Abstract
It has been proposed that during embryonic development haematopoietic cells arise from a mesodermal progenitor with both endothelial and haematopoietic potential called the haemangioblast1,2. A conflicting theory associates instead the first haematopoietic cells with a phenotypically differentiated endothelial cell with haematopoietic potential, i.e. a haemogenic endothelium3-5. Support for the haemangioblast concept was initially provided by the identification during embryonic stem (ES) cells differentiation of a clonal precursor, the blast colony-forming cell (BL-CFC), which gives rise to blast colonies with both endothelial and haematopoietic components6,7. Although recent studies have now provided evidence for the presence of this bipotential precursor in vivo8,9, the precise mechanism of generation of haematopoietic cells from the haemangioblast still remains completely unknown. Here we demonstrate that the haemangioblast generates haematopoietic cells through the formation of a haemogenic endothelium intermediate, providing the first direct link between these two precursor populations. The cell population containing the haemogenic endothelium is transiently generated during BL-CFC development. This cell population is also present in gastrulating embryos and generates haematopoietic cells upon further culture. At the molecular level, we demonstrate that the transcription factor Scl/Tal110 is indispensable for the establishment of this haemogenic endothelium population whereas the core binding factor Runx1/AML111 is critical for generation of definitive haematopoietic cells from haemogenic endothelium. Together our results merge into a single linear developmental process the two a priori conflicting theories on the origin of haematopoietic development.
The haemangioblast can be isolated from differentiated ES cells based on Flk-1 expression12 and generates a blast colony containing haematopoietic and endothelial cells after 4 days of culture6,7 (Supplementary Fig. 1). To investigate the developmental steps leading to the generation of haematopoietic cells, we followed the development of individual blast colonies by time-lapse photography. We found retrospectively that it was a sequential process divided into two stages; after 36-48 hours of culture, the haemangioblast gave rise to a tight adherent structure, then non-adherent round cells appeared and proliferated to generate a mature blast colony (Fig. 1a and Supplementary Videos 1 and 2). We observed by FACS that a high proportion of cells were positive for the endothelial marker Tie2/Tek13 after the first day (Fig. 1b and 1d). A few cells positive for CD41/αIIb integrin, which defines haematopoietic commitment both in vivo and in vitro14-16, were also detected at this stage. The percentage of CD41+ cells increased dramatically by day 2 and by day 4 most cells were CD41+Tie2- with around 30% expressing also CD45 (Fig. 1b and 1c). FACS sorting of the four cell populations defined by Tie2 and CD41 confirmed that both primitive erythroid and definitive haematopoietic potentials were confined to the CD41+ fractions (Supplementary Fig. 2). These analyses demonstrate the dynamic nature of blast colony development starting at day 1 with a subpopulation of cells expressing Tie2 to a cell population almost fully CD41+ 3 days later. Interestingly a transient Tie2hic-Kit+ cell population was detected after 48 hours of blast development (Fig. 1d) concomitant with the appearance of tight adherent structures (Fig. 1a). This Tie2hic-Kit+ population contained both CD41- and CD41+ cells (Fig. 1e). These results suggest that the Tie2hic-Kit+ cell population may represent a transitional population from which the first CD41+ cells may originate. Accordingly, the Tie2hic-Kit+CD41- fraction displayed a low but significant haematopoietic potential (Supplementary Fig. 3) suggesting that CD41+ cells may be produced from this cell population.
Figure 1. Analysis of blast colony development.
