Abstract
S100A2 and S100A6 interact with several target proteins in a Ca2+-regulated manner. However, the exact intracellular roles of the S100 proteins are unclear. In this study we identified Hsp70/Hsp90-organizing protein (Hop) and kinesin light chain (KLC) as novel targets of S100A2 and S100A6. Hop directly associates with Hsp70 and Hsp90 through the tetratricopeptide (TPR) domains and regulates Hop-Hsp70 and Hop-Hsp90 complex formation. We have found that S100A2 and S100A6 bind to the TPR domain of Hop, resulting in inhibition of the Hop-Hsp70 and Hop-Hsp90 interactions in vitro. Although endogenous Hsp70 and Hsp90 interact with Hop in resting Cos-7 cells, but not with S100A6, stimulation of these cells with ionomycin caused a Hop-S100A6 interaction, resulting in the dissociation of Hsp70 and Hsp90 from Hop. Similarly, glutathione S-transferase pulldown and co-immunoprecipitation experiments revealed that S100A6 binds to the TPR domain of KLC, resulting in inhibition of the KLC-c-Jun N-terminal kinase (JNK)-interacting protein 1 (JIP-1) interaction in vitro. The transiently expressed JIP-1 interacts with KLC in resting Cos-7 cells but not with S100A6. Stimulation of these cells with ionomycin also caused a KLC-S100A6 interaction, resulting in dissociation of JIP-1 from KLC. These results strongly suggest that the S100 proteins modulate Hsp70-Hop-Hsp90 multichaperone complex formation and KLC-cargo interaction via Ca2+-dependent S100 protein-TPR protein complex formation in vivo as well as in vitro. Moreover, we have shown that S100A2 and S100A6 interact with another TPR protein Tom70 and regulate the Tom70-ligand interaction in vitro. Thus, our findings suggest a new intracellular Ca2+-signaling pathway via S100 proteins-TPR motif interactions.
The S100 protein family consists of ∼20 low molecular weight Ca2+-binding proteins that possess two EF-hand motifs and share about 20–80% homogeneity in amino acid sequence. Unlike the prototypical EF-hand calcium sensor calmodulin, S100 proteins exhibit cell- and tissue-specific expression (1). In vivo and in vitro, the S100 proteins form hetero- or homodimers. The EF-hand proteins bind Ca2+ through their EF-hand motifs and undergo a Ca2+-dependent conformational change to expose a hydrophobic effector protein-binding surface, which in turn allows them to bind to and regulate an array of secondary effector proteins (for review, see Refs. 1–3). S100 proteins are proposed to have intracellular and extracellular roles in the regulation of many cellular processes such as contraction, cell motility and growth, cell-cycle progression, differentiation, transcription, and protein phosphorylation (1, 4). However, the precise intracellular roles of the S100 proteins are not fully understood.
Recently, we identified S100A1 to be a new member of the multichaperone complex and a potent molecular chaperone (5). To identify additional possible chaperone-interacting S100 proteins, we examined their interaction with the co-chaperone Hsp70/Hsp90-organizing protein (Hop).3 Hop is one of the co-chaperones and is able to associate directly with both Hsp70 and Hsp90 through tetratricopeptide repeat (TPR) domains (6–11). Here, we report that S100A2 and S100A6 interact with Hop-TPR domains and inhibit Hop-Hsp70 and Hop-Hsp90 complex formation in vitro and in vivo. Thus, S100A2 and S100A6 represent the first example of Ca2+-dependent regulators of the Hsp70-Hop-Hsp90 multichaperone complex.
The TPR domain is a protein-protein interaction motif that was originally identified by sequence comparisons among yeast proteins (12). The TPR domain consists of a 34-amino acid motif with a loose consensus and is present as tandem repeats in proteins with many cellular functions (13). Three-dimensional structural analyses have shown that a TPR motif contains two antiparallel α-helices such that tandem arrays of TPR motifs generate a right-handed helical structure with an amphipathic channel. The hydrophobic surface of this structure mediates protein-protein interactions between TPR- and non TPR-containing proteins (13). Hence, we hypothesized that some S100 proteins interact with other TPR-containing proteins in addition to Hop. Kinesin-1 (formerly conventional kinesin or Kif5) is a heterotetramer of two kinesin heavy chain (KHC) and two kinesin light chain (KLC) polypeptides. The N-terminal coiled-coil domain of KLC interacts with KHC, and the C-terminal TPR domain of KLC links to vesicular cargo via cytosolic adaptor proteins such as the c-Jun N-terminal kinase (JNK)-interacting protein 1 (JIP-1), JIP-2, JIP-3/Syd, JIP-4, JLP, TorsinA, Huntingtin-associated protein-1 (HAP1), and calsyntenin-1 (14–23). In the present study we also demonstrated that S100A2 and S100A6 interact with KLC-TPR domain and that S100A6 inhibits KLC-JIP-1 binding in vitro and in vivo. To further explore the relationship between S100 and TPR proteins, we focused on translocase of outer mitochondrial membrane (Tom) 70. Tom70 is a membrane-localized co-chaperone that directly associates with Hsp70 and Hsp90 through TPR motifs. We have found that S100A2 and S100A6 also interact with Tom70 and inhibit Tom70-Hsp70 and Tom70-Hsp90 complex formation in vitro. Collectively, these findings have prompted us to propose a novel physiological function for the S100 proteins and have also provided information about the regulation mechanism of TPR protein-ligand interactions.
EXPERIMENTAL PROCEDURES
Materials—Q-Sepharose, phenyl-Sepharose, glutathione-Sepharose, heparin-Sepharose, and protein G-Sepharose were purchased from GE Healthcare. Nickel-nitrilotriacetic acid-agarose was purchased from Qiagen. S100 antibodies were obtained as follows: anti-S100A1 (Sigma), anti-S100A2 (Sigma), anti-S100A6 (Sigma), anti-S100A8 (Harlan, Sussex, UK), anti-S100A9 (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-S100B (QED, San Diego, CA). Anti-S100A12 and anti-S100A13 were prepared as described previously (24). Anti-Hsp90 monoclonal antibody (AC88), anti-Hsp70 monoclonal antibody (C92F3A-5), and anti-Hop monoclonal antibody (DS14F5) were purchased from Stress-gen (Victoria, Canada). Anti-S100A6 monoclonal antibody (CACY-100) was purchased from Sigma. Anti-mouse peroxidase-conjugated secondary antibodies were purchased from GE Healthcare. Rabbit reticulocyte lysate (RRL) was purchased from Hokudo Co., Ltd (Hokkaido, Japan). Goat polyclonal antibodies to KLC (L-17) and to TorsinA, and mouse monoclonal antibody to JIP-1 (B-7) were from Santa Cruz Biotechnology. Rabbit polyclonal antibody against JIP1/2 (SH3) was from Zymed Laboratories Inc. (San Francisco, CA). Goat polyclonal antibody to KIF5B was from Abcam (Cambridge, UK). Monoclonal anti-His tag antibody was from Cell Signaling (Danvers, MA). Mouse monoclonal anti-HA antibody (12CA5) was purchased from Roche Applied Science. Lipofectamine was purchased from Invitrogen. All other reagents were of at least analytical grade.
Plasmids—Cloning of rat wild type Hop was performed as described previously (25). Briefly, a series of Hop deletion mutants (glutathione S-transferase (GST)-TPR1, residues 1–104; GST-N (1–224), residues 1–224; GST-C (225–543), residues 225–543; GST-TPR2A, residues 225–359; GST-TPR2B, residues 360–461; GST-DP2, residues 462–543) were prepared by PCR using the expression plasmid pGEX4T2-Hop wild type (WT; which encodes wild type rat Hop) as the template. Hop point mutants were generated as previously described (29). The single point mutations in the each domain were generated as follows: TPR1, Y27A and K73E; TPR2A, K301E and R305E; TPR2B, K429E and R433E. PCR products were then inserted into the BamHI/EcoRI sites of pGEX4T2 (GE Healthcare).
Human Hsp90 (full-length) and Human Hsp70 (full-length) were prepared by PCR using human Hsp90 (RIKEN number 1127) and Hsp70 (ATCC number 57494) cDNAs as templates and specific primers. PCR products were subcloned into pET-16b vector (Novagen). Human Tom70 (residues 111–608) was prepared by PCR using human Tom70 cDNA (OpenBiosystems number 6067022) as the template and specific primers. PCR products were subcloned into pGEX4T2 vector.
