Abstract
The activation of cAMP-responsive element-binding protein (CREB)-dependent gene expression is thought to be critical for the formation of different types of long-term memory. To explore the consequences of chronic enhancement of CREB function on spatial memory in mammals, we examined spatial navigation in bitransgenic mice that express in a regulated and restricted manner a constitutively active form of CREB, VP16-CREB, in forebrain neurons. We found that chronic enhancement of CREB activity delayed the acquisition of an allocentric strategy to solve the hidden platform task. The ability to turn on and off transgene expression allowed us to dissect the role of CREB in dissociable memory processes. In mice in which transgene expression was turned on during memory acquisition, turning off the transgene re-established the access to the memory trace, whereas in mice in which transgene expression was turned off during acquisition, turning on the transgene impaired memory expression in a reversible manner, indicating that CREB enhancement specifically interfered with the retrieval of spatial information. The defects on spatial navigation in mice with chronic enhancement of CREB function were not corrected by conditions that increased further CREB-dependent activation of hippocampal memory systems, such as housing in an enriched environment. These results along with previous findings in CREB-deficient mutants indicate that the relationship of CREB-mediated plasticity to spatial memory is an inverted-U function, and that optimal learning in the water maze requires accurate regulation of this pathway.
Genetic and pharmacological studies have demonstrated that the activation of the cAMP-responsive element-binding protein (CREB) pathway, important in memory storage in invertebrates, also plays an active role in memory storage in mammals (Kandel 2001; Barco et al. 2003; Josselyn and Nguyen 2005). However, critical aspects of the role of CREB in learning and memory in mammals are still unclear. In particular, the role of CREB in spatial navigation remains controversial. Numerous studies have shown that spatial memory formation is associated with increased CREB phosphorylation within the hippocampus (Mizuno et al. 2002; Colombo et al. 2003; Moncada and Viola 2006; Porte et al. 2008). Also supporting a role for CREB in hippocampal-dependent spatial memory, mice homozygous for a deletion of the α and δ isoforms of CREB were originally reported to have a specific deficit in long-term memory revealed in several memory tasks, including the Morris water maze (Bourtchuladze et al. 1994). This study represented the first evidence for a role of CREB in memory formation in rodents and was soon confirmed by experiments in rats, in which the intrahippocampal infusion of CREB antisense oligos caused deficits in spatial learning (Guzowski and McGaugh 1997). However, further analyses of the CREB hypomorphic mutants demonstrated that the spatial memory defect was sensitive to genetic background (Graves et al. 2002). Indeed, the analysis of four different strains of CREB-deficient mice (including the CREBαδ−/− hypomorphic mutant) in the water maze failed to demonstrate any specific deficit in spatial memory (Balschun et al. 2003). The apparent deficits previously described were better explained by an increment in thigmotaxis behavior rather than impaired spatial learning. Part of these discrepancies may result from compensation of CREB deficiency by up-regulation of other transcription factors belonging to the same family (Blendy et al. 1996; Mantamadiotis et al. 2002). In contrast to the most recent studies on CREB hypomorphic and brain-restricted knockout animals (Balschun et al. 2003), the induction of a dominant negative CREB mutant in the dorsal hippocampus of transgenic animals produced spatial memory deficits that could be reversed after turning the transgene off (Pittenger et al. 2002).
To further explore the role of CREB in behavior we have turned to gain-of-function approaches. These have contributed importantly to clarify the role of CREB in the consolidation of different forms of memory (Josselyn et al. 2001; Mouravlev et al. 2006; Brightwell et al. 2007; Restivo et al. 2008; Viosca et al. 2009), but have not as yet been applied to explore the role of CREB-dependent gene expression in spatial memory in the Morris water maze. To explore the consequences of chronic enhancement of CREB function on spatial memory, we examine spatial navigation in bitransgenic mice that express in a regulated and restricted manner a constitutively active form of CREB, VP16-CREB, whose strong transactivation activity is independent of the signaling cascades that regulate wild-type CREB (Barco et al. 2002). We and others have found that VP16-CREB expression up-regulated the expression of a number of CREB target genes (Barco et al. 2005), reduced the threshold for late-phase LTP (Barco et al. 2002; Marie et al. 2005; Alarcon et al. 2006), and increased neuronal excitability (Dong et al. 2006; Han et al. 2006; Lopez de Armentia et al. 2007). Here we find that the chronic enhancement of CREB activity interferes with retrieval of complex spatial associations in a reversible manner. Our results, together with previous loss-of-function studies (Bourtchuladze et al. 1994; Pittenger et al. 2002), support a role for CREB-dependent gene expression in hippocampus-dependent learning and memory, but indicate that optimal learning in the water maze requires accurate regulation of this pathway so that either prolonged or excessive overexpression of CREB activity interferes with optimal hippocampal-based memory storage and expression.
Results
Constitutive activation of CREB during learning causes spatial memory deficits
To investigate the consequences of chronic enhancement of CREB function in spatial navigation and memory, we used a bitransgenic mouse strain, VP16-CREBhigh, that expresses in a regulatable manner a constitutively active CREB variant. In this strain, strongest transgene expression occurs in pyramidal neurons of the CA1 subfield and in granular cells of the dentate gyrus, but VP16-CREB is also expressed in other forebrain regions (Barco et al. 2002).
The Morris water maze is a widely used tool to assess hippocampus-dependent learning and memory in rodents. We first tested VP16-CREBhigh mice in the visible platform task (Fig. 1A). We did not observe significant differences between genotypes in path length (Fig. 1B; F(1,54)genotype = 0.340, P = 0.562; F(1,54)genotype × session = 0.002, P = 0.968), escape latencies (Supplemental Fig. S1A), or swimming speed (Supplemental Fig. S1B), indicating normal vision, motivation, and locomotor activity and coordination. Thigmotaxis was also similar in bitransgenic mice and control littermates, indicating a similar level of anxiety (Supplemental Fig. S1C). The lack of phenotypes in the visible platform task enabled the specific assessment of spatial learning and memory using the hidden platform task, since both tasks have similar performance requirements.
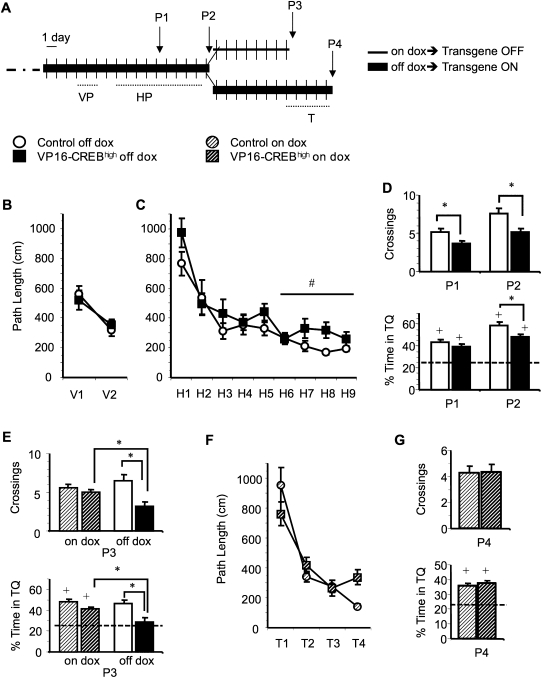
Figure 1.
