Abstract
Xenorhabdus nematophila secretes insecticidal proteins to kill its larval prey. We have isolated an ∼58-kDa GroEL homolog, secreted in the culture medium through outer membrane vesicles. The protein was orally insecticidal to the major crop pest Helicoverpa armigera with an LC50 of ∼3.6 μg/g diet. For optimal insecticidal activity all three domains of the protein, apical, intermediate, and equatorial, were necessary. The apical domain alone was able to bind to the larval gut membranes and manifest low level insecticidal activity. At equimolar concentrations, the apical domain contained approximately one-third and the apical-intermediate domain approximately one-half bioactivity of that of the full-length protein. Interaction of the protein with the larval gut membrane was specifically inhibited by N-acetylglucosamine and chito-oligosaccharides. Treatment of the larval gut membranes with chitinase abolished protein binding. Based on the three-dimensional structural model, mutational analysis demonstrated that surface-exposed residues Thr-347 and Ser-356 in the apical domain were crucial for both binding to the gut epithelium and insecticidal activity. Double mutant T347A,S356A was 80% less toxic (p < 0.001) than the wild type protein. The GroEL homolog showed α-chitin binding activity with Kd ∼ 0.64 μm and Bmax ∼ 4.68 μmol/g chitin. The variation in chitin binding activity of the mutant proteins was in good agreement with membrane binding characteristics and insecticidal activity. The less toxic double mutant XnGroEL showed an ∼8-fold increase of Kd in chitin binding assay. Our results demonstrate that X. nematophila secretes an insecticidal GroEL protein with chitin binding activity.
Xenorhabdus nematophila, a Gram-negative bacterium, resides as symbiont in the gut of a soil nematode of the genus Steinernema (1–3). The bacteria-nematode association is highly toxic to many insect species, causing rapid larval death. The bacterium has a complex life cycle, encompassing symbiotic and pathogenic stages. The symbiotic phase is spent in the nematode gut, whereas pathogenicity is manifested in the insect larval body. The bacterium is released in the insect hemocoel (3) or gut (4), where it produces a variety of effector molecules including toxic proteins to kill the prey. The larval carcass provides a nutrient-rich environment for growth and development of both the nematode and the bacteria. The bacterium alone is also able to kill the insect host when grown axenically in the laboratory medium. Earlier X. nematophila was shown to produce outer membrane vesicle (OMV)3 during growth in the broth culture (5). The OMVs contained a number of proteins and were orally toxic to neonatal larvae of Helicoverpa armigera (6). Growing concern of development of resistance in the crop pests to crystal protein toxins of Bacillus thuringiensis has initiated vigorous research world-wide to discover orally active insecticidal proteins. The proteins associated with the OMV complex of X. nematophila provided a pool of potential insecticidal molecules for investigation.
Analysis of the OMV proteins led to the identification of a ∼58-kDa GroEL homolog (XnGroEL) as a major component of the complex. The GroEL protein belongs to a highly conserved family of molecular chaperones, which facilitate folding of nascent nonnative proteins in the cell (7). The large chaperon assembly is produced by two heptameric rings of 7 identical subunits each, stacked back to back making a large double-ringed cylinder of ∼800 kDa, enclosing a central cavity (8). It requires a 10-kDa co-chaperone GroES, Mg2+, and ATP to carry out the chaperoning activity (9). The 548-amino acid-long GroEL polypeptide chain folds into three distinctive domains. A well ordered equatorial domain (residues 6–133 from the N terminus and 409–523 from the C terminus) forms a solid base around the middle of the assembly and provides most of the residues for intersubunit contacts. A considerably less ordered apical domain (residues 191–376) surrounding the opening at the ends of the central cavity shows local flexibility within the domain as well as en bloc movement around a hinge connecting it to the intermediate domain. The intermediate domain (residues 134–190 of the N terminus and 377–408 of the C terminus) is much smaller and links the equatorial domain to the apical domain (10).
Earlier, a toxic GroEL was been described in another symbiotic bacterium, Enterobacter aerogenes (11). The protein was secreted by the bacterium in the saliva of parasitic antlions, which kills its insect prey by causing paralysis. The protein was shown to paralyze cockroaches when injected in their hemocoel (11). The GroEL proteins from several pathogenic bacteria are major antigens and are highly expressed under stressful conditions (12). It is shown to be essential for growth and viability of bacterial cells (13, 14). In endosymbiotic bacteria, GroEL is expressed at a higher level under normal growth conditions compared with free living species and is reported to protect against harmful effects of accumulated mutations and preserve fitness of the species (15). In this study we describe some of the unique properties of XnGroEL, secreted by the insecticidal bacterium X. nematophila.
EXPERIMENTAL PROCEDURES
OMV Purification and Identification of XnGroEL—X. nematophila strain 19061 was grown in Luria Bertaini medium at 28 °C for 18 h with shaking at 180 rpm. OMVs were prepared from cell-free culture supernatant as described earlier (5). OMV proteins were resolved by SDS-PAGE, and a ∼58-kDa band was cut and sequenced by Edman degradation.
