Abstract
Synaptotagmin 1 (syt1) is an integral membrane protein localized on the synaptic vesicle that acts as the Ca2+ sensor for neuronal exocytosis. Synaptotagmin 1 contains two C2 domains, C2A and C2B, which bind Ca2+ ions, membranes and SNAREs. Here, site-directed-spin-labeling (SDSL) was used to determine the position and dynamics of the region that links the two C2 domains in a water soluble construct encompassing the two C2 domains (syt1C2AB). An analysis of the EPR lineshapes from this region indicates that the linker is flexible and unstructured when syt1 is in solution or bound to lipid bilayers. The ns dynamics of the linker does not change, either in the presence or absence of Ca2+, suggesting that there is no Ca2+-dependent intramolecular association between the two domains. When syt1C2AB is membrane bound, the position of the linker relative to the membrane interface was determined by measuring collision parameters for the spin-labeled syt1C2AB mutants to both soluble and membrane bound Ni(II) chelates. These data indicate that the linker does not penetrate the membrane surface, but lies approximately 7–10Å from the bilayer surface. In addition, the linker remains flexible when syt1C2AB binds to the SNARE complex, indicating that direct interactions between this linker and the SNAREs do not mediate association. These data suggest that the two C2 domains of syt1 interact independently on the membrane interface, or when bound to SNAREs.
Neurotransmitter release is the result of a highly-regulated Ca2+-triggered fusion event that is coordinated by large number of proteins within the pre-synaptic cell (1–3). Soluble N-ethylmaleimide–sensitive factor attachment receptor (SNARE1) proteins are membrane and membrane-associated proteins that assemble into a highly stable helical bundle that bridges the synaptic vesicle and the plasma membrane within the pre-synaptic cell. These SNARE proteins are essential for the fusion process, but they do not appear to act as the Ca2+-sensor in neuronal exocytosis. The available evidence suggests that synaptotagmin 1 (syt1), a protein composed of two C2 domains, acts as the Ca2+ sensor in neuronal exocytosis (4–6); however, the mechanism by which syt1 acts to trigger SNARE-mediated fusion, is not understood. For example, syt1 might directly bind to and regulate the SNAREs (7), syt1 might bind membranes and create curvature strain within the bilayer at the site of fusion (8), or syt1 might bridge across the vesicle and the plasma membrane driving these two membranes closer together (9, 10).
Synaptotagmin 1 is anchored to the synaptic vesicle through a single transmembrane segment located near its N-terminus, where the two water soluble C2 domains, C2A and C2B, are positioned on the C-terminal side of the transmembrane anchor. The two C2 domains are connected by a short linker region, and are known to bind to membranes containing negatively charged lipid in a Ca2+-dependent fashion (5, 6). When present as a water soluble fragment of syt1 containing both C2A and C2B (syt1C2AB) (see Figure 1), the two domains penetrate the bilayer surface in a Ca2+-dependent fashion so that the first and third Ca2+-binding loops of each domain are inserted into the bilayer (11). This fragment is also found to associate with the SNARE complex and there are reports that both C2A and C2B are responsible for the interactions with SNAREs. For example, NMR studies indicate that there are multiple binding sites for the syt1C2AB-SNARE interaction (12), and single molecule FRET studies imply that the C2B domain or the linker region of syt1C2AB may associate with SNAREs (13).
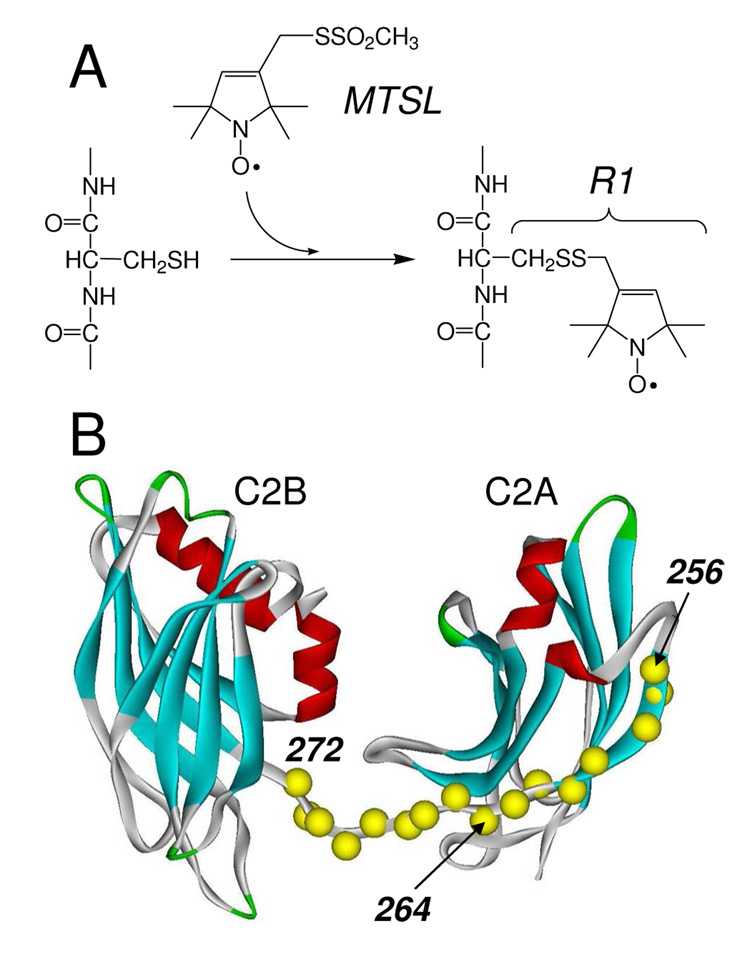
Figure 1.
A) The spin-labeled side-chain R1 is incorporated into syt1C2AB by derivatizing cysteine mutants with the MTSL. B) Model of a soluble portion of synaptotagmin 1 encompassing the two C2 domains. The first domain, C2A includes residues 140–263; the second, C2B, includes 273–418. The segment linking the two domains is formed from 264–272. Here, 16 single Cysteine mutants of a soluble fragment of syt1 (96–421) were produced (Cα positions in yellow). The C2AB model shown was built from models for the isolated C2A(PDB:1BYN) and C2B(PDB:1K5W) domains, that were connected with a linker using InsightII.
High resolution structural models for the individual C2A and C2B domains have been obtained (14–17), and recently, a Ca2+-free structure of syt1 C2AB was determined (18). In this crystal structure, the two C2 domains interact, so that the Ca2+-binding loops of C2A interact near the helix on C2B that connects strands 7 and 8. Several residues within the linker region, such as E267 and E271, also appear to make tertiary contact in this structure. Evidence for a direct interaction between the two C2 domains in syt1C2AB has also been obtained by FRET, but in this case, Ca2+ appears to induce an association of the two domains (19). In contrast to these two studies, NMR spectra of the soluble syt1C2AB fragment indicate that the two domains do not interact with each other in solution (9); however, there may be conditions, bound to membranes or bound to SNAREs, where the two domains associate.
Site-directed spin labeling (SDSL) is an EPR based method that is particularly sensitive to the local structure and dynamics at the labeled site, and SDSL can be utilized to examine proteins in solution, proteins that are bound membranes, or proteins that participate in large complexes (20–24). In the present work, we use SDSL to examine the state of the region that links C2A and C2B in the soluble fragment of synaptotagmin 1, syt1C2AB. Sixteen single cysteine mutants of syt1C2AB were expressed and derivatized with a sulfhydryl reactive spin label (Figure 1A) to incorporate the spin labeled side chain R1 into sites covering the 8th β-strand of C2A and the linker region that connects the two C2 domains in syt1C2AB (see Fig. 1B). In solution, the EPR data indicate that the linker is dynamic and unstructured, either in the presence or absence of Ca2+. Furthermore, the linker remains disordered when syt1C2AB is bound to lipid bilayers composed of POPC:POPS. When membrane bound, the collision accessibility to a surface associated Ni(II) complex indicates that the linker does not penetrate the bilayer, but lies approximately 7–10Å from a plane defined by the lipid phosphates. The flexibility of the linker is maintained when syt1C2AB binds to the soluble SNARE complex, indicating that the linker does not directly participate in the interactions with the SNAREs. Taken together, these data indicate that the two C2 domains do not associate but remain structurally independent under a wide range of conditions.