VP16-CREBhigh mice show impaired spatial memory in the water maze. (A) Schematic summary of the experimental protocol. VP16-CREBhigh (n = 35) and control mice (n = 21) were trained in the visible platform (VP) and hidden platform (HP) tasks using a four-trial per day protocol. Dox was removed from the mouse diet 3 d before starting the VP task (8 d before training in the HP task). Probe trials to assess memory acquisition were performed before training on day H5 (P1) and 24 h after concluding training in the hidden platform task (P2). After P2 (day 10 of the HP task), a randomly selected subgroup of VP16-CREBhigh mice (18) and control littermates (15) were fed again with dox food to turn off transgene expression and assess the reversal of memory deficits. The remaining 23 mice (17 VP16-CREBhigh mice and six control siblings) were kept off dox. One week later, we performed an additional probe trial (P3) and started to train the same mice using a new platform location. (T) Transfer task. A last probe trial (P4) was performed 24 h after training on T4. (B) Mice from both genotypes showed similar acquisition curves during training in the visible platform task. (C) Both genotypes also showed similar learning during the first week of training in the hidden platform task. However, VP16-CREBhigh mice failed to improve their performance as much as control littermates during the second week of training. (D) Spatial memory for the HP task was assessed in two probe trials (P1 and P2). Mice expressing constitutively active CREB spent less time in the TQ in P2 than control siblings. The analysis of annulus crossings revealed significant deficits both in P1 and in P2 (top), whereas the analysis of percentage of time in TQ only revealed significant changes in P2 (bottom). (E) The memory defect of bitransgenic mice was readily reversed by turning off transgene expression before performing a third probe trial (P3). This reversal was observed both in the number of annulus crossings (top) and percentage of time in the TQ (bottom). (Left) Mice on dox (transgene turned off); (right) mice off dox (transgene turned on). (F) Bitransgenic mice fed with dox after the second probe trial performed similarly to control littermates in the transfer task. (G) Bitransgenic animals did not show significant defects in annulus crossings (top) and time spent in the TQ (bottom) in the fourth probe trial. (#) Significant difference between genotypes in sessions H5–H9 (P < 0.05, repeated measures ANOVA); (*) significant differences between groups (P < 0.05, t-test); (+) significantly different of the chance value 25% (P < 0.05, t-test).
We therefore assessed acquisition and expression of spatial memory in VP16-CREBhigh mice using the hidden platform version of the task. Again, we did not observe any significant difference in thigmotaxis or swimming speed (Supplemental Fig. S1B,C). Both VP16-CREBhigh off dox mice and their control littermates showed significant improvement over training. Repeated measures ANOVA did not reveal significant differences on path length (Fig. 1C; F(1,54)genotype = 3.810, P = 0.056) or escape latencies (Supplemental Fig. S1A; F(1,54)genotype = 1.546, P = 0.219) over the 9 d of training. Interestingly, both genotypes showed a similar performance during the first week of training (ANOVApath length from H1 to H5: F(1,54)genotype = 0.863, P = 0.357; ANOVAlatency from H1 to H5: F(1,54) genotype = 0.132, P = 0.718), but bitransgenic animals began to drift apart during the second week (ANOVApath length from H5 to H9: F(1,54)genotype = 7.648, P = 0.008; ANOVAlatency from H5 to H9: F(1,54)genotype = 4.178, P = 0.046). Since different strategies are engaged during early and late stages of training in the hidden platform task, this result may suggest that enhanced CREB function interfered specifically with the acquisition of an allocentric spatial strategy during the second week of training. Moreover, VP16-CREBhigh mice showed significant deficits in the two probe trials (P1 and P2) performed on days 5 and 10 of training (Fig. 1D). In P1, VP16-CREBhigh mice displayed a lower number of crossings in the target annulus (control = 5.1 ± 0.4, VP16-CREBhigh = 3.6 ± 0.3, t(54) = 2.434, P = 0.018). P2 revealed an even more marked defect measured both in annulus crossings (control = 7.6 ± 0.6, VP16-CREBhigh = 5.1 ± 0.4, t(37) = 3.165, P = 0.003) and target quadrant occupancy (Fig. 1D; control = 58 ± 2%, VP16-CREBhigh = 48 ± 3%, t(54) = 2.840, P = 0.006).
These results show that chronically enhanced neuronal excitability (Lopez de Armentia et al. 2007) and a reduced threshold for L-LTP (Barco et al. 2002) do not necessarily lead to enhanced learning. In VP16-CREB mice, these traits were indeed associated with mild impairments in spatial learning and memory.
Turning off transgene expression reverses the spatial learning and memory deficits
We next assessed whether the spatial memory defect observed in VP16-CREBhigh mice was a direct consequence of the changes in CREB function and not a manifestation of altered brain development or brain damage in this mutant strain. This experiment was particularly relevant because we have previously shown that the sustained and strong enhancement of CREB function for several weeks can trigger epileptic seizures and, eventually, cause neuronal loss in the hippocampus. It should be noted, however, that our behavioral experiments were performed at a time in which no gross neuronal damage was detected (Lopez de Armentia et al. 2007).
Previous experiments have demonstrated that 1 wk of dox administration is sufficient to suppress transgene expression and reverse the electrophysiological properties of hippocampal neurons back to normal (Barco et al. 2002; Lopez de Armentia et al. 2007). To test whether the spatial memory defect was also reversed with this treatment, we fed part of the mice trained in the water maze experiment described above with dox-supplemented food (Fig. 1A). Dox administration started immediately after probe trial P2 (18 d off dox). A subset of the bitransgenic mice and control littermates were kept off dox as control for reversal. After resting in their homecages for a week without receiving any further training, the mice were tested in a third probe trial. The two groups of control mice (paired t-test: control “On/Off”: P2 = 57 ± 3%, P3 = 48 ± 4%, t(14) = 2.441, P = 0.028; control “On”: P2 = 62 ± 6%, P3 = 46 ± 4%, t(5) = 6.089, P = 0.002) and VP16-CREBhigh mice maintained off dox (paired t-test: VP16-CREBhigh “On”: P2 = 48 ± 3%, P3 = 29 ± 4%, t(16) = 6.000, P < 0.001) performed worse in P3 than in P2. In contrast, the group VP16-CREBhigh “On/Off” did not show significant forgetting of the platform location (paired t-test: VP16-CREBhigh On/Off: P2 = 48 ± 3%, P3 = 41 ± 3%, t(17) = 1.614, P = 0.125). In agreement with these observations, the comparison between genotypes revealed a recovery of the deficits in VP16-CREB “On/Off” mice (Fig. 1E, top, left: control = 48 ± 3%, VP16-CREBhigh = 41 ± 3%, t(31) =1 .368, P = 0.181; bottom, left: control = 5.6 ± 0.6 crossings, VP16-CREBhigh = 4.94 ± 0.6 crossings, t(31) = 0.730, P = 0.471), while VP16-CREB “On” mice maintained a poor performance (Fig. 1E, top, right: control = 45 ± 3%, VP16-CREBhigh = 28 ± 3%, t(21) = 2.515, P = 0.020; bottom, right: control = 6.5 ± 0.8 crossings, VP16-CREBhigh = 3.2 ± 0.5 crossings, t(21) = 3.311, P = 0.003). Furthermore, VP16-CREBhigh “On” mice showed significant deficits when compared with VP16-CREBhigh “On/Off” mice (t(33)crossings = −2.084, P = 0.045; t(33) %Time = −2.395, P = 0.022), but there was no difference between the two groups of control littermates. Two important conclusions can be drawn from this experiment: (1) The similar performance of VP16-CREB mice and control littermates fed with dox (“On/Off” groups) indicated that the memory deficits detected in the two first probe trials were a direct effect of transgene expression and could thereby be reversed; (2) the significant difference between VP16-CREB “On/Off” and VP16-CREB “On” mice in the absence of additional training suggested that either forgetting was reduced in VP16-CREB “On/Off” or that the chronic enhancement of CREB function interfered specifically with memory retrieval, rather than with memory acquisition.
We completed this water maze experiment testing whether VP16-CREB “On/Off” and control littermates were able to learn a new platform location (transfer task) to assess whether the learning deficit, like the memory impairment, could be reversed. We did not observe significant differences between genotypes either in the escape latency or path-length curves during training in the transfer task (Fig. 1F; path-length ANOVA, F(1,31)genotype = 0.128, P = 0.723; latency ANOVA, F(1,31)genotype = 0.068, P = 0.796) or in the probe trial performed after 4 d of training (Fig. 1G). These results support the view that the impairments were largely due to direct effects of VP16-CREB expression.
VP16-CREB interferes with spatial learning by mechanisms that are independent of transgene integration site and the late deleterious effects of chronic CREB activation
To strengthen the evidence indicating that the impairment of spatial learning was a direct consequence of transgene expression and confirm that transient transgene expression did not cause an irreversible impairment of hippocampal function, we carried out an independent water maze experiment in which we assessed the performance of bitransgenic mice that had expressed the transgene for 2 wk previous to training in the water maze (Fig. 2A). Under those circumstances, bitransgenic mice did not show any impairment and behaved as control littermates during training (Fig. 2B). Also, no difference was observed in the two probe trials (Fig. 2C; P1, t(12) = −0.125, P = 0.902; P2, t(12) = −2.007, P = 0.070).
