Abstract
Herein we describe the cloning, functional expression and initial characterization of ORF36 from Micromonospora carbonacae var. africana and rubN8 from Streptomyces achromogenes var. rubradiris. The purified enzymes play the same role, the double-oxidation of TDP-evernosamine to TDP-evernitrosose in the everninomycin and rubradirin pathways, respectively.
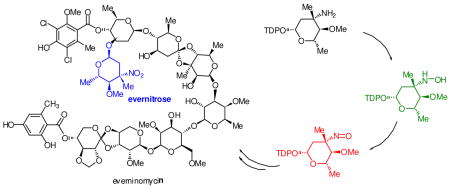
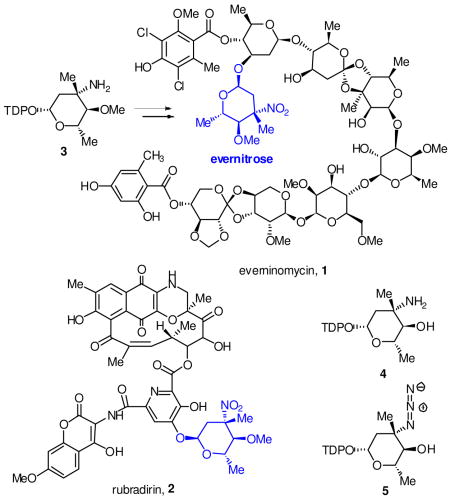
Nitrosugars decorate over 50 isolated bioactive natural products including aromatic and reduced polyketides as well as oligosaccharides.1 One of the first reported nitrosugar containing natural products is the ‘orthosomycin’ antibiotic everninomycin 1, which terminates with 2,3,6-trideoxy-3-C-methyl-4-O-methyl-3-nitro-L-ribo-aldohexo-pyranose, or evernitrose.2 Evernitrose is also present in the polyketide rubradirin 2 and is structurally related to D-kijanose from the spirotetronate polyketide antibiotic kijanimycin.3 Of note, the nitroso congener of everninomycin is a significant metabolite in fermentations of the everninomycin producer Micromonospora carbonacae var. africana.4 Moreover, ‘protorubradirin’, the nitroso analog of rubradirin is the major product of fermentation in the rubradirin producer when isolation procedures are performed in the dark, suggesting that the C-nitroso sugars are the bona fide metabolites.5
A comparative genomic analysis was previously performed for the orthosomycin antibiotics comparing two separate gene clusters encoding everninomycin octasaccharides and two gene clusters encoding avilamycins, structurally related heptasaccharides lacking the nitrosugar moiety.6 Subtractive analysis identified a cassette of genes that have translated homologs in the biochemically characterized epi-vancosamine pathway.7 ORF36 from M. carbonacae var. africana (ORF42 from var. aurantiaca) was proposed to function as a requisite oxidase in the oxidation of the 3-amino group in thymidine diphosphate TDP-evernosamine 3, a close analog of L-TDP-epi-vancosamine 4. More recently, two additional nitrosugar encoding natural product gene clusters have bolstered this hypothesis: the ansamycin rubradirin from Streptomyces achromogenese var. rubradiris and the spirotetronate polyketide kijanimicin from Actinomadura kijaniata.8,9 These clusters also contain a gene encoding a putative flavin-dependent enzyme with good overall sequence identity to ORF36/42. RubN8 is 63% identical to ORF36 and Liu and coworkers have recently proposed that KijD3, the homolog of this putative protein in the kijanimicin cluster (65% identical), is the key oxidase involved in kijanose amine oxidation.
Despite these compelling sequence-based analyses, the biochemical roles of the encoded flavin-dependent enzymes in the oxidation of TDP-evernosamine 3 and analogs have yet to be determined. Consequently, we amplified two of these putative oxidase genes from genomic DNA of M. carbonacae var. africana and S. achromogenes var. rubradiris, the everninomycin (ORF36) and rubradirin (RubN8) producing organisms respectively. PCR products were subcloned for translation as N-terminal His6-tagged fusion proteins in pET28a. Fusion proteins were overproduced in E. coli BL21(DE3) and purified to homogeneity by Ni2+-affinity chromatography.