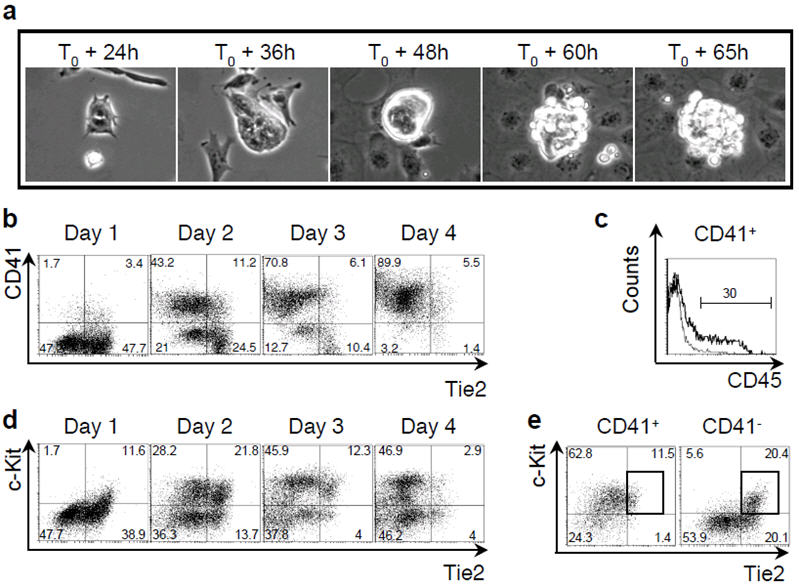
a) Phase contrast time-lapse pictures of blast colony development. b) FACS analysis of CD41 and Tie2 expression during blast colony development between day 1 and 4. c) CD45 expression of CD41+ cells at day 4 of blast colony culture. Line represents isotype control. d) FACS analysis of c-Kit and Tie2 expression during blast colony development. e) Tie2 and c-Kit expression of CD41+ (left) and CD41- (right) cells on day 2 of blast culture. Rectangles indicate Tie2hic-Kit+ population. Numbers represent percentages of respective populations.
The endothelial nature of the Tie2hic-Kit+CD41- cell population was supported by homogeneous staining for endothelial markers such as CD3117, Flk-118 and MECA3219 (Fig. 2a). Additionally these cells expressed other endothelial genes, such as CD3420, endoglin21, Flt-122 and VE-Cadherin23 (Supplementary Fig. 4a) and generated endothelial networks upon culture in matrigel plugs (Fig. 2b). Immunostaining for CD31, expressed specifically by Tie2hic-Kit+ cells at day 3 of blast development (Supplementary Fig. 4b), and for CD41 demonstrated the presence of CD31+CD41- endothelial cells corresponding to the core of tightly associated cells of blast colonies (Supplementary Fig. 4c).
Figure 2. Tie2hic-Kit+CD41- cells can generate haematopoietic progenitors.

a) FACS analyses of CD31, Flk-1 and MECA32 expression by Tie2hic-Kit+CD41- cells. b) Generation of endothelial networks in matrigel by isolated day 2 Tie2hi c-Kit+ CD41- cells. c) Tie2hic-Kit+CD41- cells at day 2 of blast development were sorted and cultured. FACS analysis of CD41 and Tie2 expression at T0 and T0 + 2 days. d) FACS analysis for the presence of Tie2+c-Kit+CD41- cells in gastrulating embryos. e) Immunostaining of gastrulating embryos for Tie2/c-Kit and (f) c-Kit/CD41. Amniotic cavity (ac), decidua (de), amnion (am), allantois (al), exocoelomic cavity (ecc), chorion (ch), and yolk sac blood islands (bi) are indicated. g) FACS analysis of CD45 and c-Kit expression (top) and May-Grunwald Giemsa staining (bottom) of cells generated by isolated E7.75 mouse embryos Tie2+c-Kit+CD41- cells co-cultured with OP9 cells. Macrophage (arrow) and mast cells (*) are indicated. Numbers indicate percentages of respective populations.