Materials and Methods
Mutagenesis, Protein Expression and Purification
DNA encoding residues for the C2A domain (96–265) and the C2A-C2B domains (96–421) of rat synaptotagmin 1 (P21707) in the vector pGEX-KG, were obtained from Carl Creutz (Pharmacology Department, University of Virginia). The single native cysteine residue at position 277 of syt1C2AB was mutated to alanine by typical polymerase chain reaction (PCR) strategies. All DNA modifications followed published protocols (25). Single-cysteine mutants of syt1C2A or syt1C2AB were prepared using either a two-step PCR or a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), and individual cysteine residues were introduced at positions 170, 176, 189, 193, 202, 217, 239, 240, 256 of the syt1C2A domain, and into positions 256, 257, 258, 260, 261, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271 and 272 of syt1C2AB (Fig. 1) All cysteine substitutions were confirmed by DNA sequencing.
The expression and purification syt1C2AB was carried out as described previously (11). Briefly, the construct was encoded in frame with glutathione S-transferase (GST), and mutant plasmids for GST-syt1C2AB were transformed into BL21(DE3)pLysS cells (Promega, Madison, WI) and grown. Following affinity purification, steps were taken to remove residual GST and thrombin, and an additional anion exchange step was added to remove nucleic acid contaminants associated with the protein as described previously (26). This fragment of syt1 consists of residues 96–421 from syt1 and 13 additional residues from the pGEX vector on the N-terminus. Protein purity and identity were confirmed by SDS-PAGE, and the purified protein was found to bind to Tb3+, using an assay described previously (11). The isolated syt1C2AB bound to POPC:POPS vesicles in a Ca2+ dependent manner as judged using a sucrose loaded vesicle sedimentation assay (27), and appeared to be correctly folded as indicated by CD spectroscopy and FTIR.
The synaptotagmin 1 C2A domain was also expressed as a GST fusion protein and was purified using GST affinity chromatography as described in a previous protocol (28). Purification of syt1C2A did not require the anion exchange step that was necessary for the purification of syt1C2AB.
The DNA of His6-tagged soluble SNARE motif fusion proteins, syntaxin 1A, SNAP-25 and synaptobrevin, encoded in the vector pET-28a were kindly provided by Reinhard Jahn and Dirk Fasshauer (Max-Plank-Institute for Biophysical Chemistry, Göttingen, Germany). The sequences covered were: syntaxin 180–262, SNAP-25 1–206 and synaptobrevin 1–96. All recombinant proteins were expressed as His6-tagged fusion proteins and purified by Ni2+-Sepharose affinity chromatography as described previously (29). All proteins were further purified by ion exchange chromatography using HiTrap Q HP or HiTrap S HP columns on an FPLC system (GE Healthcare, Piscataway, NJ). After loading, the proteins were eluted with a linear gradient of NaCl in 20 mM Tris (pH 7.4, 0–1M NaCl). The peak fractions were pooled and dialyzed against 20 mM Tris (pH 7.4) containing 50 mM NaCl. The ternary (syntaxin-SNAP-25-synaptobrevin) complex went through an initial purification step using a Q column (GE Healthcare, Piscataway, NJ) following the overnight assembly of the purified monomers. The minimal core complex was then further purified by size exclusion chromatogaphy on a Superdex 200 HiLoad 16/60 prep grade column in standard buffer containing 300 mM NaCl as described previously (29).
Spin Labeling Syt1C2AB
Single cysteine mutants of syt1C2AB were spin-labeled as described previously using the sulfhydryl reactive spin label MTSL, (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate) (Toronto Research Chemicals, North York, ON, Canada), at a 1:10 mole ratio of protein:MTSL. Excess free spin-label was removed by passing the sample through a HiPrep 26/10 desalting column (GE Healthcare, Piscataway, NJ), and the spin-labeled protein was concentrated to 20–150 µM using either Centriprep or Centricon micro concentrators (Millipore, Billerica, MA).
Large Unilamellar Vesicles
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) (Avanti Polar Lipids, Alabaster, AL) were used to prepare large unilamellar vesicles (LUVs) with a POPC:POPS ratio of 3:1 as described previously (28). Liposomes containing the Ni(II) lipid chelate 1,2-Dioleoyl-sn-Glycero-3-{[N(5-Amino-1-Carboxypentyl)iminodiAcetic Acid] Succinyl} (Nickel Salt) (DOGS-NTA-Ni(II)) (Avanti Polar Lipids, Alabaster, AL), were prepared in an identical manner with a POPC:POPS: DOGS-NTA-Ni(II) ratio of 65:25:10.
EPR Spectroscopy
EPR spectra were recorded on a Varian E-line 102 series X-band spectrometer fitted with a loop-gap resonator (Medical Advances, Milwaukee, WI). Spin-labeled samples of syt1C2AB contained protein concentrations that varied from 20–150 µM in the presence (1mM Ca2+) or absence of calcium (5mM EDTA). The spectra from membrane-bound syt1C2AB were obtained in the presence of LUVs (50mM total lipids) to ensure complete membrane binding and low surface densities of the protein. Unless otherwise noted, spectra were field sweeps of 100 G recorded using 2 mW of incident microwave power, and 100 kHz modulation with an amplitude of 1 G.
The scaled mobility, Ms, of the EPR spectrum provides a relative measure of nitroxide mobility, where a value of 1 represents the most mobile and a value of 0 represents the least mobile EPR spectra that are obtained from the R1 side chain (Fig. 1) in proteins. Ms is determined from the central linewidth of the EPR spectrum as given by Eq. 1.
| [1] |
Here, δexp is the measured central line of the EPR spectrum, and δi and δm are the corresponding values at the most immobilized and mobile sites observed in proteins. In the present study, we used values of 2.1G for δm and 8.4G for δi (30).
To determine the position of the linker region and other sites when syt1C2AB is bound to membranes, continuous-wave power saturation experiments were performed in the presence and absence of a secondary paramagnetic species as described previously (31, 32). Collision parameters, Π, for each paramagnetic reagent were also determined by normalizing the values of ΔP1/2 by the central linewidth and the P1/2 value for a solid sample of α, α’-diphenyl-β-picrylhydrazyl (DPPH) as described elsewhere (33). Two empirical depth parameters were used to estimate the position of the linker in syt1C2AB relative to the membrane interface. The first Φ1 given in Eq. 2 is based upon the ratio of oxygen and Ni(II)EDDA collision parameters.
| [2] |
The depth parameter Φ1 was previously calibrated in membranes composed of POPC:POPS (28, 34). A second depth parameter, Eq. 3, which is expected to be more sensitive to the distance variation in the aqueous phase near the membrane interface is based upon the ratio of the DOGS-NTA-Ni(II) to Ni(II)EDDA collision parameters, ΠDOGS-Ni and ΠNiEDDA, respectively.
| [3] |
In this case, the ratio of the collision parameter for DOGS-NTA-Ni(II) versus Ni(II)EDDA is taken to eliminate the effects of steric differences at the labeled sites. The dependence of the depth parameter Φ2, as a function of position is not known, and the first C2 domain of synaptotagmin 1, syt1C2A, was used to calibrate the parameter, since its orientation and position on POPC:POPS membranes is known. To make these measurements, R1 labeled mutants of the first C2 domain of synaptotagmin were produced as described previously (28) with labels that lie on the aqueous side of the membrane interface at positions 170, 176, 189, 193, 202, 217, 239, 240, and 256. The depth parameters, Φ2, were then compared with the distances from the nitrogen on the nitroxide to the plane of the lipid phosphates using the previously determined orientation and depth of C2A (28). To orient the nitroxides at individual sites, the most likely rotamers for the first three dihedral angles of the R1 side chain were selected based upon previous crystallography (35–37). Rotamers were eliminated which produced steric interference with other side chains or produced inconsistent positions of R1 with the values of the depth parameter Φ1 obtained for each site. The allowed rotations of the last two dihedral angles were used to estimate the error in the position of the side chain.