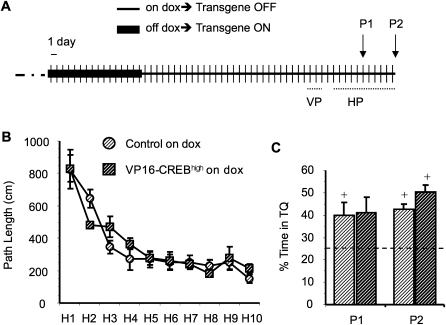
Figure 2.
Memory impairment depends on transgene expression. (A) Schematic summary of the experiment. VP16-CREBhigh (n = 6) and control mice (n = 8) were trained in the visible platform (VP, data not shown) and hidden platform (HP) tasks using a four-trial per day protocol. Both groups of mice were kept off dox for 15 d before starting the VP task. Probe trials to assess memory acquisition were performed before training on day H5 (P1) and 24 h after concluding training in the water maze (P2). (B) VP16-CREBhigh mice that had expressed the transgene for 2 wk acquired the task at a similar rate as control littermates. (C) No difference was observed in the two probe trials. (+) A percentage of time statistically different from the chance value 25% (P < 0.05, t-test).
The effective reversion of learning and memory deficits by dox together with their early onset after dox removal indicate that the strong enhancement of neuronal CREB activity in VP16-CREBhigh mice interferes with spatial learning and memory by mechanisms that are independent of the neurodegenerative process observed in this mouse strain at later times. In agreement with this view, the analysis of an independent bitransgenic strain expressing a lower level of VP16-CREB, VP16-CREBlow mice, which also showed enhanced excitability in CA1 neurons and facilitated LTP in the Schaffer-collateral pathway, but no apparent cell loss (Lopez de Armentia et al. 2007), also revealed learning impairments in the water maze, although milder than those observed in VP16-CREBhigh mice (Fig. 3). The same four-trial per day protocol described for VP16-CREBhigh mice did not reveal significant differences between VP16-CREBlow mice and their control littermates either in the path-length curve or in the two probe trials (Fig. 3B,C). However, VP16-CREBlow mice did not show a preference for the target quadrant in the first probe trial (P1 = 30.8 ± 4.9% against 25%, t(7) = 1.193, P = 0.269), whereas their control siblings already showed a significant preference (P1 = 39.4 ± 5.5% against 25%, t(7) = 2.628, P = 0.035). To confirm the existence of these mild defects in an independent experiment, VP16-CREBlow mice and control littermates were trained using a two-trial per day protocol (Fig. 3D,E). The reduced training revealed a clear delay in the path-length curve of VP16-CREBlow mice (Fig. 3D; F(1,16)genotype = 5.186, P = 0.037). Given the greater difficulty of this task, neither bitransgenic nor control mice showed significant memory for the platform location in P1. Also, no significant difference between genotypes was observed in the second probe trials (Fig. 3E). The milder spatial learning impairment of VP16-CREBlow mice correlates with the lower level of expression and weaker electrophysiological phenotypes observed in this mouse strain.
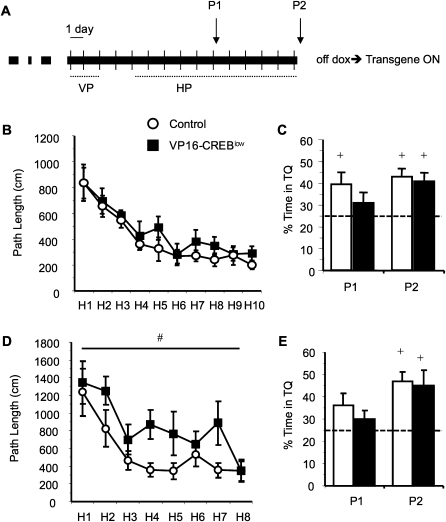
Figure 3.
VP16-CREBlow mice show milder deficits in the water maze. (A) Schematic summary of the experimental protocol: VP16-CREBlow bitransgenic mice and control littermates raised and maintained in the absence of dox were trained in the visible (VP, data not shown) and hidden platform tasks (HP). Spatial memory was assessed in two probe trials, P1 and P2. Two independent experiments, in which training consisted of either four (B,C) or two trials (D,E) per day, were performed. (B) When four training trials were given daily (eight mice per genotype), VP16-CREBlow mice showed normal learning in the HP task. (C) No significant difference between genotypes was found in the two probe trials. However, VP16-CREBlow mice did not show a preference for the target quadrant in the first probe trial, whereas their control siblings already showed a significant preference. (D) In an independent experiment, the mice (nine mice per genotype) received only two trials per day. This made the task more difficult and VP16-CREBlow mice showed a significant learning delay in the HP task. (#) Significant genotype effect in repeated measures ANOVA. (E) No significant differences were found in the probe trials. (+) A percentage of time statistically different from the chance value 25% (P < 0.05, t-test).
Chronic enhancement of CREB activity interferes with the retrieval of spatial information
We have shown that VP16-CREBhigh mice have memory deficits that disappeared when transgene expression was turned off by dox (Fig. 1, cf. D and E, left). This result argued against a pure acquisition deficit, and suggested that transgene expression interfered with memory expression rather than storage, i.e., it caused a specific deficit in memory retrieval. To confirm this result and assess whether chronic enhancement of CREB activity interfered with the retrieval of previously acquired spatial information, we carried out a new water maze experiment with VP16-CREBhigh mice, in which the mice were trained with the transgene turned off and transgene expression was switched on only during retrieval of the previously acquired spatial information (Fig. 4A). As in the experiment presented in Figure 2, bitransgenic mice kept on dox performed the visible and hidden platform task as well as their control littermates (VP: F(1,19) genotype = 0.308, P = 0.585 and HP, Fig. 4B: F(1,19) genotype = 0.438, P = 0.516). They also showed normal memory in the first probe trial (Fig. 4C: P1, transgene off). However, transgene induction and, consequently, enhancement of CREB function, interfered with the retrieval of the acquired memory for platform location during the second probe trial (Fig. 4C: P2, transgene on). The impairment was manifested in a significant worsening of the performance of VP16-CREBhigh mice during P2. Thus, bitransgenic mice did not show a significant preference for the target quadrant (% in TQ(VP16-CREB) = 28.6 ± 2.7% vs. chance value, t(12) = 1.324, P = 0.210), whereas their control littermates did (% in TQ(CONTROL) = 35.6 ± 3.3%, t(7) = 3.187, P = 0.015).
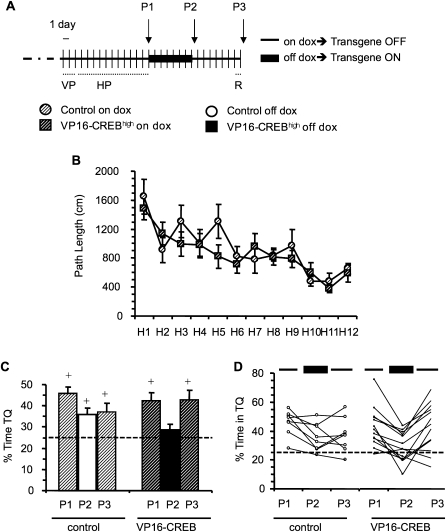
Figure 4.
Impaired retrieval. (A) Schematic summary of the experiment. VP16-CREBhigh (n = 13) and control siblings (n = 8) maintained with dox-supplemented food were trained in the visible platform (VP, data not shown) and hidden platform (HP, three trails per day) tasks. Twenty-four hours after the last training session in the HP task, a probe trial was performed (P1). The same day, dox was removed from the mouse diet. Two additional probe trials (P2 and P3) were performed in order to assess the effect of turning on and off the transgene upon memory retrieval. A single training session was performed 24 h before P3 for memory reactivation, because we had found that 2 wk after training neither genotype showed a preference for the target quadrant. Spatial memory was assessed 24 h later. (B) Control and bitransgenic mice, in which VP16-CREB expression was turned off during training, showed similar learning in the visible and hidden platform tasks. (C) Both genotypes showed similar performance in the first probe trial (gene off). VP16-CREBhigh mice had a memory defect in the second probe trial (gene on) and did not show a preference for the target quadrant, whereas their control siblings did. A third probe trial was performed one week after turning off again transgene expression (i.e., 2 wk after concluding training in the HP task); both genotypes showed again a preference for the target quadrant in P3. Repeated measures ANOVA revealed a probe × genotype interaction between P2 and P3 confirming the recovery of VP16-CREBhigh mice when the transgene was turned off. (D) The plot of individual performances across the three probes shows a clearer quadratic tendency in the group of VP16-CREBhigh mice. (+) A percentage of time statistically different from chance (P < 0.05, t-test).