The cDNA coding sequences of rat KLC1b (GenBank™ accession number M75147), amino acids 1–200 (coiled-coil domain, CC), and amino acids 167–560 (TPR) were isolated from a rat brain cDNA library. KLC1b coding sequences were amplified using PCR and subcloned into pGEX4T2, generating the constructs GST-KLC-WT, GST-KLC-CC, and GST-KLC-TPR. Likewise, the JIP-1 coding sequence (GenBank™ accession number AF109768) was amplified by PCR from a Mouse Brain Quick-clone cDNA (Clontech), Mountain View, CA and cloned into pQE30 (Qiagen), generating the construct histidine (His6)-tagged JIP (493–707), or cloned into pME18S. The TorsinA coding sequence was amplified by PCR from a verified human TorsinA cDNA clone (Origene, Rockville, MD) and subcloned into pME18S. The plasmids constructed using the pME18S vector were used for transfection into mammalian cells. The nucleotide sequences of all inserts used in this study were confirmed by a sequencer (Applied Biosystems Japan, Tokyo, Japan).
Expression and Purification of Recombinant Proteins—All recombinant S100 proteins (S100B, A1, A2, A4, A6, A10, A11, A12, and A13) were expressed in a tag-free fashion and prepared as described previously (5, 26). Briefly, S100 proteins expressed in BL21 (DE3) were extracted using TE buffer (1 mm EDTA, 10 mm Tris-HCl, pH 8.0) and purified by phenyl-Sepharose column chromatography. GST-tagged fusion proteins and His6-tagged proteins were expressed in Escherichia coli and purified according to the manufacturer's instructions.
GST Pulldown Assay and Binding Analysis—The Ca2+-binding proteins (S100 proteins or calmodulin, 25 μg each) or Hsps (Hsp70 or Hsp90, 20 μg each) and GST fusion TPR proteins (0–25 μg) were mixed in Buffer A (20 mm Tris-HCl, 0.1 m KCl, and 0.02% Tween 20, pH 7.5) with 1 mm CaCl2 or EGTA. The reaction mixtures in a total volume of 200–400 μl were incubated with 20 μl of glutathione-Sepharose beads for 60 min at 25 °C. After pelleting by microcentrifugation, the beads were subjected to replicate washes (3 × 1 ml), boiled in SDS sample buffer, and analyzed by Tricine-SDS-PAGE.
To estimate the equilibrium dissociation constant (KD), a fixed concentration of GST-Hop wild type (0.56 μm) or GST-KLCs (0.27 μm each) was mixed with increasing concentrations of purified recombinant S100 proteins (0–40 μm) in buffer A with 1 mm CaCl2. After the reaction mixtures (total volume of 200 μl) were incubated with 20 μl of glutathione-Sepharose beads for 60 min at 25 °C, the beads were washed and eluted as described above. Bound proteins were resolved by 10% Tricine-SDS-PAGE. Coomassie-stained gels were scanned, and arbitrary densitometric values for S100 proteins bound to GST-Hop or GST-KLCs were obtained. These values were converted to micromolar equivalents by normalizing them against the value for 0.4 μg of S100 proteins on the same gel. Kinetic analyses of the data to obtain the binding parameters were performed by nonlinear regression analysis using Prism software (Version 5; GraphPad Software, Inc., La Jolla, CA). The experiments were performed in triplicate.
Competition Assay in Vitro—To investigate the effects of the S100 proteins on the interactions between the TPR proteins (such as Hop, KLC, and Tom70) and their ligand proteins (such as Hsp90, Hsp70, JIP-1, TorsinA, and KHC), the GST-TPR proteins (10 or 25 μg) and the S100 proteins (0–50 μg) were mixed in buffer A with 1 mm CaCl2 or EGTA. After the reaction mixtures (total volume, 200 μl) were incubated with 20 μl of glutathione-Sepharose beads for 60 min at 25 °C, the beads were collected and washed (3 × 1 ml). Next, the beads were incubated with the appropriate recombinant ligand proteins or cell extract (2 mg of protein) in buffer A with 1 mm CaCl2 or EGTA. After the second incubation was performed for 60 min at 25 °C, the beads were washed (3 × 1 ml), boiled in SDS-sample buffer, and analyzed by Western blotting or SDS-PAGE. In some experiments 20 μl of glutathione-Sepharose was mixed with the TPR proteins, their ligand proteins, and S100 proteins simultaneously in buffer A with 1 mm CaCl2 or EGTA. After the incubation was performed for 60 min at 25 °C, the beads were treated as describe above. The details of the experimental conditions are described in the figure legends.
Immunoprecipitation of Endogenous KLC from Mouse Brain—Mouse brain extracts (2 mg of protein) supplemented with or without recombinant S100A6 (25 μg) were incubated with goat polyclonal anti-KLC antibody (2 μg) for 16 h at 4 °C in buffer A with 1 mm CaCl2 or EGTA. After the addition of protein G-Sepharose beads (50 μl), the mixture was incubated for 2 h at 4 °C. The beads were washed 3 times in buffer A with 1 mm CaCl2 or EGTA. The immunoprecipitates were analyzed by Western blotting using specific antibodies for KLC and S100A6.
Cell Culture, Transfection, Ionomycin Treatment, Cell Lysis and Immunoprecipitation—Cos-7 monkey fibroblasts were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% (v/v) fetal bovine serum (JRH Biosciences). Next, 6 × 105 cells were seeded in a 10-cm culture dish and grown until confluent. Ionomycin treatment of Cos-7 cells and immunoprecipitation were performed as described by Tomas and Moss (27). Briefly, cells were treated with or without 1 μm ionomycin for 5 min. After washing the cells with 5 ml of buffer A, 500 μl of buffer A with 1 mm CaCl2 or EGTA was added to the dishes, and the cells were collected and lysed by sonication. Lysates were centrifuged at 15,000 × g for 30 min at 4 °C, and cleared lysate was subjected to the following immunoprecipitation. Antibody resin was prepared by incubating anti-Hop, anti-KLC, or anti-HA antibody IgG (2–10 μg) with protein G-Sepharose (20 μl) in phosphate-buffered saline for 2 h at 25 °C. The antibody-conjugated resins were washed three times in buffer A. The resins were mixed with the cell lysates and rotated for 1 h at 4 °Cand washed 3 times in buffer A with 1 mm CaCl2 or EGTA. In some cases Cos-7 cells were transfected with HA-KLC, JIP-1, and S100A6 before ionomycin stimulation. The expression plasmids pME18S-HA-KLC (1.2 μg), pME18S-JIP-1 (0.05 μg), and pME18S-S100A6 (2.7 μg) were introduced into Cos-7 cells and cultured for 2 days. Transient transfections were performed using Lipofectamine reagent according to the manufacturer's instructions.
The immunoprecipitates were analyzed by Western blotting with antibodies specific for Hop, KLC, Hsp70, Hsp90, HA, JIP-1, and S100A6. Western blotting was performed as described previously (5).
Preparation of Tissue Extracts and Cell Lysates—Whole mouse brain was homogenized with 10 ml of buffer A supplemented with protease inhibitor mixture (Sigma) and centrifuged at 20,000 × g for 30 min. Expression of TorsinA in Cos-7 cells was carried out essentially as described above, using 12 μg of pME18S-TorsinA expression plasmid. After 2 days of culture, 500 μl of buffer A was added to the 10-cm dishes, and the cells were collected and lysed by sonication. Lysates were centrifuged at 15,000 × g for 30 min at 4 °C. The supernatants were stored at –30 °C until used in the immunoprecipitation or competition assays.