Determining the binding affinity of the SNARE complex to syt1C2AB
The binding of syt1C2AB to the SNARE complex was analyzed using EPR spectroscopy. The rate of exchange of syt1C2AB on and off the SNARE complex is slow relative to the EPR time scale. As a result, the EPR spectrum from a labeled site in syt1C2AB in the presence of the SNARE complex is a simple linear combination of the spectrum for unbound and bound syt1C2AB. In this case, the fraction of bound syt1C2AB, fb, can be given by:
| [4] |
Where A(0) is the amplitude of the peak-to-peak central nitroxide resonance, and Af and Ab represent the intrinsic amplitudes of the central resonance obtained for free and SNARE associated syt1C2AB, respectively.
Results
The protein segment connecting C2A and C2B is unstructured both in solution and bound to bilayers
Sixteen single labeled cysteine mutants were produced, expressed and labeled as described in Methods. As indicated above, this fragment of synaptotagmin 1 is folded and is found to bind to membranes containing POPC:POPS (3:1) in a Ca2+-dependent manner. Figure 2 shows the first derivative X-band EPR spectra obtained from these R1 mutants in solution in the presence of Ca2+ and bound to lipid vesicles composed of POPC:POPS (3:1). Residues 256 through 265 are positioned in the last β-strand of C2A and lie at the edge of the β-sandwich. Residues 266 through 271 are thought to encompass the segment that connects C2A with C2B.
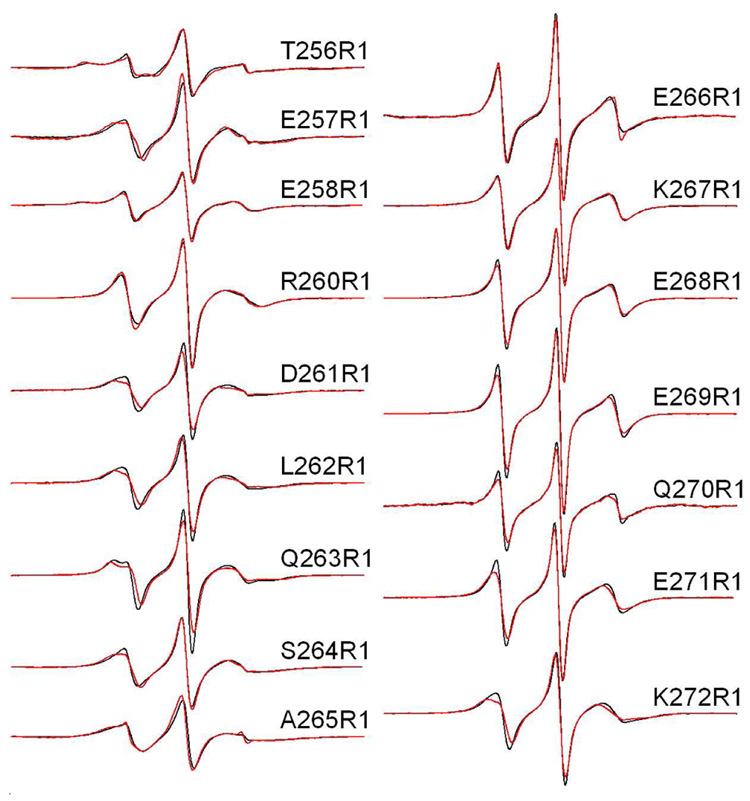
Figure 2.
X-band EPR spectra of single R1 substitutions in C2AB in the presence of Ca2+. Aqueous spectra are shown in black, spectra of C2AB completely bound to POPC:POPS (75:25) vesicles are shown in red. Spin-labeled mutants within the linker encompass residues 264–272 and mutants 256–263 cover the last β-strand in C2A. The spectra are normalized relative to each other, and the relative amplitudes provide an indication of relative nitroxide motion. The spectra are 100 Gauss scans. The spin-labeled protein concentrations in these experiments was 50 µM, with lipid concentrations of approximately 1000 fold higher (50 mM or greater). This lipid concentration is well in excess of that needed to completely bind syt1C2AB as shown previously (11).
The spectra in Figure 2 arise from labels that exhibit a range of motion, with the broadest lineshapes arising from sites in strand 8 of C2A, for example sites 256, 258, 262 and 265. These EPR lineshapes are similar to those that have been reported at other β-edge strand sites in cellular retinol binding protein (38). The most mobile lineshapes arise from residues in the linker region, 266 to 271. These spectra correspond to nitroxides that have effective correlations times of approximately 1.5 ns with highly isotropic motion. Relatively minor changes in the spectrum are detected upon membrane binding. In the absence of Ca2+, the EPR spectra obtained for the labeled sites shown in Figure 2 are unchanged from the Ca2+-bound case (supplementary data).
Shown in Figure 3 are the scaled mobilities calculated according to Eq. 4 for the sixteen residues in syt1C2AB, both in solution and bound to membranes. The scaled mobility, Ms, provides a relative measure of nitroxide motion where an Ms value of 1 represents the most mobile and a value of 0 represents the least mobile R1 side chains seen in proteins (30). This parameter is extracted from the peak-to-peak linewidth of the EPR spectrum and it is largely determined by the label correlation time (39). For residues in the linker region, the scaled mobilities are high, typically 0.8 to 0.9, indicating that they represent some of the most mobile R1 EPR lineshapes that are seen in proteins. Outside this region, the values of Ms are lower and range from about 0.5 to 0.8.
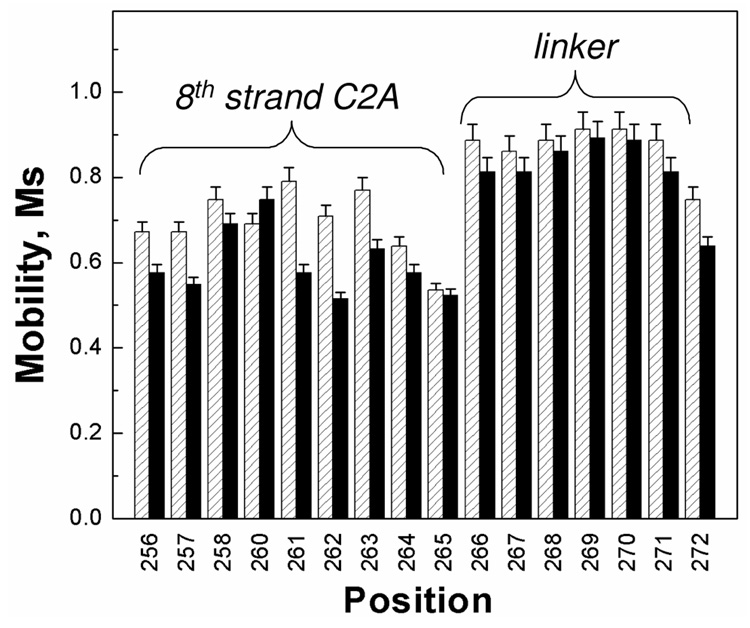
Figure 3.
The scaled mobilities, Ms, for the spin-labeled sites within syt1C2AB that are located in the linker region or in the 8th β-strand of C2A. Values of Ms were calculated from the central linewidth of the EPR spectrum using Eq. 1. Aqueous values are shown in gray, values for syt1C2AB bound to POPC:POPS (75:25) vesicles are shown in black. When bound to lipid bilayers, there is a small decrease in the scaled mobility at some sites. Some of this decrease is likely due to a decrease in protein rotation and attachment of C2A and C2B upon membrane binding. Several sites show larger changes upon binding, which may indicate changes in local structure or dynamics upon membrane association (see text).
