Abstract
L1 is a cell adhesion molecule of the immunoglobulin (Ig) superfamily, critical for central nervous system development, and involved in several neuronal biological events. It is a type I membrane glycoprotein. The L1 ectodomain, composed of six Ig-like and five fibronectin (Fn) type-III domains, is involved in homophilic binding. Here, co-immunoprecipitation studies between recombinant truncated forms of human L1 expressed and purified from insect Spodoptera frugiperda Sf9 cells, and endogenous full-length L1 from human NT2N neurons, showed that the L1 ectodomain (L1/ECD) and L1/Ig1–4 interacted homophilically in trans, contrary to mutants L1/Ig1–3 and L1/Ig2-Fn5. All mutants were correctly folded as evaluated by combination of far-UV CD and fluorescence spectroscopy. Surface plasmon resonance analysis showed comparable dissociation constants of 116 ± 2 and 130 ± 6 nm for L1/ECD-L1/ECD and L1/ECD-L1/Ig1–4, respectively, whereas deletion mutants for Ig1 or Ig4 did not interact. Accordingly, in vivo, Sf9 cells stably expressing L1 were found to adhere only to L1/ECD- and L1/Ig1–4-coated surfaces. Furthermore, only these mutants bound to HEK293 cells overexpressing L1 at the cell surface. Enhancement of neurite outgrowth, which is the consequence of signaling events caused by L1 homophilic binding, was comparable between L1/ECD and L1/Ig1–4. Altogether, these results showed that domains Ig1 to Ig4 are necessary and sufficient for L1 homophilic binding in trans, and that the rest of the molecule does not contribute to the affinity under the conditions of the current study. Furthermore, they are compatible with a cooperative interaction between modules Ig1–Ig4 in a horseshoe conformation.
The neural cell adhesion molecule L1 is a member of the immunoglobulin (Ig)3 superfamily implicated in many neuronal development steps, including neurite elongation, cell adhesion, and axon guidance and fasciculation (reviewed in Ref. 1). The multiple interactions depend on the structure of L1, which consists of a highly glycosylated extracellular region composed by six Ig-like domains (Ig1–6) and five fibronectin type III-like domains (Fn1–5), a transmembrane region and a cytoplasmic tail (Fig. 1A; reviewed in Ref. 2). L1 mediates its effects through the interaction with a variety of ligands, which include L1 (homophilic binding), other neuronal members of the Ig superfamily, integrins, and extracellular matrix components (heterophilic binding) (reviewed in Ref. 3). These interactions result in the activation of cellular signaling pathways (reviewed in Ref. 4).
FIGURE 1.
Recombinant L1 mutants expressed in insect Sf9 cells. A, schematic representation of the L1 protein and the various truncated mutants designed. L1 has six Ig domains (circles), five Fn domains (boxes), a transmembrane region (TM), and a cytoplasmic domain (Cyt). The 21 potential N-glycosylation sites (arrowheads) and the two neuronal-specific exons 2 and 27 (asterisks) are represented. Expected molecular mass of L1 truncated mutants expressed by insect Sf9 cells is shown. All mutants contained V5 followed by hexa-histidine tags at their C terminus. B, analysis of soluble L1 mutants produced by transfected Sf9 cells. Culture supernatants were sampled and analyzed by Western blot (WB), and then purified by ion metal affinity chromatography. Purified proteins were analyzed by reducing SDS-PAGE followed by Coomassie Blue G-250 staining (C) for purity and concentration assessment. L1/Ig1–4 and L1/Ig1–3 purified samples were also analyzed by non-reducing SDS-PAGE followed by Coomassie Blue G-250 staining (C) and Western blot.
Studies have been performed on domain mapping of homophilic binding, as this interaction has proven to be of fundamental importance for many of L1-mediated cellular processes, including neurite outgrowth (reviewed in Ref. 4). Siu and co-workers (5, 6) performed a series of experiments based on the use of recombinant L1 mutants produced in the Escherichia coli expression system and have shown that Ig2 is sufficient to induce homophilic interaction in trans and neurite elongation on neural retinal cells or just homophilic interaction on PC12 cells (7). More recently, Silleti and co-workers (8) showed that the bacterial-derived Fn3 domain spontaneously homomultimerizes in vitro, to form trimeric and higher order complexes suggesting that this domain could mediate L1-L1 interaction in cis.
However, studies using mammalian-derived glycosylated L1 missense mutants found in the neurological disease X-linked hydrocephalus have shown that homophilic interaction in trans is dependent on many domains, particularly the region encompassing Ig1–6 and the Fn2 domain (9). On the other hand, using Ig1–2, Ig1–3, and Ig1–4 mutants produced in HeLa cells, Haspel and co-workers (10) found that the first four Ig domains of L1 underwent homophilic binding, mediated cell adhesion, and promoted neurite outgrowth, but the whole Ig1–6 region was necessary for optimal neurite outgrowth. Accordingly, studies with L1 missense mutants expressed in COS-7 cells showed that mutations affecting the structure of domains in the Ig1–6 and Fn1–2 regions significantly reduced homophilic binding (11). Furthermore, neurons from a knock-in mouse in which Ig6 was deleted failed to attach and send out neurites on L1-coated surfaces (12).
Three-dimensional crystal structures of insect hemolin (13), chick axonin-1 (14), and its human homologue neural TAG-1 (15), which contains regions homologous to Ig1–4 from L1, showed that Ig1–4 domains adopted a horseshoe-shaped conformation, in the crystal. This suggested that a similar arrangement might occur in the Ig1–4 region of L1. A recently developed homology model of Ig1–4 from L1 further supported this possibility (16).
Previously, we have expressed the L1 ectodomain in Spodoptera frugiperda Sf9 insect cells, which was active in promoting neurite outgrowth from human NT2N neurons (17). Insect cells are adequate host systems for the expression of high amounts of recombinant glycoproteins. These cells perform glycosylation generally of the paucimannosidic-type (reviewed in Ref. 18), which is less processed than that observed for human glycoproteins, but which in many instances allows to obtain correctly folded and efficiently secreted glycoproteins.
In the present work, the homophilic interaction of the recombinant L1 ectodomain (L1/ECD) from insect cells has been observed by co-immunoprecipitation studies, and it was quantified using surface plasmon resonance analysis. Affinities between L1/ECD and L1/ECD, or L1/ECD and L1/Ig1–4 were found to be comparable, and deletion of domains Ig1 or Ig4 completely abrogated the interaction. Accordingly, cell adhesion was only detected for L1/ECD and L1/Ig1–4, and enhancement of neurite outgrowth was comparable for the two mutants.