RESULTS
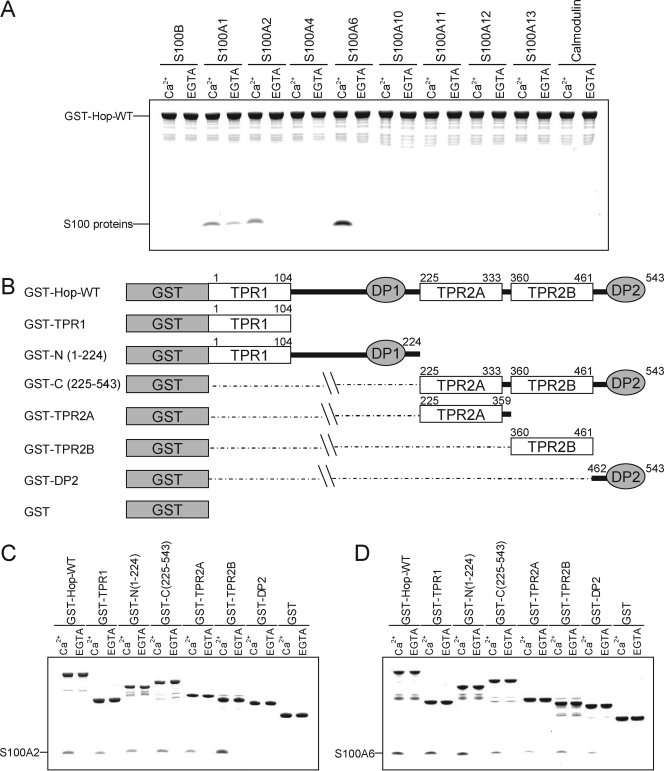
Interaction between Hop and the Members of the S100 Protein Family—Previously, we showed that S100A1 interacts with Hsp90, Hsp70, FK506-binding protein 52 (FKBP52), and cyclophilin40 (CyP40) (5). To further study the interaction of S100 proteins with molecular chaperones, we performed an in vitro binding assay using GST-Hop in the presence of 1 mm CaCl2 or 1 mm EGTA (Fig. 1A). S100A6, but not calmodulin or other S100 proteins, strongly bound to GST-Hop in the presence of Ca2+. S100A2 bound weakly to GST-Hop in a Ca2+-dependent manner, whereas S100A1 bound to Hop in a Ca2+-independent fashion. These experiments indicate that S100A6 and S100A2 exclusively interact with Hop in a Ca2+-dependent manner.
FIGURE 1.
Properties of S100 protein binding to GST-Hop-WT and deletion mutants. A, GST pulldown assays were performed using GST-Hop-WT and the S100 proteins. The S100 proteins (25 μg) and GST-Hop-WT (25 μg) were incubated with glutathione-Sepharose in buffer A with 1 mm CaCl2 (Ca2+) or EGTA (total volume 200 μl). After agitation at 25 °C for 60 min, the beads were washed and then eluted with SDS sample buffer. The eluted samples were analyzed by 10% Tricine-SDS-PAGE. The gel was stained with Coomassie Brilliant Blue. B, schematic diagrams of a series of Hop deletion mutants. The numbering refers to amino acid positions in Hop. C and D, GST pulldown assays were performed using GST-Hop, its deletion mutants, and GST alone as a control. GST-Hop fusion proteins or GST (25 μg) and S100 proteins (25 μg) were incubated with glutathione-Sepharose in buffer A with 1 mm CaCl2 (Ca2+) or EGTA (total volume 200 μl). The experiment was performed as described above.
The domain structure of Hop is shown in Fig. 1B. Hop contains three separate TPR domains, TPR1, TPR2A, and TPR2B, and two regions that contain aspartic acid-proline (DP) residue repeats; DP1 after TPR1 and DP2 after TPR2B (28, 29). The TPR1 domain of Hop co-crystallizes with a heptapeptide containing terminal EEVD residues in Hsp70, and the TPR2A domain of Hop co-crystallizes with a pentapeptide containing the terminal MEEVD residues of Hsp90 (9). To determine the S100 binding site of Hop, we performed a pulldown assay using a series of GST-Hop deletion mutants (Figs. 1, C and D). As shown in Fig. 1C, S100A2 bound to TPR2B preferentially and weakly associated with TPR1. In contrast, S100A6 exclusively interacted with TPR1 and bound weakly to TPR2A and TPR2B (Fig. 1D).
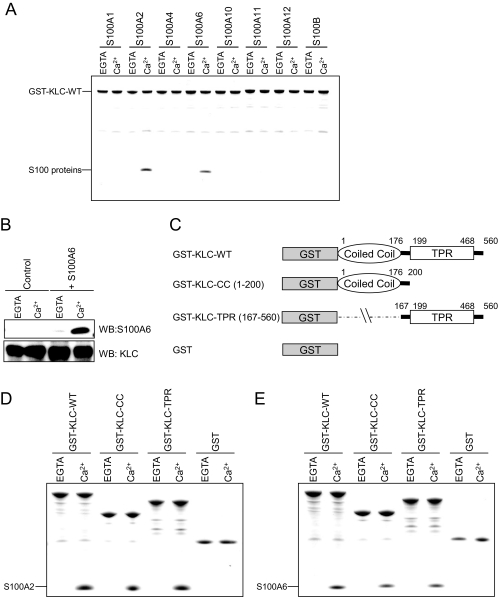
Interaction of S100A2 and S100A6 with GST-KLC—Because S100A2 and S100A6 interact with the TPR domain of Hop in a Ca2+-dependent manner, we hypothesized that some S100 proteins interact with the other TPR-containing proteins such as KLC. To investigate the interaction between KLC and the S100 proteins, we performed an in vitro binding assay using GST-KLC-WT in the presence of 1 mm CaCl2 or 1 mm EGTA. Purified GST-KLC-WT was immobilized on glutathione-Sepharose beads and then incubated with an equal amount of recombinant S100 proteins. The bound proteins were analyzed by SDS-PAGE. S100A2 and S100A6 specifically bound to GST-KLC-WT in a Ca2+-dependent manner (Fig. 2A). In contrast, the other S100 proteins such as S100A1, S100A4, S100A10, S100A11, S100A12, and S100B did not bind to GST-KLC-WT in the presence or absence of Ca2+ (Fig. 2A). Recently, Lazarov et al. (30) reported that the TPR domains in KLC tend to exhibit nonspecific binding to other proteins with suitable hydrophobic patches. They also described that this feature is a potential source of artifacts in GST pulldown assays, confirming putative interactions with a co-immunoprecipitation assay critical. Therefore, to exclude the possibility that the interaction of S100 proteins with KLC was nonspecific, we performed co-immunoprecipitation experiments. In mouse brain extracts supplemented with exogenous S100A6 (i.e. recombinant S100A6) in the presence of either Ca2+ or EGTA, endogenous KLC was immunoprecipitated with an anti-KLC polyclonal antibody. Furthermore, and solely in the presence of Ca2+, S100A6 was coprecipitated with KLC (Fig. 2B). Based on these results, the interaction of S100A6 with KLC is specific and apparently Ca2+-dependent.
FIGURE 2.
Properties of S100 protein binding to GST-KLC and deletion mutants. A, the S100 proteins (25 μg) and GST-KLC-WT (25 μg) were incubated with glutathione-Sepharose in buffer A with 1 mm CaCl2 (Ca2+) or EGTA (total volume 200 μl). The experiments were performed as described in Fig. 1A. B, mouse brain extracts (2 mg of protein) and S100A6 (0 μg (control) or 25 μg) were incubated with goat polyclonal anti-KLC antibody (2 μg) for 16 h at 4 °C in buffer B in the presence of 1 mm CaCl2 or 1 mm EGTA. After the addition of protein G-Sepharose beads (50 μl), the mixture was incubated for 2 h at 4 °C. The beads were collected by centrifugation, washed, and then eluted with the SDS-sample buffer (see “Experimental Procedures”). The immunoprecipitates were analyzed by Western blotting (WB) with antibodies specific for KLC and S100A6. C, schematic diagrams of a series of KLC deletion mutants. The numbering refers to amino acid positions in KLC. D and E, GST pulldown assays were performed using GST-KLC-WT, GST-KLC-CC, and GST-KLC-TPR (GST-KLCs) and GST alone as a control. GST-KLCs (25 μg) and S100 proteins (25 μg) were incubated with glutathione-Sepharose in buffer A with 1 mm CaCl2 (Ca2+) or EGTA (total volume 200 μl). The experiment was performed as described in the Fig. 1A legend.
The domain structure of KLC is shown in Fig. 2C. KLC consists of N-terminal heptad repeats (coiled-coil domain), which have been implicated in its binding to KHC, and a C-terminal TPR domain, which binds to cytosolic adaptor proteins such as JIP-1 and vesicular cargo proteins including TorsinA. To map the S100A2 or S100A6 binding domain in KLC, we used GST-KLC-WT, GST-KLC-CC, and GST-KLC-TPR in a GST pulldown assay (Figs. 2, D and E). The binding assay indicated that S100A2 and S100A6 bind to both the TPR and CC domains of KLC.
