Abstract
CTP:phosphocholine cytidylyltransferase (CCT), a rate-limiting enzyme in phosphatidylcholine synthesis, is regulated by reversible membrane interactions mediated by an amphipathic helical domain (M) that binds selectively to anionic lipids. CCT is a dimer; thus the functional unit has two M domains. To probe the functional contribution of each domain M we prepared a CCT heterodimer composed of one full-length subunit paired with a CCT subunit truncated before domain M that was also catalytically dead. We compared this heterodimer to the fulllength homodimer with respect to activation by anionic vesicles, vesicle binding affinities, and promotion of vesicle aggregation. Surprisingly for all three functions the dimer with just one domain M behaved similarly to the dimer with two M domains. Full activation of the wild-type subunit was not impaired by loss of one domain M in its partner. Membrane binding affinities were the same for dimers with one versus two M domains, suggesting that the two M domains of the dimer do not engage a single bilayer simultaneously. Vesicle cross-bridging was also unhindered by loss of one domain M, suggesting that another motif couples with domain M for cross-bridging anionic membranes. Mutagenesis revealed that the positively charged nuclear localization signal sequence constitutes that second motif for membrane cross-bridging. We propose that the two M domains of the CCT dimer engage a single bilayer via an alternating binding mechanism. The tethering function involves the cooperation of domain M and the nuclear localization signal sequence, each engaging separate membranes. Membrane binding of a single M domain is sufficient to fully activate the enzymatic activity of the CCT dimer while sustaining the low affinity, reversible membrane interaction required for regulation of CCT activity.
Many amphitropic proteins have multiple membrane binding domains that serve to enhance binding affinity and to enable differential regulation by various lipid ligands. Multiple binding domains may in some cases enable a stepwise process leading to full activation of the enzyme. For example several protein kinase C isoforms have a C2 domain and two C1 domains that bind acidic lipids and diacylglycerol, respectively, with full activation dependent on engagement of both types of membrane binding modules (1). Other examples include phospholipase D2, which relies on both a PH3 domain and a polybasic domain for targeting to phosphatidylinositol 4,5-bisphosphate-rich membranes (2) and phosphatidylinositol-phospholipase C β, γ, and δ, which contain C2 and PH domains for recognition of and regulation by Ca2+ and phosphatidylinositol 4,5-bisphosphate, respectively (3).
CCT, the rate-limiting enzyme in PC synthesis, is a well studied amphitropic enzyme whose weak, reversible binding is mediated by domain M, a long amphipathic helix (4). Domain M is situated just downstream of the catalytic domain in the linear sequence and is followed by a flexible, unstructured domain housing multiple phosphorylation sites. Domain M is an autoinhibitory domain as evidenced by constitutive activity resulting from deletion of domain M (5). Upon membrane engagement the autoinhibition at the active site is relieved resulting in ∼100-fold enhancement of kcat. Thus lipids activate the enzyme by entrapping an autoinhibitory domain.
The predominant CCTα isoform contains a polybasic nuclear localization signal (NLS) sequence within domain N (12RKRRK16) that directs CCT into the nucleus of many but not all cells where it translocates on and off the nuclear envelope in response to changes in the lipid composition of that membrane. Two classes of lipids have been shown to promote CCTα membrane localization in vitro and in vivo: anionic lipids and type II lipids such as unsaturated phosphatidylethanolamine and diacylglycerol (4).
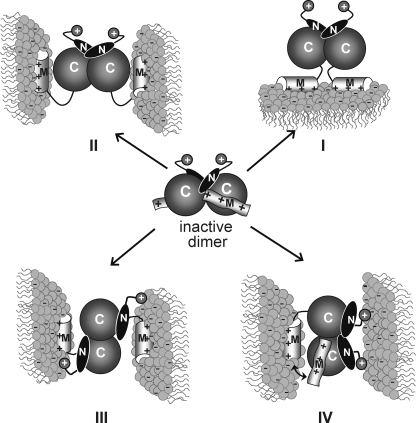
The soluble form of CCTα is a homodimer (367 amino acids per monomer) (6–8); thus the functional unit may have two M domains. The N-terminal domain consisting of domain N (residues 1 to ∼75) plus the catalytic domain C (residues 76–236) is sufficient to mediate non-covalent dimerization (5, 8). CCT236, comprised of domains N + C, is catalytically active as a dimer (5). Early models of CCT activation by membrane interaction envisioned two M domains of a dimer engaging the same bilayer in a cis mode (4, 9) (Fig. 1, Model I). Later the discovery that CCTα can cross-bridge vesicles and that domain M is required for this process suggested a trans mode in which each M domain engages a separate bilayer because of the positioning of the M domains on opposite poles of the N + C dimerization domains (Fig. 1, Model II). The trans mode of binding resulting in vesicle cross-bridging has only been detected with highly anionic vesicles (≥33 mol % anionic) (10).
FIGURE 1.
Models for CCT dimer association with membranes. The CCT dimer is inactive in the soluble form. I, the CCT dimer partitions into a single membrane using both M domains in cis. II, the CCT dimer tethers two membranes using both M domains in trans. III, the CCT dimer tethers two membranes using the two M domains in trans and two NLS motifs (+) in trans. We depict the NLS and M domains from one subunit engaging the same membrane, but they could engage separate membranes. IV, the CCT dimer tethers two membranes using a single M domain (Ma alternates with Mb) and two NLS motifs in cis. Our data offer the most support for Model IV. For clarity, domain P has been deleted from all models.
Membrane binding of CCT was associated with a loss of chemical cross-linking efficiency across the dimer interface, suggesting that membrane binding induces a rearrangement of its dimer interface (8). Although the dimer interface may reorganize, it is likely that the enzyme remains dimeric when membrane-bound and fully active for several reasons. First, it can tether two anionic vesicles as manifested by vesicle aggregation (10). Second, limited proteolysis showed that the sites within domain C that form the dimer interface remained inaccessible in the membrane form as well as the soluble form of CCT (11). If membrane binding were to dissociate the dimer into monomers the dimer interface would have become highly accessible to proteases. Third, CCT binding to and activation by lipid vesicles enriched in diacylglycerol does not reduce cross-linking efficiency.4 Lastly, a truncated form of CCT missing domains M + P is a constitutively active dimer (5), showing that the dimeric form is compatible with activity.
Why is CCT a dimer? Is the dimeric structure important for enhancement of membrane binding, for enabling membrane cross-bridging, and/or for regulation of activity by sequential binding of the M domains? Is the binding of each domain M so weak that two such domains are essential for maintaining CCT in its membrane-bound active conformation? Does membrane engagement of both M domains affect the active site of a single subunit in a way that engagement of only one domain M cannot?
To probe these questions we prepared a heterodimer composed of one subunit of the full-length CCTα (CCT367) and one subunit of a CCT truncated before domain M (CCT236). Comparing the catalytic activity of the 367 subunit in the full-length homodimer (CCT367) versus heterodimer (CCT367/236) in the presence of saturating lipid vesicles and saturating substrates would reveal the contribution of each domain M-lipid engagement to the activation process. We found that the dimer with only one domain M had a specific activity similar to that of the dimer with two M domains. The explanation for this surprising result emerged from another surprising finding that the homo- and heterodimer had the same binding affinity for PG/PC vesicles. Thus only one domain M at a time engages these membranes even when the CCT dimer has two such domains. We present arguments that activation of the two catalytic domains is highly cooperative, requiring the membrane engagement of only one M domain. Our finding that only one domain M is needed for membrane cross-bridging led us to the discovery of the N-terminal NLS as a second membrane interaction motif.
EXPERIMENTAL PROCEDURES
Materials
Egg PC and egg PG were purchased from Northern Lipids Inc. (Vancouver, British Columbia, Canada). The concentrations of phospholipid chloroform stocks were determined with a phosphorus assay (12). Phosphoryl [methyl-14C]choline, ammonium salt, the ECL Western blotting detection kit, SYPRO Orange protein gel stain, and thrombin protease were from Amersham Biosciences. Ni-NTA-agarose was purchased from Qiagen, and AcTEV protease was from Invitrogen. CTP, dithiothreitol, phenylmethylsulfonyl fluoride, Triton X-100, imidazole, SDS, and α-chymotrypsin were obtained from Sigma.
Expression and Purification of Untagged CCTα
The full-length untagged rat CCTα isoform and CCT236 (truncated at codon 236) were expressed by baculovirus infection of Trichoplusia ni cells and purified as described previously (13, 14). Purified proteins were stored in 10 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 2 mm DTT (Buffer A) at –80 °C.