EXPERIMENTAL PROCEDURES
Cell Culture—S. frugiperda Sf9 cells were grown and maintained in Sf900II medium at 27 °C and 90 rpm. Cultures were passed when they reached a cell density of about 4 × 106 cell/ml, with seeding concentration of 4 × 105 cell/ml. Human NT2N neurons were differentiated and cultured as previously described (19). Briefly, NT2– cells were maintained 5 weeks in Dulbecco's modified Eagle's medium with high glucose medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 10 μm retinoic acid, at 37 °C and 5% CO2. Cells were then plated into new flasks and maintained for 2 weeks in Dulbecco's modified Eagle's medium with high glucose medium supplemented with 5% fetal bovine serum, 1% penicillin/streptomycin, and mitotic inhibitors (1 μm cytosine arabinoside, 10 μm fluorodeoxyuridine, and 10 μm uridine). Post-mitotic human NT2N neurons were then recovered and used for functional assays. Human embryonic kidney HEK293 cells were grown at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and 1% penicillin/streptomycin.
Construction of Expression Vectors—Recombinant L1 truncated mutants comprising the entire ectodomain (L1/ECD), the Ig1 to 4 domains (L1/Ig1–4), the ectodomain without the Ig1 (L1/Ig2-Fn5), and the Ig1 to 3 domains (L1/Ig1–3) (Fig. 1A) were obtained as previously described (17), whereas L1/Ig2-Fn5 and L1/Ig1–3 mutants were constructed as previously reported (17). Briefly, they were constructed by PCR, using the pcDNA3L1A plasmid as template (Fig. 1A). The sequence corresponded to the neuronal isoform of L1 (28 exons) (GenBank™ code NM 000425). L1/Ig2-Fn5 was amplified using sense primer 5′-CATGCTAAGCTTGTGGCCAAAGGAGACAGTGAAG-3′ and the 5′-TTAATCCTCGAGCCTCAGTGGCGAAGCCAGC-3′ reverse primer. L1/Ig1–3 was amplified using the 5′-TTTGCTAAGCTTGGAGGAATATGAAGGACACCATGTG-3′ sense primer and the 5′-TTTTTCCTCGAGCAGCCTCCACGGTGACATAG-3′ reverse primer. All primers were designed to include restriction sequences for directional cloning; the forward and reverse primers had a restriction sequence for HindIII and XhoI endonucleases, respectively (first 13 bases, underscored). Fragments were cloned into the pMIB (C) expression vector and positively identified by complete automatic DNA sequencing. Standard molecular biology techniques were used.
Transfection and Production of L1 Mutants—Sf9 insect cells were transfected with pMIB-L1/Ig1–4 or pMIB-L1/Ig1–3 plasmids by the calcium phosphate method (17) and with pMIB-L1, pMIB-L1/ECD, or pMIB-L1/Ig2-Fn5 plasmids using Cellfectin (Invitrogen) according to the manufacturer's instructions. The pMIB-CAT plasmid was transfected as positive control. Transfected cells were selected with 10 mg/liter blasticidin-HCl for 2 weeks. Cell extract of Sf9-L1 cells and supernatants from the other transfected cells were analyzed by Western blot for the expression of L1 mutant proteins, and production was assessed during the days in culture. All mutant proteins were produced in shake flasks, and L1/ECD was also produced in a 2-liter bioreactor (17).
Protein Purification—Soluble recombinant L1 mutants were purified from the supernatant of transfected Sf9 cells. The purification was performed by ion metal affinity chromatography using a HiTrap Sepharose column (GE Healthcare). Briefly, 50 ml of protein-containing supernatant was run on 5 ml of Ni2+-loaded columns, washed with start buffer (0.02 m Na2HPO4, 0.5 m NaCl, 0.02 m imidazole), and eluted with elution buffer (same but with 0.5 m imidazole). Imidazole was removed from solution by washing eluate fractions with PBS, using Vivaspin concentrators (Sartorius). Purified proteins were kept at –80 °C in PBS containing 50% glycerol.
Analysis of Purified Proteins—Purified proteins were analyzed by SDS-polyacrylamide gel electrophoresis and stained with Coomassie Brilliant Blue G-250. Quantification of purified protein was performed by interpolation of the signal obtained in a calibration curve of peak area versus bovine serum albumin (BSA) mass (25–200 ng). Peak area was determined using ImageJ 1.37 gel analysis software (National Institutes of Health). Protein concentration was also determined by spectrophotometry using the extinction coefficient of L1 mutant proteins at 280 nm, calculated using the Protean version 3.11 software (DNAStar). Western blot analysis of purified proteins was performed using the mouse anti-V5 tag as primary antibody at 1:5,000 dilution; as secondary antibody an anti-mouse immunoglobulin G coupled to horseradish peroxidase was used at 1:4,000 dilution. Bands were visualized by the ECL Plus method (Amersham Biosciences).
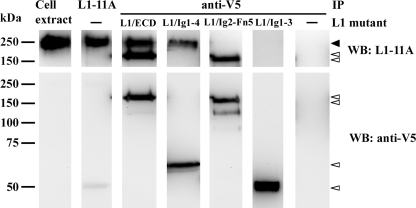
Co-immunoprecipitations—The full-length, membrane bound, native L1 protein from human NT2N neurons (hL1) was co-immunoprecipitated with each of the L1 truncated mutants. For each co-immunoprecipitation, a 20-μl aliquot of Protein A/G-agarose beads (Santa Cruz Biotechnologies) was incubated with 3 μl of the rabbit anti-V5 polyclonal antibody (serum 1712; a kind gift from Prof. Robert Doms, University of Pennsylvania) for 20 min, in a buffer containing 50 mm Tris/HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, 1% CHAPSO (Sigma), and protease inhibitors (Complete mixture, Roche). The anti-V5 antibody-coupled beads were incubated with 3 μg of the purified L1 mutants for 1 h. For each co-immunoprecipitation, the cell extract from 1 × 106 NT2N neurons was prepared with lysis buffer as described above. The lysates were then pre-cleared with Protein A/G-agarose beads, and incubated with the L1 mutant-antibody coupled beads for 4 h. The immunoprecipitates were analyzed by Western blot using, as primary antibodies, mouse anti-L1 L1–11A (a kind gift from Prof. Peter Altevogt, DKFZ, Germany), that recognizes the Fn3–5 epitope of L1 between proteolytic cleavage sites of plasmin and ADAM10 (20), and anti-V5 monoclonal antibodies.
Spectroscopic Analysis—The folding and secondary structure of L1 mutants was analyzed by circular dichroism (CD) spectroscopy. Spectra were measured using a Jasco J-815 spectropolarimeter with Peltier temperature control. Typically, 15 accumulations were recorded in the far-UV region (200–260 nm) at 20 °C using a 0.1-cm path length-polarized quartz cuvette. The lowest limit at 200 nm was imposed by high absorption below this point, which would cause distortions on the measured CD signals. CD units are expressed as mean residue extinction coefficient [Δεmrw] in m–1, cm–1, which was calculated from the relationship Δεmrw = S × mrw/(32,980 × c × d), where S represents the measured CD signal in millidegrees, c the protein concentration in mg/ml, d the path length of the cuvette in cm, and mrw the mean residue weight of each mutant. Samples were measured in 1:1 PBS/glycerol (pH 7.2) at a protein concentration of 0.1 mg/ml, in the absence or presence of 4 m guanidine hydrochloride. Data analysis was performed using the Jasco software package (Jandel Scientific). The program CDNN (Jasco) was used for deconvolution of the CD spectra and estimation of the secondary structure, from a built-in data base comprising 33 reference spectra. Tryptophan fluorescence was measured on a Varian Cary Eclipse fluorimeter equipped with Peltier temperature control. Emission spectra were recorded from 300 to 400 nm, setting the excitation at 280 nm and the emission slits at 5 nm. Samples were measured at 5 μm in 1:1 PBS/glycerol (pH 7.2). Dynamic light scattering analysis was performed on a Zetasizer Nano apparatus with temperature control. Samples were assayed at 0.3 mg/ml in PBS (pH 7.2). Diameter values were calculated using the DTSnano software (Malvern Instruments).