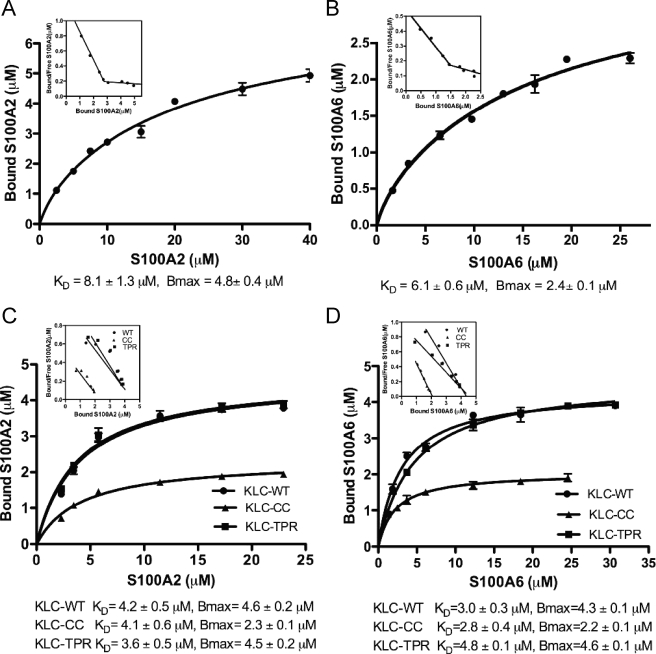
Binding Affinities of S100 Proteins to Hop and KLC—We next sought to quantify the binding affinity between Hop and the S100 proteins in vitro. We performed GST pulldown assays and analyzed the data by nonlinear regression fitting using Prism 5.0 software (Figs. 3, A and B). GST-Hop (0.56 μm) was incubated with a graded dose of S100A2 (0–40 μm) or S100A6 (0–26 μm) in the presence of Ca2+. The amounts of bound protein were measured by SDS-PAGE followed by densitometric analysis. The curves fit best to a two-site binding model, and R2 for S100A2 and S100A6 was 0.987. The KD values for S100A2 and S100A6 were 8.1 and 6.1 μm, respectively. The Scatchard plot obtained from the same experiments depicts two linear relationships that give evidence of a minimum of two different S100A2 or S100A6 binding sites (Figs. 3, A and B, insets) Although the KD values for S100A2 and S100A6 binding to Hop are almost identical, the stoichiometries of the S100 proteins binding to Hop showed a significant difference; S100A2 dimer bound to the Hop monomer in a 4:1 molar ratio, whereas the molar ratio of S100A6 dimer to Hop monomer was about 2:1.
FIGURE 3.
Binding curves of S100A2 or S100A6 to TPR-containing proteins. A and B, binding curves of S100A2 (A) and S100A6 (B) to GST-Hop-WT. A fixed amount of GST-Hop-WT (0.56 μm) was mixed with increasing amounts of S100A2 (0–40 μm) and S100A6 (0–26 μm) in the presence of 1 mm CaCl2. Coomassie-stained gels were scanned, and arbitrary densitometric values for S100 proteins bound to Hop were obtained. These values were converted to micromolar equivalents by normalizing them against the value for 0.4 μg of S100 proteins on the same gel. The KD values for S100 protein binding were determined by fitting the experimental data by non-linear regression using a two-site binding model. The mean ± S.D. value of triplicate determinations has been plotted. The inset presents the Scatchard plot from the same data. To obtain the values for free S100 proteins, the amount of bound protein was subtracted from the initial added amount. The Scatchard plot depicts two linear relationships. C and D, binding curves of S100A2 (C) and S100A6 (D) to GST-KLC and its mutants. Increasing concentrations of S100A2 (0–23 μm) or S100A6 (0–31 μm) were incubated with GST-KLC-WT, GST-KLC-CC, GST-KLC-TPR (GST-KLCs) (0.27 μm each) in the presence of 1 mm CaCl2. The experiment was performed as described above. KD was calculated by nonlinear regression analysis using a one-site binding model. The mean ± S.D. value of triplicate determinations has been plotted. The inset presents the Scatchard plots from the same data.
Similarly, we examined the binding affinity and stoichiometry between S100A2 or S100A6 and KLC followed by nonlinear regression analysis. To estimate the affinity of the interaction, GST-KLC-WT, GST-KLC-CC, and GST-KLC-TPR (0.27 μm each) were incubated with a graded dose of S100A2 (0–23 μm) or S100A6 (0–31 μm). Analysis of GST-KLC binding to S100A2 or S100A6 gave a reasonable fit with a one-site model (Figs. 3, C and D), although S100A2 or S100A6 bound to both the CC and TPR domain of KLC (Figs. 2, D and E). In addition, the Scatchard plots from the same binding data depict one linear relationship that indicates a single class of S100A2 or S100A6 binding site in each GST-KLC construct (Figs. 3, C and D, insets). A similar amount of S100A2 or S100A6 (i.e. Bmax = 4.3–4.6 μm) bound to KLC wild type and the TPR domain, and half the amount of S100A2 or S100A6 bound to the CC domain (i.e. Bmax = 2.2–2.3 μm) as shown in Figs. 3, C and D. However, the KD values for KLC wild type, the TPR domain, and the CC domain for S100A2 and S100A6 were almost identical. Based on these results, we conclude that S100A2 and S100A6 bind to both the TPR and CC domains of KLC.
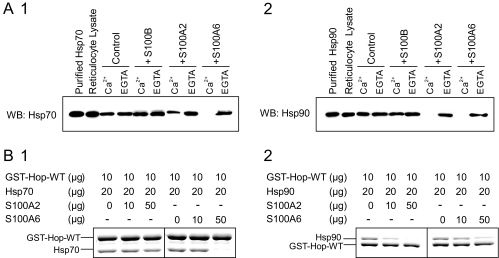
S100A2 and S100A6 Differ in Their Ability to Compete with Hsp70 and Hsp90 for Hop Binding—Hop has three TPR domains that have been implicated in its binding to Hsp70 and Hsp90. The N-terminal TPR1 domain binds to the C terminus of Hsp70, and the central TPR2 domain binds to a TPR acceptor site in the C terminus of Hsp90. Because identical structural elements (i.e. TPR1 or TPR2B) within Hop appear to be involved in the recognition of both the S100 proteins and Hsps, we next tested the influence of the S100 proteins on the Hop-Hsp70 and Hop-Hsp90 interactions. RRL was used as the source of native Hsps for the pulldown experiment. because it contains both Hsp70 (∼0.4 μg/2 mg of RRL) and Hsp90 (∼2.0 μg/2 mg of RRL) abundantly, but the S100 proteins were undetectable (Ref. 31 and data not shown). The GST-Hop-WT was incubated with recombinant S100B, S100A2, and S100A6 in the presence of 1 mm CaCl2 or EGTA, and then the resin was washed. After a brief microcentrifugation, the pelleted gel was then subjected to a second incubation with RRL. After washing the gel, retained proteins were assessed by SDS-PAGE with Coomassie Blue staining or by Western blot analysis. Fig. 4A shows representative blots that can be used to visually assess the displacement of Hsp70 and Hsp90 by the S100 proteins. Both the Hop-Hsp70 and Hop-Hsp90 interactions were completely inhibited by preincubation with S100A6 in the presence of Ca2+. Although S100A2 completely inhibited the Hop-Hsp90 interaction, the protein partially displaced Hsp70 binding. S100B did not stably bind to GST-Hop (Fig. 1A) and was not an effective competitor for Hsp70 or Hsp90 binding (Fig. 4A).
FIGURE 4.