When bound to the lipid bilayer (Fig. 3, solid bars), many labeled sites show a small decrease in label mobility. The most significant changes take place at sites 261 to 263 near the attachment of the linker region to C2A. The addition of solutes such as Ficoll, which increases solution viscosity and slows the protein correlation time (40), produces little change in the EPR spectra from syt1C2AB. This indicates that the changes seen at sites 261–263 may be due to small structural changes or changes in dynamics at this β-strand edge upon membrane binding.
Parameters that are obtained from the EPR spectrum of R1, such as the central linewidth and second moment, are an indicator of the local structure at the labeled site. A plot of the reciprocal central linewidth (ΔH0−1) versus the reciprocal of the second moment (<H2>−1) yields a map that defines regions of different label contact and environment (41, 42). These data for the syt1C2AB spectra obtained in solution are shown in Figure 4. As indicated in this plot, residues encompassing the linker region have lineshapes that are characteristic of unstructured protein segments or positions in loop segments.
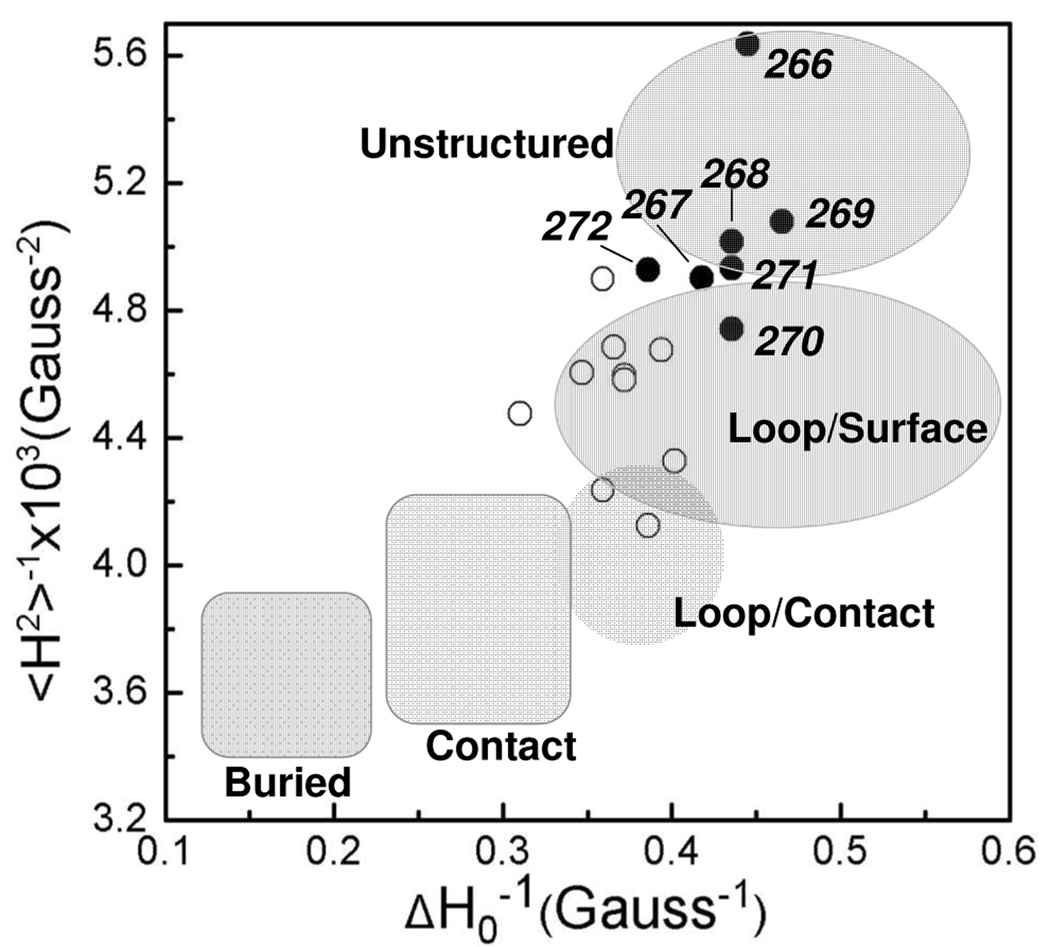
Figure 4.
Plot of the reciprocal of the central linewidth (ΔH0−1) versus the reciprocal of the second moment (<H2>−1) of the EPR spectrum for the labeled sites in syt1C2AB. Residues that make up the flexible portion of the linker, 266–272, are plotted with solid circles. The open circles represent sites outside the linker and in the 8th β-strand of C2A. Many sites within the linker have spectral characteristics of an unstructured protein segment; other sites in the linker resemble those in very dynamic loop segments. The shaded areas correspond to those defined previously, based upon EPR lineshapes from R1 at sites in proteins of known structure (42).
The spectra shown in the presence of bilayers in Figure 2 are obtained under conditions where syt1C2AB is completely membrane bound, so that the first and third Ca2+-binding loops are contacting and penetrating the bilayer interface (11). These data indicate that the linker between the two C2 domains remains flexible when the protein is membrane bound. The data suggest that the two domains are interacting independently when associated with the membrane interface. There is no evidence for a Ca2+-dependent change in the structure or dynamics of the linker region, and labels in the linker region do not show any evidence for tertiary contact. Such contact would have been expected at positions 267 and 271 if syt1C2AB assumed the conformation seen in the crystal structure (18).
Spin labels in the linker have an aqueous exposure and lie 7 to 10 Å away from the plane of the phosphates when syt1C2AB is membrane bound
To determine whether the linker region comes close to or penetrates the bilayer surface, power saturation experiments were performed to measure the exposure of R1 sites on the linker to the secondary paramagnetic species oxygen, and Ni(II)EDDA (see methods). Shown in Table 1 are the collision parameters, Πoxy and ΠNiEDDA, and the depth parameters, Φ1, for residues 262–269 covering one edge of the β-strand on C2A though the liner region. The values of Φ1 are all less than −2.0, which indicates that the labeled sites have an aqueous exposure and are greater than 5Å away from the level of the lipid phosphates (28).
Table 1.
Depth parameters, Φ1, for syt1C2A and syt1C2AB†
| Mutant | Πoxy | ΠNiEDDA | Φ1 |
|---|---|---|---|
| C2AB L262R1 | 0.13±0.01 | 1.40±0.1 | −2.4±0.1 |
| C2AB Q263R1 | 0.14±0.02 | 1.20±0.1 | −2.2±0.1 |
| C2AB S264R1 | 0.14±0.02 | 0.99±0.07 | −2.2±0.1 |
| C2AB A265R1 | 0.13±0.02 | 1.18±0.08 | −2.2±0.1 |
| C2AB Q266R1 | 0.17±0.03 | 1.17±0.10 | −2.2±0.1 |
| C2AB K267R1 | 0.16±0.02 | 2.0±0.1 | −2.5±0.1 |
| C2AB E268R1 | 0.22±0.02 | 2.1±0.1 | −2.3±0.1 |
| C2AB E269R1 | 0.22±0.03 | 2.2±0.2 | −2.3±0.1 |
Depth parameters for the single R1 labeled sites when syt1C2A or syt1C2AB are completely bound to vesicles composed of POPC:POPS (75:25). Errors are based upon fits of the data to P1/2.