Besides its clear endothelial signature, the Tie2hic-Kit+CD41- subpopulation expressed transcription factors associated with the onset of haematopoiesis (Supplementary Fig. 5a). To test if this population contained endothelial cells with haematopoietic potential i.e. a haemogenic endothelium, Tie2hic-Kit+CD41- cells were cultured in conditions supporting the generation of haematopoietic cells from the aorta-gonad-mesonephros (AGM) region of mouse embryo24. After two days, non-adherent round cells were observed (Supplementary Fig. 5b) and FACS analysis demonstrated that around 70% of the cells expressed CD41 and that some of them had down-regulated Tie2 expression (Fig. 2c). Isolation of these newly generated CD41+ cells further confirmed that haematopoietic potential was present and restricted to this fraction (Supplementary Fig. 5c). To evaluate the proportion of Tie2hic-Kit+CD41- cells becoming CD41+, we labelled them with CFSE, a fluorescent dye equally distributed following each cell division. After 24 hours of culture, about 40% of cells were CD41+ cells with a CFSE mean fluorescence 1.5 times lower than in CD41- cells (Supplementary Fig. 5d). This result suggests that CD41+ cells were produced from around 25% (i.e. 40% divided by 1.5) of the Tie2hic-Kit+CD41- cells. By limiting dilution analysis, the frequency of Tie2hic-Kit+CD41- cells generating more differentiated haematopoietic cells was found to be around 1.2 % (Supplementary Fig. 5e). This number, lower than in the CFSE analysis, reflects the fact that only a fraction of CD41+ cells are able to respond to haematopoietic growth factors. The clonality of the haemogenic endothelium was demonstrated by generation of haematopoietic cells from single Tie2hic-Kit+CD41- cells (Supplementary Fig. 5f). Finally Tie2hic-Kit+CD41- cells did not express the mesodermal marker Brachyury25 (Supplementary Fig. 6a) and were unable to generate blast colonies (Supplementary Fig. 6b) indicating that the precursors present in this cell population were clearly distinct and downstream from haemangioblasts. Overall these data demonstrate the presence of clonal haemogenic endothelial cell precursors in the Tie2hic-Kit+CD41- cell population.
A prediction from these results is that a Tie2+c-Kit+CD41- haemogenic population should be present in vivo at the onset of blood development. We were indeed able to identify Tie2+c-Kit+CD41- cells in embryos at the neural plate stage of gastrulation and their frequencies increased in subsequent stages (Fig. 2d). Immunostainings demonstrated them to be localised in developing blood islands of early headfold embryos (Fig. 2e and 2f). FACS-isolated Tie2+c-Kit+CD41- cells plated on OP9 stromal cells generated round non-adherent cells giving rise to CD45+ cells and primitive and definitive haematopoietic colonies upon replating (data not shown and Fig. 2g). Tie2+c-Kit+CD41- cells were also detected within the AGM region of E10.5 embryos in the dorsal aorta (Supplementary Fig. 7a to 7c) and these cells also had the capacity to generate haematopoietic cells (Supplementary Fig. 7d and 7e), but the origin of these Tie2+c-Kit+CD41- cells in the AGM and their precise developmental potential remain to be characterized. Altogether these data suggest that the Tie2+c-Kit+CD41- progenitor population detected in gastrulating embryos may represent an intermediate between the haemangioblast, predominantly found in the primitive streak8, and the haematopoietic precursors found in the yolk sac26.
To further investigate the molecular mechanisms implicated in the generation of the Tie2+c-Kit+CD41- population, we analyzed the effects on this cell population of knockout of two critical genes for early haematopoiesis, Runx1/AML1 and Scl/Tal1. The transcription factor Runx1 is required in vivo for the generation of definitive haematopoietic cells11 and its absence results in vitro in the generation of 20 times fewer blast colonies, with residual blast colonies restricted to a primitive haematopoietic fate27. We performed a time-lapse analysis of blast colony development from isolated Runx1-/- Flk-1+ cells. Clusters of tightly associated adherent cells were observed (Fig. 3a) but only a few of these clusters later generated blast colonies (data not shown). Cells were observed to emerge from most clusters but died instead of proliferating (Fig. 3a and Supplementary Video 3). Immunofluorescence analyses confirmed that cells within these clusters expressed CD31 but that no CD41+ cells were present (Supplementary Fig. 8). The defect in haematopoietic development was further observed by FACS analysis which showed a marked reduction in the frequency of CD41+ cells (Fig. 3b) and an increased frequency in the Tie2hic-Kit+ cell population (Fig. 3c).