Surface Plasmon Resonance Analysis—Homophilic interaction was tested in the BIAcore 2000 system using the purified recombinant L1 mutants. In this system, binding of soluble analytes to immobilized ligands is measured in arbitrary units (RU). There is a linear relationship between the mass of the analyte bound to the immobilized protein and the RU observed (1000 RU ≈ 1 ng/mm2 bound protein). L1/ECD was coupled to the CM5 chip at 1 mg/ml by the amino coupling procedure according to the manufacturer's protocol. The ligand was immobilized at ≈2.5 ng/mm2 (2500 RU). Each of the L1 mutants were diluted in Tris-buffered saline containing 2 mm MgCl2 and 2 mm CaCl2 (TBS-MC) and injected in concentrations ranging from 0 to 1000 nm. At least one concentration was used as duplicate per experiment, and three independent experiments with two independent chips were performed. Samples were injected using the KInject injection wizard for kinetic parameters determination (flow rate of 20 μl/min for 3 min at 24 °C). The complexes were dissociated by washing with TBS-MC buffer at the same flow rate for 7 min. The regeneration step was performed using 10 mm glycine-HCl (pH 2.0) for 30 s. Interaction curves were obtained after reference flow cell and blank curves subtraction, for major bulk effect correction. The association phase (10–175 s) and dissociation phase (190–360 s) regions were selected for kinetic parameter evaluation. The association rate constant (ka) and the dissociation rate constant (kd) were calculated according to BIAevaluation software version 4.1 provided by the manufacturer. Association and dissociation rate constants were calculated simultaneously using a 1:1 Langmuir binding model with drifting baseline correction to determine the fitting curve. The affinity constant (KD) was calculated from the equation KD = kd/ka.
Cell Adhesion Assay—For the cell adhesion experiments using immobilized L1 mutants, round glass coverslips (12 mm diameter) were coated with 10 mm poly-d-lysine, washed three times with water, and incubated overnight with 80-μl drops of 2 μm purified proteins (1.4 × 10–10 mol of protein/mm2 surface area). Control coverslips were coated with 2 μm BSA over poly-d-lysine. Following coating, the coverslips were washed with PBS and Sf900II medium. Sf9-CAT (mock transfected) or Sf9-L1 cells (expressing the full-length membrane bound L1) were seeded onto the coated coverslips at 5 × 104 cell/cm2 density. The binding was performed for 24 h at 27 °C, after which slides were washed three times with PBS and fixed in 4% paraformaldehyde and 4% sucrose in PBS for 20 min. Cells were subsequently permeabilized with 0.1% Triton X-100, blocked, and incubated with 1:2000 dilution mouse anti-V5 monoclonal antibody (Invitrogen), followed by 1:500 dilution goat anti-mouse Alexa 594 secondary antibody. Cell binding was quantified by counting 10 independent fields per condition, in duplicate, from three independent blind experiments. Statistical analysis was performed using the GraphPad Prism 4 software.
Soluble L1 Binding Assay—HEK cells stably overexpressing full-length L1 (21) were seeded onto poly-d-lysine-coated glass coverslips at 5 × 104 cell/cm2 density and grown in Dulbecco's modified Eagle's medium for 24 h. Cells were washed with PBS and fixed in 4% paraformaldehyde in PBS for 20 min. After washing with PBS, fixed cells were incubated with 80-μl drops of 2 μm purified L1 proteins or BSA as negative control for 24 h. Cells were washed, fixed, and blocked and permeabilized with 1% BSA and 0.1% Triton X-100 in PBS. Immunostaining was performed with 1:2000 dilution rabbit anti-V5 polyclonal antibody, followed by 1:500 dilution goat anti-rabbit Alexa 594 secondary antibody. Preparations were examined by bright-field and fluorescence microscopy, using a Leica DMRB fluorescence microscope (Leica). The experiment was performed blind three times, in duplicate.
Neurite Outgrowth Assay—The neurite outgrowth assay was performed with the human NT2N neurons cultured over glass coverslips (coating as described above) at 5 × 104 cell/cm2 density. Matrigel (0.26 mg/ml) (BD Biosciences) coating was used as positive control. Neurite outgrowth was carried out in conditioned medium with mitotic inhibitors for 24 h. Cells were observed by bright-field microscopy using a Leica DM IRB inverted microscope and images were recorded with an attached Olympus DP11 digital camera. Neurites with a length greater than one cell body diameter (≈15 μm) were measured using the NeuronJ 1.01 plugin (22) from ImageJ 1.37 software. The experiment was performed blind twice using quadruplicates for each condition. Mean neurite length was calculated by randomly measuring 250 dominant neurite processes for each condition in each experiment.
RESULTS
Expression and Purification of L1 Truncated Mutants—To study the molecular basis and kinetics of the homophilic interaction of L1, mutants corresponding to the ectodomain (L1/ECD), domains Ig1 to Ig4 (L1/Ig1–4), Ig2 to Fn5 (L1/Ig2-Fn5), and Ig1 to Ig3 (L1/Ig1–3) were designed (Fig. 1A). The PCR amplified DNA sequences were cloned into the pMIB vector that encodes the V5 and the hexa-histidine tags downstream of the cloned sequences. Sf9 insect cells were transfected with the L1 mutant vectors and selected with 10 mg/ml blasticidin-HCl. Stably transfected Sf9-L1/ECD, Sf9-L1/Ig1–4, Sf9-L1/Ig2-Fn5, and Sf9-L1/Ig1–3 cells were obtained, and corresponding mutant proteins were found to be secreted into the supernatant (Fig. 1B). All mutant proteins were produced in shaker flasks at 3, 11, 3, and 23 mg/liter concentrations, respectively. L1/ECD was further produced in a bioreactor at 13 mg/liter concentration. Mutants were purified by ion metal affinity chromatography and analyzed by SDS-PAGE and Western blot analysis (Fig. 1B), showing a high degree of purity. The L1/ECD, L1/Ig1–4, L1/Ig2-Fn5, and L1/Ig1–3 mutants had apparent molecular masses of ≈180, 65, 160, and 50 kDa, respectively. Differences between the apparent mass of the mutants and those expected for the polypeptide chains (Fig. 1A) are probably due to glycosylation, as shown in previous work (17).