To obtain a better estimate for the distance of the linker from the interface, we utilized a membrane bound paramagnetic reagent, DOGSNTA-Ni(II). In this Ni(II) lipid chelate, the metal center is believed to be extended into the aqueous phase so that the maximum concentration of the reagent resides in a layer that is approximately 14 Å above the membrane-lipid interface (43). Our expectation is that the collision rate with DOGS-NTANi(II) will drop off on either side of this layer, but may be quite broad due to dynamic motion of lipids in the bilayer interface. To account for steric differences between labeled sites and to obtain a depth parameter, the collision frequency with DOGSNTA-Ni(II) was referenced against Ni(II) EDDA, which is expected to diminish in concentration only near the interface (see Methods).
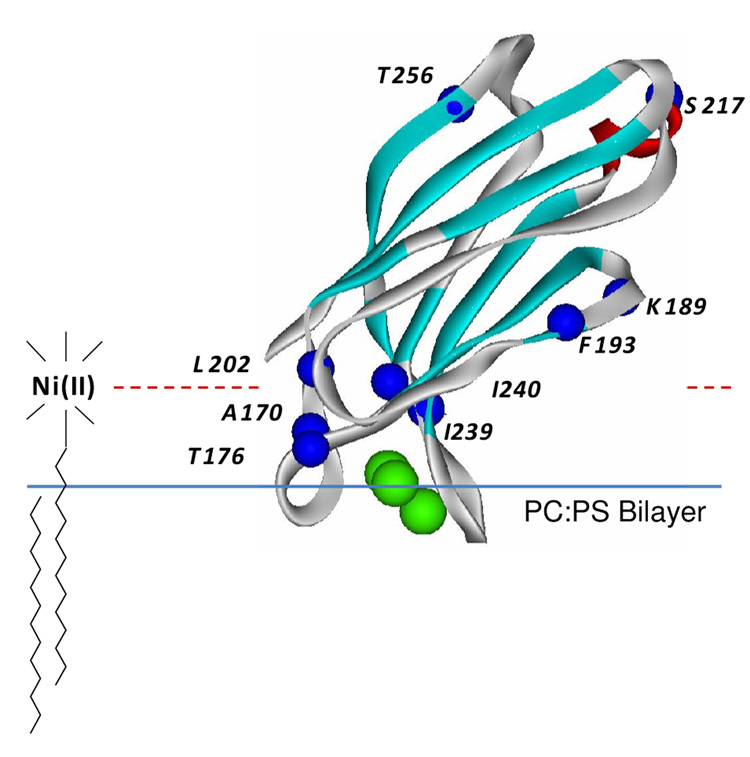
To calibrate this approach, we used the C2A domain from syt1. The orientation of this domain was previously determined on membranes composed of POPC:POPS (see Figure 5) (28) and the positions of labels placed within this domain are known relative to the membrane interface. The R1 spin label was attached to the sites indicated in Figure 5, and measurements of Πoxy, ΠNiEDDA, and ΠDOGSNi were made for 8 R1 mutants of C2A bound to extruded vesicle bilayers composed of POPC:POPS or POPC:POPS:DOGS-NTA-Ni(II) (see Methods). The data obtained in these measurements is shown in Table 2, and the dependence of the depth parameter, Φ2, upon the expected position of the R1 label relative to the membrane interface is shown in Figure 6.
Figure 5.
High resolution model for syt1C2A (PDB 1BYN) (15). The domain is shown bound to bilayers composed of POPC:POPS with a depth of penetration and orientation that were previously determined from SDSL (28). This structure was used to calibrate the position parameter Φ2 as described in Methods. This parameter is expected to be sensitive to the position of the label on the aqueous side of the bilayer interface and utilizes the lipid bound nickel chelate, DOGS-NTA-Ni(II). Single cysteine mutants were generated at the indicated residues (Cα carbons highlighted in blue) to incorporate a series of R1 labels (see Fig. 1A) at varied positions off the bilayer interface.
Table 2.
Depth parameter, Φ2, for syt1C2A and syt1C2AB.†
| Mutant | Πoxy | ΠNiEDDA | ΠDOGSNTA | Φ2 |
|---|---|---|---|---|
| C2A A170R1 | 0.27±0.03 | 3.0±0.2 | 0.18±0.02 | −2.8±0.1 |
| C2A T176R1 | 0.24±0.02 | 2.6±0.2 | 0.68±0.04 | −1.4±0.1 |
| C2A K189R1 | 0.26±0.02 | 3.3±0.2 | 0.07±0.02 | −3.9±0.3 |
| C2A F193R1 | 0.17±0.02 | 1.4±0.1 | 0.11±0.02 | −2.6±0.2 |
| C2A L202R1 | 0.30±0.02 | 4.9±0.3 | 0.41±0.03 | −2.5±0.1 |
| C2A I239R1 | 0.14±0.01 | 1.2±0.1 | 0.89±0.05 | −0.3±0.1 |
| C2A I240R1 | 0.12±0.01 | 1.6±0.1 | 0.06±0.01 | −3.3±0.2 |
| C2A S217R1 | 0.40±0.04 | 3.7±0.2 | - | - |
| C2A T256R1 | 0.26±0.02 | 3.4±0.2 | 0.004±0.01 | −6.7±2.2 |
| C2AB L262R1 | 0.13±0.01 | 1.4±0.1 | 0.1±0.01 | −2.7±0.1 |
| C2AB Q263R1 | 0.14±0.02 | 1.2±0.1 | 0.1±0.01 | −2.6±0.1 |
| C2AB K267R1 | 0.16±0.02 | 2.0±0.1 | 1.10±0.07 | −0.6±0.1 |
| C2AB E268R1 | 0.22±0.02 | 2.1±0.1 | 0.57±0.05 | −1.3±0.1 |
| C2AB E269R1 | 0.22±0.03 | 2.2±0.2 | 1.39±0.10 | −0.5±0.1 |
Depth parameters for the single R1 labeled sites when syt1C2A or syt1C2AB are completely bound to vesicles composed of POPC:POPS or POPC:POPS:DOGS-NTA-Ni(II). Errors are based upon fits of the data to P1/2.
Figure 6.
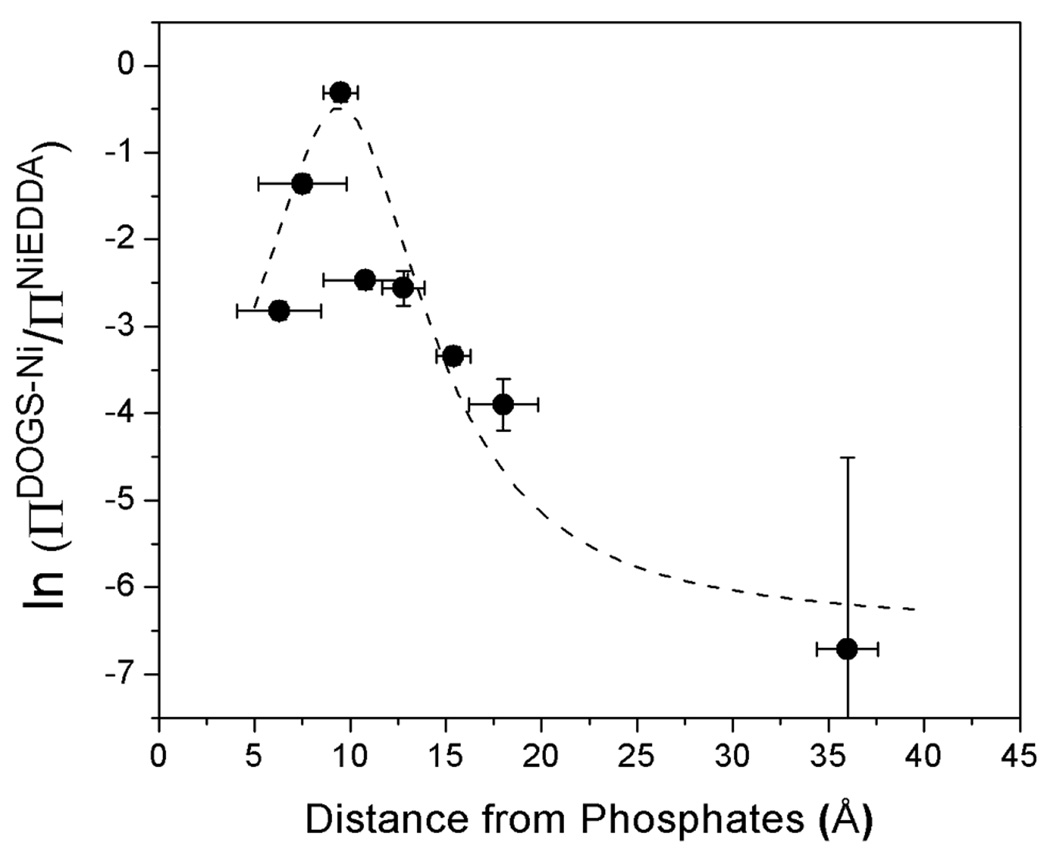
The correlation of the position or depth parameter Φ2 at sites on syt1C2A as a function of distance from the membrane surface. The parameter Φ2 was determined using ΔP1/2 values for Ni(II)EDDA and DOGS-NTA-Ni(II) as given in Eq. 3. The distances of the R1 label on C2A were extracted from a model for membrane bound syt1C2A (28) using the procedure described in Methods. The dashed line shows is a Lorentzian centered around 9.5 Å.