Construction of His-tagged CCTα Constructs
pET14b-HisCCT236(K122A)—The Lys → Ala mutation was engineered at codon 122 using the QuikChange mutagenesis method (Stratagene) and the template pVL1392-HisCCT236. The correct CCT sequence was confirmed by DNA sequencing. For expression in Escherichia coli an NdeI site was engineered just 5′ of the CCT coding region by standard PCR methods using pVL1392-HisCCT236(K122A) as a template and a 3′ primer complementary to the vector sequence. The 750-bp PCR product encompassing codons 1–236 was digested with NdeI and BamHI and ligated into NdeI/BamHI-digested pET14b to engineer an N-terminal His6 tag followed by a thrombin cleavage site.
pAX142-HisCCT(K122A)—A 261-bp EcoRV/SspI fragment from pET14b-His236(K122A) encompassing codon 122 was prepared. It was ligated into a COS cell expression vector, pAX142 (15), containing a His6 tag and a TEV protease site upstream of WT full-length CCTα after excision of the corresponding EcoRV/SspI 261-bp fragment.
pAX142-HisCCT(ΔNLS)—Codons 12–16 encoding the nuclear localization sequence were deleted by the QuikChange method (Stratagene) using a pair of complementary primers and the template HisCCT236 in pBS (KS–). The mutation was confirmed by DNA sequencing. A 292-bp MluI/SspI fragment of pBS-HisCCT(Δ12–16) containing the deleted codons was ligated with the 3849-bp Mlu1/Ssp1 fragment of pAX142-HisCCT. This HisCCT(Δ12–16) construct contained a Factor Xa cleavage site between the His tag and the CCT sequence. We later discovered, after expression, purification, and digestion of this construct, that cleavage of full-length CCTα with Factor Xa is not strictly confined to the site in the linker. Thus we replaced the Xa site with a TEV site, which cleaves faithfully only at the linker site. The 512-bp BglII/EcoRV fragment of pAX-HisCCT(Δ12–16) (CCT codons 1–168 + eight-nucleotide 5′ extension) was ligated with the 3626-bp EcoRV/BglII fragment of pBS-His-TEV site-CCT to yield pBS-His-TEV site-CCT(Δ12–16). Then the 566-bp MluI/EcoRV fragment of pBS-His-TEV site-CCT(Δ12–16), extending from 5′ of the His tag to codon 170 of CCT, was ligated with the 3599-bp MluI/EcoRV fragment of pAX-HisCCT to yield pAX-His-TEV site-CCT(ΔNLS).
pET14b-HisCCT236(K122A; ΔNLS)—This construct was made in two steps. First a 636-bp EcoRI fragment encompassing CCT codons 31–236 and the K122A mutation was obtained from pVL1392-HisCCT236(K122A). It was ligated with a 3.1-kb EcoRI fragment from pBS-His-TEV site-CCT(Δ12–16) to generate pBS-His-TEV site-CCT236(K122A; Δ12–16). In step 2 the CCT-encoding 800-bp fragment from this construct was ligated with the 5-kb vector fragment of pET14b-His-thrombin site-CCT236(K122A) using NdeI and BamHI sites just upstream of the TEV site and downstream of the stop codon, respectively.
Expression and Purification of His-tagged CCTs and Cleavage of Tag
Expression of the pET14b-CCT236 constructs used the Rosetta strain of E. coli (derived from BL-21). Rosetta cells transformed with these plasmids (250 ml) were cultured to an optical density of ≈0.8. CCT proteins were expressed by induction with 0.4 mm isopropylthio-β-d-galactoside for 4 h at 28 °C. To purify His-tagged CCT236 proteins from E. coli cells, the cell pellet from a 250-ml cell culture was incubated for 5 min at 37 °C in phosphate-buffered saline, pH 7.4, 1 mm DTT, 0.2 mg/ml lysozyme, 10 μg/ml deoxyribonuclease I, six protease inhibitors (16) and lysed by sonication on ice 4 × 15 s with a 20-s rest between bursts. To the lysate we added 0.1 volume of 10× binding buffer (50 mm Na2PO4, 5 m NaCl, 150 mm imidazole) with mixing. The lysate was centrifuged at 13,000 rpm for 30 min at 4 °C, and the supernatant containing ∼18 mg of protein was applied to a Ni-NTA-agarose column (1.5-ml bed volume). The nickel resin was washed, and the His-tagged CCTs were eluted as described previously (8).
Full-length CCTs in the pAX142 vector were expressed transiently in COS-1 cells by transfection using DEAE-dextran (17). Transfection times were 64 h for pAX-HisCCT(K122A) and 48 h for pAX-HisCCT(ΔNLS). Longer expression times for the latter led to higher protein expression but also decreased solubility. To purify His-tagged CCT367 proteins from COS cells, cells from 10 15-cm dishes were harvested with phosphate-buffered saline, 2.5 mm EDTA, and the cell pellet was homogenized by sonication on ice in 20 mm phosphate buffer, pH 7.4, 1% Triton X-100, six protease inhibitors (16). To the cell lysate (∼8 mg of cell protein in 7.5 ml) we added 0.1 volume of 10× binding buffer, and the lysate was centrifuged at 13,000 rpm for 10 min at 4 °C. The supernatant (∼7 mg of protein) was applied to a Ni-NTA-agarose column (1.2-ml bed volume). The nickel resin was washed, and the His-tagged CCTs were eluted as described previously (8).
The eluted HisCCTs were dialyzed against 500 volumes of 20 mm phosphate buffer, pH 7.4, 100 mm NaCl, 0.15 mm Triton X-100, 2 mm DTT (HisCCT236 proteins) or 10 mm Tris, pH 7.4, 100 mm NaCl, 0.15 mm Triton X-100, 2 mm DTT (HisCCT367 proteins) for 3 h at 4°C with one buffer change to remove the imidazole and stored at –80 °C. The concentrations of the pure enzymes were determined by the method of Bradford (18) using bovine serum albumin as standard.
For some experiments the His tag of HisCCT236(K122A) was removed by digestion with thrombin protease (1 unit/35 μg of His-tagged protein) at 4 °C for ∼16 h. The cleavage reaction in phosphate-buffered saline, pH 7.4, 0.15 mm Triton X-100, 2 mm DTT (PBST Buffer) was quenched with a mixture of protease inhibitors (1 μg/ml antipain, 2 μg/ml chymostatin, 2.5 μg/ml leupeptin), and the sample was dialyzed for 3 h at 4 °C against the same buffer to remove the His peptide. The His tag of His-TEV-CCT(ΔNLS) was cleaved by digestion with AcTEV protease (1 unit/1.4 μg of His-tagged protein) at 4 °C for ∼16 h in 10 mm Tris, pH 7.4, 150 mm NaCl, 0.25 mm Triton X-100, 2 mm DTT. At the end of the cleavage reaction imidazole (15 mm) was added to block non-specific binding. Ni-NTA-agarose beads were added (∼0.1 μl/μg of CCT), and the sample was incubated for 1 h at 4 °C. After centrifugation at 5,000 × g for 1 min CCT(ΔNLS) was recovered in the supernatant; the His-AcTEV protease and traces of uncleaved His-TEV-CCT(ΔNLS) remained bound to the resin.
Preparation and Quantification of CCT367/His236(K122A) Heterodimers in Vitro
Native dimers of CCT367 and catalytically inactive HisCCT236(K122A) were dissociated into monomers by incubation in Buffer A with 1 mm SDS at a 70:1 SDS/protein molar ratio at 37 °C for 15 min. Disruption of dimers was indicated by loss of glutaraldehyde-mediated covalent cross-linking of the two subunits as assessed by SDS-PAGE (data not shown). The CCT367/His236(K122A) heterodimer was formed by mixing the SDS-treated CCT367 and HisCCT236(K122A) in a 1:1 molar ratio followed by a 5-min incubation at 37 °C. Spontaneous reassociation of the protein monomers was initiated by 1:1 dilution in Buffer A followed by removal of the SDS by overnight dialysis at 4 °C. Sample containing the three reconstituted CCT dimers was applied to a Ni-NTA-agarose column and processed as described above. The untagged CCT367 homodimer was recovered in the flow-through fraction; CCT367/His236(K122A) and HisCCT236(K122A) dimers co-eluted with 350 mm imidazole, pH 8.0. Because the HisCCT236(K122A) homodimer is catalytically inactive it should not affect activity measurements with the heterodimer. The CCT dimers in the flow-through and eluted fractions were dialyzed for 3 h at 4°C against PBST Buffer and stored at –80 °C.
The concentration of the active CCT367 subunit in the CCT367/His236(K122A) heterodimer was determined as follows. First, the two proteins were separated by SDS-PAGE, quantitatively stained with SYPRO Orange, and imaged with a Typhoon 9410 imager. The ratio, R = [CCT367]/[HisCCT236(K122A)], was determined by densitometry (Image Quant). Second, the total protein concentration, [CCT367] + [HisCCT236(K122A)], was determined (18), and the two equations were used to calculate [CCT367] and [HisCCT236(K122A)] in the mixture. The same procedures were used to make and quantify a CCT367/His367(K122A) heterodimer, which co-eluted with the catalytically inactive HisCCT367(K122A) homodimer.
