Abstract
Radiation hybrid (RH) mapping has become one of the most well established techniques for economically and efficiently navigating genomes of interest. The success of the technique relies on random chromosome breakage of a target genome, which is then captured by recipient cells missing a pre-selected marker. Selection for hybrid cells that have DNA fragments bearing the marker of choice, plus a random set of DNA fragments from the initial irradiation, generates a set of cell lines that recapitulates the genome of the target organism several-fold. Markers or genes of interest are analyzed by PCR using DNA isolated from each cell line. Statistical tools are applied to determine both the linear order of markers on each chromosome, and the confidence of each placement. The resolution of the resulting map relies on many factors, most notably the degree of breakage from the initial radiation as well as the number of hybrid clones and mean retention value.
A high resolution RH map of a genome derived from low pass or survey sequencing (coverage from 1 to 2x) can provide essentially the same comparative data on gene order that is derived from high-coverage (greater than 7x) genome sequencing. When combined with Fluorescence in Situ Hybridization (FISH), RH maps are complete and ordered blueprints for each chromosome. They give information about the relative order and spacing of genes and markers, and allow investigators to move between target and reference genomes, such as those of mouse or human, with ease although the approach is not limited to mammal genomes.
Keywords: Animals, Dogs, Genetic Markers, Genotype, Male, Radiation Hybrid Mapping, methods, Sequence Analysis, DNA, methods, Sequence Homology, Nucleic Acid
1. Introduction
Genome maps are essential for identifying disease genes or loci controlling traits of interest. Development of maps for the dog genome began in the late 1990s with the production of both meiotic linkage (1) and radiation hybrid maps (2). Analysis of the same sets of markers on both reference families and RH panels (3) allowed the meiotic linkage and RH maps to be quickly integrated (4). While such maps were useful for the identification of several disease loci (5,6), the lack of an agreed upon nomenclature for the dog’s 38 chromosomes until the late 1990s (7) made discussion of the data problematic. Most dog chromosomes are small and acrocentric. Thus, only those having expertise with techniques like fluorescence in situ hybridization (FISH) could readily identify specific dog chromosomes. The availability of a set of flow-sorted canine chromosomes (8,9), from which DNA fragments could be both FISH and RH mapped (10 11,), as well as used to develop microsatellite markers for meiotic linkage mapping, proved invaluable to the canine community. It allowed the first fully integrated maps to be developed in which most linkage groups were assigned to named chromosomes (12). In addition, it allowed researchers to orient linkage groups on chromosomes, thus facilitating the first studies of comparative genomics between the dog and human (13).
Since 2001 the community has advanced rapidly, generating new maps every 12 to 18 months (13,14,15,16). Particular emphasis has been placed on maps that have ordered microsatellite markers, genes sequences, and sequenced BAC ends that can facilitate positional cloning efforts. In addition, researchers have focused on increased understanding of the relationship between the dog, mouse and human genomes, initially defining about 85 conserved segments (13). As map resolution improved and researchers moved from a mapping panel with 600 Kb resolution (17) to 200 Kb resolution, the number of conserved ordered fragments increased to 264 (16).
Key to the success of the field has been the development of new bioinformatics tools that allow more precise positioning of markers then was possible with existing programs (18), together with the assignment of scores reflecting the relative confidence of local map order (19,20).
In 2003, The Institute for Genomic Research (TIGR) released 1.5x sequence coverage of a Standard Poodle genome (21). The 6.22 million sequence reads contained canine gene fragments that were orthologous to 84% of annotated human genes, and this was put to immediate use by researchers interested in cloning genes associated with disease and morphology traits. We subsequently opted to use the 1.5x sequence, or survey sequence as it has become termed, as a resource for constructing a very dense canine RH map, composed primarily of canine gene fragments.