When we turned off transgene expression for an additional week and tested for memory retention 2 wk after training, we found that neither control nor VP16-CREBhigh mice showed a significant preference for the target quadrant (results not shown). However, both genotypes showed similar reactivated memory after a single training session (Fig. 4C: P3, transgene off). Notably, repeated measures analysis revealed a significant genotype × probe trial interaction (F(2,38) = 3.578, P = 0.038), indicating that both groups behaved differentially during the probe trials. Interestingly, this analysis showed that the performance of VP16-CREB mice across the three probes followed a clear quadratic tendency (F(1,12) = 24.616, P < 0.001). In control mice the trend was revealed between quadratic (F(1,7) = 5.591, P = 0.050) and linear (F(1,7) = 4.340, P = 0.070). The comparison between P2 and P3 (F(1,19) = 5.886, P = 0.025) revealed a significant genotype × probe trial interaction, suggesting that the memory trace was preserved in mutant mice despite the expression memory deficit observed in P2 when retrieval was evocated with the transgene turned on. These differences in the performance across probe trials between genotypes are especially evident if we observe the behavior of individual mice (Fig. 4D).
Environmental enrichment does not correct the learning deficits of VP16-CREB mice
Environmental enrichment (EE) augments CREB immunoreactivity in the hippocampus (Williams et al. 2001; Huang et al. 2006, 2007), enables LTP reinforcement, increases intrinsic neuronal excitability, and reduces AHP in CA1 neurons (Artola et al. 2006; Irvine et al. 2006; Kumar and Foster 2007), changes which are also observed in VP16-CREBhigh mice readily after transgene induction (Barco et al. 2002; Lopez de Armentia et al. 2007). We speculated that housing in an enriched environment could accentuate the cognitive impairments associated with chronic enhancement of CREB function by further increasing CREB activity. Worsening of memory defects by environmental enrichment would be, however, paradoxical, because it is well known that laboratory rodents housed in an EE perform better in cognitive task than those housed in standard cages (SC).
To test this idea, we examined the performance of VP16-CREBhigh mice and control littermates housed in an enriched environment in the water maze (Fig. 5A). Similar to the experiment described in Figure 1, we did not observe significant differences between genotypes in the visible platform task or during the first week of the hidden platform task, but bitransgenic mice showed delayed learning during the second week of training (ANOVA analysis H5-H8: path length: F(1,69)genotype = 8.409, P = 0.005; escape latency: F(1,69)genotype = 5.672, P = 0.020). ANOVA did not reveal a general effect of housing in either parameter (path length: F(1,69)housing = 0.257, P = 0.614; escape latency: F(1,69)housing = 1.570, P = 0.214), maybe because housing had a differential effect in each genotype. In agreement with this view, we found that whereas the performance of control mice improved, as expected, with environmental enrichment (path length: F(1,34)housing = 4.344, P = 0.045, Fig. 5B; escape latency: F(1,34)housing = 6.173, P = 0.018), this condition did not ameliorate the learning impairment of VP16-CREBhigh mice (path length: F(1,35)housing = 0.256, P = 0.616, Fig. 5C; escape latency: F(1,35)housing = 0.003, P = 0.954).
Figure 5.
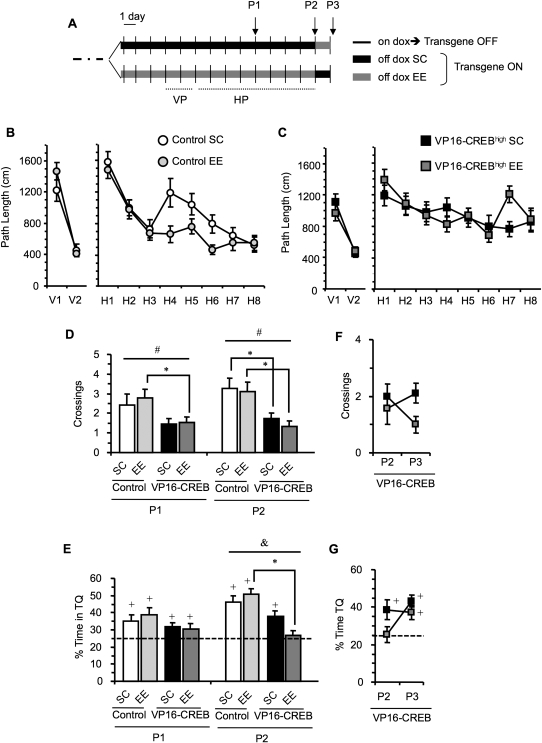
Effect of environmental enrichment in spatial memory. (A) Scheme of the protocol used. VP16-CREBhigh mice (n = 39) and control siblings (n = 38) were trained in the visible platform (VP) and hidden platform tasks (HP). Three days before starting training in the VP task, dox was removed from the mouse diet and half of the mice were housed in enriched environments (EE: 19 VP16-CREBhigh and 19 control littermates), while the other half remained in standard cages (SC: 20 VP16-CREBhigh and 19 control littermates). Spatial memory was assessed in three probe trials: P1 was performed before training on day H5, P2 was performed 24 h after finishing the 8-d training protocol in the HP task. In a subset of the animals (19 VP16-CREBhigh and 19 control littermates), the housing conditions were switched after P2 (VP16-CREBhigh SC→EE = 10, VP16-CREBhigh EE→SC = 9, control SC→EE = 9, and control EE→SC = 10). A third probe trial (P3) was performed 20 h later. (B) Environmental enrichment improved learning in control mice. (C) Both groups of bitransgenic mice showed poor learning. (D) VP16-CREBhigh mice showed reduced number of annulus crossings in P1 and P2. This deficit was especially significant in bitransgenic mice housed in an EE, although bitransgenics housed in SC also showed deficits in P2. (E) The analysis of time spent in the TQ revealed a significant genotype × housing interaction, indicating that environmental enrichment had a differential effect on the performance of control and VP16-CREB mice. Mutant mice housed in an EE, in addition, did not show memory for the platform location in P2. (F) Housing in an EE for 20 h reduced the number of annulus crossings in the VP16-CREB SC→EE group (dark square in P2, gray square in P3). (G) Placing the mice in standard cages ameliorated the deficit observed in %Time in TQ in the VP16-CREBhigh EE→SC group (gray square in P2, dark square in P3). Asterisks indicate statistical differences between groups. (*) P < 0.05; (&) a significant genotype × housing interaction; (#) a general effect of genotype; (+) a percentage of time statistically different from 25% (chance).
Also consistent with previous experiments (Fig. 1), we found that although VP16-CREBhigh mice showed some memory for the platform location, they performed worse than control mice in both probe trials, P1 (annulus crossings: F(1,69)genotype = 7.883, P = 0.006; Fig. 5D), and P2 (annulus crossings: F(1,69)genotype = 15.819, P < 0.001; Fig. 5D; %Time in TQ: F(1,69)genotype = 23.457, P < 0.001; Fig. 5E). Interestingly, we found a significant genotype × housing interaction in P2 (F(1,69) = 6.008, P = 0.017), indicating that memory expression is differentially affected by environmental enrichment in control and VP16-CREBhigh mice (Fig. 5E). A deleterious effect of environmental enrichment on VP16-CREBhigh mice was suggested by the results of the comparison between genotypes for a given housing condition (Fig. 5D,E). Bitransgenic mice housed in standard cages (SC) showed a lower number of annulus crossings in P2 than control littermates maintained in the same conditions (F(1,35) = 6.349, P = 0.016). The memory deficits were even more pronounced in the group of animals housed in an enriched environment (EE) (annulus crossings in P1: F(1,34) = 5.780, P = 0.022; annulus crossings in P2: F(1,34) = 9.713, P = 0.004; %Time in TQ in P2: F(1,34) = 36.384, P < 0.001).