Initially we tried to make a heterodimer in vitro from GST-tagged CCT367 and HisCCT236(K122A) homodimers using the same procedure of dissociation/reassociation of the dimers followed by two-step affinity chromatography on Ni-NTA and glutathione-Sepharose. However we could not find conditions appropriate to dissociate both the CCT and GST dimers: 1–3 mm SDS used to dissociate the CCT dimers was not sufficient to dissociate the GST dimers, and some of the treatments used to achieve monomerization of GST, e.g. 15–30% acetonitrile (19), failed to dissociate the CCT dimers.
Glutaraldehyde Cross-linking and Limited Proteolysis
Protein samples (∼20 μg/ml) in PBST Buffer were incubated with 1 mm glutaraldehyde at 37 °C for 15 min. The cross-linking reaction was quenched with ethanolamine (0.1 m final concentration). The cross-linked samples were analyzed by SDS-PAGE (10% gels) and silver staining (20) or by Western analysis using antibodies against residues 164–176 of the catalytic domain (8) or residues 256–288 of domain M (21). The tertiary structure of native and reconstituted CCT dimers was assessed by limited chymotrypsin digestion (9, 14). Samples contained 2.0 μg of CCT in Buffer A and a 1:220 weight ratio of chymotrypsin/CCT. Reactions were quenched after various times at 37 °C with 2 mm phenylmethylsulfonyl fluoride. The fragments were separated by SDS-PAGE (12% gels) and visualized with silver stain (20).
Enzyme Activity Assay
CCT activity was determined essentially as described previously (22). A standard reaction mixture contained 20 mm Tris, pH 7.4, 10 mm DTT, 88 mm NaCl, 12 mm MgCl2, 16 mm CTP, 2.5 mm [14C]phosphocholine (specific activity, 1 mCi/mmol), enzyme dimer concentration of 14 nm, and 0.2 mm PC/PG (1:1, mol/mol) small unilamellar vesicles (SUVs). SUVs were prepared as described previously (23). Reactions were at 37 °C for 10 min. To analyze the response of CCT dimers to variable concentrations of PG vesicles, enzyme velocity measurements were conducted using the standard assay conditions, but the concentration of lipid was varied from 0.1 to 50 μm. For this analysis LUVs were prepared (13) by extrusion of multilamellar vesicles at room temperature through two 100-nm polycarbonate membranes (Lipofast Micro-extruder, Avestin, Ontario, Canada). LUVs were in 10 mm Tris, pH 7.4, 1 mm EDTA, 2 mm DTT and contained trace amounts of 1,2-[3H]dipalmitoyl-sn-glycerophosphocholine to allow quantification of recovery after extrusion. Data were analyzed using GraphPad Prism 4 software by non-linear regression fit to a one-site binding equation: V = Vmax[L]/(K½ + [L]) where [L] is the molar concentration of accessible lipid. The maximal activity Vmax and the K½, an apparent dissociation constant that measures the strength of binding between lipid and protein (24, 25), were determined. The K½ is equivalent to the reciprocal of Kp, which is described below.
The kinetic parameters Vmax, kcat, and Km with respect to phosphocholine and CTP were determined using primary plots of the enzyme velocity versus substrate concentration ([S]). Phosphocholine was varied from 0.1 to 3.5 mm in the presence of 16 mm CTP, and CTP was varied from 0.2 to 15 mm in the presence of 2.5 mm phosphocholine. The velocity versus [S] data were analyzed using GraphPad Prism 4 software and were fit to the Michaelis-Menten equation: V = Vmax[S]/(Km + [S]) where Vmax is the velocity extrapolated to infinite substrate concentration and Km is the Michaelis constant.
Membrane Binding Assay
Binding of CCT dimers to sucrose-loaded large unilamellar vesicles (SLVs) was measured as described previously (10, 26). Briefly multilamellar vesicles containing trace amounts of 1,2-[3H]dipalmitoyl-sn-glycerophosphocholine were prepared in 20 mm HEPES, pH 7.5, 170 mm sucrose, 10 mm DTT; extruded through two 100-nm polycarbonate filters; diluted 5-fold in 100 mm NaCl, 20 mm HEPES, pH 7.5, 10 mm DTT; and sedimented by ultracentrifugation at 100,000 × g for 30 min. CCT dimers (∼0.3 μm) were equilibrated with the SLVs for 20 min at room temperature, and the vesicle-bound protein was separated from free protein by centrifugation at 25 °C as above. In the same experiment a protein sample in the absence of added SLVs was treated similarly to determine the vesicle-independent protein sedimentation. The quantity of CCT in the supernatant and pellet was determined by SDS-PAGE using a Tricine gel system (27). Gels were stained with SYPRO Orange and visualized as described above. The percentage of bound protein was calculated from the equation: % Bound = 100 P/(S + P) where P and S are the fluorescence intensities of the bands for the pellet and supernatant, respectively. The intensity of the pellet band was corrected for contamination by supernatant as well as protein sedimented in the absence of SLVs (26). To ensure that the lipid/protein ratio was not limiting in these analyses using 0.3 μm CCT dimers and PC/PG (1:1) SLVs, we repeated the binding analysis using 0.07 μm CCT dimers and found a variance in the Kp values of only 2-fold between the two determinations.
The percentage of protein bound was plotted as a function of accessible lipid concentration (the concentration of lipid in the outer leaflet of the membrane ([L]access = 0.5[L]tot), and the data were fit to the equation: %Bound protein = 100Kp[L]access/(1 + Kp[L]access) where Kp is the molar partition coefficient. The molar partition coefficient is the reciprocal of the accessible lipid concentration required to bind 50% of the protein (28).
Vesicle Aggregation Assay
CCT-induced vesicle aggregation was assayed at 20 °C by measuring the increase in the apparent absorbance at 400 nm of SUVs or LUVs after addition of protein (14). The absorbance due to vesicles (0.1 mm lipid in 10 mm Tris, pH 7.4 or 8.5, 150 mm NaCl, 1 mm EDTA, 2 mm DTT) was measured. Then variable amounts of CCT were added from a concentrated stock while mixing, and the sample absorbance increase was recorded every 30 s for 5 min. The absorbance at the plateau of the hyperbolic ΔA400 versus time curve was plotted for each CCT concentration trial.
RESULTS
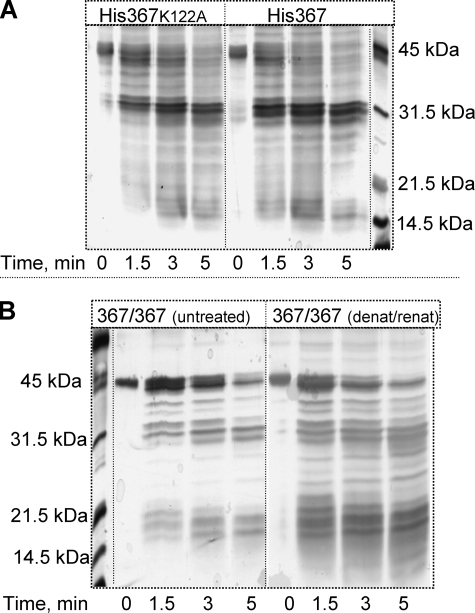
A Functional CCT Heterodimer Composed of One Full-length Subunit and One Truncated Subunit Can Be Formed in Vitro—Our goal was to compare the specific activity of the CCT367 catalytic domain when present in a dimer with one versus two M domains (CCT367/236 versus CCT367). Because CCT236 is constitutively active (active in the absence of lipids) it would complicate the analysis of the effects of membrane engagement of the CCT367 subunit on the activity of the heterodimer. Therefore, we used a catalytically dead mutant of CCT236 in which lysine 122, a critical catalytic residue (29), is substituted with alanine. This mutation does not impact the tertiary conformation of CCT as evidenced by a nearly identical limited proteolysis fragmentation pattern compared with WT enzyme (Fig. 2A). This mutant displayed no detectable activity in our standard enzyme assay in which the lower limit of detection is ∼0.03 units/nmol of CCT active site. Thus the CCT367/236(K122A) heterodimer pairs a subunit containing functional catalytic and membrane binding domains with a subunit that is catalytically non-functional and lacks a domain M-membrane interaction.
FIGURE 2.
Tertiary structure of CCT dimers probed by limited proteolysis. A, effect of the K122A mutation. HisCCT367 or HisCCT367(K122A) was digested with chymotrypsin using a mass ratio of 220:1 (CCT:chymotrypsin). Proteolysis was quenched at the indicated times with 2 mm phenylmethylsulfonyl fluoride. CCT fragments were separated by 12% SDS-PAGE and stained with silver. The His-tagged CCT367 parent species run as doublets on SDS gels likely because of heterogeneous phosphorylation. Fragments containing the phosphorylated domain P in the 36–42-kDa region are thus “fuzzy.” The bands in the 25–30-kDa region are derived from domains N and C (11) and are sharper. B, effect of the denaturation/renaturation protocol used to prepare heterodimers. CCT367 was denatured with 1 mm SDS and renatured as described under “Experimental Procedures” (367/367 (denat/renat)). This CCT and a native CCT control (367/367 (untreated)) were digested with chymotrypsin as above and analyzed on 12% SDS-PAGE gels and silver-stained.