Figure 3. Runx1 requirement in blast colony development.
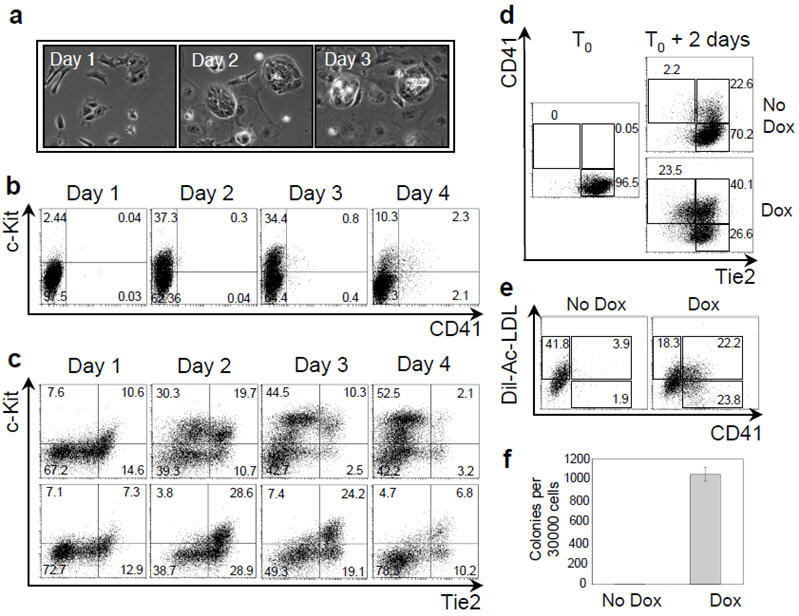
a) Phase contrast time-lapse photographs of Runx1-/- blast colony development. b) FACS analysis of CD41 and c-Kit expression during Runx1-/- blast colony development between day 1 and 4. c) FACS analysis of Tie2 and c-Kit expression during Runx1+/+ (top) and Runx1-/- (bottom) cells blast colony development. d) FACS analysis of Tie2/CD41 expression after 2 days of culture of iRunx1 Runx1-/- Tie2hic-Kit+CD41- cells in absence or presence of doxycycline (dox) (0.1 μg/ml). e) Dil-Ac-LDL and CD41 expression analysis of iRunx1 Runx1-/- Tie2hic-Kit+CD41- cells were evaluated in absence (left) and presence of doxycycline (right). Numbers indicate the percentage of the respective populations. f) Numbers of definitive haematopoietic colonies generated in methylcellulose by iRunx1 Runx1-/- cells harvested after 2 days of culture with or without doxycycline. Error bars indicate standard deviation of the mean (n=3).
To further investigate the requirement for Runx1, we generated a Runx1-/- ES cell line in which Runx1 expression can be induced by addition of doxycycline (iRunx1 Runx1-/-). When doxycycline was added at day 2 of blast development, at least a 10-fold increase in CD41+ cell frequency was observed after 24 hours, associated with downregulation of Tie2 expression (Supplementary Fig. 9a) and induction of expression of genes involved in myeloid cell development (Supplementary Fig. 9b). To establish if the developmental block observed in the absence of Runx1 was at the level of the Tie2hic-Kit+CD41- cell population, we isolated these cells and cultured them in the presence of doxycycline. After 48 hours, around 60% of the cells expressed CD41 and one third of these cells down-regulated Tie2 expression (Fig. 3d). Additionally the CD41+ cells incorporated acetylated LDL, which is taken up by endothelial cells28, further supporting their endothelial origin (Fig. 3e). Finally definitive haematopoietic precursors were exclusively detected in colony assays in cultures induced with doxycycline (Fig. 3f). Altogether these results demonstrate that the Tie2hic-Kit+CD41- haemogenic endothelial cell population is generated in absence of Runx1 but that Runx1 is indispensable for the generation of definitive haematopoietic cells from this population.