We concluded this experiment by switching overnight the housing conditions of a subset of the animals in the four experimental groups and performing an additional probe trial (P3). The animals did not receive further training between P2 and P3. The change of housing conditions had no effect in the two groups of control mice (data not shown), but affected the performance of transgenic mice (Fig. 5F,G). The elimination of environmental enrichment (EE→SC) reversed the stronger deficit observed in the VP16-CREBhigh EE group to the significant, but milder deficits previously observed in the VP16-CREBhigh SC group. Repeated measures analysis of the performance of both bitransgenic groups in P2 and P3 revealed a significant probe × group interaction (target quadrant occupancy: F(1,17) = 7.974, P = 0.012, annulus crossings: F(1,17) = 5.379, P = 0.033), indicating that the switching of housing conditions had a differential effect in the two groups of VP16-CREBhigh mice. Strikingly, the group of VP16-CREBhigh mice EE→SC improved their performance in P3 and now showed a significant preference for the target quadrant that was absent the day before, in P2, when the mice were housed in the enriched environment (P2 = 25.4 ± 4.2% against 25%, t(8) = 0.099, P = 0.923; P3 = 43.1 ± 3.1% against 25%, t(8) = 5.857, P < 0.001; Fig. 5G). Whereas VP16-CREBhigh SC→EE showed a relatively stronger deficit in P3 (VP16-CREBhigh SC→EE = 1 ± 0.2 crossings vs. control SC→EE = 3.7 ± 0.5 crossings, F(1,17) = 22.549, P < 0.001) than in P2 (Fig. 5D, right: VP16-CREBhigh SC = 1.7 ± 0.2 crossings, control SC = 3.2 ± 0.5 crossings, F(1,35) = 6.349, P = 0.016). Nevertheless, mutant mice still underperformed their control siblings in P3 (annulus crossings: F(1,34)genotype = 23.611, P < 0.001; %Time in TQ: F(1,34)genotype = 8.363, P = 0.007). Overall, these results indicate that environmental enrichment did not correct the memory deficits observed in VP16-CREBhigh mice. On the contrary, some parameters even suggested that the expression memory impairment was accentuated rather than ameliorated by environmental enrichment.
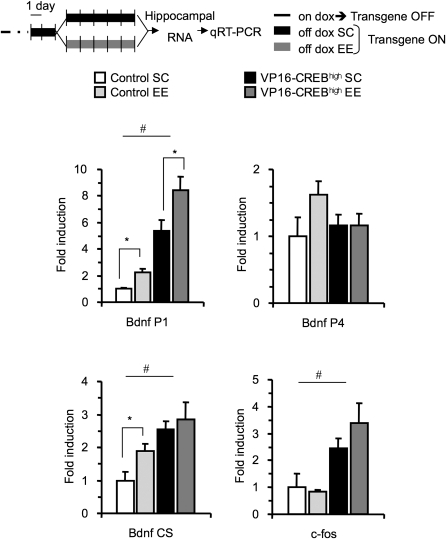
Both environmental enrichment and CREB enhancement have been proposed to modulate hippocampal gene expression, in some cases acting on similar gene programs (Barco et al. 2005; Nithianantharajah and Hannan 2006). To demonstrate the synergistic action of environmental enrichment and VP16-CREB on gene expression, we investigated the expression of the neurotrophin BDNF, which has been found to be one of the main effectors of both environmental enrichment (Rossi et al. 2006) and VP16-CREB (Barco et al. 2005) effects, in VP16-CREBhigh mice and control littermates housed in an enriched environment. The genomic structure of the BDNF gene is unusually complex since it contains multiple promoters that may be differentially regulated (Aid et al. 2007). Experiments in rat neuronal cultures have found that membrane depolarization induces the transcription from promoters P1 and PIV (PIII in the rat), which both contain CRE sites (Tao et al. 2002). We have previously found that VP16-CREB was very effective in driving BDNF expression from the CRE located at PI, whereas PIV was strikingly not affected, despite the presence of a functional CRE (Barco et al. 2005). We examined BDNF induction by enriched environment in VP16-CREBhigh bitransgenic animals and control littermates and found that, as expected, both conditions increased BDNF expression. The effects of transgene expression and environmental enrichment on PI transcriptional activity were additive (Fig. 6). Environmental enrichment enhanced PI-driven transcription in both control (t(2.7) = −4.198, P = 0.028) and VP16-CREBhigh mice (t(6.8) = −2.307, P = 0.055). In contrast, PIV transcriptional activity appeared to be modulated only by environmental enrichment (Fig. 6). In other loci, the interaction may be more complex; for example, in the case of c-fos, we observed a chronic elevation of c-fos transcripts in VP16-CREBhigh mice, whereas its expression was only transiently up-regulated in control mice housed in an enriched environment in response to novelty (Fig. 6) (results not shown).
Figure 6.
Synergistic effect of VP16-CREB and environmental enrichment on gene expression. VP16-CREBhigh and control mice were housed in an enriched environment (EE: five VP16-CREBhigh and three control littermates) or stayed in their standard cages (SC: four VP16-CREBhigh and four control littermates) for 5 d and were then sacrificed. Dox was removed from the mouse diet 2 d before enrichment. Hippocampal tissue was collected for analysis of gene expression by qRT-PCR using primer pairs targeted to bdnf and c-fos genes. Transcript levels of BDNF and c-fos were increased in VP16-CREB mice. BDNF expression was also enhanced by environmental enrichment. In the case of BDNF, specific primers were used for detecting the transcripts produced from promoter I (BDNF PI) and IV (BDNF P4), as well as primers targeted to a coding sequence common to all transcript (BDNF CS). (*) Significant differences between groups (P < 0.05, t-test); (#) overall differences between genotypes (two-way ANOVA).
Discussion
The behavioral analysis of bitransgenic mice, in which it is possible to increase CREB activity in forebrain neurons, particularly in pyramidal and granular neurons of the hippocampus, in a regulatable manner, has allowed us to gain novel insight into the role of CREB-mediated activity in spatial memory, as well as into the possible consequences of chronic enhancement of this signaling pathway. We have shown that mice expressing a constitutively active CREB variant have state-dependent memory deficits in the water maze. The early onset and reversibility of these deficits, together with the detection of similar traits in a second bitransgenic strain and the absence of appreciable cell loss at the times in which the mice underwent water maze training, demonstrate that the spatial memory impairment is a direct consequence of the chronic enhancement of CREB function and, likely, of the concomitant changes in the physiology of hippocampal neurons rather than an indirect consequence of cellular damage or abnormal brain development.
The ability to turn on and off transgene expression with dox has allowed us to gain novel insight into the mechanism by which chronic enhancement of CREB function interferes with learning and to define the memory stage affected by this molecular manipulation. In mice in which transgene expression was turned on during memory acquisition, turning off the transgene re-established the access to the memory trace, whereas in mice in which transgene expression was turned off during acquisition, turning on the transgene impaired memory expression in a reversible manner. These results suggest that spatial memory can still be acquired under high CREB activity, but chronic enhancement of CREB function can cause retrograde amnesia by interfering with the retrieval of complex spatial information. Interestingly, the learning deficit of VP16-CREBhigh mice was only manifested in the escape latency and path-length curves during the second week of training (as observed in the experiments described in Figs. 1 and 5). Different strategies are engaged during early and late stages of training in the hidden platform task. After a first phase of habituation to the pool, the mice switch from a random search strategy to locate the pool to an allocentric spatial strategy, in which the spatial map is refined. Retrieval of spatial information is likely more important in the second phase of the task, which is what may explain that bitransgenic mice only expressed a deficit during the second week of training. Although the identity of the molecular alterations underlying VP16-CREB-induced retrieval defects remains unknown, the synergistic interaction between environmental enrichment and CREB enhancement suggests that the relationship of CREB-mediated plasticity to memory retrieval is a steep inverted-U function. Whereas the shift in neural plasticity caused by environmental enrichment facilitated learning in control animals, it interfered further with retrieval in VP16-CREB mutants. The interaction at the level of BDNF PI promoter, which may also likely occur at other loci, provides a possible molecular substrate for this synergistic action. Thus, it has been shown that overexpression of BDNF can cause learning impairments (Croll et al. 1999; Pietropaolo et al. 2007).