Effects of the S100 proteins on the Hop-Hsp70 and Hop-Hsp90 interactions in vitro. A, GST-Hop-WT (25 μg) and S100 proteins (0 μg (control) or 25 μg of S100B, S100A2, and S100A6) were incubated with glutathione-Sepharose in buffer A with 1 mm CaCl2 (Ca2+) or EGTA (total volume 200 μl). After agitation at 25 °C for 60 min, the beads were collected, washed, and then mixed with RRL (2 mg of protein) in buffer A with 1 mm CaCl (Ca2+) or 1 mm EGTA (total volume, 1 ml). After the second incubation (see “Experimental Procedures”), the resulting samples were subjected to Western blotting (WB) using antibody against Hsp70 (panel 1) or Hsp90 (panel 2). B, the protein amounts of each reaction are indicated on the top of the panels. GST-Hop-WT (10μg) and S100 proteins (0, 10, and 50 μg) were incubated with 20 μl of glutathione-Sepharose in buffer A with 1 mm CaCl2 (total volume 200 μl). After agitation at 25 °C for 60 min, the beads were collected, washed, and then mixed with Hsp70 (20 μg) or Hsp90 (10 μg) in buffer A with 1 mm CaCl2 (total volume 200 μl). After the second incubation, the resulting samples were analyzed by 7.5% Laemmli-SDS-PAGE. The gel was stained with Coomassie Brilliant Blue.
To confirm these results, we performed a GST-Hop pulldown assay using recombinant Hsp90 and Hsp70. Fig. 4B shows that preincubation with increasing amounts of S100A2 or S100A6 was able to effectively inhibit the Hop-Hsp90 interaction (Fig. 4B, 2). Under similar binding conditions S100A6 also effectively inhibited the Hop-Hsp70 interaction; however, S100A2 did not affect the Hop-Hsp70 interaction (Fig. 4B, 1).
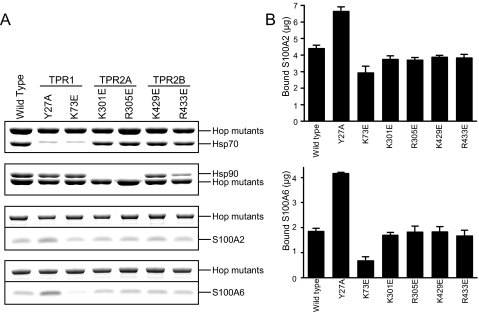
Point Mutations of the Hsp Binding Site within Hop Had Little Effect on S100 Protein Binding—The C-terminal regions of Hsp70 and Hsp90, both of which terminate with the amino acid sequence EEVD, are involved in the interaction with the Hop TPR domains. Crystallographic studies (9) have shown that EEVD interacts with basic amino acids (called calboxylate clamp) in Hop TPR binding pockets, and point mutations altering these calboxylate clamp residues to acidic amino acids can disrupt Hsp binding (29). To further explore the interaction between Hop and S100 proteins, we performed GST pulldown assays using Hop mutants of the calboxylate clamp residue in each TPR domain. After the method of Carrigan et al. (29), the single point mutations in Hop were generated as follows: TPR1, Y27A and K73E; TPR2A, K301E and R305E; TPR2B, K429E and R433E. In a qualitative experiment, mutants of TPR1 (Y27A and K73E) had greatly reduced binding to Hsp70 (Fig. 5A, uppermost panel). TPR2A mutants (K301E and R305E) also failed to bind Hsp90 (Fig. 5A, top panel). These observations are consistent with earlier reports (29). Fig. 5B shows the amount of binding of S100A2 (upper panel) or S100A6 (lower panel) to the GST-Hop mutants. Unexpectedly, Y27A (TPR1) had slightly increased binding to both S100A2 and S100A6. On the other hand, K73E (TPR1) had reduced binding to S100A2 and S100A6. The binding of the other mutants (K301E, R305E, K429E, and R433E) to S100A2 and S100A6 was unchanged. Based on these findings (Figs. 5, A and B), we conclude that the point mutations of calboxylate clamps within Hop had little effect on S100 protein binding.
FIGURE 5.
Interaction of Hop point mutants with Hsp70, Hsp90, S100A2, and S100A6. A, GST pulldown assays were performed using GST-Hop point mutants. GST-Hop point mutants (10 μg) were incubated with Hsp70 (20 μg), Hsp90 (20 μg), S100A2 (50 μg), and S100A6 (50 μg) in buffer A with 1 mm CaCl2 (total volume 200 μl). The experiments were performed as described in the legend of Fig. 1A. The samples were analyzed by SDS-PAGE. To separate the proteins clearly, we used both Laemmli (for Hsp70 and Hsp90) and Tricine (for S100A2 and S100A6) SDS-PAGE systems. The regions of the Coomassie-stained bands containing GST-Hop point mutants, Hsp70, Hsp90, S100A2, and S100A6 are shown. B, the Coomassie-stained bands from triplicate experiments were quantified by densitometry, and arbitrary densitometric values for S100 proteins bound to Hop were converted to micrograms by normalizing them against the value for 0.4 μg of S100 proteins running on the same gel. These values were converted to the amount of the bound S100 proteins to 10 μg of GST-Hop point mutants. The mean ± S.E. value of triplicate determinations has been plotted.
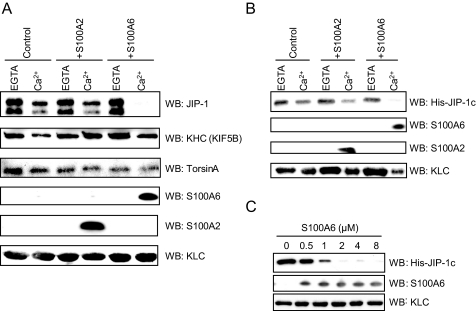
Effects of S100A6 on the Interactions between KLC and KLC-interacting Proteins—Next, we tested the effects of S100A2 and S100A6 on KLC ability to bind to KLC-interacting proteins. KLC interacts with a number of potential cargo molecules. KLC contains an N-terminal coiled-coil domain, termed the heptad repeats, that binds to KHC. However, the most important region for cargo binding is thought to be a C-terminal region of KLC that contains six TPRs which bind to a wide range of proteins such as JIP-1 and TorsinA (14, 15, 16, 20, and 21). First we examined the binding specificity of KLC-interacting proteins for KLC using a GST pulldown assay in which GST or GSTKLC was incubated with mouse brain extracts containing endogenous JIP-1 and KHC or TorsinA-transfected Cos-7 lysates. GST-KLC was precipitated with these KLC-interacting proteins, but GST alone did not precipitate (data not shown). Similarly, GST pulldown assays, in which GST-KLC was pretreated with S100A2 or S100A6, were performed in the presence or absence of Ca2+ to analyze the effects of S100A2 and S100A6 on the binding of KLC to KLC-interacting proteins. Although S100A2 or S100A6 bound to KLC in the presence of Ca2+, KHC and TorsinA binding to KLC was not inhibited by the S100A2 or S100A6 binding (Fig. 6A). In contrast, the KLC-JIP-1 interaction was completely inhibited by preincubation with S100A6 in the presence of Ca2+ but not in the absence of Ca2+. However, S100A2 did not inhibit the interaction between JIP-1 and KLC in the presence or absence of Ca2+. To confirm the findings of the KLC-JIP-1 interaction, we performed a GST-KLC pulldown assay using a bacterially expressed truncation mutant of His-JIP-1c, which includes the binding site to the TPR domain of KLC (15, 16). Fig. 6B shows that preincubation with S100A6 in the presence of Ca2+ effectively inhibited the KLC-JIP-1c interaction, whereas is-JIP-1c bound to KLC in the presence of EGTA. We further tested the replacement experiments using increasing concentrations of S100A6. S100A6 inhibited JIP-1c binding to KLC in a dose-dependent fashion (Fig. 6C). From these results we propose that S100A6 directly inhibits the interaction between KLC and JIP-1 in a Ca2+-dependent manner.
FIGURE 6.
Effects of S100A2 or S100A6 on the interaction between KLC and KLC-associating proteins in vitro. A and B, GST-KLC-WT (25 μg) and S100 proteins (0 μg (control) or 25 μg of S100A2 and S100A6) were incubated with 20 μl of glutathione-Sepharose in buffer A with 1 mm CaCl2 (Ca2+) or EGTA (total volume 200 μl). After agitation at 25 °C for 60 min, the beads were collected, washed, and then mixed with 2 mg of mouse brain extract (as the source of JIP-1 and KHC) (A) or 25 μg of recombinant His-JIP-1c (B) in buffer A with 1 mm CaCl2 (Ca2+) or 1 mm EGTA (total volume 400 μl). After the second incubation (see “Experimental Procedures”), the resulting samples were analyzed by Western blotting (WB) using the specific antibodies against JIP-1, KHC, S100A2, S100A6, and KLC. 500 μg of Cos-7 cell lysates (TorsinA transiently expressed) were used for the TorsinA binding to GST-KLC-WT, and the blot was stained using a specific antibody against TorsinA (A). C, effects of a graded dose of S100A6 on the KLC-JIP-1 interaction. GST-KLC-WT (0.27 μm) and a graded dose of S100A6 (0–8 μm) were incubated with 20 μl of glutathione-Sepharose beads in buffer A with 1 mm CaCl2 (total volume 200 μl). After agitation at 25 °C for 60 min, the beads were collected, washed, and then mixed with His-JIP-1c (0.2 μm) in the presence of 1 mm CaCl2 (total volume 200μl). The bound proteins were detected by Western blotting (WB) using the antibodies against KLC, His tag, and S100A6.