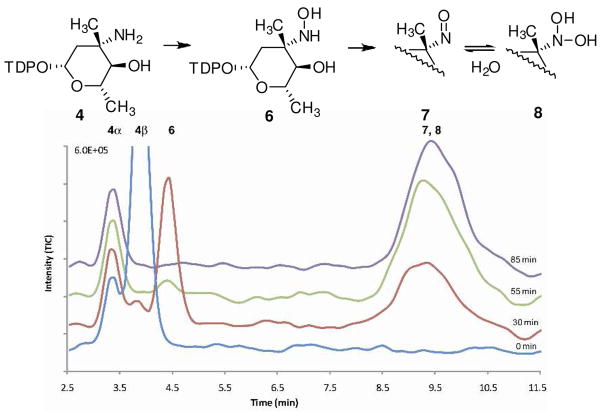
A close substrate analog of 3, L-TDP-epi-vancosamine 4 (4-O-desmethyl L-TDP-evernosamine) was prepared by in situ Staudinger reduction of a synthetic azide congener 5(5:1 β/α)10 with tris(2-carboxyethyl)phosphine. L-TDP-epi-vancosamine 4 was incubated separately with the putative oxidases, FAD, NADPH and flavin reductase from Vibrio fischeri. Product formation was assayed by electrospray HPLC/MS using a porous graphitic matrix column11 in negative ion mode, which effectively separated all TDP products. As shown in Figure 1, time course assay revealed rapid disappearance of amino sugar 4 (m/z = 544), transient appearance of intermediary hydroxylamine 6 (m/z = 560) and ultimate formation of a nitroso compound TDP-epi-vancosonitrose 7/8 (m/z = 558 and its hydrate at 576). Tandem MS fragmentation (Figure S6) of selected product ions showed diagnostic fragmentation patterns of 7/8 including loss of TDP anion (m/z = 401) and thymidine (m/z = 432). Of note, only the β– anomer of 4 was oxidized and no formation of nitro sugar was observed under any reaction conditions.
Figure 1.
Time course HPLC/MS of product formation in RubN8 oxidation reaction. Further details can be found in Figure S4 for RubN8 and Figure S7 for ORF36 in Supporting Information.
Control reactions omitting either ORF36/RubN8, FAD or NADPH produced no detectable oxidation products. In the absence of flavin reductase, hydroxylamine 6 was detected, but at very low rates. V. fischeri flavin reductase is known to stimulate hydrogen peroxide and superoxide production (as they are FADH2 aerobic decomposition byproducts).12,13 Considering the possibility that these reactive oxygen species (ROS) may oxidize epi-vancosamine, all enzymatic reactions were also performed in the presence of a catalytic excess of catalase and superoxide dismutase. Under these conditions, no substantial changes in the pattern of oxidation of L-TDP-epi-vancosamine to the corresponding nitroso sugar were observed.