The resulting resource, discussed in detail here advanced our knowledge of the canine genome in several ways. First, the dense map, composed of portions of nearly 10,000 independent genes detailed nearly all canine/human conserved ordered segments (16). This meant that, for the first time, we could move freely between the dog map and reference human and mouse genome assembled sequences. Second, the ordered presentation of over half of what are now known to be 19,000 canine genes offered an opportunity to verify and edit the 7.5x assembly of the Boxer sequence (22) and a way to check data related to the orientation of contigs. Finally, the work suggests a new paradigm for analysis of the many genomes currently being sequenced at only 2x coverage. These include a variety of mammalian genomes that are being sequenced primarily to identify conserved genomic elements (e.g. elephant, armadillo, cat, tenrec, sloth, hedgehog, shrew, squirrel, rabbit, bat and bushbaby; http://www.genome.gov/10002154). If high-density RH maps were constructed to accompany each survey sequence, researchers in the relevant fields would have essentially the same mapping resources for comparative genomics as those who benefit from 7–8x sequencing. We thus present the detailed methodology used for construction and utilization of a 10,000 gene RH map.
2. Materials
2.1. Marker identification and primer sequence design
Computer with Unix operating system
Primer design software “Primer3” (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi)
DNA sequence Alignment Programs BLAST, BLAT, MEGABLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
2.2. Marker genotyping
PCR Thermocycler
Electrophoresis apparatus and gel casting systems
Hot start PCR kits with high fidelity Taq enzyme and master mixes
Electrophoresis buffer. TE, Ph 7.5 10mM Tris-HCl, 1mM EDTA and 2% agarose gels for typing
Genotyping acquisition data -software (see Note 1)
DNA gel or capillary sequencer for sequencing and typing
Sequence analysis software
2.3. Map Construction
Radiation hybrid mapping analysis software (see Note 2)
Database with a portable relational database management system
3. Methods
A key limitation of survey-sequencing is the fragmentary nature of the data. The absence of long-range continuity greatly restricts the ability to predict gene and marker order in the surveyed genome. RH mapping can maximize the value of survey-sequencing through the localization of thousands of markers derived from the 1–2x sequence data. This has been tested with the canine 1.5x coverage (21) for which fragments of orthologs for 18,473 of 24,567 annotated human genes were identified (see Note 3).
3.1. Survey sequencing
Canine genomic sequence was obtained by end-sequencing of plasmid clones (2 kb and 10 kb inserts) prepared from genomic DNA of a male Standard Poodle. After trimming to remove vector or poor-quality sequence data, 6.22 million reads (mean 576 bases) provides approximately 1.5x coverage of the 2.5 Gb haploid canine genome. The assembled data consists of 1.2 million contigs (mean 1414 bases) and 0.4 million singletons. (21).
3.2. Orthologous Sequence Identification Strategy
All contigs and singletons from the survey sequence assembly are searched against an annotated reference genome that is closely related to the surveyed genome (e.g. human) using the Blastn alignment program (23). Sequences can also be searched across multiples species to ensure the selection of sequences that are highly conserved through evolution (24).
3.3. Orthologous Sequence Identification Criteria
-
Selected sequences have mutually-best alignments.
Blastn comparison of the test (dog) and reference (human) genome sequences yields many alignments of short segments, termed high scoring pairs (HSPs). HSPs are defined as mutually-best when the segment of aligned test sequence has no higher-scoring hit elsewhere on the reference genome and the segment of aligned reference sequence has no higher-scoring alignment with another test sequence.
The test/reference alignment overlaps at least one exon of the reference genome (as defined by the Ensembl annotation of 24,567 human genes; version 13.31.1)
Ensure that the test sequence is not a processed pseudogene. Where multiple HSPs are separated by <25 bases on a test sequence, but >300 bases on the reference sequence, the test sequence is considered as a potential pseudogene, and should be eliminated.
Ensure that the test sequence does not align end-to-end with the reference genome. A test sequence for which an alignment extends over its complete length may not permit the design of primers that are specific for the test sequence, and should be eliminated.