Accordingly Runx1-/- Flk-1+ cells generated significantly more colonies of tightly associated cells than wild type cells in BL-CFC clonogenic assays (Supplementary Fig. 10). Their number was inversely correlated with the number of blast colonies observed with wild type cells (Supplementary Fig. 11a). The presence of haemogenic endothelium in these tight structures was supported by detection of a large proportion of CD31+ or Tie2+c-Kit+ cells in individual colonies by immunostaining (Supplementary Fig. 8 and 11b respectively) and of Tie2hic-Kit+CD41- cells by FACS analysis on pooled colonies (Supplementary Fig. 11c). Additionally around 75% of individual colonies (26 out of 34) were able upon Runx1 re-expression to give rise to haematopoietic cells (Supplementary Fig. 11d). These data support the notion that these tight colonies generated from the haemangioblast contain cells with haemogenic endothelium potential that are unable to initiate haematopoiesis in the absence of Runx1.
Scl/Tal1 is another critical regulator of haematopoiesis as Scl-/- cells are unable to generate either primitive or definitive haematopoiesis10. Plating of Scl-/- Flk-1+ cells resulted, as previously shown29, in the absence of blast colonies but also in a complete lack of clusters of tightly associated adherent cells (Fig. 4a). We were unable to detect any CD41+ cells (Fig. 4b). Although Tie2 was expressed at levels similar to Scl+/+ cells, only a very small fraction of cells co-expressed Tie2 and c-Kit (Fig. 4c) and none of these cells expressed CD31, Flk-1 or MECA32 (data not shown). These data indicate that Scl is critical for the generation of Tie2hic-Kit+CD41- haemogenic endothelium population and place its role in haematopoietic specification prior to Runx1 requirement.
Figure 4. SCL requirement during blast colony development.
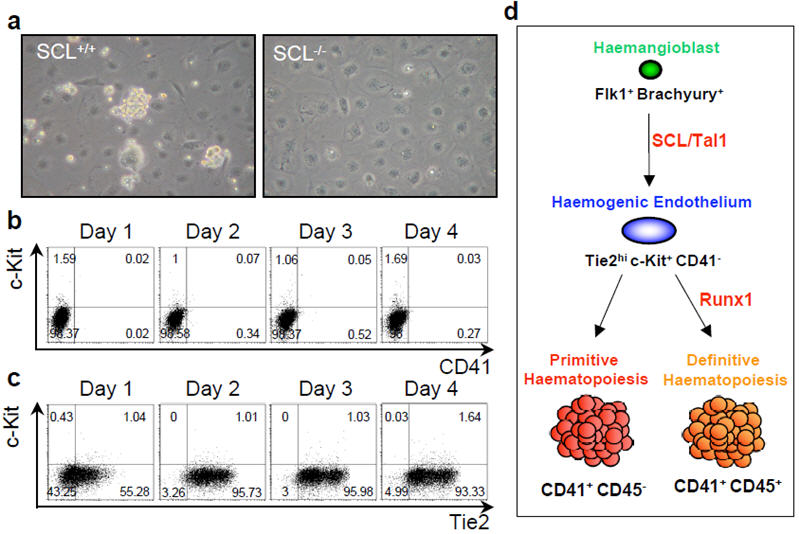
a) Phase contrast pictures of day 3 blast colony culture from Scl+/+ and Scl-/- ES cells. b) FACS analysis of CD41 and c-Kit expression during Scl-/- blast colony development. Numbers indicate the percentage of the respective populations. c) FACS analysis of Tie2 and c-Kit expression during Scl-/- blast colony development. d) Model of haemangioblast differentiation toward haematopoiesis in the yolk sac.
In summary, our data provide the first evidence for a direct link between the haemangioblast and a downstream haemogenic endothelium. The generation of this cell population is characterised by the up-regulation of c-Kit in a Tie2hi population and this process is Scl/Tal1 dependent (Fig. 4d). The consecutive generation of definitive haematopoietic cells, characterised by CD41 expression and Tie2 down-regulation requires the transcription factor Runx1 whereas primitive haematopoiesis is Runx1 independent. Identification of these discrete developmental steps will provide the opportunity to further explore the molecular regulation of haematopoietic development.