The flavin-dependent double oxidation described in this work contrasts with previously reported amine oxidases in nitroaromatic and deoxysugar metabolite pathways. AurF, from aureothin biosynthesis, a [2Fe-2S] Rieske non-heme iron oxygenase,14–16 and PrnD, a binuclear metalloenzyme from pyrrolnitrin biosynthesis, have been implicated in aromatic nitro group formation.17, 18 In the former case, evidence for a transient hydroxylamino intermediate has been obtained. The flavin dependent mechanism of ORF36/RubN8 is also likely distinct from CalE10 a P450 N-oxidase involved in hydroxylamino deoxysugar formation in calicheamicin biosynthesis.19, 20
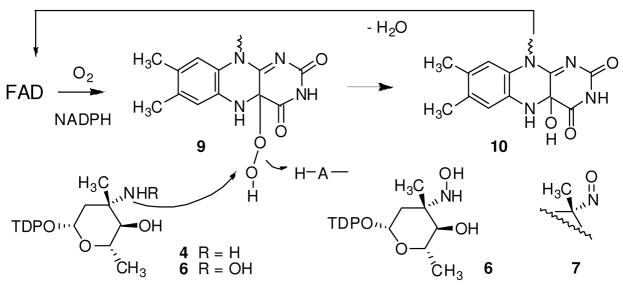
The four electron flavin-mediated oxidation of an amine to a nitroso functional group likely involves a two step process in which the amino sugar 4 is first oxidized to the corresponding hydroxylamine 6 via a flavin monooxygenase mechanism in analogy with L-ornithine N5-oxygenase from pyoverdin biosynthesis21 and p-hydroxybenzoate hydroxylase22 (Scheme 1). Correspondingly, we propose that bound FADH2, generated by flavin reductase, reacts with molecular oxygen to form flavin 4a-hydroperoxide 9 which is the electrophilic oxidizing species in the formation of the hydroxylamino sugar. The elimination of the resultant 4-a-hydroxide 10 regenerates FAD. In the second step, oxidation of the hydroxylamino sugar 6 to nitroso sugar 7/8 may be affected by an iterative oxidative process in which FAD would again be reduced by flavin reductase/NADPH prior to an additional round of oxidation. To confirm FAD recycling, reactions were performed with stoichiometrically limiting NAD(P)H. The formation of hydroxylamine 6 was observed (Figure S5, Supporting Information) but no further oxidation products appeared until additional NAD(P)H was added to the reaction, suggesting that four electrons are supplied for the formation of the FAD-4a-hydroperoxide via NAD(P)H.
Scheme 1.
Proposed pathway for nitrososynthase oxygenation reaction.
This study employs the 4-O-desmethyl analog of L-TDP-evernosamine, and the possibility exists that the methylation state may affect subsequent oxidation reactions at the 3-position. However our data are in accord with previous observations in which the nitroso compound is the predominant product produced under aphotic fermentation conditions. Taken together, these data suggest that the metastable nitroso congener of evernitrose is the likely product of the deoxysugar biosynthetic pathway. Nitrososugar metabolites may be subsequently transformed to the nitro oxidation state via a latent (photo)chemical process or endogenous ROS. Protorubradirin and its methanolysis derived nitroso sugar methyl glycoside are reportedly converted to the nitro form upon exposure to ambient light and air.5
These results speak to the high utility of comparative genomic analysis of similar gene clusters in elucidating biosynthetic pathways. Limited sequence similarity is also observed for ORF36/RubN8 to characterized oxidases in secondary metabolism. Dibenzothiophene oxidase dszC (35% similarity) catalyzes an analogous net 4-electron oxidation of a sulfide functional group to a sulfoxide23 and isobutylamine oxidase vlmH (34% similarity) converts isobutyl amine to isobutyl N-hydroxylamine an intermediate in pathway for the azoxy functional natural product valanimycin.24
To the best of our knowledge, this study provides the first biochemical evidence for a new class of flavin-dependent monooxygenase that is utilized in the pathways for the preponderance of alkylnitro functional natural products described to date. The observed activity of ORF36 and RubN8 provide direct biochemical evidence for the everninomycin and rubradirin gene clusters respectively. Future studies will investigate the detailed kinetics and mechanism of the flavin catalyzed enzymatic double N-oxidation reaction and the subsequent nonenzymatic formation of the nitro natural products.
Supplementary Material
Experimental data on protein expression, purification and biochemical assays are available. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by the Office of Naval Research N000140610144 and NIH 1R01GM077189-01.