We have earlier found a chronic reduction of the threshold for L-LTP and increased intrinsic excitability of CA1 neurons in the hippocampus of VP16-CREBhigh mice (Barco et al. 2002; Lopez de Armentia et al. 2007). This might cause an excessive number of synapses within the hippocampal network of transgenic mice to become potentiated over the several days of training in the water maze, which, in turn, might interfere with the access to stored memories. Consistent with this view, post-training LTP induction can also cause retrograde amnesia, possibly by interfering with the activation of recently formed hippocampal memory networks (Martin and Morris 2002; Diamond et al. 2004). Interestingly, we have also observed that the expression of VP16-CREB bypassed the requirement for de novo gene expression associated with long-term memory formation in contextual and cued fear conditioning tasks, suggesting that CREB-dependent gene expression can be sufficient for fear memory consolidation (Viosca et al. 2009). The effects in fear memory consolidation and spatial memory retrieval observed in mice with enhanced CREB activity might have a physiological correlate in normal behaving animals. Memory reinforcement and retroactive amnesia are two features associated with the formation of flashbulb memory during highly stressful experiences (Diamond et al. 2004, 2006). During such events a number of neuromodulator molecules are widespread released within several forebrain areas. These neuromodulatory inputs have been shown to trigger CREB activation and de novo gene expression, inhibit afterhyperpolarization, and reduce the threshold for L-LTP (Sah and Bekkers 1996; Cohen et al. 1999; Haug and Storm 2000; Fuenzalida et al. 2007). All of these alterations are observed in the hippocampus of VP16-CREBhigh mice shortly after transgene induction. Therefore, VP16-CREB expression might mimic, at the cellular level, the chronic activation of neuromodulatory afferent inputs. Retrograde amnesia and interference with the acquisition of complex tasks may be a necessary downside effect of robust LTP and enhanced intrinsic plasticity.
Genetic enhancement of learning has been achieved through manipulations that provided a net increase in the signal-to-noise ratio of neuronal networks (Tang et al. 1999; Malleret et al. 2001; Genoux et al. 2002; Chen et al. 2003; Jeon et al. 2003; Nolan et al. 2004; Wang et al. 2004; Kushner et al. 2005). In contrast, chronic activation of plasticity-related signaling pathways may disrupt memory formation by increasing the noise, therefore reducing instead of enhancing the signal-to-noise ratio (Gerlai et al. 1998; Migaud et al. 1998; Uetani et al. 2000; Pineda et al. 2004; Bourtchouladze et al. 2006). The situation in VP16-CREBhigh mice may be similar to that described for flies with the dunce mutation, both VP16-CREBhigh mice and dunce flies show an enhancement of the cAMP pathway, both show an enhanced response to tetanic stimulation (Kuromi and Kidokoro 2000), and both perform worse than wild-type animals in behavioral tasks (Dudai et al. 1976), indicating that from flies to mammals, accurate regulation of the cAMP-pathway is required for learning.
Our results support a role of CREB-dependent gene expression in spatial learning and memory and encourage the ongoing effort in the identification of drug compounds targeted to this signaling pathway to enhance memory formation. However, they also suggest that caution is required in such studies, because only pharmaceuticals that enhance CREB-mediated gene induction within a reasonable range and do not elevate excessively the uninduced, basal level of transcription can restore or enhance complex forms of learning and memory storage. Molecular constraints of memory storage, such as phosphodiesterases (PDE), phosphatases, or transcriptional repressors may prove to be the targets of choice for pharmacological and therapeutic manipulation of memory.
Materials and Methods
Transgenic mice
VP16-CREBhigh and VP16-CREBlow mice have been described before (Barco et al. 2002; Lopez de Armentia et al. 2007). We used as control littermate mice carrying either pCaMKII-tTA, tetO-VP16-CREB, or no transgene. For VP16-CREBhigh mice, dox was administrated in the food at 40 mg/Kg and removed or added at the indicated times during experimentation to induce or repress, respectively, transgene expression (Mayford et al. 1996). VP16-CREBlow mice were raised in the absence of dox. The genetic background of all mice was C57BL6/J. Mice were maintained and bred according to animal-care standards established by the Institutional Animal Care and Use Committees and national guidelines. The mice were group housed in single-sex cages on a light:dark cycle (12/12 h) with food and water available ad libitum. In all experiments, we used bitransgenic mice and control littermates with the same sex and age range. In the experiments described in Figures 1–3, we used 3–5-mo-old males. When we first tackled the environmental enrichment experiment, we found that males started to fight when housed in an enriched environment. To avoid this situation, we performed the experiment described in Figure 5 with adult (3–5 mo) females. The genotype effect observed in both experiments (Figs. 1 and 5) indicates that the impairment in VP16-CREBhigh mice is sex independent. The experiment described in Figure 4 was also performed with adult females. For environmental enrichment, the mice were housed in 50 × 50 × 30 cm white acrylic glass boxes with diverse kinds of bedding, tubing, objects, nesting material, and complete pet food for guinea pigs. The spatial configuration within the enriched boxes was changed every day.
Water maze experiments
The basic water maze experiment was divided in two phases: (1) visible platform (VP) task, in which the animal is forced to swim in a pool filled with opaque water and learns that in order to escape the pool it must reach a transparent platform submerged 1 cm under the water and cued with a black bar. The platform location changed every trial. Mice failing to find the platform after 120 sec were gently guided to it by hand and allowed to remain on it for 15 sec. Mice received four trials of 120 sec maximum, unless otherwise indicated, separated by a 60–120 min intertrial interval every day. (2) In the hidden platform (HP) task, the basic procedures were the same as in the visible platform task, but the submerged platform was not cued and its position did not change during the task. Therefore, in this version of the task, the animal is forced to remember the position of the hidden escape platform using distal surrounding cues. In all of our experiments, the HP task was performed after the VP task so that mice were habituated to the pool. In the experiment described in Figure 1, we extended the water maze experiment one additional week, performing 5 d of transfer task, in which the hidden platform was moved to the opposite corner of the pool. Probe trials of 60 sec, in which the platform was removed, were performed on specific days during training to assess memory formation. After 60 sec, mice were gently guided to the original platform location and allowed to remain in this position for 15 sec to avoid extinction (the experimenter held the platform in his hand). In some experiments, additional probe trials were performed at specific times after acquisition of the task to evaluate long-term retention of previously acquired spatial information. For the analysis of probe trials, we calculated quadrant occupancy (percent of time), number of crossings in an annulus (area double than that of platform) located at platform position (TQ, target quadrant), and the average of crossings in same sized annuli at equivalent positions in the other three quadrants of the pool. Experiments in Figures 1–3 were performed in a 122-cm pool using HVS Image Analysis System (HVS Image Ltd.). Experiments in Figures 4 and 5 were carried out in a 170-cm pool using SMART software (S.L. Panlab), which made the task more difficult. The confirmation of the same deficits in two different laboratories using different equipment supports the robustness of the phenotype (Crabbe et al. 1999). All behavioral procedures were conducted during the light phase of the light cycle. Experimenters were blind to genotypes. The result of the PCR-based genotyping was provided as a factor for statistical analysis of the behavioral data after task conclusion.
Quantitative RT-PCR
Total mRNA was extracted from the hippocampus of bitransgenic and control mice. qRT-PCRs were performed in an Applied Biosystems 7300 real-time PCR unit using SYBR GreenER mix (Invitrogen) and primers specific for c-fos, for the BDNF transcripts transcribed from promoter PI and PIV (Barco et al. 2005), and for GADPH. Each independent sample was assayed in duplicate and normalized using GAPDH levels.
Statistical methods
Training curves were analyzed using repeated measures ANOVAs including session as the within-subject factor and genotype as the between-subject factor. Housing was included as a second between-subject factor in Figure 5. In probe trials, comparisons between groups used t-tests and ANOVAs. Within-group comparisons were performed using paired t-tests. Significant preferences toward the target quadrant were determined within each group using single t-tests (against the chance value: 25%). Repeated measures ANOVAs including probe trial as the within-subject factors were applied in Figures 4 and 5. The results presented in Figures 1 and 5 and their description in the text correspond to two duplicate experiments with similar results. “Experiment” was introduced as an additional between-subject factor in the ANOVA analysis of these data. There was no significant “Experiment” effect or interaction. Type III sum of squares was used in all ANOVA models. Data in all figures are presented as mean ± SEM.