The method that we developed to prepare CCT367/236 heterodimers for the present study generated a species with native folding and high recovery of activity. Untagged full-length wild-type enzyme (CCT367) and a His-tagged version of the truncated, catalytically dead enzyme (HisCCT236(K122A)) were separately denatured with SDS, mixed together, renatured by dialysis of the SDS, and repurified. Table 1 shows that this procedure resulted in 80–90% recovery of the enzyme activity of CCT367 and CCT236 with the wild-type lysine at position 122. The proteolytic cleavage pattern of the CCT367 dimer was also very similar before and after the denaturation/renaturation protocol (Fig. 2B), suggesting recovery of a native fold following renaturation.
TABLE 1.
Specific molar activities of various CCT dimers
CCT activity was assayed with 2.5 mm phosphocholine, 16 mm CTP, and 0.2 mm lipid. Activities are in units/nmol of active catalytic site. 1 unit = 1 nmol of CDP-choline formed per min. Data are means ± range for two independent determinations or means ± S.D. for three or more independent determinations. When n = 1, the data are means ± range of independent duplicate assays. ND, not determined.
|
CCT dimer
|
Denature, renature, and repurify
|
Specific molar activity per active catalytic site
|
|||
|---|---|---|---|---|---|
| No lipid (n) | PG/PC (1:1) SUVs (n) | 100% PG LUVs (n) | |||
| 1 | 236/236 | - | ND | 294 ± 27 (2) | ND |
| 2 | 236/236 | + | ND | 235 ± 14 (1) | ND |
| 3 | 367/367 | - | 22 ± 4a (5) | 745 ± 13 (3) | ND |
| 4 | 367/367 | + | 34 ± 6 (4) | 680 ± 46 (4) | 673 ± 15 (2) |
| 5 | 367/236(K122A) | + | 108 ± 29b (6) | 454 ± 15b (3) | 487 ± 13 (2) |
| 6 | 367/His367(K122A) | + | 20 ± 7 (2) | 466 ± 56b (3) | 539 ± 11 (1) |
| 7 | 367/His236(K122A) | + | 60 ± 17a (4) | 445 ± 18b (3) | 438 ± 11 (1) |
For data sets with n > 2, t test analyses showed that the probability of being the same value as the CCT367/367 dimer on row 4 is ≤0.03a. Other sets were not significantly different from the 367/367 dimer.
For data sets with n > 2, t test analyses showed that the probability of being the same value as the CCT367/367 dimer on row 4 is ≤0.003. Other sets were not significantly different from the 367/367 dimer.
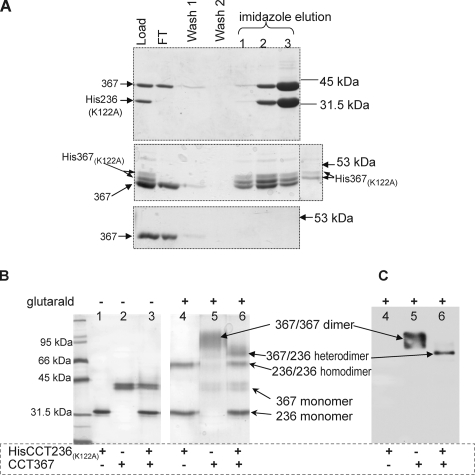
This protocol generated a mixture of three dimers (CCT367, CCT367/His236(K122A), and HisCCT236(K122A)) in a ratio of ∼1:2:1. The full-length homodimer (CCT367) was separated from a mixture of the HisCCT236(K122A) homodimer and the CCT367/His236(K122A) heterodimer by nickel affinity chromatography because only the 236 species was His-tagged. Evidence for formation of a CCT367/His236(K122A) heterodimer is shown in the top panel of Fig. 3A where imidazole co-elutes a 42-kDa species (CCT367) along with HisCCT236(K122A). Moreover glutaraldehyde cross-linking of this preparation generated a new species with a mass of ∼70 kDa, the sum of CCT367 monomer (42 kDa) plus the HisCCT236(K122A) (28.5 kDa) (Fig. 3B). This species scored positive for reaction with anti-domain M antibody (Fig. 3C) and with an antibody against the catalytic domain of CCT (data not shown). Although the preparation of the CCT367/His236(K122A) heterodimer was contaminated with the HisCCT236(K122A) homodimer, this latter species did not contribute to enzyme activity measurements because it was catalytically dead. It also did not interfere with analysis of the membrane binding of the 367 species in the heterodimer because the CCT367 species was readily separated from the CCT236 species on the gels used for monitoring membrane partitioning (see “Experimental Procedures”). Importantly the preparation of heterodimer was not contaminated with the full-length wild-type CCT367 dimer; i.e. there was no species migrating at the position of the CCT367 cross-linked homodimer either in the silver-stained gel or the anti-domain M blot (Fig. 3, B and C). Moreover once reconstituted, the heterodimers were stable because no CCT367 homodimer reappeared in a preparation of CCT367/His236(K122A) that was stored frozen for 6 months, thawed, and reanalyzed by cross-linking (data not shown). As a matched control for the CCT367/His236(K122A) dimer we prepared a CCT367/His367(K122A) heterodimer using the same approach. Fig. 3A shows the separation of this heterodimer from the CCT367 homodimer. CCT367 and HisCCT367(K122A) subunits co-eluted with imidazole (Fig. 3A, middle panel), whereas the untagged CCT367 dimer did not; the latter eluted in the flow-through fraction (Fig. 3A, lower panel). Thus none of the untagged CCT367 homodimer contaminated the CCT367/His367(K122A) heterodimer.
FIGURE 3.
Preparation of CCT heterodimers. Homodimers of CCT367 and either HisCCT236(K122A) or HisCCT367(K122A) were denatured with 1 mm SDS, mixed together, and renatured as described under “Experimental Procedures.” The renatured heterodimer and inactive His-tagged homodimer with the K122A mutation were separated from the full-length CCT367 on a Ni-NTA-agarose column. A, isolation of heterodimers by Ni-NTA-agarose. Top panel, resolution of renatured CCT367/His236(K122A) and His236(K122A) from CCT367. Middle panel, resolution of renatured CCT367/His-367(K122A) and HisCCT367(K122A) from CCT367. The lane on the far right contains purified HisCCT367(K122A) as a marker. Bottom panel, behavior of renatured CCT367 homodimer on the Ni-NTA-agarose column. FT, flow-through fraction. Fractions from the nickel column were evaluated by 10% SDS-PAGE (top panel) or 9% PAGE (middle and bottom panels) and stained with Coomassie Blue. B and C, evidence for formation of a CCT367/His236(K122A) heterodimer by glutaraldehyde cross-linking. Samples containing either HisCCT236(K122A), CCT367 homodimers, or CCT367/His236(K122A) heterodimer (lanes 3 and 6) were cross-linked with 1 mm glutaraldehyde for 15 min. Ethanolamine quencher was added at 0 min to samples in lanes 1–3 in B. Samples were analyzed by 10% SDS-PAGE and stained with silver (B) or visualized by Western blot with antibody against domain M (C).
A CCT Dimer with a Single Domain M Can Be Activated by Lipids to the Same Extent as a CCT Dimer with Two M Domains—To assess the contribution of each domain M to the activation by lipids we determined the enzyme activity under conditions of lipid and substrate saturation for CCT dimers with two versus one domain M (CCT367 versus CCT367/236(K122A)). There are at least four formal possibilities for the effect of membrane binding of domains Ma and Mb on the activity of the dimeric enzyme (where Ma and Mb refer to the M domains of subunits A and B in the homodimer, respectively). (i) Domains Ma and Mb contribute equally and independently to the activation such that the activity of subunit A is not influenced by membrane engagement of Mb, (ii) domains Ma and Mb contribute cooperatively to the activation such that the activity of subunit A increases upon membrane binding of Mb, (iii) the M domains contribute negatively to the activation such that the activity of subunit A decreases upon membrane binding of Mb, or (iv) full activation requires engagement of either Ma or Mb such that activity is maximal when either Ma or Mb alone are engaged. Our analysis initially compared the specific molar activity of the wild-type CCT367 subunit when paired with an identical subunit or with a catalytically dead CCT236 lacking domain M (CCT236(K122A)). If each domain M contributed equally and independently to the activation we would expect the specific activity of the CCT367 subunit to be the same regardless of whether its partner was active or dead. However, the specific molar activity of the CCT367 subunit in the CCT367/236(K122A) species was 67 ± 2% of the CCT367 subunit in the CCT367 homodimer (Table 1, compare rows 4 and 5; activity in the presence of PG/PC SUVs). This finding suggested that full activation might require cooperative effects of two M domains or alternatively cooperative effects between active sites, only one of which was working in the heterodimer.