Purification of the Native XnGroEL from Culture Supernatant (Extracellular) and Cell Lysate (Intracellular)—XnGroEL was purified from both the pellet and supernatant of X. nematophila culture. The proteins in the culture supernatant were precipitated with 70% ammonium sulfate and resolved on a 25-ml Q-Sepharose column. XnGroEL was eluted with 100 ml of NaCl gradient (0.3–1.0 m). The partially purified protein was concentrated and further purified by Superose 12 gel filtration column in a fast protein liquid chromatography system. The cell pellet was used for isolation of intracellular XnGroEL. Cells were disrupted by sonication, and the cell lysate was further purified as described earlier for extracellular protein. The proteins were resolved by SDS-PAGE, and purity was checked by silver staining of the gel. For purification of native GroEL protein from Escherichia coli, cells from 5 liters of LB culture were subjected to heat stress at 42 °C for 6 h and further treated as described above for intracellular protein of X. nematophila.
Cellular Fractionation and Localization of XnGroEL and XnGroES Proteins—Different cellular fractions of X. nematophila were prepared as described earlier (5, 16), and the proteins were resolved by SDS-PAGE and detected with antibodies of GroEL or GroES (Stressgen).
Pronase Treatment of OMV Proteins—OMVs were incubated with 1 unit of Pronase (Roche Applied Science) at 37 °C for 30 min in buffer A (50 mm sodium phosphate, pH 7.2). The reaction was stopped by 1× protease inhibitor mixture (Roche Applied Science), and the proteins were subjected to SDS-PAGE.
Biochemical Characterization of XnGroEL—ATPase activity and in vitro porcine lactate dehydrogenase refolding assay was performed as described previously (17, 18).
Construction of Genomic DNA Library and Cloning of groEL Gene—Genomic DNA was digested with different restriction enzymes and probed with a 45-bp nucleotide fragment, derived from the N-terminal amino acid sequence of the protein. A DNA fragment reacting with the probe in the EcoRI-digested DNA was cloned in pUC18 cloning vector (pMJ plasmid) and transformed in E. coli DH5α cells producing the MJ strain. Using the cloned fragment as template and primers from the 5′ and 3′ ends of the groEL coding sequence, a 1.7-kb DNA was amplified by PCR. The amplified DNA was cloned in pGEMTeasy vector producing pMJ1 plasmid. The sequences encoding apical domain (558 bp) and apical-intermediate domains (876 bp) of XnGroEL and GroES protein were PCR-amplified using specific primers, and the products were cloned in pGEMTeasy vector producing pMJ2, pMJ3, and pMJ4 plasmids, respectively.
Expression and Purification of Recombinant XnGroEL and Domain Proteins—The 1.7-kb fragment from pMJ1 plasmid was ligated in pET28a expression vector (pMJ5) and transformed in the E. coli BL21DE3-producing MJ5 strain. MJ5 cells were grown in LB medium containing 50 μg/ml kanamycin at 37 °C to exponential phase and induced with 1 mm isopropyl thiogalactopyranoside for 3–4 h. The cells were washed and lysed by sonication, and the cell-free supernatant was purified by nickel-nitrilotriacetic acid-agarose affinity matrix in cold using standard protocol. The recombinant apical domain (∼21 kDa), apical-intermediate domain (∼30 kDa), and GroES protein (∼10 kDa) were also expressed and purified as above, producing plasmids pMJ6, pMJ7, and pMJ8, respectively.
Site-directed Mutagenesis—Point mutations in the full-length XnGroEL and apical domain were done using cloned pMJ5 and pMJ6 constructs as template with the site-directed mutagenesis kit (Stratagene) as per the manufacturer's instructions. Five polar substitutions on the outer surface of the apical domain (Tyr-219, Ser-244, Asn-297, Thr-347, and Ser-356) and four on the outer surface of the equatorial domain (Ser-126, Lys-133, Asn-474, and Thr-481) were mutated to alanine. The mutated proteins were expressed and purified as the wild type proteins. CD spectra of all the proteins were recorded to compare their secondary structure.
Evaluation of Oral Insecticidal Activity—The insect bioassay was carried out in 12- or 24-well flat-bottom plates (NUNC). Different concentrations of the proteins were diluted in 10 mm sodium phosphate buffer, pH 7.5, and each group contained 24–30 neonates. The proteins were applied on the surface of artificial diet and allowed to percolate down. One neonatal larva (24 h old) of H. armigera was released on the surface of the diet in each well (19), and the plates were incubated at 25 °C (16-h-day-length periods) with 80% relative humidity. Mortality and larval weight were recorded periodically over the entire larval period. The dose of protein shown in the results was the amount of protein added to the diet. The bioassays were performed more than three times. Heat-inactivated XnGroEL, bovine serum albumin, GroEL homologue from E. coli K-12, and buffer were used as controls. Contribution of different domains of XnGroEL was evaluated by linear regression analysis of percent mortality at equimolar protein concentrations. The 50% lethal concentration (LC50) was determined by Probit analysis, and statistical analysis of the data was done using R package, a web-based tool for statistical computing.
