oral Microbiol Immunol. 2007;22(5):347–51. doi: 10.1111/j.1399-302X.2007.00369.x. [DOI] [PubMed] [Google Scholar]
- 26.Botero JE, Vidal C, Contreras A, Parra B. Comparison of nested polymerase chain reaction (PCR), real-time PCR and viral culture for the detection of cytomegalovirus in subgingival samples. Oral Microbiol Immunol. 2008;23(3):239–244. doi: 10.1111/j.1399-302X.2007.00418.x. [DOI] [PubMed] [Google Scholar]
- 27.Contreras A, Slots J. Active cytomegalovirus infection in human periodontitis. Oral Microbiol Immunol. 1998;13(4):225–230. doi: 10.1111/j.1399-302x.1998.tb00700.x. [DOI] [PubMed] [Google Scholar]
- 28.Cohen JI. Clinical aspects of Epstein-Barr virus infection. In: Robertson E, editor. Epstein-Barr Virus. Norfolk: Calister Academic Press; 2005. pp. 33–54. [Google Scholar]
- 29.Gouarin S, Vabret A, Scieux C, Agbalika F, Cherot J, Mengelle C, Deback C, Petitjean J, Dina J, Freymuth F. Multicentric evaluation of a new commercial cytomegalovirus real-time PCR quantitation assay. J Virol Meth. 2007;146(12):147–154. doi: 10.1016/j.jviromet.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 30.Hakim H, Gibson C, Pan J, Srivastava K, Gu Z, Bankowski MJ, Hayden RT. Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood. J Clin Microbiol. 2007;45(7):2151–2155. doi: 10.1128/JCM.02308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grinde B. Human circoviruses: Clincal and applied perspectives. Curr Top Virol. 2003;3:81–90. [Google Scholar]
- 32.Hafstrom CA, Wikstrom MB, Renvert SN, Dahlen GG. Effect of treatment on some periodontopathogens and their antibody levels in periodontal abscesses. J Periodontol. 1994;65(11):1022–1028. doi: 10.1902/jop.1994.65.11.1022. [DOI] [PubMed] [Google Scholar]