After applying criteria 1–4, a single test sequence for each reference gene is selected. When application of these criteria gives several hits, the sequence having the highest Blastn score is selected.
With the objective to map more than 10,000 gene sequences, selection of ~ 12,000 sequences is recommended to anticipate failures at each step of the mapping. The reference genome is divided into the smallest number of equivalently-sized segments that permits one or more genes to be located in each of 12,000 distinct segments. Here, this entails dividing the human genome into 40,000 “bins” of 75 kb, 11,818 of which contains a gene target.
Select a test sequence for each of the 75 kb bins that contain a gene. If the bin contains multiple genes, select the sequence providing the highest Blastn score.
Selection of test sequences is determined in two rounds from alternate bins. Processing in two rounds permits first-round failures to be retried during the second round, thereby minimizing the size of gaps in the final map.
3.4. Orthologous Sequence Content Criteria
-
Gene-based sequence selection.
In order to characterize conserved synteny between a test and a reference genome, fragments of orthologous genes are first predicted using mutually-best Blastn alignments. Although any unique conserved sequences can be used to anchor regions of conserved synteny, there is particular interest in the comparison of protein-coding genes, and analyses have therefore focused on identification of orthologous exons.
-
Microsatellite sequences within genes (see Note 4).
In the course of exon-based sequence selection, short tandem repeats (in flanking introns) were retrieved for 10% of the 11,818 selected sequences (see Note 5). Such sequences are an additional resource for genetic linkage and cloning studies and depending upon the ultimate goal, they could be specifically selected to be included within the marker sequence (25).
-
Non-coding conserved sequence selection.
Genome-wide sequence alignments lead to an estimate that 5% of a mammalian genome is under negative selection and thus potentially functional (22,26). This is about three times higher than the portion coding for proteins. Selection of such non-coding sequences, expected to be transcribed into non-coding RNA, or be involved in gene expression regulation, can also be selected for large-scale genomic comparative analysis. These can be particularly useful for comparative mapping of large genomic regions that lack protein-coding genes.
3.5. Gene-based Marker Design for Mapping
Selected genomic survey-sequences (300–1000 bp) have mutual best alignments with fragments of annotated human genes. For each sequence, the boundary between aligned and non-aligned segments is identified. In order to limit cross-amplification of the carrier hamster DNA present in RH DNA, primers should be designed in segments of the survey sequence that flank the aligned region. Alternatively, one primer can be selected from a flanking segment and the other from an aligned region. Markers derived from the sequence are amplified using two primers preferentially selected to be 25 bp in length and to work under a single optimal set of PCR conditions (salt, Tm, Mg+2, etc.) generating PCR products of 100–400 bp. Primers are selected for mapping using a standard selection program, i.e. Primer3 software, within non-repetitive sequences.
3.6. Radiation Hybrid Panel Characteristics
Radiation hybrid cells are constructed by fusing host cells that have been gamma-irradiated using a dose of 9,000 to 12,000 rads (17,27). This ensures a resolving power sufficient to map 10–12,000 markers at individual positions.
Radiation hybrid cell lines should be selected to contain 15–30% of the host genome with a standard deviation as low as possible. Approximately 200 markers distributed randomly on all chromosomes should be tested to determine consistent retention values. Selection of 90–95 hybrid cell lines (from a stock in our case of 387 cell lines) is required to constitute a complete RH panel with appropriate statistical power.
3.7. Marker Genotyping
Genotype markers as follows: all reactions are performed using a 96- well or 384-well format in a volume of 10–15 microliters. An initial screen using 50 ng dog DNA, 50 ng hamster DNA, and a 1:3 mix of dog/hamster DNA (50 ng) is used to select primers that under PCR amplification produced a clear DNA band with the host sample and no band with hamster DNA.
PCR amplifications are done using 50 ng of RH DNA and a touchdown program: 8 min 95°C, followed by 20 cycles of 30 sec 94°C, 30 sec 63°C decreasing of 0.5°C per cycle, 1 min 72 °C and 15 cycles of 30 sec 94°C, 30 sec 53°C, 1 min 72°C and final extension of 2 min 72°C.