To test whether the apparent cooperativity could be explained by coupled effects at the two active sites rather than cooperative contributions of the two M domains, we prepared a heterodimer composed of one wild-type CCT367 subunit paired with HisCCT367(K122A). This construct makes a more closely matched control for the CCT367/His236(K122A) heterodimer because the only difference is the loss of one C-terminal tail. When the CCT367/His236(K122A) heterodimer and the CCT367/His367(K122A) heterodimer were assayed in parallel, the lipid-stimulated specific molar activities of the active 367 subunit were nearly the same (466 versus 445 units/nmol of CCT367; p = 0.58; Table 1, rows 6 and 7). The 33% reduction in activity for these His-tagged heterodimers in comparison with the CCT367 homodimer is not due to the His tag because the activity of CCT367/236(K122A) was the same with or without the tag (p = 0.68; Table 1, rows 5 and 7). Thus the functioning of the active site of one subunit has a small but significant impact on the functioning of the active site in the partnering subunit (compare the lipid-stimulated specific molar activity of CCT367 versus CCT367/His367(K122A); Table 1, rows 4 and 6; p < 0.005). However, when we accounted for this effect, we found that a dimer containing two M domains has activity nearly identical to that of a dimer containing one M domain.
To examine the basis of the cooperation between subunit active sites we compared the kinetic parameters of the CCT367 dimer containing two functioning active sites with CCT367/His367(K122A) or CCT367/His236(K122A), both of which contain one debilitated and one functioning active site (Table 2). First, the procedure to generate heterodimers did not impair the kinetic parameters of the enzyme; kcat and Km values were the same for the full-length wild-type enzyme with or without denaturation/renaturation (Table 2, row 3 versus row 4). Second, pairing a dead active site with a functioning one reduced kcat for the functioning subunit by ∼50% (Table 2, row 2 versus row 3). It had no effect on the CTP Km and slightly decreased the phosphocholine Km. Thus the subunit with a mutationally disabled active site can reduce the kcat of its wild-type partner. Lastly, the kcat values for CCT367/His367(K122A) and CCT367/His236(K122A) were the same (p = 0.84; Table 2, row 1 versus row 2) in agreement with the analysis shown in Table 1. The 3-fold increase in the CTP Km for the 367/236 heterodimer may be a consequence of the pairing of the active CCT367 subunit with the CCT236 truncation, which is known to have a very high CTP Km of 4.5 mm (5). This Km effect also highlights the interdependence of the two active sites of the CCT dimer.
TABLE 2.
Kinetic constants for CCT dimers
CCT activity was assayed in the presence of 0.2 mm PG/PC (1:1) SUVs as described under “Experimental Procedures.” Plots of activity versus [phosphocholine] or versus [CTP] were fit to the Michaelis-Menten equation (V = Vmax[S]/(Km + [S]), using GraphPad Prism 4 software, to generate the kinetic parameters kcat and Km. The r2 values for the fit of the data to this equation were ≥0.96 for each trial. Km and kcat are derived from the best fit to this equation ±S.E. of the fit. n is the number of independent determinations. The S.E. values associated with the kcat values were obtained by error propagation of the S.E. of the kcat values for individual substrates (CTP and phosphocholine (PCho)).
|
CCT dimer
|
kcat (n)
|
Km (n)
|
|||
|---|---|---|---|---|---|
| PCho | CTP | ||||
| min-1 | mm | ||||
| 1 | 367/His236(K122A) | 454 ± 11a (4) | 0.48 ± 0.07 (2) | 1.82 ± 0.16 (2) | |
| 2 | 367/His367(K122A) | 474 ± 10a (5) | 0.28 ± 0.03 (3) | 0.61 ± 0.08 (2) | |
| 3 | 367/367 (denatured/renatured) | 861 ± 32 (4) | 0.41 ± 0.10 (2) | 0.72 ± 0.14 (2) | |
| 4 | 367/367 (untreated) | 885 ± 35 (2) | 0.46 ± 0.12 (2) | 0.64 ± 0.10 (2) | |
For data sets with n > 2, t test analyses showed that the probability of being the same as the CCT367/367 dimer on row 3 is <0.005.
These results eliminate scenarios ii and iii above, the positively and negatively cooperative models for the effect of domain M-lipid binding on activity, but do not distinguish between scenarios i and iv. To further evaluate and potentially distinguish between these models we needed to assess whether both M domains of the 367 homodimer engage the PC/PG (1:1) vesicles with equivalent binding strength.
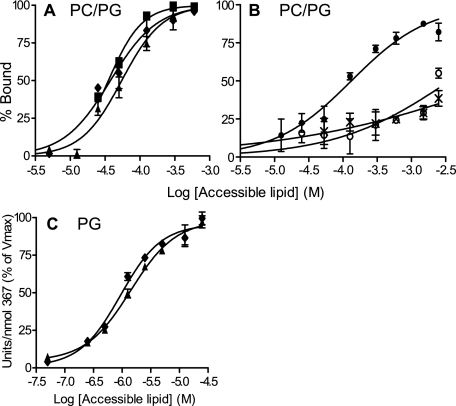
A CCT Dimer with Only One Domain M Binds as Tightly to Anionic Vesicles as a CCT Dimer with Two M Domains—To assess the contribution of each domain M of the CCT dimer to membrane binding we measured the membrane partitioning of the CCT367/236(K122A) heterodimer and a similarly prepared CCT367 homodimer using SLVs. These analyses used untagged CCTs. Fig. 4A shows binding of constructs to PC/PG (1:1) SLVs. If both M domains of the CCT367 homodimer engage the membrane, we would expect its binding curve to be shifted far to the left (lower [L]) of the binding curve for the heterodimer with only one M domain; however, the binding curves were remarkably similar. The partition coefficients, calculated as described under “Experimental Procedures,” are shown in Table 3. The construct with two M domains had a Kp only ∼1.5-fold higher than the CCT dimer with one domain M; this translates into a ΔΔGbind of only 0.2 kcal/mol. This surprising result suggests that the M domains experience a large negative cooperativity in binding.
FIGURE 4.
Membrane binding affinity of CCT dimers with two, one, or zero M domains. Membrane binding was assayed using SLVs composed of PC/PG (1:1) and sedimentation analysis at 25 °C (A and B) or PG LUVs and activity measurements at 37 °C (C). The binding curves are compiled from two to three independent experiments. Using Prism 4 GraphPad software, the curves were fit to the equation: %Bound protein = 100Kp[L]/(1 + Kp[L]) where [L] is the concentration of accessible lipid and Kp is the partition coefficient. The data points are means of two to three independent experiments; error bars show range (n = 2) or S.D. (n = 3). See Table 3 for the partition coefficients extracted from these binding curves. Note the different x axis scales in A, B, and C. CCT constructs are as follows: CCT367 (untreated) (▪), CCT367 (denatured and renatured) (♦); CCT367/236(K122A) (▴); CCT236 (×), HisCCT236(K122A) at pH 7.5 (•); HisCCT236(K122A) at pH 8.5 (○). All binding curves were at pH 7.5 and used untagged constructs unless indicated.
TABLE 3.
Partition coefficients and binding free energies for CCT dimers
Membrane binding was assayed using PC/PG (1:1) SLVs and an ultracentrifugation technique (25 °C) (rows 1–3) or PG LUVs and activity analysis (37 °C) (rows 4 and 5). The binding data in Fig. 4, A and C, were fit by non-linear regression to the equation %Bound protein = 100Kp[L]access/(1 + Kp[L]access) using GraphPad Prism 4 software. Kp = 1/[L] when CCT is 50% bound (28). The error is plus or minus the 95% confidence interval with respect to the best fit Kp value. The ΔG values were derived from ΔG = – RT ln Kp[H2O].
| CCT dimer | Denature, renature, and repurify | Phospholipid | n | Partition coefficient Kp | Binding free energy ΔG | |
|---|---|---|---|---|---|---|
| ×104m-1 | kcal/mol | |||||
| 1 | 367/367 | - | PG/PC (1:1) | 3 | 2.8 ± 0.3 | -8.4 ± 0.1 |
| 2 | 367/367 | + | PG/PC (1:1) | 3 | 2.7 ± 1.1 | -8.4 ± 0.2 |
| 3 | 367/236(K122A) | + | PG/PC (1:1) | 2 | 1.8 ± 0.3 | -8.2 ± 0.1 |
| 4 | 367/367 | + | 100% PG | 2 | 107 ± 27 | -11.0 ± 0.2 |
| 5 | 367/236(K122A) | + | 100% PG | 2 | 71 ± 21 | -10.8 ± 0.2 |
CCT can bind vesicles with a high anionic lipid content in a cross-bridging manner that we previously interpreted to mean that the two M domains are positioned on opposite sides of the dimerization domain (Ref. 10 and see Fig. 1, Model II). Could the apparent non-binding function of the second M domain of the dimer be explained by the idea that the M domains are not engaged with the same bilayer? To explore this idea we examined the binding of the CCT dimers to 100% PG vesicles, which have a higher tendency for CCT-induced aggregation than do 50% PG vesicles (Ref. 10 and Fig. 5A) and would potentially have a greater proportion of both M domains of the CCT367 homodimer engaged with membranes in the cross-bridging mode. If the two M domains engage the same bilayer with positive cooperativity the binding energies should be roughly additive, but if the two M domains engage two separate membranes the binding energy would be much less than additive because of entropic penalties associated with constraining two vesicles.