References
- 1.Buckingham J. Dictionary of Natural Products. CRC Press; Boca Raton, FL: 2007. [Google Scholar]
- 2.Ganguly AK, Sarre OZ, Reimann H. J Am Chem Soc. 1968;90:7129. doi: 10.1021/ja01027a047. [DOI] [PubMed] [Google Scholar]
- 3.Waitz JA, Horan AC, Kalyanpur M, Lee BK, Loebenberg D, Marquez JA, Miller G, Patel MG. J Antibiot. 1981;34:1101. doi: 10.7164/antibiotics.34.1101. [DOI] [PubMed] [Google Scholar]
- 4.Alroy R, Blaidell J, Morenberg A, Eugene S. US 6,320,044 2001
- 5.Bannister B, Zapotocky BA. J Antibiot. 1992;45:1313. doi: 10.7164/antibiotics.45.1313. [DOI] [PubMed] [Google Scholar]
- 6.Farnet, C. M.; Zazopoulos, E.; Staffa, A. CA 2002-CA432, 2002.
- 7.Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Proc Natl Acad Sci. 2000;97:11942. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CG, Lamichhane J, Song KI, Nguyen VD, Kim DH, Jeong TS, Kang SH, Kim KW, Maharjan J, Hong YS, Kang JS, Yoo JC, Lee JJ, Oh TJ, Liou K, Sohng JK. Arch microbiol. 2008;189:463. doi: 10.1007/s00203-007-0337-3. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, White-Phillip JA, Melancon CE, 3rd, Kwon HJ, Yu WL, Liu HW. J Am Chem Soc. 2007;129:14670. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberthur M, Leimkuhler C, Kahne D. Org Lett. 2004;6:2873–76. doi: 10.1021/ol049187f. [DOI] [PubMed] [Google Scholar]
- 11.Xing J, Apedo A, Tymiak A, Zhao N. Rapid Commun Mass Spectrom. 2004;18:1599. doi: 10.1002/rcm.1524. [DOI] [PubMed] [Google Scholar]
- 12.Gaudu P, Touati D, Niviere V, Fontecave M. Journal Biol Chem. 1994;269:8182. [PubMed] [Google Scholar]
- 13.Inouye S. FEBS letters. 1994;347:163. doi: 10.1016/0014-5793(94)00528-1. [DOI] [PubMed] [Google Scholar]
- 14.He J, Hertweck C. J Am Chem Soc. 2004;126:3694–5. doi: 10.1021/ja039328t. [DOI] [PubMed] [Google Scholar]
- 15.Winkler R, Hertweck C. Angew Chem Int Ed. 2005;44:4083. doi: 10.1002/anie.200500365. [DOI] [PubMed] [Google Scholar]
- 16.Winkler R, Hertweck C. Chembiochem. 2007;8:973. doi: 10.1002/cbic.200700042. [DOI] [PubMed] [Google Scholar]
- 17.Kirner S, Hammer PE, Hill DS, Altmann A, Fischer I, Weislo LJ, Lanahan M, van Pee KH, Ligon JM. J Bacteriol. 1998;180:1939. doi: 10.1128/jb.180.7.1939-1943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Simurdiak M, Zhao H. J Biol Chem. 2005;280:36719. doi: 10.1074/jbc.M505334200. [DOI] [PubMed] [Google Scholar]
- 19.Gao Q, Thorson JS. FEMS Microbiol Lett. 2008;282:105. doi: 10.1111/j.1574-6968.2008.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmons SC, Thorson JS. Curr Opin Chem Biol. 2008;12:297. doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meneely KM, Lamb AL. Biochemistry. 2007;46:11930. doi: 10.1021/bi700932q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alfieri A, Fersini F, Ruangchan N, Prongjit M, Chaiyen P, Mattevi A. Proc Natl Acad Sci. 2007;104:1177. doi: 10.1073/pnas.0608381104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei B, Tu SC. J Bacteriol. 1996;178:5699. doi: 10.1128/jb.178.19.5699-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parry RJ, Li W, Cooper HN. J Bacteriol. 1997;179:409. doi: 10.1128/jb.179.2.409-416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental data on protein expression, purification and biochemical assays are available. This material is available free of charge via the Internet at http://pubs.acs.org.