Preparation of Brush Border Membrane Vesicles (BBMV) and Binding of XnGroEL—BBMV were prepared from dissected gut of fourth and fifth instar larvae by MgCl2 precipitation, as described previously (20). For binding assay, 20 μg of BBMV protein was incubated with 10 μg of XnGroEL or variant proteins in a total volume of 30 μl and incubated at 4 °C for 30 min followed by centrifugation at 12,000 × g for 5 min in cold to remove the unbound protein. The pellet was washed with 50 μl of 1× PBS twice and resuspended in 20 μl of 1× PBS. The samples were boiled in Laemmli sample dye for 5 min and resolved by SDS-PAGE. The proteins were transferred on nitrocellulose membrane and blotted with anti-GroEL antibodies diluted 1:20,000. The Western blots were scanned, and the integrated density value of each band representing bound protein was determined. Different substances like soluble chitin (21), chitosan, and crystalline cellulose were used to test protein binding to BBMV. XnGroEL was incubated with different concentrations of the above compound at 4° for 30 min and centrifuged, and the supernatant containing the unbound protein was added to BBMV and incubated as above, and the membrane-bound protein was estimated as above. To determine the specificity of binding of XnGroEL, competitive inhibition by sugar derivatives like GalNAc, N-acetylneuraminic acid, GlcNAc, N-acetyllactosamine, glucose, mannose, chito-oligosaccharides-N,N′-diacetyl chitobiose, N,N′,N′-triacetyl chitotriose, and hexa-N-acetyl chitohexaose (Dextra Laboratories) were also tested in the binding assay. The sugars were preincubated with the proteins at 4 °C for 30 min followed by incubation with BBMV as described above. To investigate the nature of binding of XnGroEL with the gut epithelium, the BBMVs were treated with different proteases or chitinase from Serratia marcescens (Sigma) at 37 °C for 20 min followed by binding and detection of the protein as described above. Chitinase digestion was also carried out in the presence of 1× protease inhibitor mixture (Roche Applied chemicals). Activity of aminopeptidase N, a protein exposed on the surface of the epithelial membrane, was measured (22) to evaluate the effect of chitinase treatment on the membrane surface.
Detection of Binding of XnGroEL by Immunofluorescence—Fourth to fifth instar larvae of H. armigera were starved for 12 h and dissected to take out the gut. The latter were washed in 1× MET buffer (50 mm Tris-HCl, pH 7.0, 100 mm mannitol, 1× protease inhibitor mixture, and 1 mm EGTA) and fixed in fixing solution (1% formaldehyde + 50 mm phosphate buffer, pH 7.0) for 16 h at 4 °C. The samples were embedded in paraffin blocks, and 6-μm-thick sections were cut and placed on glycerol-coated slides. For detection of protein binding, the sections were deparaffinated with xylene for 10 min at room temperature followed by sequential washing with ethanol (100, 80, 70, 50, and 20%), distilled water, and 1× PBS. Twenty μg of the proteins (XnGroEL or double mutant) were laid over the gut sections and incubated for 2 h at 4 °C. The slides were washed 3 times with wash buffer (1× PBS + 0.01% Tween 20) followed by blocking with 3% bovine serum albumin in 1× PBS for 2 h at 4 °C. The sections were incubated with anti-XnGroEL antibodies (1:10,000) for 16 h at 4 °C followed by extensive washing with wash buffer. The sections were incubated with anti-rabbit ALEXA 488 secondary antibodies (Molecular Probes, OR) at a dilution of 1:1500 and incubated for 2 h at 4°C. The samples were washed extensively with wash buffer, and coverslips were placed on the sections with anti-fade agent (Bio-Rad) and viewed in a Fluorescence microscope (Nikon ECLIPSE TE 2000-U) under blue light at a magnification of 20×.
Chitin Binding Assay—Binding of the wild type and mutant proteins with α-chitin (from Crab shells, Sigma Aldrich) was evaluated as described earlier (23). A 20 mg/ml stock suspension of the substrate α-chitin was prepared in buffer (50 mm Tris-HCl, pH 7.0). For studying the time course of binding, a 500-μl reaction mixture contained 0.5 mg of substrate and 50 μg of protein in the above buffer. The tubes were incubated on a rotary shaker at room temperature, and samples were taken at intervals (5, 10, 15, 30, and 60 min). The suspension was centrifuged for 5 min at 13,000 × g, and optical density of the supernatant was measured at 280 nm to determine the amount of unbound protein.