Resolve PCR products on 1.8% or 2% agarose gels, electrophoresed for 30 minutes using standard electrophoresis gel apparatus (2,12).
View PCR bands under UV light after ethidium bromide staining, and record image.
Submit images to manual or automated software for data acquisition.
3.8. Radiation Hybrid Map Construction
Score images of PCR products resolved on agarose gels for the absence or presence of a band as ‘0’ and ‘1’, respectively, in plain text format (ambiguous data are scored as ‘2’). Typically a genotype corresponds to a string of ‘0s’ and ‘1s’ distributed in 90–100 data points. A complete RH data, termed RH vector, comprises the Marker Id and its retention pattern as: Marker_Id_1 0000011010000110010101000010……001010000010000100
-
Computational analysis.
The complete set of RH vectors (each RH vector characterize one marker) is clustered into RH groups using pairwise calculations with two-points linkage analysis, using dedicated RH software (18,19,20). The Lod score statistical test is applied to the whole dataset in order to assign markers to RH groups. On average, when a map of this resolution is constructed, each chromosome will be represented by one to three individuals RH groups. Lod score threshold, while empirically determined by users, relies on several factors, notably the degree of breakage from the initial radiation (ie the RH panel resolution) and the number of markers that cluster. A conventional threshold of 6.0 to 8.0 is commonly accepted as significant. RH groups are sets of markers that are linked to at least one another marker at a Lod score higher than the Lod score threshold.
Ordering markers within RH groups is then carried out using multipoint linkage analysis. This step computes the final map order and delivers inter-marker distances expressed in centiRay (cR). Resulting maps are either comprehensive maps where all markers are placed and ordered respectively to other markers or framework map when only a subset of selected markers is analyzed, the other markers being inserted between the framework markers (see Note 6).
3.9. Comparative Map Construction
Comparative genomics aims to describe the structural organization between genomes at both large-scale and micro-rearrangement level. Gene-based markers in a genome having orthologs unambiguously identified in a second genome are informative anchor sites between genomes (28). A RH map encompassing 10,000 gene-based markers (one every 250–300 kb on average) orthologous to genes mapped in reference genomes is a powerful tool for constructing comparative maps. About 90% of the human genome is in large blocks of homology with the genomes of dog and/or other mammals sequenced to date. The regions of conserved synteny reveal many genes from canine chromosomes that match blocks of genes in human chromosomes defining conserved segments (CS). Large sub-blocks that have their markers co-linearly arranged in the test RH map and in the reference sequence are termed conserved segments ordered (CSO) (28).
Compare the marker positions in the test RH map with the positions of their orthologs in the reference sequence to display chromosomal rearrangements such as inversions, translocations and duplications. The comparison of 10,000 independent gene positions between genomes will identify nearly all test/reference conserved segments greater than 500 kb(16,22).
Beyond the description of conserved segments between genomes, the RH map comprised of survey-sequencing data can be used to identify evolutionary breakpoints between CS and CSO.
Careful identification of synteny breakpoints defines regions of inter-chromosomal and intra-chromosomal rearrangement and can pinpoint genomic loci prone to chromosomal breakage and fusion.
Identify sites of rearrangement that tend to occur in regions containing duplicated sequences and gene family members. Categorization of evolutionary breakpoints classified as either lineage-specific or shared through species evolution permit derivation of ancestral genomes (29).
Orthologs corresponding to gene-based markers from new species can be searched after construction of the RH map with the alignment program Blastn. This allows integration data from new genomes for further multi-species comparative genomics analysis.