Acknowledgments
We thank the members of the Barco's laboratory and Eloisa Herrera for critical reading of the manuscript and helpful comments. We also thank Bing Liu and Román Olivares for technical assistance on mice genotyping. J.V. holds a fellowship from the Generalitat Valenciana (BFPI06/316), and E.B. holds a fellowship from the Gobierno Vasco. A.B. and J.V. are supported by the European Commission grant MEXT-CT-2003-509550, the Spanish MEC Grants BFU2005-00286 and CSD2007-00023, and grants from Fundació La Marató de TV3 and Fundación Ramón Areces. E.R.K, G.M., and S.V. were supported by the Howard Hughes Medical Institute and the Kavli Institute for Brain Sciences.
Footnotes
[Supplemental material is available online at www.learnmem.org.]
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.1220309.
References
- Aid T., Kazantseva A., Piirsoo M., Palm K., Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J. Neurosci. Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon J.M., Barco A., Kandel E.R. Capture of the late phase of long-term potentiation within and across the apical and basilar dendritic compartments of CA1 pyramidal neurons: Synaptic tagging is compartment restricted. J. Neurosci. 2006;26:256–264. doi: 10.1523/JNEUROSCI.3196-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artola A., von Frijtag J.C., Fermont P.C., Gispen W.H., Schrama L.H., Kamal A., Spruijt B.M. Long-lasting modulation of the induction of LTD and LTP in rat hippocampal CA1 by behavioural stress and environmental enrichment. Eur. J. Neurosci. 2006;23:261–272. doi: 10.1111/j.1460-9568.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Balschun D., Wolfer D.P., Gass P., Mantamadiotis T., Welzl H., Schutz G., Frey J.U., Lipp H.P. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J. Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A., Alarcon J.M., Kandel E.R. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. doi: 10.1016/s0092-8674(02)00657-8. [DOI] [PubMed] [Google Scholar]
- Barco A., Pittenger C., Kandel E.R. CREB, memory enhancement and the treatment of memory disorders: Promises, pitfalls and prospects. Expert Opin. Ther. Targets. 2003;7:101–114. doi: 10.1517/14728222.7.1.101. [DOI] [PubMed] [Google Scholar]
- Barco A., Patterson S., Alarcon J.M., Gromova P., Mata-Roig M., Morozov A., Kandel E.R. Gene expression profiling of facilitated L-LTP in VP16-CREB mice reveals that BDNF is critical for the maintenance of LTP and its synaptic capture. Neuron. 2005;48:123–137. doi: 10.1016/j.neuron.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Blendy J.A., Kaestner K.H., Schmid W., Gass P., Schutz G. Targeting of the CREB gene leads to up-regulation of a novel CREB mRNA isoform. EMBO J. 1996;15:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- Bourtchuladze R., Frenguelli B., Blendy J., Cioffi D., Schutz G., Silva A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R., Patterson S.L., Kelly M.P., Kreibich A., Kandel E.R., Abel T. Chronically increased Gsα signaling disrupts associative and spatial learning. Learn. Mem. 2006;13:745–752. doi: 10.1101/lm.354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightwell J.J., Smith C.A., Neve R.L., Colombo P.J. Long-term memory for place learning is facilitated by expression of cAMP response element-binding protein in the dorsal hippocampus. Learn. Mem. 2007;14:195–199. doi: 10.1101/lm.395407. [DOI] [PubMed] [Google Scholar]
- Chen A., Muzzio I.A., Malleret G., Bartsch D., Verbitsky M., Pavlidis P., Yonan A.L., Vronskaya S., Grody M.B., Cepeda I., et al. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–669. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- Cohen A.S., Coussens C.M., Raymond C.R., Abraham W.C. Long-lasting increase in cellular excitability associated with the priming of LTP induction in rat hippocampus. J. Neurophysiol. 1999;82:3139–3148. doi: 10.1152/jn.1999.82.6.3139. [DOI] [PubMed] [Google Scholar]
- Colombo P.J., Brightwell J.J., Countryman R.A. Cognitive strategy-specific increases in phosphorylated cAMP response element-binding protein and c-Fos in the hippocampus and dorsal striatum. J. Neurosci. 2003;23:3547–3554. doi: 10.1523/JNEUROSCI.23-08-03547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe J.C., Wahlsten D., Dudek B.C. Genetics of mouse behavior: Interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- Croll S.D., Suri C., Compton D.L., Simmons M.V., Yancopoulos G.D., Lindsay R.M., Wiegand S.J., Rudge J.S., Scharfman H.E. Brain-derived neurotrophic factor transgenic mice exhibit passive avoidance deficits, increased seizure severity and in vitro hyperexcitability in the hippocampus and entorhinal cortex. Neuroscience. 1999;93:1491–1506. doi: 10.1016/s0306-4522(99)00296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond D.M., Park C.R., Woodson J.C. Stress generates emotional memories and retrograde amnesia by inducing an endogenous form of hippocampal LTP. Hippocampus. 2004;14:281–291. doi: 10.1002/hipo.10186. [DOI] [PubMed] [Google Scholar]
- Diamond D.M., Campbell A.M., Park C.R., Woodson J.C., Conrad C.D., Bachstetter A.D., Mervis R.F. Influence of predator stress on the consolidation versus retrieval of long-term spatial memory and hippocampal spinogenesis. Hippocampus. 2006;16:571–576. doi: 10.1002/hipo.20188. [DOI] [PubMed] [Google Scholar]
- Dong Y., Green T., Saal D., Marie H., Neve R., Nestler E.J., Malenka R.C. CREB modulates excitability of nucleus accumbens neurons. Nat. Neurosci. 2006;9:475–477. doi: 10.1038/nn1661. [DOI] [PubMed] [Google Scholar]
- Dudai Y., Jan Y.N., Byers D., Quinn W.G., Benzer S. dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. 1976;73:1684–1688. doi: 10.1073/pnas.73.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuenzalida M., Fernandez de Sevilla D., Buno W. Changes of the EPSP waveform regulate the temporal window for spike-timing-dependent plasticity. J. Neurosci. 2007;27:11940–11948. doi: 10.1523/JNEUROSCI.0900-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoux D., Haditsch U., Knobloch M., Michalon A., Storm D., Mansuy I.M. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418:970–975. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- Gerlai R., Henderson J.T., Roder J.C., Jia Z. Multiple behavioral anomalies in GluR2 mutant mice exhibiting enhanced LTP. Behav. Brain Res. 1998;95:37–45. doi: 10.1016/s0166-4328(98)00002-3. [DOI] [PubMed] [Google Scholar]
- Graves L., Dalvi A., Lucki I., Blendy J.A., Abel T. Behavioral analysis of CREB αδ mutation on a B6/129 F1 hybrid background. Hippocampus. 2002;12:18–26. doi: 10.1002/hipo.10003. [DOI] [PubMed] [Google Scholar]
- Guzowski J.F., McGaugh J.L. Antisense oligodeoxynucleotide-mediated disruption of hippocampal cAMP response element binding protein levels impairs consolidation of memory for water maze training. Proc. Natl. Acad. Sci. 1997;94:2693–2698. doi: 10.1073/pnas.94.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.H., Bolanos C.A., Green T.A., Olson V.G., Neve R.L., Liu R.J., Aghajanian G.K., Nestler E.J. Role of cAMP response element-binding protein in the rat locus ceruleus: Regulation of neuronal activity and opiate withdrawal behaviors. J. Neurosci. 2006;26:4624–4629. doi: 10.1523/JNEUROSCI.4701-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug T., Storm J.F. Protein kinase A mediates the modulation of the slow Ca2+-dependent K+ current, IsAHP, by the neuropeptides CRF, VIP, and CGRP in hippocampal pyramidal neurons. J. Neurophysiol. 2000;83:2071–2079. doi: 10.1152/jn.2000.83.4.2071. [DOI] [PubMed] [Google Scholar]
- Huang F.L., Huang K.P., Wu J., Boucheron C. Environmental enrichment enhances neurogranin expression and hippocampal learning and memory but fails to rescue the impairments of neurogranin null mutant mice. J. Neurosci. 2006;26:6230–6237. doi: 10.1523/JNEUROSCI.1182-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.L., Huang K.P., Boucheron C. Long-term enrichment enhances the cognitive behavior of the aging neurogranin null mice without affecting their hippocampal LTP. Learn. Mem. 2007;14:512–519. doi: 10.1101/lm.636107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine G.I., Logan B., Eckert M., Abraham W.C. Enriched environment exposure regulates excitability, synaptic transmission, and LTP dentate gyrus of freely moving rates. Hippocampus. 2006;16:149–160. doi: 10.1002/hipo.20142. [DOI] [PubMed] [Google Scholar]