To determine the binding constants of XnGroEL variant proteins, the assay procedure was as follows; each protein variant was diluted to a range of concentrations (10–300 μg/ml) in 50 mm Tris-HCl, pH 7.0, from a stock solution of known strength, and A280 of each dilution was measured to create a standard curve of the individual variants. For chitin binding assay, 0.5 mg/ml α-chitin was added to different concentrations of XnGroEL variants (10, 20, 50, 100, 200, and 300 μg/ml), in a total volume of 1 ml, and the tubes were mixed gently on a rotary shaker at 60 rpm for 16 h at room temperature. Subsequently the samples were centrifuged at 13,000 × g for 5 min, A280 of the supernatants was measured, and protein concentrations were calculated from the standard curves. All the values below 20 μg/ml were verified by protein estimation by Bradford reagent. A suitable blank containing 0.5 mg/ml α-chitin in buffer was used in all the experiments, performed in triplicate. The dissociation constant Kd and substrate binding capacities Bmax were determined by fitting the binding isotherms to the one-site binding equation, Pbound = Bmax[Pfree]/Kd + [Pfree], where Pbound denotes protein specifically bound to the substrate and Pfree is unbound protein in the supernatant, by nonlinear regression using the Graph Pad Prism software 3.0 (San Diego, CA). To analyze the nature of molecular interactions between XnGroEL and α-chitin, the binding assay was also performed in the presence of 10–100 mm sodium chloride.
Sequence and Structure Analysis—NCBI BLAST (24) was used to find out the closest homologous sequence to XnGroEL. Global alignment of X. nematophila GroEL sequence with E. coli GroEL was done using Needle program from EMBOSS (25). SWISS-MODEL (26) was used to generate the three-dimensional structure, which was viewed and analyzed with CHIMERA (27). Energy calculations of proteins were done using NAMD and VMD (28). Multiple sequence alignment and phylogenetic analysis of proteins were done using ClustalW2 (29).
Nucleotide Sequence and Accession Number—GroESL operon sequence has been submitted to GenBank™ (accession number AY184491).
RESULTS
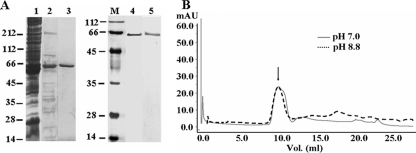
Identification and Purification of Native XnGroEL Protein—The SDS-PAGE profile of OMV proteins prepared from culture supernatant of X. nematophila contained multiple proteins ranging from 10 to 200 kDa (5). The N-terminal sequence of a predominant protein band of ∼58 kDa identified it as a homolog of E. coli heat shock protein GroEL. The XnGroEL protein eluting between 0.5 and 0.7 mm NaCl from the ion-exchange column contained minor impurities (Fig. 1A, lane 2). The partially purified protein was passed through Superose-12 size fractionation column, and XnGroEL was eluted in the void volume in oligomeric form. The void volume fractions contained pure homogeneous native XnGroEL protein (Fig. 1A, lane 3), also seen by silver staining (Fig. 1A, lane 4). No lipopolysaccharide was found associated with the protein (data not shown). The final yield of purified protein was ∼2 mg per liter of culture supernatant. EcGroEL protein was also purified from 5 liters of cell lysate, and homogeneity was examined by silver staining.
FIGURE 1.
Purification of XnGroEL from X. nematophila culture supernatant. A, SDS-PAGE gel stained with Coomassie Blue. Lane 1, ammonium sulfate-precipitated culture supernatant proteins; lane 2, Q-Sepharose-purified proteins; lane 3, purified protein from Superose-12 gel filtration column; lanes 4 and 5, purified native and recombinant XnGroEL respectively, after silver staining. B, elution profile of recombinant XnGroEL from Superdex-200 gel filtration column; the arrow indicates void volume peak containing XnGroEL at pH 7.0 and 8.8. mAb, milliabsorbance units.
Biochemical Characterization of XnGroEL Protein—Elution of XnGroEL in the void volume of Superdex-200 column indicated that the purified protein existed as high molecular (>600 kDa) oligomer (data not shown), as reported for EcGroEL. XnGroEL was able to hydrolyze γ-P32-labeled ATP with the release of inorganic phosphate. Synthesis of the protein by X. nematophila was higher compared with E. coli at 28 °C and was further enhanced when subjected to heat shock at 37 °C. The protein was able to function as chaperone in an in vitro lactate dehydrogenase folding system (data of the above experiments not shown).
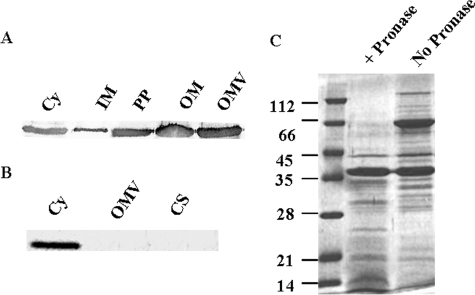
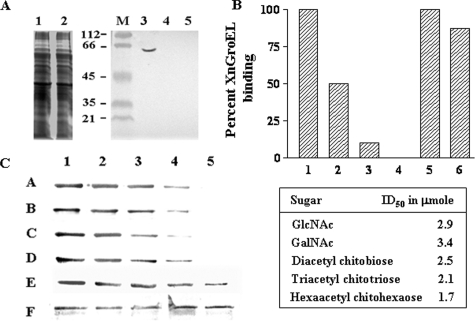
Export of XnGroEL Protein—Because the protein was found in culture supernatant, we examined whether the protein was actually secreted or was present due to cell lysis. Different cellular fractions, e.g. cytosolic, inner membrane (IM), periplasmic, outer membrane (OM), and extracellular proteins, were subjected to SDS-PAGE analysis followed by Western blotting (Fig. 2A). The presence of XnGroEL was observed in all the above fractions, whereas HNS (histone-like nucleoid structuring protein), a cytosolic protein, or DsbA, a periplasmic marker protein, was not detected in the culture supernatant (data not shown), suggesting a specific secretion pathway for export of XnGroEL outside the cell. GroES, the canonical co-chaperone, was present in the cytoplasmic fraction but absent in culture supernatant or OMVs of X. nematophila (Fig. 2B). Treatment of OMV preparations with Pronase degraded XnGroEL completely, indicating its location on the surface, possibly in association with the outer membrane of the bacteria (Fig. 2C).