Spin labels on C2A at positions 176, 202 and 239 are located approximately 8–10Å above the membrane surface, and they have the highest collision parameters, ΠDOGSNi, and the least negative depth parameters, Φ2. The remaining sites are above or below 8–10 Å, and they have lower values of ΠDOGSNi and lower values of Φ2. As seen in Figure 6, the value of Φ2 appears to undergo a maximum as expected, and the position of this maximum lies at approximately 8–10Å in these bilayers composed of POPC/POPS/DOGS-NTA-Ni(II). In addition to the change in saturation behavior, the high collision rate between R1 and Ni(II) leads to a slight broadening in the EPR spectra. This effect is most notable for the spectra obtained from sites 176, 202 and 239 (see supplemental data).
Depth measurements using DOGSNTA-Ni(II) were made for several sites in the central linker for membrane bound syt1C2AB. As shown in Table 2, the values of ΠDOGSNi are very high for labels in the linker (267–269), and the values of Φ2 for the linker residues 267–269 in C2AB lie between the values obtained for sites 176 and 239 on C2A. These data indicate that the central linker is located at approximately the same distance as these residues in C2A, which is 7–10Å above the membrane surface and near the region where the concentration of Ni(II) is a maximum for the DOGSNTA-Ni(II) containing liposomes. The EPR lineshapes from the linker sites also indicate the linker is close to this Ni(II) layer, and the spectra from these sites show significant broadening due to collisional exchange with Ni(II) (see Supplemental).
The linker between C2A and C2B remains flexible when syt1C2AB binds to the core SNARE complex
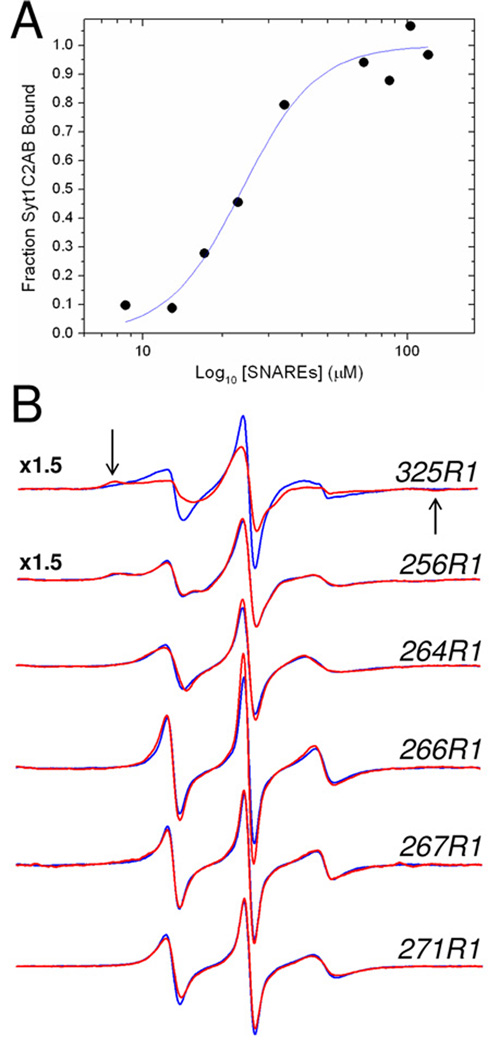
To determine whether the linker in syt1 remains flexible when syt1C2AB binds to SNAREs, or is directly involved in mediating interactions with SNAREs, EPR spectra were obtained from spin-labeled sites within the linker under conditions where syt1C2AB is bound to the core SNARE complex. The binding affinity of syt1C2AB was estimated by placing a spin label at position 325 in the C2B domain of syt1C2AB, which resides within the polybasic region of C2B. This region thought to make up a region of C2B that interacts with the SNARE complex (12). The spectra shown in Figure 7 for 325R1 were taken in the absence and presence of the SNARE complex. This EPR spectrum broadens dramatically in the presence of the SNAREs, indicating that label at this site undergoes a decrease in motion upon SNARE binding. The appearance of the broad hyperfine extrema in this spectrum of 325R1 (arrows Fig. 7B) indicates that the change in the EPR spectrum results from tertiary contact at the labeled site. Similar changes are seen in other resides within the polybasic region of the C2B domain (data not shown) and are due to tertiary contact of the SNARE proteins at this site. Using this labeled site, the spectrum of 325R1 was recorded as a function of the concentration of the core SNARE complex, and the fraction of syt1C2A bound to the SNAREs, which was calculated according to Eq. 4, is presented in Fig. 7A. The fit to these data yields an affinity of the soluble SNAREs for syt1C2AB of approximately 25 µM.
Figure 7.
A) Titration of the fraction of syt1C2AB bound to SNAREs as a function of the concentration of added soluble SNARE complex. The fraction of bound label was determined using Eq. 4. The fit to the data was made using a standard Hill Equation, and yield an affinity of 24 µM. A slight apparent cooperativity is found in this binding with n=2. In this titration, the concentration of syt1C2AB was held constant at 23 µM with a Ca2+ concentration of 1 mM. B) EPR spectra of single R1 substitutions in the absence (blue trace) or presence (red trace) of 120 M SNARE complex in the presence of 1 mM Ca2+. Site 325 is located in the C2B domain and is likely at a site that is involved in tertiary contact with the SNAREs. The arrows indicate the position of the hyperfine extrema in this spectrum. Site 256 lies within the 8th β-strand in the C2A domain, and sites 264, 266, 267 and 271 lie within the linker connecting C2A and C2B. The amplitudes of these EPR spectra are normalized against the second integral of the EPR spectrum and are expanded by a factor of 1.5 for 325R1 and 256R1.
Shown in Figure 7B are EPR spectra recorded for site 256 in the 8th β-sheet of the C2A domain, and four sites in the linker in the absence and presence of 120 µM SNARE complex. This concentration that is sufficient to bind virtually all syt1C2AB according to the data obtained in Fig. 7A. These spectra show essentially no change as a function of SNARE addition, and indicate that the dynamics of the linker on the ns time scale are not altered by the binding of syt1C2AB with the SNAREs. Furthermore, the lack of change in these spectra indicates that tertiary contact between these labeled sites in the linker, or nearby side chains in the linker, with the SNAREs is not taking place upon SNARE binding. Thus, sites within the linker connecting C2A and C2B do not directly contact the SNAREs.