FIGURE 5.
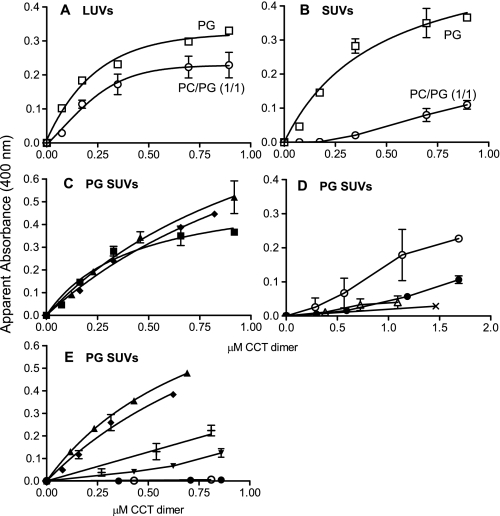
Aggregation of vesicles induced by various CCT dimers. The absorbance due to vesicles was adjusted to 0, and the protein-induced increase was monitored for 5 min. The plateau value is plotted for each CCT concentration. The assay pH was 7.4 in A–D and 8.5 in E. Data represent means ± S.D. of three or range of two independent determinations. CCT constructs are as follows: A and B, CCT367 untreated; C, CCT367 untreated (▪) or denatured/renatured (♦) and CCT367/236(K122A) (▴); D, HisCCT236 (○), HisCCT236ΔNLS (•), CCT367ΔNLS (▵), and CCT236 (×); E, CCT367/His236(K122A) (▴), CCT367 denatured/renatured (♦), CCT367ΔNLS/His236(K122A) (+), CCT367/His236ΔNLS(K122A) (▾), HisCCT236ΔNLS (•), and HisCCT236 (○).
We were unable to evaluate the binding of the CCT dimers to 100% PG vesicles by sedimentation and gel analysis because the binding was so strong that even at the lowest protein concentration that would allow detection of the CCT (0.14 μm CCT monomer) the lipid/protein ratio became limiting for the binding reaction. Instead we evaluated the lipid concentration dependence for enzyme activation by PG vesicles using conditions where [L] was in excess of [P], the total protein concentration, even at the low range of [L] and used these plots to estimate the binding constants for the two dimers (Table 3, rows 4 and 5). Fig. 4C shows that the binding of CCT367 versus CCT367/236(K122A) by 100% PG LUVs was nearly identical. The Kp values computed from these curves were approximately the same for the two dimers (Table 3, rows 4 and 5) and were nearly 2 orders of magnitude higher than for binding to PG/PC 1:1 SLVs. This finding, that CCT367 binds PG vesicles with the same apparent affinity as CCT367/236(K122A), is consistent with two distinct possibilities. (i) Each domain M of the CCT dimer binds a separate membrane, or (ii) the two M domains could bind the same bilayer but with a large negative cooperativity. To differentiate between these we measured the contribution of each domain M to vesicle aggregation.
A CCT Dimer with Only One Domain M Induces Anionic Vesicle Cross-bridging as Effectively as a CCT Dimer with Two M Domains—Fig. 5, A and B, show that full-length CCT induces the aggregation of LUVs and SUVs composed of 100% PG and is less effective toward either type of vesicle composed of PG/PC (1:1). To assess the contribution of each domain M to vesicle tethering we compared the concentration of CCT367 and CCT367/236(K122A) needed to aggregate 100% PG SUVs. We found that the heterodimer with only one domain M was as effective as the homodimer in promoting the aggregation of the 100% PG vesicles (Fig. 5C). This finding contradicted our original model (Fig. 1, Model II) where two M domains are required for cross-bridging. Another membrane binding motif that mediates vesicle cross-bridging could explain this quandary. This second motif would be present in both full-length CCT and the truncated CCT236.
Deletion of the Nuclear Localization Signal from the CCT Dimer Eliminates Anionic Vesicle Cross-bridging Function—The polybasic nuclear localization signal near the N terminus of domain N was considered as a potential electrostatic membrane binding motif. Deletion of residues 12RKRRK16 (CCT367(ΔNLS)) obliterated the induction of vesicle aggregation (Fig. 5D), indicating a requirement of this motif for the cross-bridging mode of vesicle interaction. It seemed unlikely that the NLS motif on its own could function in membrane tethering, and in keeping with this idea we could barely detect the binding of the CCT236 homodimer to SLVs of PG/PC (1:1) (Fig. 4B) nor could we detect induction of PG vesicle aggregation by CCT236 (Fig. 5D). However, augmenting the positive charge at the N terminus of CCT236 with a His6 tag did induce vesicle aggregation (Fig. 5D) albeit much more weakly than did the full-length CCT or the CCT367/236(K122A) heterodimer (Fig. 5C). In SLV binding assays we also observed an enhancement of membrane binding (yielding a Kp value of ∼7,000 m–1) when a His tag was placed proximal to the NLS on CCT236 (Fig. 4B). Deletion of the NLS from the His-tagged CCT236 diminished the cross-bridging action (Fig. 5D), suggesting that the combination of the two proximal polybasic domains (His tag and the NLS) are required in this construct to forge sufficiently strong interactions to tether two membranes. In keeping with this, the His-tagged CCT236 did not promote vesicle aggregation at pH 8.5, i.e. when the charge on the His motif would be neutral (Fig. 5E). The binding affinity of HisCCT236(K122A) to SLVs was also too weak to measure at pH 8.5 (Fig. 4B).
To assess the contribution of each NLS to vesicle cross-bridging we prepared two additional heterodimers by the SDS denaturation/renaturation method: CCT367(ΔNLS)/His236(K122A) and CCT367/His236(ΔNLS; K122A). Vesicle aggregation was assayed at pH 8.5 to neutralize the charge of the His tag. A switch of pH from 7.4 to 8.5 had very little effect on domain M participation in vesicle tethering (compare CCT367 in Fig. 5, C and E). Deletion of the NLS from either the 367 or the 236 subunit of the heterodimers reduced the vesicle aggregation activity but did not eliminate it (Fig. 5E). Thus although the pairing of two polybasic NLS motifs per dimer strengthens the tethering action, the orientation of the CCT dimers is such that some tethering is obtained with only one NLS per dimer. This finding helps to differentiate between models for CCT cross-bridging (Fig. 1) as discussed below.
To assess whether the NLS-dependent cross-bridging mode of membrane binding impacts the activity of CCT, we compared the activity of full-length WT CCT367 versus CCT367(ΔNLS) in the presence of saturating 100% PG SUVs. The WT CCT367 should tether these vesicles, whereas the ΔNLS mutant should not. We found that the specific activities were nearly identical (WT CCT367, 508 ± 56 units/nmol CCT; CCT367ΔNLS, 493 ± 31 units/nmol CCT for two independent analyses), suggesting that the cross-bridging ability does not impact the activity of the enzyme.
DISCUSSION
The experiments in this study were driven by the following central question: how does the dimeric structure of CCT contribute to its activity, membrane binding, and membrane cross-bridging? To answer this question we compared a CCT dimer with one versus two membrane binding domains (domain M) with respect to these three functions.
The CCT Dimer Uses Only One M Domain at a Time to Bind Membranes—In the simplest case (Fig. 1, Model I), both M domains of the CCT dimer engage the same bilayer upon membrane binding, with positive cooperation between M domains, such that Kp(dimer) ≈ [Kp(monomer)]2 and ΔG(dimer) = 2[ΔG(monomer)] (30). According to this model, the dimer with two M domains would have up to twice the binding strength of the dimer with only one M domain. Our finding that the binding strength for 50 or 100% PG vesicles was equal for CCT dimers with one or two M domains can be interpreted in various ways. Initially we supposed that the lack of a contribution of the second M domain reflected its role in cross-bridging. In this scenario, engagement of each M domain with separate vesicles would be characterized by lower binding energy than for their simultaneous engagement with a single bilayer because of entropic penalties associated with immobilization of two vesicles. According to this model, both M domains would be required for binding two vesicles in trans (Fig. 1, Model II). However, the vesicle cross-bridging function required only a single M domain because CCT367 and CCT367/236 were equally effective at inducing vesicle aggregation. These data, along with the discovery of an NLS requirement for membrane cross-bridging, are more consistent with a model where the M domains alternately engage the same bilayer (Fig. 1, Model IV). Simultaneous membrane engagement may be precluded by steric interference of the two M domains; i.e. inability to simultaneously position both M domains on the same plane as the membrane surface.
Comparative analysis of the membrane affinities of protein kinase C isoforms and their multiple C1 and C2 membrane binding modules has likewise revealed that although the two domains do show some cooperative binding the measured binding affinity of the holoenzyme is much less than the product of the individual module binding affinities (1). Moreover the protein kinase Cδ isoform utilizes only one of its available membrane binding modules (its C1 domain) to engage anionic membranes. Its C2 membrane binding module appears to be incapable of binding anionic vesicles when the holoenzyme is analyzed (31). Thus it may be common in amphitropic proteins that steric interference of individual modules limits the membrane binding strength of the protein so that binding and activity can be regulated by small variations in the content of regulatory lipids.