FIGURE 2.
Export of XnGroEL. A, proteins in subcellular fractions immunoblotted with antibodies to XnGroEL (1:20,000); Cy, cytosolic; IM, inner membrane; PP, periplasmic proteins; OM, outer membrane. B, cellular fractions blotted with antibodies of GroES (1:5000). CS, culture supernatant proteins. C, SDS-PAGE analysis of Pronase digested OMV proteins stained with Coomassie Blue.
Cloning, Expression, and Purification of Recombinant Proteins—Southern blot analysis of genomic DNA after restriction digestion identified a ∼4-kb fragment containing the gene in the EcoRI digest. The partial library produced with 3–5 kb of EcoRI-digested DNA fragments in pUC18 vectors led to the isolation of a positive clone with an ∼3.8-kb insert (pMJ1). The sequence of the 3.8-kb DNA fragment showed that it contained GroESL operon spanning ∼2.4 kb preceded by two hypothetical open reading frames and part of aspartate ammonium lyase A. The nucleotide sequence of GroESL operon of X. nematophila was very similar to closely related Photorhabdus spp. (85%) and E. coli (84%). The translated amino acid sequence of XnGroEL was (89%) similar to the EcGroEL sequence. The recombinant XnGroEL protein was obtained from the pET28a construct (pMJ2); it was purified to homogeneity by nickel-nitrilotriacetic acid column (Fig. 1A, lane 5). The purified recombinant XnGroEL was obtained as oligomeric complex, which remained stable at pH 8.8 (Fig. 1B). The protein showed ATPase activity and in vitro chaperoning activity like native XnGroEL. The 21-kDa apical domain and the 30-kDa apical-intermediate domains existed as monomer and had no ATPase activity (data not shown).
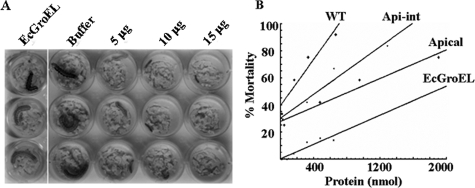
Toxicity of XnGroEL in H. armigera Larvae—Both the native and recombinant XnGroEL proteins retarded growth of H. armigera neonates at 1–20 μg/g diet when fed orally. Reduction in the average weight of the surviving larvae was 50–55% on sixth day of the larval period. A dose-dependent effect on larval mortality was observed at concentrations 5–15 μg/g (Fig. 3A). Heat-inactivated XnGroEL, EcGroEL, and bovine serum albumin used as control, caused low mortality (∼5%) of the insect larvae. The 50% lethal concentration (LC50) of native protein purified from cell lysate (intracellular) and culture supernatant (extracellular) were ∼3.8 μg/g, and ∼3.6 μg/g, respectively. The recombinant full-length protein was toxic with LC50 ∼ 4.8 μg/g. The apical domain of XnGroEL was also toxic to insect neonates when fed orally in a dose-dependent manner (LC50 ∼ 5.8 μg/g). The relationship between larval death and protein concentration was evaluated by linear regression analysis. The values of the slope (m) representing toxicity were 0.006, 0.01, and 0.02 for apical domain, apical-intermediate domain, and full-length protein, respectively, indicating that the apical domain retained ∼1/3 that of the toxicity of the full-length protein. The toxicity was increased to ∼1/2 that of the full-length protein by the addition of intermediate domain (apical-intermediate domain) (analysis of variance, p < 0.001) (Fig. 3B). These results suggested that although the apical domain of XnGroEL alone was able to manifest toxicity, the intermediate and the equatorial domains too have some role in toxicity of the protein. Reduced toxicity of the domains could also be attributed to their inability to oligomerize, which could be necessary for providing strength by cooperative binding.
FIGURE 3.
Insecticidal activity of XnGroEL on H. armigera larvae. A, purified proteins XnGroEL (5–15 μg/g diet) and EcGroEL at a concentration of 20 μg/g diet were added to the diet of 1-day-old neonates, and growth and mortality were recorded during 6–12 days of larval period. B, plot of percent mortality against protein concentration. Each dot in the plot represents the average of three individual experiments. The range of protein concentration with high statistical significance (p < 0.001) was selected for linear regression analysis; concentration range shown in the plot: XnGroEL (∼16–400 nmol), apical-intermediate (∼30–1300 nmol), apical (∼40–2000 nmol), and EcGroEL (∼16–700 nmol). The parameters calculated for each line are as follows: slope (m) 0.006, 0.011, 0.021, and 0.006, intercept (c) 6.638, 7.207, 9.430, and 0.011 for apical, apical-intermediate, full-length XnGroEL, and EcGroEL proteins, respectively. WT, wild type.