Discussion
Site-directed spin labeling is a powerful approach to obtain information regarding backbone dynamics and tertiary contact at the labeled site. In this work, we used SDSL to examine the protein segment that links the first and second C2 domains in syt1. The data indicate that the region connecting the two C2 domains is highly dynamic and unstructured, and there is no evidence for tertiary interactions of spin-labeled side chains in this linker with other regions of the protein. The flexibility of the linker is maintained either in solution (with or without Ca2+) or when the two C2 domains of syt1 are bound to bilayers of POPC:POPS. Furthermore the linker remains dynamic and unstructured when syt1C2AB is bound to the core SNARE complex, and the sites examined within the linker do not directly interact with the SNARE complex.
These data also suggest that the two C2 domains in syt1 do not directly interact. A direct interaction of the two C2 domains, as indicated in the crystal structure (18), should restrain the linker and bring certain side-chains within the linker into tertiary contact with other regions of syt1. However, these interactions are not seen, and there is no evidence for a significant change in local ordering within the linker with Ca2+ addition. These results are consistent with NMR data showing that C2A and C2B have the same chemical shifts and do not interact in solution (9). However, the data obtained here also suggest that the two C2 domains function independently when syt1C2AB is bound membranes or to SNAREs, since the dynamics of the linker are unchanged upon membrane or SNARE association.
The interactions of syt1 with the SNAREs have been reported to be mediated by the two C2 domains of syt1 (for example, (12, 13, 44)), and models where the linker between C2A and C2B straddle the bilayer have been proposed (4, 45). The measurements made here indicate that the flexibility of the linker is maintained when syt1C2AB is bound to the SNAREs. This result is consistent with a report indicating that syt1-SNARE interactions are largely mediated by C2B (12, 46), and that there may be multiple binding modes for the syt1-SNARE interaction (12). Previous data obtained from single molecule FRET indicated that sites within the linker were in close proximity to the SNARE complex when syt1 was bound (13). The results obtained here indicate that while residues within the linker might be close to the SNAREs when syt1 is bound, they do not directly interact with the SNAREs. However, the measurements made here are not entirely analogous to these FRET measurements, since they have not yet been carried out in the presence of membrane bound SNAREs.
At the present time, the mechanism by which syt1 triggers neuronal exocytosis is not known. Synaptotagmin 1 appears to interact both with SNAREs and with membranes (45, 47), and it is likely that both these interactions are necessary for the action of syt1 (12, 48). Many models for syt1-regulated membrane fusion are consistent with the results found here, and require that the two C2 domains interact independently. For example, it has been proposed that C2A and C2B bind to the vesicle and target (plasma) membrane respectively, thereby assuming a trans-configuration. This requires that protein segment connecting the two domains be flexible (10). C2A and C2B are also reported to have different lipid specificities, where C2B appears to have a higher affinity towards PI(4,5)P2, which is found in the plasma membrane (49). Based upon this finding, it has been suggested that the two C2 domains of syt1 may target different membranes under different conditions, again requiring that the linker be flexible. However, the length of the linker appears to be important. Previous work demonstrated that lengthening the linker abolishes the interaction between syt1C2AB and the SNAREs and decreases the membrane fusion efficiency (50). The results imply that while the two domains may be flexibly linked, the relative position or separation between the two domains is important.
The data shown in Figure 7 indicate that the affinity of the core SNARE complex for syt1C2AB in solution is approximately 25 µM. This is three orders of magnitude weaker than the apparent affinity measured for syt1C2AB towards the membrane bound SNARE complex (51). This dramatic discrepancy and the increased affinity of syt1C2AB towards membrane bound SNAREs is expected if there are multiple binding sites for syt1; for example, if syt1C2AB makes simultaneous interactions with both the membrane interface and the SNAREs. Indeed, previous work suggests that there is a complex formed between syt1, SNAREs and the membrane interface (12). In this case the free energies of interaction of syt1C2AB towards SNAREs and the lipid bilayer should be roughly additive.
Previous work using site-directed spin labeling indicated that both the first and third Ca2+-binding loops of the C2A and C2B domains of syt1 penetrated the bilayer when sytC2AB is membrane bound (11). This model was based upon depth data from SDSL, and the high-resolution solution structures of the individual C2A and C2B domains. In this model, the extensions of the C-terminal end of C2A and the N-terminal end of C2B are projected towards the bilayer interface, but no data on the separation between domains or the position of the linker were obtained. The collision accessibility data obtained here using Ni(II)EDDA or DOGS-NTA-Ni(II) indicate that the linker does not penetrate the bilayer but resides on the aqueous side of the membrane-solution interface. Presently, we are using data on the linker, depth constraints, and distance constraints obtained from double electron-electron resonance, to generate a more refined structural model for membrane associated syt1C2AB.
Information on the position of protein segments lying on the aqueous side of the membrane solution interface can be obtained using gradients of charged paramagnetic regents, such as chromium oxalate (52, 53). In this approach, high surface charge densities and low ion strength are typically utilized to establish a concentration gradient of the charged reagent. The lipid chelate DOGS-NTA-Ni(II) was used previously to identify spin-labeled sites on KcsA that lie near the membrane interface (43). The present work demonstrates that this lipid-Ni(II) chelate can also be used to localize spin labels on proteins at positions off the membrane interface, and that the Ni(II) bound to DOGS-NTA has a maximum concentration in a region that lies 8 to 10 Å from the layer of the phosphates on the membrane interface. The collision parameters for DOGS-NTA-Ni(II) will be sensitive to the relative diffusion rates of DOGS-NTA-Ni(II) within different bilayers or different membrane regions. This may require different calibrations on different membrane surfaces, and might lead to interesting effects in membrane proteins if there are regions of immobilized lipid near labeled sites.
In summary, the position and conformation of the region that links C2A and C2B in syt1 was determined by site-directed spin labeling. This region of syt1C2AB is flexible and largely unstructured in solution, with or without Ca2+ and when bound to the membrane interface. When syt1C2AB is bound to membranes, residues within the linker reside approximately 7–10Å on the aqueous side of the lipid phosphates. The linker remains flexible and unstructured when syt1C2AB is bound to the SNARE complex, and it does not directly participate in syt1/SNARE interactions. These results are consistent with models for membrane fusion where the two C2 domains interact independently, either on membrane or protein sites.
Supplementary Material
Supporting information includes additional EPR spectra obtained for syt1C2AB in solution in the presence and absence of Ca2+, EPR spectra for syt1C2A, and EPR spectra for bound syt1C2AB in the presence of DOGS-NTA-Ni(II). This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
We would like to thank Drs. Reinhard Jahn and Dirk Fasshauer (Max-Planck-Institute, Gottingen, Germany) for providing DNA for the His-tagged SNARE proteins, Drs. Wayne Hubbell, and Christian Altenbach (UCLA) for LabView software used in the EPR data analysis, and members of Dr. Lukas Tamm’s lab for assistance with the assembly of the SNARE complexes.
Footnotes
This work was supported by grants from the National Institutes of Health, GM 62305 and GM 072694.
Abbreviations used: DOGS-NTA-Ni(II), 1,2-Dioleoyl-sn-Glycero-3-{[N(5-Amino-1-Carboxypentyl) iminodiAcetic Acid] Succinyl} (Nickel Salt); EPR, electron paramagnetic resonance spectroscopy; LUV, large unilamellar vesicle; MTSL, (1-oxy-2,2,5,6-tetramethylpyrroline-3-methyl) methanethiosulfonate; PI(4,5)P2, phosphatidylinositol 4–5 bisphosphate; POPC, palmitoyloleoylphosphatidycholine; POPS, palmitoyloleoylphosphatidylserine; R1, spin-labeled side chain produced by derivatization of a cysteine with MTSL; SDSL, site-directed spin labeling; SNARE, soluble N-ethylmaleimide–sensitive factor attachment receptor; syt1, synaptotagmin 1; syt1C2AB, soluble fragment of syaptotagmin 1 containing the C2A and C2B domains; syn1C2A, C2A domain of synaptotagmin 1.