4. Notes
Acquisition software is a computer application which allows researchers to accurately and quickly score electrophoresis data from a Radiation Hybrid gel image. It displays the image of the gel on the screen, detects PCR products automatically using a specifically designed pattern recognition algorithm. Once all of the desired bands have been selected, acquisition programs such as AutoScore (available upon request to the corresponding author) allows users to save the scored data in a text file. The AutoScore application is written in Java. Therefore it will run on any hardware platform that supports Java. AutoScore is able to read and import gel images in GIF or JPEG formats. It allows researchers to overlay a resizable grid onto the displayed image. The grid can be manipulated so that the grid cells align with the bands in the image. The image is automatically rotated at an angle of 90° when it is loaded from disk. AutoScore allows the user to “click” on any grid cell in order to select the band intensity. The cell will be highlighted at three different levels (0=off (black), l=on (white), 2=maybe (gray)). Successive clicks will toggle through these three values. The resulting array of scores can be saved in a text file, including the image name.
-
RH package softwares description and distribution
TSP/CONCORDE: An approach to construction of RH maps that uses the CONCORDE software for solving the Travelling Salesman Problem can efficiently map large numbers of markers and can construct maps combining two RH panels (19). http://www.tsp.gatech.edu/concorde.html
RHMAP at the University of Michigan: Written in Fortran, multiple retention models and mapping approaches : http://csg.sph.umich.edu/boehnke/rhmap.php (30).
MultiMap at Rutgers University: Written in CLISP and C++, highly automated, equal retention model. http://compgen.rutgers.edu/multimap/multimapdist.html (18).
RHMAPPER at the Whitehead Institute/MIT Center for Genome Research: Written in C and PERL, automated, equal retention model: Z-extensions developed at the Sanger Centre provides additional functionality to RHMAPPER: ftp.sanger.ac.uk/pub/zmapper (30)
CarthaGene at INRA Toulouse France: Program based on a maximum likelihood multipoint RH and genetic data mapping tool. Haploid/diploid equal retention models with a very fast EM algorithm. Can build maps combining RH/genetic datasets. Automated C++/Tcl/Tk multi-platform (Windows/Unix) program with a rich graphic user interface. http://www.inra.fr/Internet/Departements/MIA/T/CarthaGene/index.html (32)
-
Gene-based orthologous sequence selection:
If the primary objective of a sequencing project is to generate gene-based markers for RH-mapping, 1x sequence coverage of a genome offers several advantages over large collections of ESTs. Unlike cDNA libraries, the representation of genes is unaffected by cellular expression levels, and identification of orthologous exons is not biased by the length of 3′ untranslated mRNA. In addition, the low but significant conservation of intronic sequences between species is useful for distinguishing between paralogous sequences that share substantial sequence identity within exons.
Microsatellites are tandem arrays made up of many copies of a short repeating unit typically 1–8 bp in length (25). Microsatellites have been found in 1054 gene-marker sequences to date, that have been identified by screening the 10,000 marker sequences using the sputnik program http://espressosoftware.com/pages/sputnik.jsp.
Microsatellites selected for their highly repeated motif number and the absence of point mutation that disrupt the periodic pattern have a higher chance to present allelic polymorphism. Analyses should therefore focus on such microsatellites.
-
Comprehensive and framework maps:
A comprehensive RH map is a map in which all markers are ordered using, for example, the state of the art CONCORDE algorithm. The RH map is produced using a global method that searches for local improvement, starting with an initial tour (map) defined by the nearest neighbor. The heuristics used in the CONCORDE chained Lin-Kernighan algorithm allows random “kick” to the tour (map) and reruns the tour, providing an improved solution when possible or returning back to the tour to continue on local improvement. The different algorithms used by TSP/CONCORDE package are independent in terms of computation principle, in that combinatorial and maximum likelihood approaches are both used in the analysis. Within the two approaches, variations are made to incorporate unknown entries to reduce the effect of unknowns in the quality of map produced by TSP.