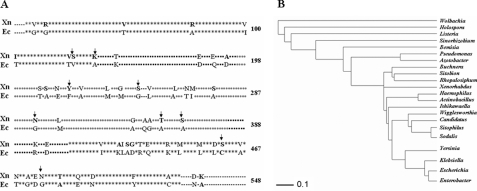
Protein Sequence and Structural Analysis—To investigate the basis of interaction of XnGroEL at molecular levels, pairwise alignment of primary amino acid sequence with non-toxic E. coli protein was performed (Fig. 4A). In addition, the GroEL sequences of free-living and symbiotic bacteria were compared to examine the degree of similarity in the protein sequences of the two bacterial groups and also identify substitutions that were conserved by natural selection (data not shown). A phylogenetic tree was generated from the data (Fig. 4B).
FIGURE 4.
Structural analysis of XnGroEL protein. A, comparison of protein sequences of XnGroEL (AY184491) and EcGroEL (NC_000913) showing the three domains. Star, equatorial domain; solid squares, intermediate domain; open squares, apical domain; dash, undefined. The 59 substitutions are shown by amino acids symbols. The mutations performed in the study are shown by arrows. XN, X. nematophila. EC, E. coli. B, phylogenetic tree was constructed by PHYLIP using ClustalW web server. Wolbachia (AAW52271), Holospora obtusa (BAA14046), Listeria welshimeri (CAK21507), Sinorhizobium meliloti (NP_435641), bacterial isolate from Bemisia tabaci (AAR23105), Pseudomonas aeruginosa (AAB34346), Azotobacter vinelandii (AAL25964), Buchnera aphidicola (AAO33050), bacterial isolate from Sitobion avenae (AAB47483), bacterial isolate from Rhopalosiphum padi (AAB47482), X. nematophila (AY184491), Haemophilus influenzae (NP_438701), Actinobacillus actinomycetemcomitans (AAM88539), Ishikawaella (BAF44092), Wigglesworthia glossinidia (AAK07427), Candidatus Blochmannia (AAZ40717), bacterial isolate from Sitophilus oryzae (AB97670), Sodalis glossinidius (AAK92204), Yersinia pestis (YP_653837), Klebsiella pneumoniae (ABY64753), E. coli (NC_000913), and E. aerogenes (AAL09389).
Attribution of toxicity in XnGroEL in contrast to EcGroEL notwithstanding 89% homology in their primary sequence (GenBank™ accession number NC_000913) suggested that substitutions acquired by XnGroEL (GenBank™ accession number AY184491) might be responsible for its activity. Sequence analysis showed that XnGroEL has 59 substitutions (Fig. 4A) compared with EcGroEL, scattered all across the protein. A three-dimensional homology model of XnGroEL demonstrated that of a total of 59 substitutions, 21 were localized in the apical domain, 6 in the intermediate domain and 32 in the equatorial domain. Polar character of the apical and equatorial domains was increased, as seven (Ser-212, Tyr-219, Ser-244, Ser-275, Asn-297, Thr-347 and Ser-356), and nine (Arg-13, Arg-78, Ser-126, Lys-133, Ser-427, Asp-456, Ser-458, Asn-474, and Thr-481) substitutions, respectively, were from hydrophobic to polar in nature. The intermediate domain had no polar substitution. In contrast, substitutions of polar to hydrophobic residues were one (Ala-343) in apical, one (Ala-188) in the intermediate, and five (Val-48, Ile-101, Val-125, Ala-425, and Gly-428) in the equatorial domain. These substitutions led to a net increase in hydrophilic character of the protein compared with E. coli (Eisenberg scale 3.175 and 6.434, respectively). Structural analysis of the apical domain showed that five of seven polar residues (Tyr-219, Ser-244, Asn-297, Thr-347, and Ser-356) were exposed on the outer surface, whereas two were buried in the core of the protein. In the equatorial domain Ser-126, Lys-133, Asn-474, Thr-481 were on the outer surface, and Arg-13 and Asp-456 were on the inner surface (facing the cavity) whereas Arg-78, Ser-427, and Ser-458 were located on the interface between the two heptameric rings. To examine the role of surface-exposed polar substitutions in XnGroEL, the latter were mutated to alanine (results are described under “Discussion”).