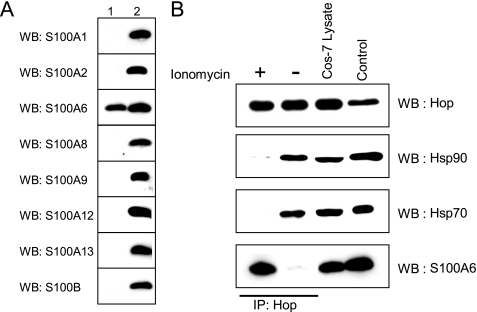
In Vivo Interaction of S100 Proteins with Hop—To examine the in vivo Ca2+-dependent interaction of the S100 proteins with Hop, we performed co-immunoprecipitation experiments using anti-Hop antibody. We used Cos-7 cells, because they are enriched in endogenous Hop, Hsp70, Hsp90, and S100A6. Cos-7 is an ideal cell line for investigation of the effects of S100A6 since the other S100 proteins, such as S100B, S100A1, S100A2, S100A8, S100A9, S100A12, and S100A13 were not detected by Western blot using appropriate anti-S100 antibodies (Fig. 7A). We stimulated Cos-7 cells with 1 μm ionomycin for 5 min, and then the whole cell lysates were immunoprecipitated using an anti-Hop antibody (Fig. 7B). When Cos-7 cells were stimulated with ionomycin, the amounts of Hsp70 and Hsp90 bound to Hop were greatly decreased, and S100A6 was co-immunoprecipitated with Hop. Comparative studies using non-stimulated cell extracts confirmed the interaction of Hsp70 and Hsp90 with Hop, but no interaction of S100A6 with Hop was detected. These results suggest that S100A6 binds to Hop, resulting in inhibition of the interactions between Hsp70 and Hop and Hsp90 and Hop in vivo.
FIGURE 7.
S100A6 modulates the interaction of Hop with Hsp90 and Hsp70 in vivo. A, Western blot analysis of the S100 protein distribution in Cos-7 cells. Equal amounts of total protein (40 μg/lane) from Cos-7 cells were resolved by Tricine-SDS-PAGE. Western blotting (WB) was performed using antibodies specific to the S100 proteins indicated on the left. Lane 1, Cos-7 lysates (40 μg/lane); lane 2, purified recombinant S100 proteins (50 ng/lane). B, Cos-7 cells (contain endogenous Hop, Hsp70, Hsp90, and S100A6) were treated with (+) or without (–)1 μm ionomycin for 5 min. After treatment, the cells were washed with buffer A, and 500 μl of buffer A with 1 mm CaCl2 was added to the dishes, and the cells were collected and lysed by sonication. Lysates were centrifuged at 15,000 × g for 30 min at 4 °C. The Hop-interacting protein complexes were immunoprecipitated (IP) by anti-Hop mouse monoclonal DS14F5 antibody. The resulting immunoprecipitates were analyzed by Western blotting (WB) using antibodies against Hop, Hsp90, Hsp70, and S100A6. Authentic Hop (25 ng), Hsp90 (100 ng), Hsp70 (50 ng) and S100A6 (50 ng) were electrophoresed and analyzed by Western blotting (control).
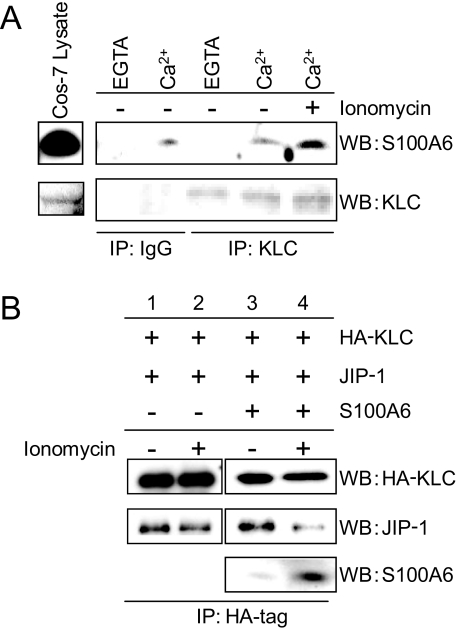
Effects of S100A6 on the KLC-JIP-1 Interaction in Cultured Cells—Similarly, to examine the Ca2+-dependent interactions between S100A6 and KLC in cultured cells, we performed KLC immunoprecipitation of Cos-7 cells treated with or without 1 μm ionomycin for 5 min. After the treatment, cell lysates were prepared and immunoprecipitated with an anti-KLC antibody or anti-goat IgG. When resting Cos-7 cells were lysed with a Ca2+-containing buffer, but not with an EGTA-containing buffer, endogenous S100A6 was co-immunoprecipitated with endogenous KLC (Fig. 8A). When Cos-7 cells were stimulated with ionomycin, the amount of S100A6 bound to KLC was greatly increased compared with non-stimulated cells (Fig. 8A, right-most lane). To study the effect of S100A6 on the KLC-JIP-1 interaction, we carried out co-immunoprecipitation experiments using double or triple-transfected Cos-7 cells with HA-KLC, S100A6, and/or JIP-1 (Fig. 8B). In the HA-KLC and JIP-1 double-transfected cells, the amount of co-precipitated JIP-1 did not change with or without ionomycin treatment. When the triple-transfected (i.e. HA-KLC, JIP-1, and S100A6 cotransfected) Cos-7 cells were stimulated with ionomycin, the amount of JIP-1 bound to HA-KLC was greatly decreased, and S100A6 was co-precipitated together with HA-KLC (Fig. 8B, fourth lane). Using non-stimulated cells, the anti-HA antibody precipitated KLC together with JIP-1 but not S100A6 (Fig. 8B, third lane). These results suggest that S100A6 binds to KLC, resulting in inhibition of the interaction of KLC with JIP-1 in intact cells.
FIGURE 8.
S100A6 regulates the KLC-JIP-1 interaction in cultured cells. A, Cos-7 cells (contain endogenous KLC and S100A6) were treated with (+) or without (–)1 μm ionomycin for 5 min. Cell lysates were immunoprecipitated (IP) with a normal goat IgG or an anti-KLC antibody in the presence of 1 mm CaCl2 (Ca2+) or EGTA (EG). The experimental details are as described in the legend of Fig. 7B. The resulting immunoprecipitates were analyzed by Western blotting (WB) using antibodies against KLC and S100A6. B, Cos-7 cells were transfected with HA-KLC and JIP-1 in the presence or absence of S100A6. The transfected components of each dish are indicated on the top panels. The transfected Cos-7 cells were treated with (+) or without (–)1 μm ionomycin for 5 min. Cell lysates were immunoprecipitated with anti-HA antibody. The experimental details are as described in the legend of Fig. 7B. The resulting immunoprecipitates were analyzed by Western blotting (WB) using antibodies against HA, JIP-1, and S100A6.