Structural Analysis of L1 Mutants—To investigate the conformation and folding of the produced L1 mutants, the purified proteins were analyzed by CD spectroscopy. CD spectra were recorded in the far-UV region (200–260 nm), which is informative regarding the various types of secondary structure (Fig. 2A, Table 1). The spectrum of L1/ECD is typical of a folded protein, with a single negative band centered at ≈216 nm, denoting a predominant β-sheet structure. This observation is in agreement with that predicted for the extracellular part of L1, because the six Ig and five Fn domains are formed by β-sheets (2). L1/ECD unfolded upon incubation with 4 m guanidine hydrochloride, a strong denaturing agent, which led to the disruption of the secondary structure (Fig. 2A). This data are in agreement with the results obtained by Hall and co-workers (23) for mouse L1 mutants expressed in mammalian cells, suggesting that the overall fold of Sf9-expressed L1 mutants is similar to that of mammalian-derived L1. The far-UV CD spectra of the L1/Ig1–4, L1/Ig2-Fn5, and L1/Ig1–3, which are characterized by a weak negative band (≈1.5 m–1 × cm–1) with minima at 210–220 nm, denote that the truncated mutants are folded, and also β-sheet proteins (Table 1). In fact, both the Ig and Fn domains are all β proteins, although a recent report has identified a small, novel α-helix within human Fn1 (24). Overall, far-UV CD analysis shows that the purified mutants correspond to structured and folded proteins.
FIGURE 2.
Structural and conformational analysis of L1 mutants. A, far-UV CD spectra of L1 mutants, and of L1/ECD in the presence of 4 m guanidine hydrochloride (Gdn/HCl). Protein concentration was 0.1 mg/ml in PBS/glycerol (1:1) (pH 7.2). B, Trp emission spectra of L1/Ig1–4 (solid lines) and L1/Ig1–3 mutants (dashed lines) recorded at 20 °C (folded state, F) or 90 °C (thermally unfolded state, U). Protein concentration was 5 μm in PBS/glycerol (1:1) (pH 7.2).
TABLE 1.
Estimated secondary structure content for L1 variants determined by far-UV CD
|
Protein
|
Secondary structure content
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| α-Helix | Antiparallel β | Parallel β | β Turn | Random coil | |||||
| % | |||||||||
| L1/ECD | 10 | 32 | 6 | 18 | 35 | ||||
| L1/Ig1-4 | 9 | 33 | 5 | 20 | 35 | ||||
| L1/Ig1-3 | 9 | 33 | 5 | 20 | 35 | ||||
| L1/Ig2-Fn5 | 10 | 30 | 6 | 20 | 35 | ||||
The structural conformation of L1/Ig1–4 and L1/Ig1–3 was further analyzed, to investigate if the Ig1–4 region of L1 could adopt a U-shaped structure similar to the one found in TAG-1 protein (15). Such a conformation would involve interactions between the Ig1 and Ig4 domains. L1/Ig1–4 and L1/Ig1–3 contain, respectively, six and four tryptophan residues, whose intrinsic fluorescence is an excellent probe to monitor differences in the conformation of the two variants. Upon excitation at 280 nm, the emission spectra of L1/Ig1–4 and L1/Ig1–3 exhibited maxima at 330 and 335 nm, respectively (Fig. 2B, letter F). The red-shifted emission maximum of L1/Ig1–3 suggests that the emitting residues in this variant are in a more solvent-exposed conformation (25). When thermally denatured, the tryptophan moieties, which were inaccessible to the solvent in the folded state, become solvent exposed, and the emission maxima shifts to 350 nm (Fig. 2B, letter U). In an attempt to discriminate between conformations with differences in compactness and hydrodynamic diameter, these mutants were also analyzed by dynamic light scattering. However, no clear difference was determined in the diameters of L1/Ig1–4 (12.4 ± 0.7 nm) and L1/Ig1–3 (12.8 ± 0.6 nm).
It was previously shown that the Ig1–3 region, expressed as a recombinant protein in HEK293 cells, was able to dimerize by intermolecular disulfide bonds (20). This dimerization was suggested to be due to improper folding of the truncated mutant, with consequent loss of function. To verify if L1/Ig1–3 was able to dimerize, the purified protein was analyzed by non-reducing SDS-PAGE followed by Western blot analysis and Coomassie staining (Fig. 1B, right). L1/Ig1–3 appeared as a major band at ≈40 kDa, by Coomassie staining and Western blot, but no other bands at a higher molecular mass were detected. L1/Ig1–4 mutant was also analyzed for comparison, and it appeared at a molecular mass of ≈52 kDa (Fig. 1B).
In Vitro L1 Homophilic Interaction by Co-Immunoprecipitation Assay—To test the homophilic interaction for the L1 mutants, a co-immunoprecipitation assay was designed using the native L1 protein from human NT2N neurons (hL1). In this assay, purified, V5-tagged L1 truncated mutants were incubated with rabbit anti-V5 antibody previously coupled to Protein A/G-agarose beads. The bead-adsorbed L1 mutants were then incubated with cell extracts from NT2N neurons. A positive control was performed, using anti-L1 (L1–11A) antibody to immunoprecipitate hL1. The hL1 was detected as a band of ≈220 kDa that specifically co-immunoprecipitated with L1/ECD or L1/Ig1–4 (Fig. 3, closed arrowhead). On the contrary, no hL1 was detected in L1/Ig2-Fn5 or L1/Ig1–3 co-immunoprecipitated samples. As the L1–11A antibody targets the Fn3–5 epitope (see “Experimental Procedures”), the L1 truncated mutants were detected using the mouse anti-V5 antibody (Fig. 3, lower panel). These results showed that L1/ECD and L1/Ig1–4 mutants interacted with hL1, contrary to L1/Ig2-Fn5 and L1/Ig1–3, in the conditions assayed. This indicates that the complete Ig1–4 region is sufficient for homophilic interaction with full-length hL1.
FIGURE 3.
Co-immunoprecipitation (IP) of native hL1 from NT2N neurons with L1 truncated mutants. L1 and V5 tagged mutants were analyzed by Western blot (WB) using as primary antibodies L1–11A and anti-V5, respectively. The hL1 protein from NT2N neurons is indicated with a closed arrowhead, and the truncation mutants with open arrowheads.