Methods Summary
Time lapse analyses were performed in a Solent Scientific environmental chamber kept at 37°C for the duration of the analysis and observed under a Zeiss Axiovert 200M microscope. Brachyury-GFP ES25, Scl-/-10, Ainv1830, Ainv18 Runx1+/-, Runx1-/- and iRunx1 Runx1-/- ES cell lines were used. Flow cytometry and cell sorting were performed on a FACSCalibur (Becton Dickinson) and FACSVantage or FACSAria cell sorters (Becton Dickinson). For immunostainings, frozen sections were fixed, blocked, incubated with primary Tie2, c-Kit and CD41 antibodies, then with secondary antibodies, mounted and observed under a Zeiss Axiovert 200M microscope.
Full methods accompany this paper.
Methods
Embryonic stem cell growth and differentiation
Brachyury-ES25, Scl-/-10, Ainv1830, Ainv18 Runx1+/-, Runx1-/- and iRunx1 Runx1-/- ES cell lines (data not shown) were used. Growth and differentiation of ES cells were performed as described previously25.
Embryo generation
Time matings of ICR mice were set up and the morning of vaginal plug detection was considered day 0.5. Gastrulating embryos were staged by morphological landmarks. All animal work was performed under regulation in accordance with the United Kingdom’s Animal Scientific Procedures Act (ASPA) 1986.
Adherent BL-CFC and haemogenic endothelium cultures
Flk-1+ EB cells were plated on gelatin in the BL-CFC media described previously25. For haemogenic endothelium culture, Tie2+c-Kit+CD41- cells were cultured either on gelatin (cells isolated from day 2 or day 3 BL-CFC cultures) or on OP9 stromal cells (cells isolated from embryos) in conditions described before24. Cultures were maintained in a humidified chamber in a normal or low O2 (5%) 5% CO2-air mixture at 37°C.
Matrigel plug assays
Tie2+c-Kit+CD41- cells isolated from day 2 BL-CFC liquid culture were transferred to 50 μl Matrigel plugs in 96-well plates. Matrigel (BD Biosciences) was diluted 1:1 with IMDM supplemented with 10% FCS, 50 ng/ml VEGF, 5 ng/ml bFGF and allowed to solidify at 37°C before the addition of cells. Cultures were maintained at 37°C, 5% CO2, 5% O2 for 5 days.
Flow cytometry and cell sorting
Staining was done as described previously25 and analyses were performed with a FACSCalibur (Becton Dickinson). Sorts were performed with a FACSVantage or a FACSAria (Becton Dickinson). Monoclonal antibodies and streptavidin used were Flk-1-bio25, CD31-bio (MEC13.3, BD Bioscience), MECA32-bio (BD Bioscience), Tie2-bio (TEK4, eBioscience), Tie2-PE (TEK4, eBioscience), c-Kit-APC (2B8, eBioscience), CD41-FITC (MWReg30, BD Bioscience), Strep-PECy5 (BD Bioscience) and Strep-PECy7 (eBioscience). Tie2+c-Kit+CD41- cells from iRunx1 Runx1-/- day 3 blast culture were incubated with Dil-Ac-LDL (10μg/ml; Invitrogen Molecular Probes) for 3 hours, washed twice to remove all Dil-Ac-LDL and Runx1 expression was induced with doxycycline. FACS analysis was performed two days later.
Carboxyfluorescein succinimidyl ester (CFSE) labelling
Tie2+c-Kit+CD41- cells were resuspended in PBS 5% FCS at 0.5×106 cells/ml and CFSE was added to 5 μM final concentration. Cells were incubated for 5 min at room temperature, washed three times and put back in cultures.