Framework 1000:1 maps such as produced by RHMAPPER, Carthagene or MultiMap are not true framework maps. All alternative orders have a log likelihood within the initial map larger than an “adding threshold”. This is a weakness of all simple heuristic procedures that are based on iterative insertion processes. Furthermore, so-called framework maps place only subsets (<50%) of markers. The remaining markers are then inserted between best neighbor markers
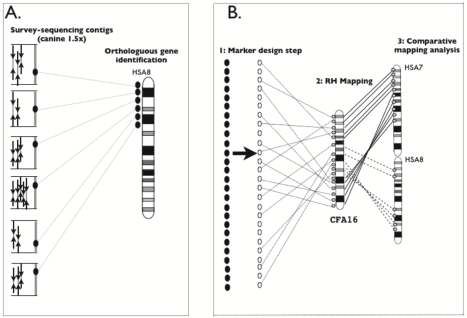
Figure. Strategy to construct comparative maps using survey-sequencing combined with dense RH gene maps.
-A- End-sequencing (black arrows) of clones from 2kb or 10 kb inserts are assembled into contigs with on average five reads per contig as represented on the left of the figure. Gene fragments, symbolized by black circles, are identified by sequence-similarity search and selected based on an even distribution on a reference genome as schematized by human chromosome 8 on the right of the figure.
-B- (1): Gene-based markers (white circles) are derived from gene fragment sequences (black circles). (2): The RH mapping analysis clusters and orders markers (represented by small grey circles) along chromosomes, here canine chromosome 16 (CFA16) shown in the middle part of panelB. (3): A human-canine comparative map for CFA16 is constructed using orthologous markers that serve as comparative anchors between the two species as shown on the right part of the figure. The comparative map identifies blocks of conserved synteny between CFA16 and HSA17 and HSA8 as well as conserved ordered segments (CSO) that contain adjacent markers in the same order and orientation.
Acknowledgments
We acknowledge all our collaborators involved in this work and for their contribution to this chapter. We acknowledge the American Kennel Club Canine Health Foundation, U.S. Army Grant DAAD19-01-1-0658 (E.A.O. and F.G.) and NIH R01CA-92167 (E.A.O, E.K. and F.G.). FG and CH are supported by the French Centre National de la Recherche Scientifique (CNRS) and by the Conseil Regional de Bretagne. This research was also supported [in part] by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.”
References
- 1.Mellersh CS, Langston AA, Acland GM, et al. A linkage map of the canine genome. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- 2.Priat C, Hitte C, Vignaux F, et al. A whole-genome radiation hybrid map of the dog genome. Genomics. 1998;54:361–378. doi: 10.1006/geno.1998.5602. [DOI] [PubMed] [Google Scholar]
- 3.Vignaux F, Hitte C, Priat C, Chuat JC, Andre C, Galibert F. Construction and optimization of a dog whole-genome radiation hybrid panel. Mamm Genome. 1999;10:888–894. doi: 10.1007/s003359901109. [DOI] [PubMed] [Google Scholar]
- 4.Mellersh CS, Hitte C, Richman M, et al. An integrated linkage-radiation hybrid map of the canine genome. Mamm Genome. 2000;11:120–130. doi: 10.1007/s003350010024. [DOI] [PubMed] [Google Scholar]
- 5.Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005;15:1831–1837. doi: 10.1101/gr.3736605. [DOI] [PubMed] [Google Scholar]
- 6.Parker HG, Ostrander EA. Canine Genomics and Genetics: Running with the Pack. PLoS Genet. 2005;1(5) doi: 10.1371/journal.pgen.0010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breen M, Bullerdiek J, Langford CF. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res. 1999;7:401–406. doi: 10.1023/a:1009224232134. [DOI] [PubMed] [Google Scholar]
- 8.Breen M, Thomas R, Binns MM, Carter NP, Langford CF. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 1999;61:145–155. doi: 10.1006/geno.1999.5947. [DOI] [PubMed] [Google Scholar]
- 9.Yang F, O’Brien PC, Milne BS, et al. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Graphodatsky AS, O’Brien PC, et al. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res. 2000;8:393–404. doi: 10.1023/a:1009210803123. [DOI] [PubMed] [Google Scholar]