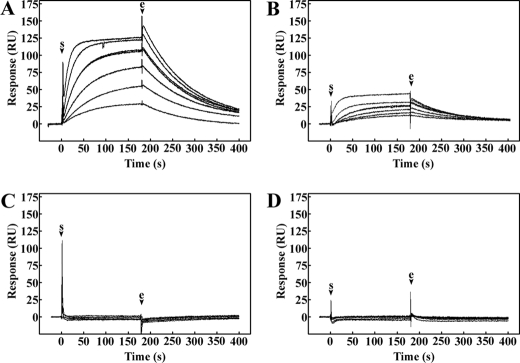
Analysis of L1 Homophilic Interaction Kinetics Using the BIAcore System—To investigate the homophilic interaction kinetics of L1, the purified L1 mutants were assayed in the BIAcore system. With this high-sensitivity system, kinetic constants for the association and dissociation rates of isolated molecules can be obtained, and from these parameters the dissociation equilibrium constant can be calculated. L1/ECD mutant was directly immobilized into the CM5 sensor chip as the ligand, and the binding of soluble L1/ECD or L1/Ig1–4 was measured using concentrations of the analyte between 31.25 and 1000 nm (Fig. 4, A and B). The interaction observed was concentration-dependent and indicated single-phase association and dissociation kinetics. The fitting curve was determined using a 1:1 Langmuir binding model with drifting baseline correction and an acceptable fit was obtained (χ2 = 1.03 ± 1.13 and 0.39 ± 0.18 for L1/ECD and L1/Ig1–4, respectively). The apparent dissociation constant (KD) calculated for L1/ECD and L1/Ig1–4 were 1.16 ± 0.02 × 10–7 m and 1.30 ± 0.06 × 10–7 m, respectively (Table 2). The small difference in the KD values for these two mutants to immobilized L1/ECD may be due to the stability of the larger L1/ECD when compared with L1/Ig1–4. Nevertheless, this difference does not suggest a considerable impairment of L1/Ig1–4 binding. Differences between Rmax values for L1/ECD and L1/Ig1–4 (151 ± 21 and 39 ± 6, respectively) probably result from differences in molecular weight of the truncated proteins. Mutants L1/Ig2-Fn5 and L1/Ig1–3 showed no interaction with immobilized L1/ECD (Fig. 4C). Taken together, these results showed that deletion of either Ig1 or Ig4 domains impaired L1 homophilic interaction.
FIGURE 4.
Sensorgrams of L1 mutants binding to L1 ectodomain. L1/ECD (A), L1/Ig1–4 (B), L1/Ig2-Fn5 (C), or L1/Ig1–3 (D) were passed over a CM5 chip coated with L1/ECD. Protein concentrations of 31.25, 62.5, 125, 250, 500, and 1000 nm were used. Arrows indicate the start (s) and the end (e) of the injection. Sensorgrams shown are representative of three independent experiments.
TABLE 2.
KD values calculated from kinetics for L1 homophilic interaction between immobilized L1 ectodomain and soluble L1 mutants measured in the BIAcore system
Results are the mean ± S.D. values of three independent experiments one of which considered triplicates of protein concentration.
| Injected analyte | kd | ka | KD |
|---|---|---|---|
| ×10-3/s | ×104/m s | ×10-7m | |
| L1/ECD | 9.97 ± 0.15 | 8.57 ± 0.28 | 1.16 ± 0.02 |
| L1/Ig1-4 | 9.96 ± 0.39 | 7.71 ± 0.42 | 1.30 ± 0.06 |
Homophilic Binding between Membrane-bound L1 and Truncated L1 Mutants—To investigate the homophilic binding in trans involving L1, two binding adhesion assays were developed. First, the adhesion of Sf9 cells stably expressing L1 to monolayers of L1/ECD and mutants was monitored. BSA was used as negative control for immobilized protein, and mock-transfected Sf9 cells (Sf9-CAT, expressing the V5 tag) were used as negative control for the cells. Cells were seeded and, after 24 h in culture, they were fixed and detected with anti-V5 antibody. Results showed that Sf9-L1 cells seeded over L1/ECD and L1/Ig1–4-coated surfaces adhered in higher numbers (168 ± 34 and 153 ± 23 cells/field, respectively) when compared with those seeded over L1/Ig2-Fn5, L1/Ig1–3, and BSA surfaces (24 ± 17, 18 ± 12, and 17 ± 4 cells/field, respectively) (Fig. 5). This suggested that membrane-bound L1 expressed by Sf9 cells interacted homophilically with L1/ECD and L1/Ig1–4 thus promoting cell adhesion, contrary to mutants that did not contain domains Ig1 or Ig4. This interaction was specific, as no differences in Sf9-CAT cell adhesion were observed to the different mutants (Fig. 5).
FIGURE 5.
Adhesion of Sf9-L1 cells to L1 mutant-coated surfaces. Insect Sf9 cells expressing membrane-bound, full-length L1 (Sf9-L1: A and C–F) or mock transfected (Sf9-pCAT: B) were seeded over glass coverslips coated with 2 μm BSA (A), L1/ECD (B and C), L1/Ig1–4 (D), L1/Ig2-Fn5 (E), or L1/Ig1–3 (F). Anti-V5 tag primary antibody was used in immunofluorescence microscopy. G, number of cells from each of 10 independent fields per glass slide. Countings were blind, from three independent assays (n = 3) in duplicate. Mean ± S.D. was calculated, and statistical analysis was performed by one-way analysis of variance followed by a Dunnett's post hoc comparison test. Asterisks indicate significant difference (p < 0.01) from mean number of attached Sf9-L1 cells over BSA. Bar = 50 μm.
Second, binding of the L1 mutants to HEK cells stably transfected with full-length L1 (HEK-L1) was analyzed. HEK-L1 cells seeded over poly-d-lysine-coated coverslips were incubated for 24 h with the L1 mutants, and detected by immunofluorescence microscopy with anti-V5 antibody. BSA was used as negative control. Results showed that HEK-L1 cells incubated with L1/ECD or L1/Ig1–4 were stained, whereas those incubated with L1/Ig2-Fn5, L1/Ig1–3, and BSA were not (Fig. 6).
FIGURE 6.
Homophilic binding of L1 mutants to full-length L1 from HEK-L1 cells. Anti-V5 tag primary antibody was used in immunofluorescence microscopy (upper panel). The presence of cells on the surface was confirmed by bright-field microscopy (lower panel). This assay was performed three independent times in duplicate. Bar = 50 μm.
These results showed that both L1/ECD and L1/Ig1–4 bound L1 from intact cells via homophilic interaction in trans. Furthermore, this interaction was abolished for mutants without domains Ig1 or Ig4.