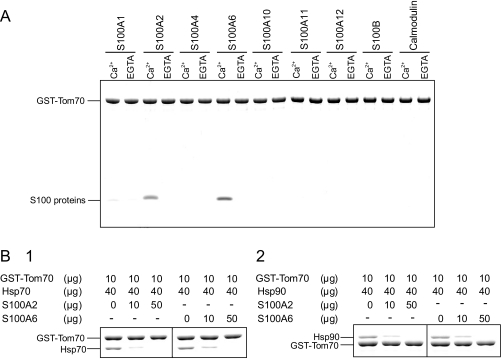
S100A2 and S100A6 Regulation of the Tom70-Hsp70 and Tom70-Hsp90 Interactions in Vitro—We have shown that S100A2 and S100A6 interact with the TPR domains of Hop and KLC in a Ca2+-dependent manner, resulting in regulation of the TPR protein-ligand interactions. More than 30 TPR-containing proteins have so far been identified (13). We speculated that the other TPR-containing proteins interact with particular S100 proteins and that their functions are regulated. To further confirm our hypothesis, we investigated whether the interaction of another TPR protein Tom70, with its specific ligand, is regulated by S100 proteins in a Ca2+-dependent manner. Tom70 is localized in the outer mitochondrial membrane and is involved in the transportation of cytosolic proteins into mitochondria. Tom70 contains seven TPR motifs, and interacts directly with Hsp70 and Hsp90 (32). We fused the cytosolic segment of Tom70 (residues of 111–608) to GST and then tested its binding to the S100 proteins using a GST pulldown approach. As expected, S100A2 and S100A6 interacted with Tom70 in a Ca2+-dependent manner (Fig. 9A). To explore whether the interactions of Tom70-Hsp90 and Tom70-Hsp70 are regulated by the S100 proteins, we performed a competition assay using GST-Tom70, Hsp90, Hsp70, and S100 proteins in the presence of Ca2+. Fig. 9B shows that the Tom70-Hsp70 interaction was effectively inhibited by increasing the amount of S100A2 and S100A6 (Fig. 9B, 1). Under similar binding conditions, the interaction of Tom70-Hsp90 was also effectively inhibited by S100A2 and S100A6 (Fig. 9B, 2). These data suggest that S100A2 and S100A6 bind to Tom70 and regulate Tom70-Hsps complex formation in a Ca2+-dependent manner in vitro.
FIGURE 9.
The effects of S100 proteins on Tom70-Hsp70 and Tom70-Hsp90 interactions. A, GST pulldown assays were performed using GST-Tom70. GST-Tom70 (25 μg) and S100 proteins (25 μg) were incubated with 20 μl of glutathione-Sepharose in buffer A with 1 mm CaCl2 (Ca2+) or EGTA (total volume 200 μl). The experiments were performed as described in the legend of Fig. 1A. The Coomassie-stained gel is shown. B, GST-Tom70 (10 μg), the S100 proteins (0–50 μg), and Hsps (40 μg) were incubated with 20 μl of glutathione-Sepharose in buffer A supplemented with 1 mm CaCl2 (total volume, 200 μl). The protein amounts in each reaction are indicated on the top of the panels. After agitation at 25 °C for 1 h, the beads were collected, washed, and then eluted with SDS-sample buffer. The eluted samples were analyzed by 7.5% Laemmli-SDS-PAGE. The gel was stained with Coomassie Brilliant Blue.
DISCUSSION
We have demonstrated that S100A2 and S100A6, but not calmodulin, specifically interact with the TPR domains of Hop and KLC in a Ca2+-dependent manner, resulting in regulation of the TPR protein-ligand interactions in vitro. Initially, we identified Hop as a novel target for S100A2 and S100A6. Hop directly associates with Hsp70 and Hsp90 through the TPR domain and regulates Hsp70-Hop-Hsp90 heterocomplex formation. The KD values for S100A2 (KD = 8.1 μm) and S100A6 (KD = 6.1 μm) are in the micromolar range, and similar KD values for Hsp70 and Hsp90 binding to Hop have been reported, i.e. 1.3 μm and 0.24 ∼ 1.5 μm, respectively (11, 33, 34). Therefore, we speculated that these S100 proteins are able to compete with Hsp binding to Hop in intact cells. In fact, S100A6 and Hop interact in ionomycin-stimulated Cos-7 cells, and this interaction inhibits Hsp70 and Hsp90 binding to Hop. These results suggest that endogenous S100A6 modulates Hsp70-Hop-Hsp90 heterocomplex assembly with an increased intracellular Ca2+ level in vivo. The S100 proteins are expressed in a strictly tissue- and cell-specific fashion (1). Therefore, the interaction between Hop and the S100 proteins will be regulated by the availability of the S100 proteins in a given tissue and cell type. These findings have prompted us to propose that Ca2+ regulates the TPR protein-ligand interaction through the S100 proteins.
The mechanism of the Hop-Hsp interactions has been well established. The highly conserved basic side chains inside the TPR domains form salt bridges with acidic residues of the Hsp EEVD regions. Furthermore, point mutations of the calboxylate clamp diminish the binding of Hop to the Hsps. We assessed the effects of mutations of the calboxylate clamp within Hop TPR domains on S100 protein binding (Fig. 5). Our results suggested that the point mutations of the calboxylate clamps within Hop had little effect on S100 protein binding and that the mode of interaction of the S100 proteins with Hop is different from the Hop-Hsp electrostatic interaction. In general, S100 proteins undergo a Ca2+-dependent conformational change to expose a hydrophobic effector protein binding surface, resulting in binding to the target protein. Moreover, our results have shown that substituting tyrosine 27 with alanine (Y27A) increases binding for S100 proteins. Thus, we speculate that the mode of the interaction between Hop and the S100 proteins depends on a hydrophobic interaction.
Based on the above results, we hypothesized that both S100A2 and S100A6 bind to the TPR-containing proteins and block binding of the protein to the partner proteins. To verify this hypothesis, we examined the S100A2 and S100A6 Ca2+-dependent interactions with KLC, which contains six TPR motifs. Among the S100 proteins tested, S100A2 and S100A6 specifically bound to both the CC and TPR domains of KLC in a Ca2+-dependent manner. Next, the binding kinetics of S100A6 binding to KLC wild type, isolated CC domain, and TPR domain were determined using the GST pulldown method. Unexpectedly, S100A2 and S100A6 bound to the CC and TPR domains with KD values in a similar micromolar range. Although the stoichiometries of S100A2 and S100A6 binding to wild type KLC (S100A2 or S100A6 dimer:KLC monomer = 2:1) and isolated TPR domain (2:1) are similar, S100A2 or S100A6 dimer binds to the CC domain in a 1:1 molar ratio. Thus, the sum of the bound S100A2 or S100A6 to isolated CC domain and TPR domain is out of accord with the stoichiometry of S100 binding to KLC wild type. Because there are no structural data for the KLC-S100A2 or KLC-S100A6 interactions, the basis of this discrepancy is unclear at present. However, it is perhaps not surprising that truncations of KLC are associated with either loss or gain of S100A2 or S100A6 binding activity.
The fact that S100A2 or S100A6 bind both the CC domain, a domain that mediates KHC interaction, and the TPR domain of KLC, a domain that mediates multiple KLC-ligand interactions, raises the interesting possibility that S100A2 or S100A6 might play a role in the KHC-KLC and KLC-cargo interactions. In this regard, we wanted to determine whether S100A2 or S100A6 could compete with KHC, JIP-1, and TorsinA binding to KLC. Indeed, S100A6, but not S100A2, inhibited JIP-1 binding to KLC in the presence of Ca2+ in vitro. Furthermore, JIP-1 was not found to be associated with KLC when Cos-7 cells were stimulated with ionomycin. Thus, S100A6 appears to inhibit the KLC-JIP-1 interaction via Ca2+-dependent S100A6·KLC complex formation.
Although the precise binding site(s) of the S100 proteins has not yet been determined, as the TPR domain of KLC is relatively large (six times repeat), we speculate that the binding site of S100A2 in the TPR domain might be different from that of S100A6. In striking contrast to JIP-1, S100A6 failed to compete with TorsinA and KHC binding to KLC in vitro. At present, the functional significance of the interaction between S100A6 and the CC domain is unclear as S100A6 has no effect on KLC-KHC complex formation. The precise S100A6 binding site in the CC domain needs to be elucidated. Although the majority of TorsinA is localized in the lumen of the endoplasmic reticulum, Kamm et al. (20) identified KLC (a cytoplasmic protein) as a TorsinA-interacting protein through a yeast two-hybrid screen, co-immunoprecipitation, and fractionation studies. In accordance with that report, in the present study TorsinA seemed to bind to GST-KLC in the pulldown assay. However, because S100A6 does not compete against TorsinA binding to KLC, the functional role of S100A6 in the KLC-TorsinA interaction is unclear at present. The TPR domain of KLC has been shown to associate with many of its key binding partners, including JIP-1 and TorsinA, but the precise binding site within the TPR domain that contributes to the association has not yet been identified (14–23). The precise mapping of the binding sites for these KLC binding partners and S100A6 will shed light on the mechanism of Ca2+/S100A6 dependent kinesin motor regulation.