Limiting dilution and single cell analyses
Defined numbers (400, 200, 100 and 50 cells) of Tie2hic-Kit+CD41- cells were plated on OP9 stromal cells using a FACSAria equipped with an automatic cell deposition unit (Becton Dickinson). For each cell number a minimum of 24 wells were seeded. The cells were first cultured for 2 days in conditions supporting the transition of haemogenic endothelial cells to haematopoietic progenitors24 and subsequently in conditions supporting the differentiation to mature haematopoietic cells. The wells were scored for the presence of haematopoietic cells two days later. The haematopoietic nature of the cells was confirmed by May-Grunwald Giemsa staining. The fraction of wells negative for haematopoietic cells were plotted against each dilution and the linear regression was calculated with the Origin software (OriginLab). To obtain the frequency of Tie2hic-Kit+CD41- cells able to generate haematopoietic cells we used the limdil function of the statistical package R (http://bioinf.wehi.edu.au/software/limdil/index.html). For single cell analysis, single Tie2hic-Kit+CD41- cells were sorted on 96 well-plates coated with gelatin. The culture conditions were as described above and the wells were scored for the presence of haematopoietic cells after 8 days. The haematopoietic nature of the cells was confirmed by May-Grunwald Giemsa staining.
Immuno-histochemistry
7-10 μm sections were cut with a Leica CM3050S cryostat from dissected frozen embryos embedded in OCT (Tissue Tek). Sections were fixed with acetone, incubated with 3% H2O2, blocked with 10% goat serum (DAKO) and Avidin/biotin blocking kit (Vector). Primary antibodies, Tie2-Bio (TEK4, eBioscience), c-Kit (ACK2, eBioscience) and CD41-Bio (MWReg30, eBioscience) were incubated overnight at 4°C. Incubations with secondary Streptavidin-AlexaFluor555 (Invitrogen) and goat anti-rat IgG2b-HRP (ABD Serotec) were followed by AlexaFluor488 tyramide signal amplification (Invitrogen). The sections were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen), analysed using Zeiss Axiovert 200M microscope and processed with Metamorph software (Universal Imaging).
Time-lapse photography
An Axiovert 200M microscope equipped with a Zeiss x10 objective lens, a Roper Coolsnap HQ Camera and Solent Scientific environmental chamber kept at 37°C for the duration of the experiment was used for time-lapse analysis. Phase contrast images were taken every minute (supplementary videos 1 and 3) or every 5 minutes (supplementary video 2) with the image capture and processing software MetaMorph (Universal Imaging). Video animations were made using the software Imaris (Bitplane). The movies were edited and compressed using Final Cut Studio 2 (Apple).
Gene Expression Analysis
For gene specific PCR, total RNA was extracted from each sample with an RNeasy mini kit and treated with RNAse-free DNase (Qiagen). Five hundred to 1000 ng of total RNA were reverse-transcribed into cDNA with random hexamer using an Omniscript RT kit (Qiagen). The PCR reactions were performed using GoTaq (Promega) and 0.2 μM of each primer. Cycling conditions were as follows; 94°C for 5 min followed by 30-35 cycles of amplification (94°C denaturation for 30 sec, 60°C annealing for 30 sec, 72°C elongation for 60 sec) with a final incubation at 72°C for 10 min. The list of primers is available upon request.