The distribution of CCT dimers in cells is largely non-membranous by both biochemical fractionation and fluorescence imaging and is observed to partition into the membrane only when the content of regulatory lipids is experimentally augmented (17, 32–34). For CCT to be effectively regulated by small variations in the mol percent anionic lipid in the physiological range of 10–30 mol % (35) the Kd for membrane dissociation must be in the range of the molar concentration of anionic lipid (calculated by assuming that the lipid uniformly takes up the volume of the cell). This requirement is met if the Kd is in the 0.1–1 mm range. If the Kd value for the CCT dimer were in the sub-μm range, as in the case where the dimer Kd is the square of the monomer Kd, the protein would be constitutively membrane-bound. Our measurements showed that even for 50 mol % anionic vesicles the Kd for a WT CCT dimer is ∼100 μm and would be even higher for membranes with lower anionic lipid contents.
The Nuclear Localization Signal Sequence Is Required for Membrane Cross-bridging—We have shown that membrane tethering requires one domain M (but not two) and at least one NLS. A role for the NLS in vesicle tethering is also supported by the finding that the CCTβ2 isoform (which lacks the NLS) cannot induce vesicle cross-bridging, and replacing domain N of CCTβ2 with the corresponding region of CCTα results in a gain of tethering function.5 The anionic vesicle binding and tethering ability of HisCCT236 argues that the NLS itself constitutes a weak membrane binding domain rather than acting as a domain that merely modulates the membrane affinity of domain M. Analysis of CCT367/His236(K122A) heterodimers with the NLS deleted from the 367 subunit or the 236 subunit showed that cross-bridging activity is impaired but not obliterated by loss of one NLS. Thus although the pairing of two polybasic NLS motifs per dimer strengthens the tethering action, the orientation of the CCT dimers is such that some tethering is obtained with only one NLS per dimer. Vesicle aggregation induced by a CCT dimer with only a single NLS motif and one M domain per dimer supports a membrane cross-bridging model for WT CCT dimers in which the two M domains alternately bind to vesicle 1 and the two NLS motifs engage vesicle 2 (Fig. 1, Model IV) and eliminates Model III with the M and NLS motifs of one subunit engaging the same bilayer. Vesicle cross-bridging by CCT367(ΔNLS)/His236(K122A) also eliminates a variant of Model III in which the NLS and domain M of separate subunits engage the same bilayer. That the His-tagged CCT236 homodimer, which lacks any M domains, can weakly induce vesicle cross-bridging might be better rationalized with Fig. 1, Model III. However, the weak tethering by CCT236 with a His6 tag may result from increased orientational possibilities of a CCT dimer lacking domain M (where tethering via domain M would restrict tumbling of the dimer) so that the two NLS + His6 extensions could engage two separate membranes in trans. (As a word of caution, researchers should pay heed to the potential membrane binding enhancement of His tags placed adjacent to polybasic sequences.)
The role of the N-terminal NLS in CCTα in nuclear import has been clearly demonstrated. Its deletion results in exclusion of CCTα from the nucleus (36) and explains the cytoplasmic localization of CCTβ isoforms (37), which have domain N sequences divergent from the α isoform. Our discovery of a membrane binding function for the CCT NLS is novel for CCT but has some precedence. There are a few reports that allude to NLS motif doubling as membrane binding motifs in other proteins (38–40). Mid1p, a protein that helps establish the position of the cytokinetic ring in fission yeast, requires both a short amphipathic helix and an adjacent NLS for association with the nuclear or plasma membrane (40). Heo et al. (38) noticed that a polybasic plasma membrane-targeting motif in Rit or Rin GTPases could be converted into a nuclear targeting signal by replacing a single tryptophan with alanine. NLS motifs bind tightly to importin α until delivery into the nucleoplasm (41) where dissociation is promoted by Ran·GTP. As polybasic motifs, the free NLS motif could then interact electrostatically with anionic membrane surfaces. The polybasic motif-membrane interaction is well described for peripheral membrane proteins such as myristoylated alanine-rich C kinase substrate, Ras family GTPases, or the Src kinases, which use this motif in combination with lipid anchors or hydrophobic amino acids to bind the plasma membrane or internal membranes (28, 30, 39, 42). NLS involvement in membrane cross-bridging is without precedent.
Whether CCT and its NLS participate in membrane tethering interactions in cells is not known. Our in vitro analysis suggests that supraphysiological contents of anionic lipids are required for NLS function in tethering. However, there are hints that membrane tethering can be promoted by CCT in cells. Overexpression of a full-length CCT domain M mutant with enhanced membrane affinity in Chinese hamster ovary cell nuclei induced the formation of stacked intranuclear tubules invaginating from the inner nuclear membrane that were decorated with CCT as imaged with immunogold (34). These membrane stacks may have been tethered by the CCT. Lung epithelial cells express CCTα in the cytoplasm rather than the nucleus (43). These cells generate membrane stacks-lamellar bodies, the precursors to secreted lung surfactant. The stacked morphology was disrupted in cells with a CCTα gene deletion (44), suggesting a role for CCTα in bilayer adhesion. The role of the NLS in these processes remains to be explored, but an interesting idea is that, in lung cells, the NLS of CCT is involved in membrane tethering; thus it is not available for capture and importation into the nucleus.
Only One M Domain Is Required for Full Activation of the CCT Dimer—We found equivalent kcat values for the WT subunit of CCT in CCT367/His236(K122A) and CCT367/ His367(K122A) in the presence of 50 or 100% PG vesicles (Tables 1 and 2). These two dimers differ only in the number of M + P domains; the catalytic domains are identical. The equivalent activities for a CCT dimer with one versus two M domains can be rationalized with their equivalent membrane binding affinities. If we accept the conclusion that only one domain M at a time is involved in the binding (see above), then only one at a time contributes to activation. Two possibilities must be evaluated: (i) independent activation of individual subunits where binding of Mb can only activate its own catalytic domain and has no influence on the activity of the active site of the partnering subunit A and (ii) fully cooperative activation where binding of Ma or Mb is sufficient for full activation of the CCT dimer.
The critical data we use to differentiate between these models are the equivalent specific molar activity (kcat) values for the WT subunit of CCT in the CCT367/His367(K122A) and CCT367/His236(K122A) dimers (∼450/min). The activity analysis utilized lipid concentrations in which the CCT dimers were fully bound to vesicles. The CCT367/His236(K122A) dimer can only bind membranes via Ma because there is no Mb associated with the 236 subunit. Thus 450/min reflects the kcat of a single active CCT367 subunit. The CCT367/His367(K122A) dimer with two M domains can bind vesicles using either Ma or Mb with equal probability, and each dimer orientation contributes 50% to the kcat value. When Mb is engaged this will not activate catalytic domain B because the debilitating K122A mutation is in subunit B. If binding of Mb in CCT367/His367(K122A) could only influence its own catalytic domain then the activity of this CCT dimer when bound via Mb would be negligible; i.e. half of the population of CCT367/His367(K122A) would be dead. Thus the specific molar activity of the WT subunit in CCT367/His367(K122A) should be half that of CCT367/His236(K122A). If, on the other hand, the catalytic domain of subunit A is fully activated by the binding of Mb, then the specific molar activity of CCT367/His367(K122A) will be 450/min regardless of whether Ma or Mb is engaged with the membrane, and the specific molar activities of the WT 367 subunit should be the same for the two heterodimers, which is what we observed. If the two M domains of CCT interact with each other, one can envision how the binding of one M domain to a membrane might displace the second M domain even though only one M domain inserts into the bilayer, leading to derepression of both active sites of the dimer.
The CCT367 homodimer had a specific molar activity of ∼900/min per monomer, twice that of the heterodimers. We propose that the reduced activity upon pairing CCT367 with HisCCT367(K122A) is due to a coupling of active sites. How might the active site functioning of subunit A influence that of subunit B? A portion of the active site of CCT is situated near the dimer interface based on a homology model of the CCT catalytic domain (11) with the related CTP:glycerol-3-phosphate cytidylyltransferase (45). Two separate solved structures of glycerol-3-phosphate cytidylyltransferase with bound substrate (45) or product (46) indicate movement of the helix B-loop 2 segment at the base of the active site during a catalytic cycle that likely serves to orient key catalytic lysines residing on this loop (Lys44 and Lys46 in glycerol-3-phosphate cytidylyltransferase and Lys122 in CCT (29, 46)). Optimal restructuring of both active sites during turnover may require synchronized movements. If the Lys122 → Ala mutation impairs movements in catalytic site B this coordinated motion may be impeded, thus impacting the facility with which subunit A induces the transition state or releases product.