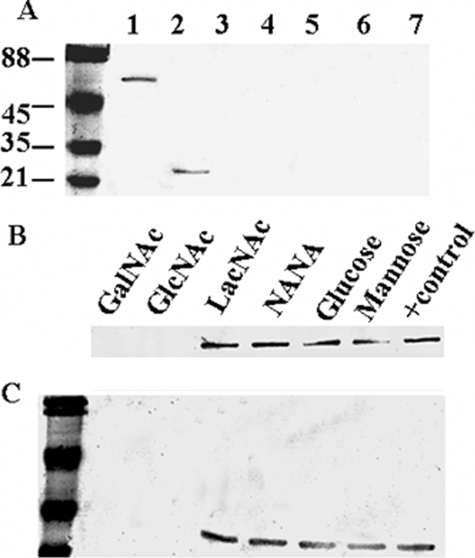
Interaction of XnGroEL with Larval BBMs and Competitive Inhibition by Sugars—In contrast to EcGroEL, wild type XnGroEL and the apical domain protein were able to bind with BBMVs prepared from H. armigera larval gut (Fig. 5A, lanes 1, 2, and 4). Heating the proteins at 60 °C for 20 min abolished their ability to bind with the membrane (Fig. 5A, lane 3 and 5). Among the sugars tested, GalNAc and GlcNAc inhibited binding of protein with BBMV, whereas other sugars like glucose, mannose, LacNAc, and N-acetylneuraminic acid had no effect (Fig. 5, B and C). The membrane binding behavior of the apical domain was identical to the full-length protein (Fig. 5, B and C).
FIGURE 5.
Interaction of XnGroEL with larval gut BBMV. The proteins were incubated with BBMVs, washed, and resolved by SDS-PAGE. Membrane-bound protein was detected by immunoblotting with antibody (1:20,000) of XnGroEL. A, lane 1, full-length protein; lane 2, apical domain protein; lane 3, heated full-length protein; lane 4, EcGroEL; lane 5, heated apical domain; lane 6, membranes alone; lane 7, bovine serum albumin. B and C, inhibition of binding of XnGroEL and apical domain respectively by sugars, 300 mm each. Protein binding was detected as above. NANA, N-acetylneuraminic acid.
Chitinase treatment of the BBMV abolished binding of XnGroEL completely (Fig. 6A), indicating that chitin in the peritrophic membrane may be the primary target of XnGroEL binding. Chitinase treatment of BBMV in the presence of protease inhibitor also abolished protein binding, showing no contaminating protease was present in the chitinase. Different forms of chitin were used as the control in the BBMV binding assay (Fig. 6B). Soluble chitin showed dose-dependent inhibition of protein binding between 10–100 μg. Chitosan (85% deacetylated) reduced binding by ∼18%, whereas crystalline cellulose (Avicel, Sigma) had no effect on binding of protein to the larval membrane. GalNAc and GlcNAc inhibited protein binding to BBMV with an ID50 of 3.4 and 2.9 μmol, respectively (Fig. 6C). Among the chito-oligosaccharides, the inhibitory concentration was inversely proportional to the size of the molecule, and hexa-N-acetyl chitohexaose was most efficient, with an ID50 of 1.7 μmol, whereas the ID50 values of N,N′,N″-triacetyl chitotriose and N,N′-diacetyl chitobiose were 2.1 and 2.5 μmol, respectively (Fig. 6C). Furthermore, no significant change in the aminopeptidase N activity of the BBMV was observed after chitinase treatment, indicating that the membrane surface was not affected (data not shown). No binding was observed when the membranes were treated with varying concentrations and time (0.1–20 μg) of proteinase K and Pronase for 1–10 min. However, because all the proteins were completely digested (data not shown), involvement of any specific binding protein could not be ascertained. Treatment of BBMV with 0.1–25 μg of trypsin showed neither any effect on protein profile nor on the binding of XnGroEL to BBMV (data not shown).
FIGURE 6.
Inhibition of XnGroEL binding to BBMV by different chitin derivatives. A, detection of XnGroEL binding to BBMV after chitinase treatment. The BBMV were incubated with chitinase followed by incubation with XnGroEL, and the membrane proteins were processed and resolved by SDS-PAGE as above. The gel was stained with Coomassie Blue; lane 1, untreated BBMV proteins; lane 2, treated with chitinase. Samples in lanes 1 and 2 were blotted with anti-GroEL antibodies after Western transfer; M, prestained markers; lane 3, untreated BBMV; lane 4, BBMV with chitinase and protease inhibitor; lane 5, with chitinase, no protease inhibitor. B, the binding assay mixture contained 20 μg of BBMV, 10 μg of XnGroEL, and the competing compounds. 1, no inhibitory compound; 2, 3, and 4, 0.05, 0.1, and 0.5 mg of solubilized chitin, respectively; 5, 1.0 mg of cellulose; 6, 1.0 mg of chitosan. C, XnGroEL binding in the presence of lanes 1–5, 0, 50, 100, 150, and 200 mm GalNAc, respectively (A); lanes 1–5, 0, 50, 100, 150, and 200 mm GlcNAc, respectively (B), lanes 1–5, 0, 50, 100, 150, and 200 mm diacetyl chitobiose, respectively (C), lanes 1–5, 0, 50, 75, 100, and 125 mm triacetyl chitotriose, respectively (D); lanes 1–5, 0, 20, 40, 60 and 80 mm hexaacetyl chitohexaose, respectively (E); loading control, BBM blotted with antibody against N-aminopeptidase (F). The table shows ID50 values calculated by plotting the integrated density values of the bands shown in panel C. The experiments were repeated three times, and data from one of the representative experiments are presented.