- Jeon D., Yang Y.M., Jeong M.J., Philipson K.D., Rhim H., Shin H.S. Enhanced learning and memory in mice lacking Na+/Ca2+ exchanger 2. Neuron. 2003;38:965–976. doi: 10.1016/s0896-6273(03)00334-9. [DOI] [PubMed] [Google Scholar]
- Josselyn S.A., Nguyen P.V. CREB, synapses and memory disorders: Past progress and future challenges. Curr. Drug Target. CNS Neurol. Disord. 2005;4:481–497. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- Josselyn S.A., Shi C., Carlezon W.A., Jr, Neve R.L., Nestler E.J., Davis M. Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. J. Neurosci. 2001;21:2404–2412. doi: 10.1523/JNEUROSCI.21-07-02404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel E.R. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kumar A., Foster T. Environmental enrichment decreases the afterhyperpolarization in senescent rats. Brain Res. 2007;1130:103–107. doi: 10.1016/j.brainres.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromi H., Kidokoro Y. Tetanic stimulation recruits vesicles from reserve pool via a cAMP-mediated process in Drosophila synapses. Neuron. 2000;27:133–143. doi: 10.1016/s0896-6273(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Kushner S.A., Elgersma Y., Murphy G.G., Jaarsma D., van Woerden G.M., Hojjati M.R., Cui Y., LeBoutillier J.C., Marrone D.F., Choi E.S., et al. Modulation of presynaptic plasticity and learning by the H-ras/extracellular signal-regulated kinase/synapsin I signaling pathway. J. Neurosci. 2005;25:9721–9734. doi: 10.1523/JNEUROSCI.2836-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Armentia M., Jancic D., Olivares R., Alarcon J.M., Kandel E.R., Barco A. cAMP response element-binding protein-mediated gene expression increases the intrinsic excitability of CA1 pyramidal neurons. J. Neurosci. 2007;27:13909–13918. doi: 10.1523/JNEUROSCI.3850-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret G., Haditsch U., Genoux D., Jones M.W., Bliss T.V., Vanhoose A.M., Weitlauf C., Kandel E.R., Winder D.G., Mansuy I.M. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T., Lemberger T., Bleckmann S.C., Kern H., Kretz O., Martin Villalba A., Tronche F., Kellendonk C., Gau D., Kapfhammer J., et al. Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- Marie H., Morishita W., Yu X., Calakos N., Malenka R.C. Generation of silent synapses by acute in vivo expression of CaMKIV and CREB. Neuron. 2005;45:741–752. doi: 10.1016/j.neuron.2005.01.039. [DOI] [PubMed] [Google Scholar]
- Martin S.J., Morris R.G. New life in an old idea: The synaptic plasticity and memory hypothesis revisited. Hippocampus. 2002;12:609–636. doi: 10.1002/hipo.10107. [DOI] [PubMed] [Google Scholar]
- Mayford M., Bach M.E., Huang Y.Y., Wang L., Hawkins R.D., Kandel E.R. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Migaud M., Charlesworth P., Dempster M., Webster L.C., Watabe A.M., Makhinson M., He Y., Ramsay M.F., Morris R.G., Morrison J.H., et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein [see comments] Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- Mizuno M., Yamada K., Maekawa N., Saito K., Seishima M., Nabeshima T. CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav. Brain Res. 2002;133:135–141. doi: 10.1016/s0166-4328(01)00470-3. [DOI] [PubMed] [Google Scholar]
- Moncada D., Viola H. Phosphorylation state of CREB in the rat hippocampus: A molecular switch between spatial novelty and spatial familiarity? Neurobiol. Learn. Mem. 2006;86:9–18. doi: 10.1016/j.nlm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Mouravlev A., Dunning J., Young D., During M.J. Somatic gene transfer of cAMP response element-binding protein attenuates memory impairment in aging rats. Proc. Natl. Acad. Sci. 2006;103:4705–4710. doi: 10.1073/pnas.0506137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantharajah J., Hannan A.J. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat. Rev. Neurosci. 2006;7:697–709. doi: 10.1038/nrn1970. [DOI] [PubMed] [Google Scholar]
- Nolan M.F., Malleret G., Dudman J.T., Buhl D.L., Santoro B., Gibbs E., Vronskaya S., Buzsaki G., Siegelbaum S.A., Kandel E.R., et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119:719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Pietropaolo S., Paterna J.C., Bueler H., Feldon J., Yee B.K. Bidirectional changes in water-maze learning following recombinant adenovirus-associated viral vector (rAAV)-mediated brain-derived neurotrophic factor expression in the rat hippocampus. Behav. Pharmacol. 2007;18:533–547. doi: 10.1097/FBP.0b013e3282da0bf6. [DOI] [PubMed] [Google Scholar]
- Pineda V.V., Athos J.I., Wang H., Celver J., Ippolito D., Boulay G., Birnbaumer L., Storm D.R. Removal of Giα1 constraints on adenylyl cyclase in the hippocampus enhances LTP and impairs memory formation. Neuron. 2004;41:153–163. doi: 10.1016/s0896-6273(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Pittenger C., Huang Y.Y., Paletzki R.F., Bourtchouladze R., Scanlin H., Vronskaya S., Kandel E.R. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34:447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- Porte Y., Buhot M.C., Mons N. Alteration of CREB phosphorylation and spatial memory deficits in aged 129T2/Sv mice. Neurobiol. Aging. 2008;29:1533–1546. doi: 10.1016/j.neurobiolaging.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Restivo L., Tafi E., Ammassari-Teule M., Marie H. Viral-mediated expression of a constitutively active form of CREB in hippocampal neurons increases memory. Hippocampus. 2008 doi: 10.1002/hipo.20527. [DOI] [PubMed] [Google Scholar]
- Rossi C., Angelucci A., Costantin L., Braschi C., Mazzantini M., Babbini F., Fabbri M.E., Tessarollo L., Maffei L., Berardi N., et al. Brain-derived neurotrophic factor (BDNF) is required for the enhancement of hippocampal neurogenesis following environmental enrichment. Eur. J. Neurosci. 2006;24:1850–1856. doi: 10.1111/j.1460-9568.2006.05059.x. [DOI] [PubMed] [Google Scholar]
- Sah P., Bekkers J.M. Apical dendritic location of slow afterhyperpolarization current in hippocampal pyramidal neurons: Implications for the integration of long-term potentiation. J. Neurosci. 1996;16:4537–4542. doi: 10.1523/JNEUROSCI.16-15-04537.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.P., Shimizu E., Dube G.R., Rampon C., Kerchner G.A., Zhuo M., Liu G., Tsien J.Z. Genetic enhancement of learning and memory in mice [see comments] Nature. 1999;401:63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- Tao X., West A.E., Chen W.G., Corfas G., Greenberg M.E. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33:383–395. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- Uetani N., Kato K., Ogura H., Mizuno K., Kawano K., Mikoshiba K., Yakura H., Asano M., Iwakura Y. Impaired learning with enhanced hippocampal long-term potentiation in PTPδ-deficient mice. EMBO J. 2000;19:2775–2785. doi: 10.1093/emboj/19.12.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viosca J., Lopez de Armentia M., Jancic D., Barco A. Enhanced CREB-dependent gene expression increases the excitability of neurons in the basal amygdala and primes the consolidation of contextual and cued fear memory. Learn. Mem. 2009;16 doi: 10.1101/lm1254209. (this issue). [DOI] [PubMed] [Google Scholar]
- Wang H., Ferguson G.D., Pineda V.V., Cundiff P.E., Storm D.R. Overexpression of type-1 adenylyl cyclase in mouse forebrain enhances recognition memory and LTP. Nat. Neurosci. 2004;7:635–642. doi: 10.1038/nn1248. [DOI] [PubMed] [Google Scholar]
- Williams B.M., Luo Y., Ward C., Redd K., Gibson R., Kuczaj S.A., McCoy J.G. Environmental enrichment: Effects on spatial memory and hippocampal CREB immunoreactivity. Physiol. Behav. 2001;73:649–658. doi: 10.1016/s0031-9384(01)00543-1. [DOI] [PubMed] [Google Scholar]