References
- 1.Jahn R, Lang T, Suhof TC. Membrane Fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 2.Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 3.Sudhof TC. The synaptic vesicle cycle. Annual Review of Neuroscience. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 4.Koh TW, Bellen HJ. Synaptotagmin I, a Ca2+ sensor for neurotransmitter release. Trends in Neurosciences. 2003;26:413–422. doi: 10.1016/S0166-2236(03)00195-4. [DOI] [PubMed] [Google Scholar]
- 5.Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 7.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Ca(2+)-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 8.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 9.Arac D, Chen X, Khant HA, Ubach J, Ludtke SJ, Kikkawa M, Johnson AE, Chiu W, Sudhof TC, Rizo J. Close membrane-membrane proximity induced by Ca2+-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 10.Stein A, Radhakrishnan A, Riedel D, Fasshauer D, Jahn R. Synaptotagmin activates membrane fusion through a Ca2+-dependent trans interaction with phospholipids. Nat Struct Mol Biol. 2007;14:904–911. doi: 10.1038/nsmb1305. [DOI] [PubMed] [Google Scholar]
- 11.Herrick DZ, Sterbling S, Rasch KA, Hinderliter A, Cafiso DS. Position of synaptotagmin I at the membrane interface: cooperative interactions of tandem C2 domains. Biochemistry. 2006;45:9668–9674. doi: 10.1021/bi060874j. [DOI] [PubMed] [Google Scholar]
- 12.Dai H, Shen N, Arac D, Rizo J. A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release. Journal of Molecular Biology. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen ME, Weninger K, Ernst J, Chu S, Brunger AT. Single-molecule studies of synaptotagmin and complexin binding to the SNARE complex. Biophys J. 2005;89:690–702. doi: 10.1529/biophysj.104.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 15.Shao X, Fernandez I, Sudhof TC, Rizo J. Solution structures of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: does Ca2+ induce a conformational change? Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez I, Arac D, Ubach J, Gerber SH, Shin O, Gao Y, Anderson RG, Sudhof TC, Rizo J. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Sequeira SM, Malinina L, Tereshko V, Sollner TH, Patel DJ. Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain. Protein Sci. 2004;13:2665–2672. doi: 10.1110/ps.04832604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuson KL, Montes M, Robert JJ, Sutton RB. Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association. Biochemistry. 2007;46:13041–13048. doi: 10.1021/bi701651k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia RA, Forde CE, Godwin HA. Calcium triggers an intramolecular association of the C2 domains in synaptotagmin. Proc Natl Acad Sci U S A. 2000;97:5883–5888. doi: 10.1073/pnas.100127197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 21.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 22.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 23.Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Klug CS, Feix JB. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol. 2008;84:617–658. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 26.Rufener E, Frazier AA, Wieser CM, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 27.Buser CA, Sigal CT, Resh MD, McLaughlin S. Membrane binding of myristylated peptides corresponding to the NH2 terminus of Src. Biochemistry. 1994;33:13093–13101. doi: 10.1021/bi00248a019. [DOI] [PubMed] [Google Scholar]
- 28.Frazier AA, Roller CR, Havelka JJ, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42:96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- 29.Fasshauer D, Eliason WK, Brunger AT, Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998;37:10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- 30.Langen R, Cai K, Altenbach C, Khorana HG, Hubbell WL. Structural features of the C-terminal domain of bovine rhodopsin: a site-directed spin-labeling study. Biochemistry. 1999;38:7918–7924. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 31.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient-method to determine the immersion depth of nitroxides in lipid bilayers. Application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Victor K, Cafiso DS. Structure and position of the N-terminal binding domain of pp60src at the membrane interface. Biochemistry. 1998;37:3402–3410. doi: 10.1021/bi9721501. [DOI] [PubMed] [Google Scholar]
- 33.Farahbakhsh ZT, Altenbach C, Hubbell WL. Spin-labeled cysteines as sensors for protein-lipid interaction and conformation in rhodopsin. Photochemistry and Photobiology. 1992;56:1019–1033. doi: 10.1111/j.1751-1097.1992.tb09725.x. [DOI] [PubMed] [Google Scholar]
- 34.Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, Falke JJ, Cafiso DS. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 35.Langen R, Oh KJ, Cascio D, Hubbell WL. Crystal structures of spin labeled T4 lysozyme mutants: implications for the interpretation of EPR spectra in terms of structure. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 36.Guo Z, Cascio D, Hideg K, Kalai T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16:1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17:228–239. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lietzow MA, Hubbell WL. Motion of spin label side chains in cellular retinol-binding protein: correlation with structure and nearest-neighbor interactions in an antiparallel beta-sheet. Biochemistry. 2004;43:3137–3151. doi: 10.1021/bi0360962. [DOI] [PubMed] [Google Scholar]
- 39.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4–58 bZip free and bound to DNA. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 40.Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in a-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 41.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 42.Isas JM, Langen R, Haigler HT, Hubbell WL. Structure and dynamics of a helical hairpin and loop region in annexin 12: a site-directed spin labeling study. Biochemistry. 2002;41:1464–1473. doi: 10.1021/bi011856z. [DOI] [PubMed] [Google Scholar]
- 43.Gross A, Hubbell WL. Identification of protein side chains near the membrane-aqueous interface: a site-directed spin labeling study of KcsA. Biochemistry. 2002;41:1123–1128. doi: 10.1021/bi015828s. [DOI] [PubMed] [Google Scholar]
- 44.Chapman ER, Hanson PI, An S, Jahn R. Ca2+ regulates the interaction between synaptotagmin and syntaxin 1. J Biol Chem. 1995;270:23667–23671. doi: 10.1074/jbc.270.40.23667. [DOI] [PubMed] [Google Scholar]
- 45.Bai J, Chapman ER. The C2 domains of synaptotagmin--partners in exocytosis. Trends Biochem Sci. 2004;29:143–151. doi: 10.1016/j.tibs.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Rickman C, Archer DA, Meunier FA, Craxton M, Fukuda M, Burgoyne RD, Davletov B. Synaptotagmin Interaction with the Syntaxin/SNAP-25 Dimer Is Mediated by an Evolutionarily Conserved Motif and Is Sensitive to Inositol Hexakisphosphate. J Biol Chem. 2004;279:12574–12579. doi: 10.1074/jbc.M310710200. [DOI] [PubMed] [Google Scholar]
- 47.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 48.Lynch KL, Gerona RRL, Larsen EC, Marcia RF, Mitchell JC, Martin TFJ. Synaptotagmin C2A loop 2 mediates Ca2+-dependent SNARE interactions essential for Ca2+-triggered vesicle exocytosis. Mol Biol Cell. 2007;18:4957–4968. doi: 10.1091/mbc.E07-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai J, Tucker WC, Chapman ER. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat Struct Mol Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 50.Bai J, Wang CT, Richards DA, Jackson MB, Chapman ER. Fusion pore dynamics are regulated by synaptotagmin-t-SNARE interactions. Neuron. 2004;41:929–942. doi: 10.1016/s0896-6273(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 51.Tang J, Maximov A, Shin OH, Dai H, Rizo J, Sudhof TC. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 52.Lin Y, Nielsen R, Murray D, Hubbell WL, Mailer C, Robinson BH, Gelb MH. Docking phospholipase A2 on membranes using electrostatic potential-modulated spin relaxation magnetic resonance. Science. 1998;279:1925–1929. doi: 10.1126/science.279.5358.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ball A, Nielsen R, Gelb MH, Robinson BH. Interfacial membrane docking of cytosolic phospholipase A2 C2 domain using electrostatic potential-modulated spin relaxation magnetic resonance. Proc Natl Acad Sci U S A. 1999;96:6637–6642. doi: 10.1073/pnas.96.12.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information includes additional EPR spectra obtained for syt1C2AB in solution in the presence and absence of Ca2+, EPR spectra for syt1C2A, and EPR spectra for bound syt1C2AB in the presence of DOGS-NTA-Ni(II). This material is available free of charge via the Internet at http://pubs.acs.org.