Supplementary Material
Acknowledgements
We thank Karim Labib, Crispin Miller and Tim Somervaille for critical reading of the manuscript, Jeff Barry and Michael Hughes for cell sorting, Steve Bagley for help with the time-lapse photography, Garry Ashton for help with preparation of sections and Laetitia Gautreau for advice with the OP9 cultures. Cancer Research UK supported this work.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Sabin FR. Studies on the origin of blood vessels and of red corpuscules as seen in the living blastoderm of the chick during the second day of incubation: contributions to embryology. Contrib Embryol. 1920;9:213–262. [Google Scholar]
- 2.Murray PDF. The development in vitro of the blood of the early chick embryo. Proc R Soc London. 1932;11:497–521. [Google Scholar]
- 3.Jaffredo T, Gautier R, Eichmann A, Dieterlen-Lievre F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 1998;125:4575–4583. doi: 10.1242/dev.125.22.4575. [DOI] [PubMed] [Google Scholar]
- 4.Nishikawa SI, et al. In vitro generation of lymphohematopoietic cells from endothelial cells purified from murine embryos. Immunity. 1998;8:761–769. doi: 10.1016/s1074-7613(00)80581-6. [DOI] [PubMed] [Google Scholar]
- 5.North TE, et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy M, et al. A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature. 1997;386:488–493. doi: 10.1038/386488a0. [DOI] [PubMed] [Google Scholar]
- 7.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 8.Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- 9.Vogeli KM, Jin SW, Martin GR, Stainier DY. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature. 2006;443:337–339. doi: 10.1038/nature05045. [DOI] [PubMed] [Google Scholar]
- 10.Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 11.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 12.Faloon P, et al. Basic fibroblast growth factor positively regulates hematopoietic development. Development. 2000;127:1931–1941. doi: 10.1242/dev.127.9.1931. [DOI] [PubMed] [Google Scholar]
- 13.Dumont DJ, Yamaguchi TP, Conlon RA, Rossant J, Breitman ML. tek, a novel tyrosine kinase gene located on mouse chromosome 4, is expressed in endothelial cells and their presumptive precursors. Oncogene. 1992;7:1471–1480. [PubMed] [Google Scholar]
- 14.Ferkowicz MJ, et al. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130:4393–4403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- 15.Mikkola HK, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–516. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Ferkowicz MJ, Johnson SA, Shelley WC, Yoder MC. Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem Cells Dev. 2005;14:44–54. doi: 10.1089/scd.2005.14.44. [DOI] [PubMed] [Google Scholar]
- 17.Newman PJ, et al. PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science. 1990;247:1219–1222. doi: 10.1126/science.1690453. [DOI] [PubMed] [Google Scholar]
- 18.Hirai H, et al. Hemogenic and nonhemogenic endothelium can be distinguished by the activity of fetal liver kinase (Flk)-1 promoter/enhancer during mouse embryogenesis. Blood. 2003;101:886–893. doi: 10.1182/blood-2002-02-0655. [DOI] [PubMed] [Google Scholar]
- 19.Hallmann R, Mayer DN, Berg EL, Broermann R, Butcher EC. Novel mouse endothelial cell surface marker is suppressed during differentiation of the blood brain barrier. Dev Dyn. 1995;202:325–332. doi: 10.1002/aja.1002020402. [DOI] [PubMed] [Google Scholar]
- 20.Young PE, Baumhueter S, Lasky LA. The sialomucin CD34 is expressed on hematopoietic cells and blood vessels during murine development. Blood. 1995;85:96–105. [PubMed] [Google Scholar]
- 21.Li DY, et al. Defective angiogenesis in mice lacking endoglin. Science. 1999;284:1534–1537. doi: 10.1126/science.284.5419.1534. [DOI] [PubMed] [Google Scholar]
- 22.Fong GH, Rossant J, Gertsenstein M, Breitman ML. Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature. 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 23.Breier G, et al. Molecular cloning and expression of murine vascular endothelial-cadherin in early stage development of cardiovascular system. Blood. 1996;87:630–641. [PubMed] [Google Scholar]
- 24.Mukouyama Y, et al. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta-gonadmesonephros region. Dev Biol. 2000;220:27–36. doi: 10.1006/dbio.2000.9617. [DOI] [PubMed] [Google Scholar]
- 25.Fehling HJ, et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 26.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 27.Lacaud G, et al. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood. 2002;100:458–466. doi: 10.1182/blood-2001-12-0321. [DOI] [PubMed] [Google Scholar]
- 28.Voyta JC, Via DP, Butterfield CE, Zetter BR. Identification and isolation of endothelial cells based on their increased uptake of acetylated-low density lipoprotein. J Cell Biol. 1984;99:2034–2040. doi: 10.1083/jcb.99.6.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Souza SL, Elefanty AG, Keller G. SCL/Tal-1 is essential for hematopoietic commitment of the hemangioblast but not for its development. Blood. 2005;105:3862–3870. doi: 10.1182/blood-2004-09-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