In conclusion, our results strongly suggest that the Ca2+-dependent interactions of the S100 proteins with the TPR proteins such as Hop and KLC regulate the TPR protein-specific ligand interaction in vivo as well as in vitro. Moreover, we have found that the Tom70-Hsps interactions are regulated by the S100 proteins in a Ca2+-dependent manner in vitro. TPR domains generally mediate protein-protein interactions, and different TPR domains have different protein binding specificities. This structural property enables TPR-containing proteins to work as scaffold proteins and allows them to be involved in a variety of cellular functions. A common regulatory factor for the TPR domain-ligand interaction has yet to be identified. Therefore, our findings represent the first example of Ca2+-dependent regulation of the TPR domain-ligand interactions, namely via the S100 proteins. More than 30 TPR-containing proteins have so far been identified (for review, see Ref. 13). An important question that arises is whether the other TPR-containing proteins interact with particular S100 proteins and how their functions are regulated. Future experiments will be necessary to address this question.
Acknowledgments
We thank Dr. Takashi Hatakeyama (Akita University) for kindly providing the KLC plasmids. We are grateful to Naoko Uematsu and Takaki Nimura (Kagawa University) for excellent technical assistance.
This work was supported in part by grants-in aid for Scientific Research 18300123 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Hop, Hsp70/Hsp90-organizing protein; Hsp, heat shock protein; TPR, tetratricopeptide repeat; GST, glutathione S-transferase; RRL, rabbit reticulocyte lysate; KLC, kinesin light chain; KHC, kinesin heavy chain; JIP-1, c-Jun N-terminal kinase (JNK)-interacting protein 1; Tom, translocase of outer mitochondrial membrane; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; HA, hemagglutinin; WT, wild type; CC, coiled-coil domain.
References
- 1.Donato, R. (1999) Biochim. Biophys. Acta 1450 191–231 [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya, S., Bunick, C. G., and Chazin, W. J. (2004) Biochim. Biophys. Acta 1742 69–79 [DOI] [PubMed] [Google Scholar]
- 3.Santamaria-Kisiel, L., Rintala-Dempsey, A. C., and Shaw, G. S. (2006) Biochem. J. 396 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donato, R. (2003) Microsc. Res. Tech. 6 540–551 [DOI] [PubMed] [Google Scholar]
- 5.Okada, M., Hatakeyama, T., Itoh, H., Tokuta, N., Tokumitsu, H., and Kobayashi, R. (2004) J. Biol. Chem. 279 4221–4233 [DOI] [PubMed] [Google Scholar]
- 6.Smith, D. F., Sullivan, W. P., Marion, T. N., Zaitsu, K., Madden, B., MaCormick, D. J., and Toft, D. O. (1993) Mol. Cell. Biol. 13 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassle, M., Blatch, G. L., Kundra, V., Takatori, T., and Zetter, B. R. (1998) J. Biol. Chem. 273 3679–3683 [DOI] [PubMed] [Google Scholar]
- 8.Prodromou, C., Siligardi, G., O'Brien, R., Woolfson, D. N., Regan, L., Panaretou, B., Ladbury, J. E., Piper, P. W., and Pearl, L. H. (1999) EMBO J. 18 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sceufler, C., Brinker, A., Bourenkov, G., Pegoraro, S., Moroder, L., Bartunik, H., Hartl F. U., and Moarefi, I. (2000) Cell 101 199–210 [DOI] [PubMed] [Google Scholar]
- 10.Van Der Spuy, J., Kana, B. D., Dirr, H. W., and Blatch, G. L. (2000) Biochem. J. 345 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odunuga, O. O., Hornby, J. A., Bies, C., Zimmermann, R., Pugh, D. V., and Blatch, G. L. (2003) J. Biol. Chem. 278 6896–6904 [DOI] [PubMed] [Google Scholar]
- 12.Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L. Y., and Patterson, C. (1999) Mol. Cell. Biol. 19 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blatch, G. L., and Lassle, M. (1999) BioEssays 11 932–939 [DOI] [PubMed] [Google Scholar]
- 14.Verhey, K. J., and Rapoport, T. A. (2001) Trends Biochem. Sci. 9 545–550 [DOI] [PubMed] [Google Scholar]
- 15.Verhey, K. J., Meyer, D., Deehan, R., Blenis, J., Schnapp, B. J., Rapoport, T. A., and Margolis, B. (2001) J. Cell Biol. 152 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda, S., Matsuda, Y., and D'Adamio, L. (2003) J. Biol. Chem. 278 38601–38606 [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa, N., and Takemura, R. (2005) Nat. Rev. Neurosci. 6 201–214 [DOI] [PubMed] [Google Scholar]
- 18.Kelkar, N., Standen, C. L., and Davis, R. J. (2005) Mol. Cell. Biol. 25 2733–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen, Q., Lee, C. M., Le, A., and Reddy, E. P. (2005) J. Biol. Chem. 280 30185–30191 [DOI] [PubMed] [Google Scholar]
- 20.Kamm, C., Boston, H., Hewett, J., Wilbur, J., Corey, D. P., Hanson, P. I., Ramesh, V., and Breakefield, X. O. (2004) J. Biol. Chem. 19 19882–19892 [DOI] [PubMed] [Google Scholar]
- 21.Adio, S., Reth, J., Bathe, F., and Woehlke, G. (2006) J. Muscle Res. Cell Motil. 27 153–160 [DOI] [PubMed] [Google Scholar]
- 22.McGuire, J. R., Rong, J., Li, S. H., and Li, X. J. (2006) J. Biol. Chem. 281 3552–3559 [DOI] [PubMed] [Google Scholar]
- 23.Konecna, A., Frischknecht, R., Kinter, J., Ludwig, A., Steuble, M., Meskenaite, V., Indermuhle, M., Engel, M., Cen, C., Mateos, J. M., Streit, P., and Sonderegger, P. (2006) Mol. Biol. Cell 17 3651–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada, M., Tokumitsu, H., Kubota, Y., and Kobayashi, R. (2002) Biochem. Biophys. Res. Commun. 292 1023–1030 [DOI] [PubMed] [Google Scholar]
- 25.Okada, M., Itoh, H., Hatakeyama, T., Tokumitsu, H., and Kobayashi, R. (2003) Biochem. J. 374 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita, K., Oyama, Y., Shishibori, T., Matsushita, O., Okabe, A., and Kobayashi, R. (1999) Protein Expression Purif. 16 47–52 [DOI] [PubMed] [Google Scholar]
- 27.Tomas, A., and Moss, S. E. (2003) J. Biol. Chem. 278 20210–20216 [DOI] [PubMed] [Google Scholar]
- 28.Chen, S., and Smith, D. F. (1998) J. Biol. Chem. 273 35194–35200 [DOI] [PubMed] [Google Scholar]
- 29.Carrigan, P. E., Nelson, G. M., Roberts, P. J., Stoffer, J., Riggs, D. L., and Smith, D. F. (2004) J. Biol. Chem. 279 16185–16193 [DOI] [PubMed] [Google Scholar]
- 30.Lazarov, O., Morfini, G. A., Lee, E. B., Farah, M. H., Szodorai, A., DeBoe;r, S. R., Koliatsos, V. E., Kins, S., Lee, V. M., Wong, P. C., Price, D. L., Brady, S. T., and Sisodia, S. S. (2005) J. Neurosci. 9 2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, P. J. M., Kanelakis, K. C., Galigniana, M. D., Morishima, Y., and Pratt, W. B. (2001) J. Biol. Chem. 276 30092–30098 [DOI] [PubMed] [Google Scholar]
- 32.Young J. C., Hoogenraad, N. J., and Hartl, F. U. (2000) Cell 112 41–50 [DOI] [PubMed] [Google Scholar]
- 33.Hernandez, M. P., Sullivan, W. P., and Toft, D. O. (2002) J. Biol. Chem. 277 38294–38304 [DOI] [PubMed] [Google Scholar]
- 34.Siligardi, G., Hu, B., Panaretou, B., Piper, P. W., Pearl, L. H., and Prodromou, C. (2004) J. Biol. Chem. 279 51989–51998 [DOI] [PubMed] [Google Scholar]