In the soluble form of CCT, is each M domain able to independently suppress the catalytic activity of the dimer, or do the M domains cooperatively repress catalysis? Although our data show that CCT367/236(K122A) had much lower activity in the absence than in the presence of lipids, it had ∼3-fold higher lipid-independent activity than WT CCT367 (Table 1). Thus full repression of catalytic activity may require input from both M domains of the dimer (cooperative suppression).
In this work we sought to answer how the dimeric structure of CCTα conveys an advantage to the enzyme. We discovered that the dimer does not enhance membrane affinity via dual engagement of domain M, but rather the dimeric structure appears to enable (i) cooperation between active sites and (ii) cooperation between the NLS and domain M to facilitate membrane cross-bridging. In the process we obtained valuable information regarding the orientation of domains M and N within the CCT dimer. The stage is now set to unravel the contribution of the CCT NLS to membrane binding and tethering in cells.
Acknowledgments
We thank Joseph Lee for assistance with expression of His-CCT236 in bacteria and Dr. Neale Ridgway for valuable comments on our manuscript.
This work was supported by Canadian Institutes of Health Research Grant 12134. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PH, pleckstrin homology; CCT, α isoform of rat; CTP:phosphocholine cytidylyltransferase; GST, glutathione S-transferase; NLS, nuclear localization signal; PC, l-α-phosphatidylcholine (from egg); PG, l-α-phosphatidylglycerol (from egg); Ni-NTA, nickel-nitrilotriacetic acid; SUVs, sonicated unilamellar vesicles; LUVs, large unilamellar vesicles; SLV, sucrose-loaded vesicle; TEV, tobacco etch virus; DTT, dithiothreitol; WT, wild-type; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
R. B. Cornell, unpublished data.
M. K. Dennis, S. Taneva, and R. B. Cornell, unpublished data.
References
- 1.Johnson, J. E., Giorgione, J., and Newton, A. C. (2000) Biochemistry 39 11360–11369 [DOI] [PubMed] [Google Scholar]
- 2.Sciorra, V. A., Rudge, S. A., Wang, J., McLaughlin, S., Engebrecht, J., and Morris, A. J. (2002) J. Cell Biol. 159 1039–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee, S. G. (2001) Annu. Rev. Biochem. 70 281–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornell, R. B., and Northwood, I. C. (2000) Trends Biochem. Sci. 25 441–447 [DOI] [PubMed] [Google Scholar]
- 5.Friesen, J. A., Campbell, H. A., and Kent, C. (1999) J. Biol. Chem. 274 13384–13389 [DOI] [PubMed] [Google Scholar]
- 6.Weinhold, P. A., Rounsifer, M. E., Charles, L., and Feldman, D. A. (1989) Biochim. Biophys. Acta 1006 299–310 [DOI] [PubMed] [Google Scholar]
- 7.Cornell, R. B. (1989) J. Biol. Chem. 264 9077–9082 [PubMed] [Google Scholar]
- 8.Xie, M., Smith, J. L., Ding, Z., Zhang, D., and Cornell, R. B. (2004) J. Biol. Chem. 279 28817–28825 [DOI] [PubMed] [Google Scholar]
- 9.Craig, L., Johnson, J. E., and Cornell, R. B. (1994) J. Biol. Chem. 269 3311–3317 [PubMed] [Google Scholar]
- 10.Taneva, S. G., Patty, P. J., Frisken, B. J., and Cornell, R. B. (2005) Biochemistry 44 9382–9393 [DOI] [PubMed] [Google Scholar]
- 11.Bogan, M. J., Agnes, G. R., Pio, F., and Cornell, R. B. (2005) J. Biol. Chem. 280 19613–19624 [DOI] [PubMed] [Google Scholar]
- 12.Bartlett, G. R. (1959) J. Biol. Chem. 234 466–468 [PubMed] [Google Scholar]
- 13.Davies, S. M., Epand, R. M., Kraayenhof, R., and Cornell, R. B. (2001) Biochemistry 40 10522–10531 [DOI] [PubMed] [Google Scholar]
- 14.Taneva, S., Johnson, J. E., and Cornell, R. B. (2003) Biochemistry 42 11768–11776 [DOI] [PubMed] [Google Scholar]
- 15.Walkey, C. J., Kalmar, G. B., and Cornell, R. B. (1994) J. Biol. Chem. 269 5742–5749 [PubMed] [Google Scholar]
- 16.MacDonald, J. I., and Kent, C. (1993) Protein Expr. Purif. 4 1–7 [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. E., Xie, M., Singh, L. M., Edge, R., and Cornell, R. B. (2003) J. Biol. Chem. 278 514–522 [DOI] [PubMed] [Google Scholar]
- 18.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 19.Vargo, M. A., and Colman, R. F. (2004) Protein Sci. 13 1586–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poehling, H.-M., and Neuhoff, V. (1981) Electrophoresis 2 141–147 [Google Scholar]
- 21.Johnson, J. E., Aebersold, R., and Cornell, R. B. (1997) Biochim. Biophys. Acta 1324 273–284 [DOI] [PubMed] [Google Scholar]
- 22.Sohal, P. S., and Cornell, R. B. (1990) J. Biol. Chem. 265 11746–11750 [PubMed] [Google Scholar]
- 23.Cornell, R. B. (1991) Biochemistry 30 5881–5888 [DOI] [PubMed] [Google Scholar]
- 24.Yang, W., and Jackowski, S. (1995) J. Biol. Chem. 270 16503–16506 [DOI] [PubMed] [Google Scholar]
- 25.Helmink, B. A., and Friesen, J. A. (2004) Biochim. Biophys. Acta 1683 78–88 [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. E., Goulding, R. E., Ding, Z., Partovi, A., Anthony, K. V., Beaulieu, N., Tazmini, G., Cornell, R. B., and Kay, R. J. (2007) Biochem. J. 406 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schagger, H., and von Jagow, G. (1987) Anal. Biochem. 166 368–379 [DOI] [PubMed] [Google Scholar]
- 28.Murray, D., Hermida-Matsumoto, L., Buser, C. A., Tsang, J., Sigal, C. T., Ben-Tal, N., Honig, B., Resh, M. D., and McLaughlin, S. (1998) Biochemistry 37 2145–2159 [DOI] [PubMed] [Google Scholar]
- 29.Helmink, B. A., Braker, J. D., Kent, C., and Friesen, J. A. (2003) Biochemistry 42 5043–5051 [DOI] [PubMed] [Google Scholar]
- 30.Buser, C. A., Sigal, C. T., Resh, M. D., and McLaughlin, S. (1994) Biochemistry 33 13093–13101 [DOI] [PubMed] [Google Scholar]
- 31.Giorgione, J. R., Lin, J. H., McCammon, J. A., and Newton, A. C. (2006) J. Biol. Chem. 281 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Y., MacDonald, J. I., and Kent, C. (1993) J. Biol. Chem. 268 5512–5518 [PubMed] [Google Scholar]
- 33.Jamil, H., Hatch, G. M., and Vance, D. E. (1993) Biochem. J. 291 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gehrig, K., Cornell, R. B., and Ridgway, N. D. (2008) Mol. Biol. Cell 19 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vance, J. E., and Steenbergen, R. (2005) Prog. Lipid Res. 44 207–234 [DOI] [PubMed] [Google Scholar]
- 36.Wang, Y., MacDonald, J. I., and Kent, C. (1995) J. Biol. Chem. 270 354–360 [DOI] [PubMed] [Google Scholar]
- 37.Lykidis, A., Baburina, I., and Jackowski, S. (1999) J. Biol. Chem. 274 26992–27001 [DOI] [PubMed] [Google Scholar]
- 38.Heo, W. D., Inoue, T., Park, W. S., Kim, M. L., Park, B. O., Wandless, T. J., and Meyer, T. (2006) Science 314 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams, C. L. (2003) Cell. Signal. 15 1071–1080 [DOI] [PubMed] [Google Scholar]
- 40.Celton-Morizur, S., Bordes, N., Fraisier, V., Tran, P. T., and Paoletti, A. (2004) Mol. Cell. Biol. 24 10621–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodel, M. R., Corbett, A. H., and Hodel, A. E. (2001) J. Biol. Chem. 276 1317–1325 [DOI] [PubMed] [Google Scholar]
- 42.McLaughlin, S., and Murray, D. (2005) Nature 438 605–611 [DOI] [PubMed] [Google Scholar]
- 43.Ridsdale, R., Tseu, I., Wang, J., and Post, M. (2001) J. Biol. Chem. 276 49148–49155 [DOI] [PubMed] [Google Scholar]
- 44.Tian, Y., Zhou, R., Rehg, J. E., and Jackowski, S. (2007) Mol. Cell. Biol. 27 975–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber, C. H., Park, Y. S., Sanker, S., Kent, C., and Ludwig, M. L. (1999) Struct. Fold. Des. 7 1113–1124 [DOI] [PubMed] [Google Scholar]
- 46.Pattridge, K. A., Weber, C. H., Friesen, J. A., Sanker, S., Kent, C., and Ludwig, M. L. (2003) J. Biol. Chem. 278 51863–51871 [DOI] [PubMed] [Google Scholar]