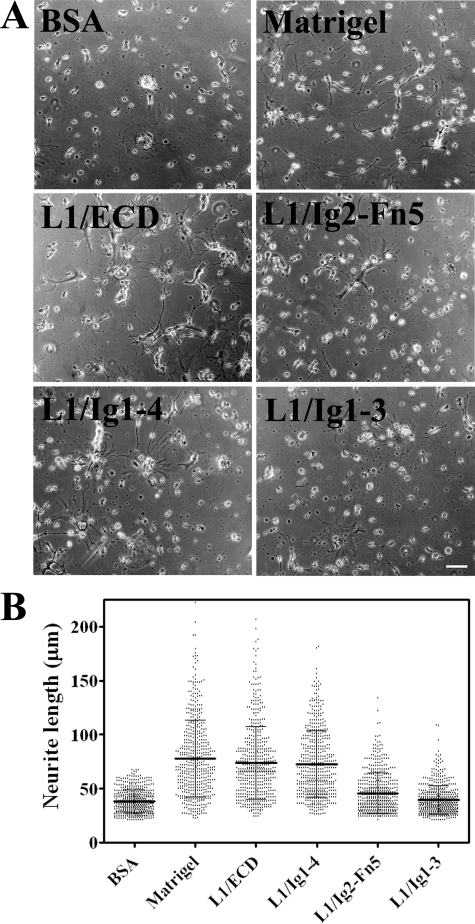
Enhancement of Neurite Outgrowth Activity of the L1 Mutants—To test the neuritogenic activity of L1 mutants, human NT2N neurons were seeded onto 2 μm L1/ECD, L1/Ig1–4, L1/Ig2-Fn5, or L1/Ig1–3-coated coverslips. BSA was used as negative control and Matrigel as positive control. Matrigel is able to promote neurite outgrowth due to the interaction of its component laminin with integrins from NT2N cells (26). Long neurites were formed from NT2N neurons cultured on Matrigel (78 ± 36 μm), L1/ECD (74 ± 33 μm), and L1/Ig1–4 (73 ± 31 μm) contrary to those grown on L1/Ig1–3 (39 ± 13 μm) or BSA (38 ± 11 μm) (Fig. 7). L1/Ig2-Fn5 had a small effect on the enhancement of neurite outgrowth (46 ± 19 μm). Mean lengths of the neurites extending on L1/ECD or L1/Ig1–4-coated surfaces were statistically different from those on BSA (p < 0.001), but did not differ from each other, as evaluated by the one-way analysis of variance analysis with a Tukey's post hoc multiple comparison test, and the corresponding neurite outgrowth enhancement was ≈1.9-fold. Neurite length on L1/Ig2-Fn5 was ≈0.6-fold lower than on L1/ECD or L1/Ig1–4 (p < 0.001); however, it was 1.2-fold higher than on BSA (p < 0.001) or L1/Ig1–3 (p < 0.01). Therefore, L1/Ig1–4 was able to induce neurite outgrowth from NT2N neurons to a similar extent as L1/ECD.
FIGURE 7.
Effect of immobilized L1 mutants on neurite outgrowth from NT2N neurons. A, bright-field micrographs of NT2N neurons cultured on different immobilized proteins. Bar = 50 μm. B, neurite length of NT2N neurons cultured on different coatings. Mean ± S.D. values were calculated from two independent blind experiments.
DISCUSSION
In this study we have investigated the kinetics of L1-L1 binding and its implications on homophilic interaction mechanisms. For the purpose we produced several L1 truncated mutants in stably transfected Sf9 insect cells. This expression system was used for high-throughput expression of soluble, glycosylated L1 mutants, which upon purification corresponded to folded, predominantly β-sheet proteins, as expected for the native L1 (2, 23). Homophilic interaction between these mutants was tested by co-immunoprecipitation analysis and quantified by surface plasmon resonance. The correlation with biological processes, namely cell adhesion, binding to cells and inducement of neurite outgrowth, has also been studied.
L1 molecules mediate cell-cell adhesion and other cellular processes via homophilic binding. Initial studies using bacteria-expressed L1 mutants proposed that the Ig2 domain solely accounted for homophilic binding (5, 6). However, studies using mammalian-derived L1 mutants showed that domains Ig1–Ig4 would be the minimum unit required for the binding, and Ig5–Ig6 and Fn2 domains also contributed to optimal interaction (9–11). Accordingly, in this work, recombinant L1/Ig1–4 from a eukaryotic source capable of performing N-glycosylation was found to mediate the homophilic interaction, contrary to mutants L1/Ig2-Fn5 or L1/Ig1–3, which contained the Ig2 domain. The characteristics of the host system used for the production of L1 mutants probably account for the differences observed. In eukaryotes, secretory N-linked glycoproteins undergo N-glycosylation and folding in the endoplasmic reticulum assisted by chaperones that include BiP and the lectin calnexin, and only leave the endoplasmic reticulum for subsequent secretion after they have been correctly folded. The quality control of glycoproteins in the endoplasmic reticulum is dependent on protein N-glycosylation (27). On the other hand, in bacteria, the non-glycosylated forms of L1 mutants have been accumulated in inclusion bodies, and were further recovered after solubilization with chaotropic agents, and refolding in vitro. It is probable that proteins from bacteria underwent a non-native pathway of folding that led to artificial homophilic interactions (discussed in Ref. 3), whereas the eukaryotic proteins either from insect cells (present work) or mammalian cells (9–11) were glycosylated and folded via pathways more similar to that found in human cells. These aspects are particularly relevant for L1 functionality because it is a heavily glycosylated protein.
Based on the kinetic analysis of the interaction between L1/ECD and the different mutants analyzed, the calculated KD for L1/ECD-L1/ECD interaction was found to be 116 ± 2 nm, which was comparable with that found for the L1/ECD-L1/Ig1–4 interaction (130 ± 6 nm). Therefore, Ig1–4 is the minimum part of the protein required for homophilic binding that exhibits kinetic properties similar to the full ectodomain. The additional importance of other domains Ig5–Ig6 and Fn2 (9, 11) and Ig6 (12) for the interaction found by others, might be associated with providing additional stability to the conformation of the L1/Ig1–4 domains, and may account for the slightly lower value of KD calculated for the L1/ECD-L1/ECD interaction. The dissociation constant of L1 homophilic interaction was approximately twice than that calculated for another molecule of the same family, the neural cell adhesion molecule (64 nm) (28), which indicated a comparatively lower affinity of L1 homophilic binding.
Following the crystal structure of Ig1–4 domains of insect hemolin (13), chick axonin-1 (14), or its human ortholog TAG-1 (15), two main mechanisms were proposed to explain the homophilic interaction. The first supported by the crystal structure of hemolin, and the second supported by the crystal structure of axonin-1 (14), TAG-1 (15), and the model of L1 Ig1–4 domains (16). The first mechanism proposes that Ig1–4 exists in an equilibrium between the extended form and a horseshoe conformation and that homophilic interaction in trans is mediated by the extended conformation (13). Accordingly, Ig domains would interact in an antiparallel orientation (i.e. Ig1–Ig4′, Ig2–Ig3′, Ig3–Ig2′, and Ig4–Ig1′). On the other hand, the second mechanism proposes that the horseshoe conformation is the active form in mediating homophilic interaction. In this model, compact horseshoe structures of opposing L1 molecules do not open up, but rather interact with one another. The study on the structure of chick axonin-1 proposes that the interaction is mediated by non-covalent bonds between the FG loop of Ig2 of one protein with the CE loop of Ig3 of the opposing L1 molecule (14). Alternatively, for the human ortholog of axonin-1, TAG-1, the interaction site would be restricted to the FG loop of Ig2 of opposable L1 molecules (15). This is a cooperative model that has found support in studies using rotary shadowing electron microscopy (23) or gradient sedimentation and negative stain electron microscopy techniques (29). Other studies involving the L1-neurocan binding site have also considered this model to explain neurocan-induced inhibition of L1 homophilic binding (30).