- 11.Breen M, Langford CF, Carter NP, et al. FISH mapping and identification of canine chromosomes. J Hered. 1999;90:27–30. doi: 10.1093/jhered/90.1.27. [DOI] [PubMed] [Google Scholar]
- 12.Breen M, Jouquand S, Renier C, et al. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 2001;11:1784–1795. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guyon R, Lorentzen TD, Hitte C, et al. A 1-Mb resolution radiation hybrid map of the canine genome. Proc Natl Acad Sci USA. 2003;100:5296–5301. doi: 10.1073/pnas.0831002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guyon R, Kirkness EF, Lorentzen TD, et al. Building comparative maps using 1.5x sequence coverage: human chromosome Ip and the canine genome. Cold Spring Harb Symp Quant Biol. 2003;68:171–177. doi: 10.1101/sqb.2003.68.171. [DOI] [PubMed] [Google Scholar]
- 15.Breen M, Hitte C, Lorentzen TD, et al. An integrated 4249 marker FTSH/RH map of the canine genome. BMC Genomics. 2004;5:1–11. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hitte C, Madeoy J, Kirkness EF, et al. Survey sequencing combined with dense radiation hybrid gene mapping facilitates genome navigation. Nat Rev Genet. 2005;6:643–648. doi: 10.1038/nrg1658. [DOI] [PubMed] [Google Scholar]
- 17.Vignaux F, Priat C, Jouquand S, et al. Toward a dog radiation hybrid map. J Hered. 1999;90:62–67. doi: 10.1093/jhered/90.1.62. [DOI] [PubMed] [Google Scholar]
- 18.Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- 19.Agarwala R, Applegate DL, Maglott D, Schuler GD, Schaffer AA. A fast and scalable radiation hybrid map construction and integration strategy. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitte C, Lorentzen T, Guyon R, et al. Comparison of the MultiMap and TSP/CONCORDE packages for constructing radiation hybrid maps. J Hered. 2003;94:9–13. doi: 10.1093/jhered/esg012. [DOI] [PubMed] [Google Scholar]
- 21.Kirkness EF, Bafna V, Halpern AL, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- 22.Lindblad-Toh K, Wade CM, Mikkelsen T, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margulies EH, Maduro VV, Thomas PJ, et al. Comparative sequencing provides insights about the structure and conservation of marsupial and monotreme genomes. Proc Natl Acad Sci USA. 2005;102:3354–3359. doi: 10.1073/pnas.0408539102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas EE. Short, local duplications in eukaryotic genomes. Curr Opin Genet Dev. 2005;15:640–644. doi: 10.1016/j.gde.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Hardison RC. Comparative genomics. PLoS Biol. 2003;1(2) doi: 10.1371/journal.pbio.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senger F, Priat C, Hitte C, et al. The first radiation hybrid map of a perch-like fish: The gilthead seabream (Sparus aurataL) Genomics. 2006;87:793–800. doi: 10.1016/j.ygeno.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien SJ, Womack JE, Lyons LA, et al. Anchored reference loci for comparative genome mapping in mammals. Nat Genet. 1993;3:103–112. doi: 10.1038/ng0293-103. [DOI] [PubMed] [Google Scholar]
- 29.Murphy WJ, Larkin DM, Everts-van der Wind A, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 30.Boehnke M, Lange K, Cox DR. Statistical methods for multipoint radiation hybrid mapping. Am J Hum Genet. 1991;49:1174–1188. [PMC free article] [PubMed] [Google Scholar]
- 31.Soderlund C, Lau T, Deloukas P. Z extensions to the RHMAPPER package. Bioinformatics. 1998;14:538–539. doi: 10.1093/bioinformatics/14.6.538. [DOI] [PubMed] [Google Scholar]
- 32.Schiex T, Gaspin C. CARTHAGENE: constructing and joining maximum likelihood genetic maps. Proc Int Conflntell Syst Mol Biol. 1997;5:258–267. [PubMed] [Google Scholar]