In this work, it was shown that deletion of the Ig1 or Ig4 domains was sufficient to impair interaction. These results clearly support the latter mechanism and suggest that deletion of either the Ig1 or Ig4 domains would disrupt the closed horseshoe fold and impair L1-L1 interaction. The results could not be explained by the first model because deletion of either Ig1 or Ig4 from one of the binding partners would eliminate just one of the four domain pairs of the interaction site with a consequent decrease of affinity of L1 interaction, but not impair it. In agreement, the analysis of the kinetics of neural cell adhesion molecule homophilic interaction (mediated by the double-reciprocal interaction of IgI–IgII to IgII′–IgI′ from the extended conformation of two molecules), has shown that the affinity of the single interaction between individual IgI and IgII′ domains (KD 5.5 ± 1.6 × 10–5 m) was approximately 3 orders of magnitude lower than the affinity of the double-reciprocal interaction (estimated to be KD 3.3 ± 1.8 × 10–9 m) (31). In this perspective, the deletion of either L1 Ig1 or Ig4 domains would result in a lower but still measurable KD value for either L1/ECD-L1/Ig2-Fn5 or L1/ECD-L1/Ig1–3 interaction. Therefore, the surface plasmon resonance results obtained in the present work favor that the binding occurred between the horseshoe structures of two opposable molecules.
The L1/Ig1–4 functional unit is thought to be stabilized through non-covalent bonds between Ig1–Ig4 and Ig2–Ig3 (14, 15, 16). However, the diameters calculated by DLS for L1/Ig1–4 and L1/Ig1–3 were not sufficiently different to discriminate between a closed or open conformation. Nevertheless, the calculated diameter value of the L1/Ig1–3 mutant is in agreement with the value expected for the extended molecule considering 4 nm as the length per L1 Ig domain (29). The calculated diameter value of L1/Ig1–4 was smaller than that expected, and this observation raises the possibility that a horseshoe-shaped L1/Ig1–4 mutant could be in equilibrium with an open, elongated conformation.
The homophilic binding activity of L1-truncated mutants was found to be well correlated with their participation in cell adhesion. L1/ECD and L1/Ig1–4, which presented homophilic binding in trans, also supported the adhesion of Sf9 cells expressing the full-length, membrane-bound form of L1, contrary to L1/Ig2-Fn5 and L1/Ig1–3. The same was true for the binding assay, where HEK cells overexpressing the full-length L1 were found to bind L1/ECD and L1/Ig1–4 but not L1/Ig2-Fn5 nor L1/Ig1–3. Similar results were observed in previous studies, where L1 deletion mutants were used for homophilic interaction site mapping. There, the Ig1–Ig3 mutant did not mediate homophilic interaction either in a coated microsphere aggregation assay (11) or as a coating molecule for rat cerebellar cell adhesion (10).
The correlation between homophilic binding and neuritogenic potential was also investigated for all truncated mutants. The relation between these two processes has been examined by others using domain-deletion (5, 7, 10) and pathological, missense (11, 32) mutants. In those studies, eukaryotic-derived L1 mutants indicated that both homophilic interaction and neuritogenic activity depended on, at least, the first four Ig domains. Accordingly, our results showed that L1/ECD and L1/Ig1–4 were able to promote neurite outgrowth from human NT2N neurons contrary to L1/Ig1–3. This is in contrast with that reported before (12) for cerebellar neurons from a knock-in mouse with deleted Ig6 that failed to attach to recombinant human L1-Fc from COS cells and to send out neurites. The discrepancy might be due to the different biological materials used: here we use postmitotic NT2N neurons that have characteristics of primary human neurons (19) attached to recombinant human L1 from Sf9 cells. The use of human neurons might present an advantage over mouse neurons for the study of homophilic interaction of human L1.
The L1/Ig2-Fn5 mutant promoted neurite outgrowth, although to a low extent. In a previous deletion mutant study, Jacob and co-workers (33) showed that the deletion of the neuronal-specific exon 2 sequence was sufficient to impair homophilic binding and to reduce neuritogenic activity of L1. The sequence corresponding to this exon is located immediately N-terminal to the Ig1 domain, and its role could be in the stabilization of the closed, horseshoe fold of Ig1–4 through interaction with the junction between Ig4 and Ig5 (32, 33). It is possible that the exon 2 deletion mutant presents a similar effect to that of L1/Ig2-Fn5. L1-mediated neuritogenesis was also reported to depend on the RGD motif present in the Ig6 domain, through interaction with integrins (7, 17, 34, 35). Therefore, the effect is probably due to such interaction.
Several studies have reported a number of downstream signaling pathways implicated in L1-triggered neuritogenesis, namely the FGFR pathway (36). Additionally, the mitogen-activated protein kinase pathway was found to be implicated in L1-integrin-mediated neurite outgrowth in PC12 cells (7). It would be interesting to investigate the role of these pathways in the L1-induced neurite outgrowth here described for the human NT2N cells. This characterization would allow a better understanding of L1-mediated signaling and its roles in NT2N neuritogenesis and brain plasticity. This could be of considerable clinical interest, as NT2N neurons have been used for transplantation therapies (37).
In summary, milligram amounts of truncated mutants of L1 have been produced and purified from insect cells that are folded and glycosylated and, therefore, suitable for functional and structural studies. L1/ECD and L1/Ig1–4 were shown to be active in homophilic binding in trans, with domains Ig1–4 exhibiting a dissociation constant comparable with that of the entire ectodomain. Deletion of Ig1 or Ig4 completely abrogated the homophilic interaction, therefore, providing evidence that supports the cooperative model proposing a horseshoe conformation of domains Ig1–Ig4 to underlie homophilic binding. L1/Ig1–4 was also shown to be the minimum unit required for cell adhesion and inducement of neurite outgrowth.
Acknowledgments
We gratefully acknowledge Prof. Peter Altevogt, DKFZ, Heidelberg, Germany, for plasmid pcDNA3 L1A (3.9 kb) and the L1–11A antibody; Prof. Robert Doms, University of Pennsylvania, for the anti-V5 antibody; Prof. Adriano Henriques, Instituto de Tecnologia Química e Biológica (ITQB), for the use of the Biacore 2000 equipment; Dr. Christophe Quétard, BIAcore AB, for advice with surface plasmon resonance instrumentation and data rationalization; Cristina Escrevente, ITQB, for help with the HEK-L1 cells; and Hugo Botelho, ITQB, for assistance during DLS measurements.
This work was supported in part by CellPROM number 500039-2 and Signalling & Traffic number LSHG-CT-2004-503228 from the European Commission. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Ig, immunoglobulin; BSA, bovine serum albumin; Fn, fibronectin type III; NT2-, NTERA-2 cl.D1 pluripotent human embryonal carcinoma; CHAPSO, 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate; PBS, phosphate-buffered saline; RU, response unit; HEK, human embryonic kidney.
References
- 1.Burden-Gulley, S. M., Pendergast, M., and Lemmon, V. (1997) Cell Tissue Res. 290 415–422 [DOI] [PubMed] [Google Scholar]
- 2.Bateman, A., Jouet, M., MacFarlane, J., Du, J. S., Kenwrick, S., and Chothia, C. (1996) EMBO J. 15 6050–6059 [PMC free article] [PubMed] [Google Scholar]
- 3.Haspel, J., and Grumet, M. (2003) Front. Biosci. 8 s1210–s1225 [DOI] [PubMed] [Google Scholar]
- 4.Kenwrick, S., Watkins, A., and De Angelis, E. (2000) Hum. Mol. Genet. 9 879–886 [DOI] [PubMed] [Google Scholar]
- 5.Zhao, X., and Siu, C. H. (1995) J. Biol. Chem. 270 29413–29421 [DOI] [PubMed] [Google Scholar]
- 6.Zhao, X., Yip, P. M., and Siu, C. H. (1998) J. Neurochem. 71 960–971 [DOI] [PubMed] [Google Scholar]
- 7.Yip, P. M., and Siu, C. H. (2001) J. Neurochem. 76 1552–1564 [DOI] [PubMed] [Google Scholar]
- 8.Silletti, S., Mei, F., Sheppard, D., and Montgomery, A. M. (2000) J. Cell Biol. 149 1485–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Angelis, E., MacFarlane, J., Du, J. S., Yeo, G., Hicks, R., Rathjen, F. G., Kenwrick, S., and Brummendorf, T. (1999) EMBO J. 18 4744–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haspel, J., Friedlander, D. R., Ivgy-May, N., Chickramane, S., Roonprapunt, C., Chen, S., Schachner, M., and Grumet, M. (2000) J. Neurobiol. 42 287–302 [PubMed] [Google Scholar]
- 11.De Angelis, E., Watkins, A., Schafer, M., Brummendorf, T., and Kenwrick, S. (2002) Hum. Mol. Genet. 11 1–12 [DOI] [PubMed] [Google Scholar]
- 12.Itoh, K., Cheng, L., Kamei, Y., Fushiki, S., Kamiguchi, H., Gutwein, P., Stoeck, A., Arnold, B., Altevogt, P., and Lemmon, V. (2004) J. Cell Biol. 165 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su, X. D., Gastinel, L. N., Vaughn, D. E., Faye, I., Poon, P., and Bjorkman, P. J. (1998) Science 281 991–995 [DOI] [PubMed] [Google Scholar]
- 14.Freigang, J., Proba, K., Leder, L., Diederichs, K., Sonderegger, P., and Welte, W. (2000) Cell 101 425–433 [DOI] [PubMed] [Google Scholar]
- 15.Mortl, M., Sonderegger, P., Diederichs, K., and Welte, W. (2007) Protein Sci. 16 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arevalo, E., Shanmugasundararaj, S., Wilkemeyer, M. F., Dou, X., Chen, S., Charness, M. E., and Miller, K. W. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouveia, R. M., Morais, V. A., Peixoto, C., Sousa, M., Regalla, M., Alves, P. M., and Costa, J. (2007) Protein Expression Purif. 52 182–193 [DOI] [PubMed] [Google Scholar]
- 18.Altmann, F., Staudacher, E., Wilson, I. B., and Marz, L. (1999) Glycoconj. J. 16 109–123 [DOI] [PubMed] [Google Scholar]
- 19.Brito, C., Escrevente, C., Reis, C. A., Lee, V. M., Trojanowski, J. Q., and Costa, J. (2007) J. Neurosci. Res. 85 1260–1270 [DOI] [PubMed] [Google Scholar]
- 20.Mechtersheimer, S., Gutwein, P., Agmon-Levin, N., Stoeck, A., Oleszewski, M., Riedle, S., Postina, R., Fahrenholz, F., Fogel, M., Lemmon, V., and Altevogt, P. (2001) J. Cell Biol. 155 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escrevente, C., Morais, V. A., Keller, S., Soares, C. M., Altevogt, P., and Costa, J. (2008) Biochim. Biophys. Acta 1780 905–913 [DOI] [PubMed] [Google Scholar]
- 22.Meijering, E., Jacob, M., Sarria, J. C., Steiner, P., Hirling, H., and Unser, M. (2004) Cytometry A 58 167–176 [DOI] [PubMed] [Google Scholar]
- 23.Hall, H., Bozic, D., Fauser, C., and Engel, J. (2000) J. Neurochem. 75 336–346 [DOI] [PubMed] [Google Scholar]
- 24.Mendiratta, S. S., Sekulic, N., Hernandez-Guzman, F. G., Close, B. E., Lavie, A., and Colley, K. J. (2006) J. Biol. Chem. 281 36052–36059 [DOI] [PubMed] [Google Scholar]
- 25.Lakowicz, J. R. (1999) Principles of Fluorescence Spectroscopy, Kluwer Academic/Plenum Publishers, New York
- 26.Stipp, C. S., and Hemler, M. E. (2000) J. Cell Sci. 113 1871–1882 [DOI] [PubMed] [Google Scholar]
- 27.Helenius, A., and Aebi, M. (2004) Annu. Rev. Biochem. 73 1019–1049 [DOI] [PubMed] [Google Scholar]
- 28.Retzler, C., Gohring, W., and Rauch, U. (1996) J. Biol. Chem. 271 27304–27310 [DOI] [PubMed] [Google Scholar]
- 29.Schurmann, G., Haspel, J., Grumet, M., and Erickson, H. P. (2001) Mol. Biol. Cell 12 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oleszewski, M., Gutwein, P., von der Lieth, W., Rauch, U., and Altevogt, P. (2000) J. Biol. Chem. 275 34478–34485 [DOI] [PubMed] [Google Scholar]
- 31.Kiselyov, V. V., Berezin, V., Maar, T. E., Soroka, V., Edvardsen, K., Schousboe, A., and Bock, E. (1997) J. Biol. Chem. 272 10125–10134 [DOI] [PubMed] [Google Scholar]
- 32.De Angelis, E., Brummendorf, T., Cheng, L., Lemmon, V., and Kenwrick, S. (2001) J. Biol. Chem. 276 32738–32742 [DOI] [PubMed] [Google Scholar]
- 33.Jacob, J., Haspel, J., Kane-Goldsmith, N., and Grumet, M. (2002) J. Neurobiol. 51 177–189 [DOI] [PubMed] [Google Scholar]
- 34.Yip, P. M., Zhao, X., Montgomery, A. M., and Siu, C. H. (1998) Mol. Biol. Cell 9 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaess, S., Kammerer, R. A., and Hall, H. (1998) J. Neurochem. 71 2615–2625 [DOI] [PubMed] [Google Scholar]
- 36.Loers, G., Chen, S., Grumet, M., and Schachner, M. (2005) J. Neurochem. 92 1463–1476 [DOI] [PubMed] [Google Scholar]
- 37.Hara, K., Yasuhara, T., Maki, M., Matsukawa, N., Masuda, T., Yu, S. J., Ali, M., Yu, G., Xu, L., Kim, S. U., Hess, D. C., and Borlongan, C. V. (2008) Prog. Neurobiol. 85 318–334 [DOI] [PubMed] [Google Scholar]