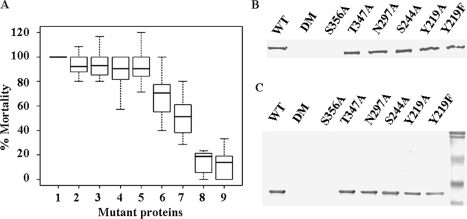
Effect of Mutations on Bioactivity of XnGroEL—The role of surface-exposed polar residues in insecticidal activity of the protein was analyzed by mutating these residues with alanine. The mutations Y219A, Y219F, S244A, and N297A did not alter insecticidal activity of the protein significantly (p > 0.4), whereas T347A and S356A led to an ∼30% (p < 0.001) and ∼50% (p < 0.001) loss of insecticidal activity, respectively. A double mutant, T347A,S356A, resulted in loss of insecticidal activity by ∼80% (p < 0.001) (Fig. 7A). The above data show that residues Thr-347 and Ser-356 together play an important role in the toxicity of XnGroEL. Binding of the mutated proteins to BBMVs was in agreement with their insecticidal activity. There was no effect on the binding of mutated proteins Y219A, Y219F, S244A, N297A, and T347A, but mutation S356A abolished binding of the protein with the gut membrane. Similarly, the double mutant also did not bind to the gut membrane (Fig. 7B). Corresponding mutations in the apical domain showed similar behavior in membrane binding assay (Fig. 7C).
FIGURE 7.
Effect of mutations on bioactivity of XnGroEL. A, box plot of percent toxicity (y axis), normalized relative to wild type scores. 1, wild type; 2, S244A; 3, N297A; 4, Y219A; 5, Y219F; 6, T347A; 7, S356A; 8, double mutant; 9, EcGroEL. The thick bar represents percent medians for different proteins. The box includes range of scores falling into the middle 50% of the distribution (interquartile range (IQR) = 75th–25th percentile), and the whiskers are the minimum and maximum scores in the distribution, mathematically defined as (±1.5 IQR) upper and lower fences. B, binding of XnGroEL and mutated proteins to BBMV. WT, wild type; DM, double mutant. C, binding and detection of apical domain and corresponding domain variants. Protein binding was detected as described in Fig. 5.
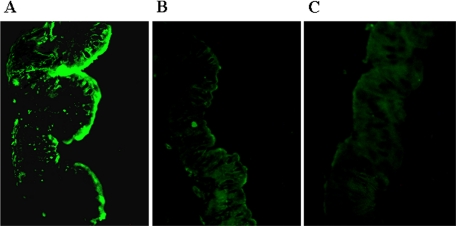
Detection of Protein Binding by Immunohistochemistry—The results are shown in Fig. 8, A–C, demonstrating localization of the protein on the lumen side of the epithelial membrane. A diffused and homogenous binding of the wild type protein was observed, suggesting distribution of the binding moiety uniformly on the apical face of the membrane. The double mutant protein was not able to bind to BBMV, as observed in earlier studies.
FIGURE 8.
Detection of XnGroEL binding to gut epithelial membrane by immunohistochemistry. The gut was dissected from the fourth to fifth instar larvae of H. armigera, washed and fixed in fixing solution. 6-μm-thick sections were incubated with proteins (XnGroEL or double mutant) and stained with anti-XnGroEL antibodies (1:10,000). The sections were treated with anti-rabbit ALEXA 488 secondary antibodies at a dilution of 1:1500 and viewed in a fluorescence microscope (Nikon ECLIPSE TE 2000-U) under blue light at a magnification of 20×. A, wild type XnGroEL; B, double mutant; C, negative control, no protein.
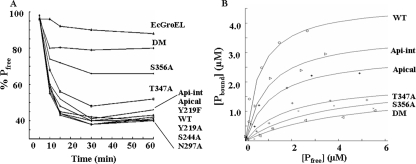
Binding of XnGroEL Variants to α-Chitin—To investigate the possibility that XnGroEL is a chitin-binding protein, binding constants of interaction were determined. The time course of binding showed that most of the protein binding was completed within 1 h of the start of incubation (Fig. 9A). More than 60% of wild type protein was bound to chitin after 1 h when saturation occurred. The initial rate of binding of the apical domain at equimolar concentrations was slightly lower than the full-length protein. EcGroEL was not able to bind to α-chitin, as the percentage of free protein remained close to 95% till the end of incubation. The mutations Y219A, Y219F, S244A, and N297A showed no effect on binding as observed earlier with the BBMVs. Partial disruption in binding was observed in the mutations T347A and S356A. The double mutant T347A,S356A showed drastic inhibition of binding as more than 90% of the protein remained free at the end of incubation (Fig. 9A). These results are in good agreement with the earlier results of interaction with gut membranes. Protein binding to chitin was not significantly affected at lower concentrations, but 100 mm sodium chloride completely disrupted binding of XnGroEL to α-chitin (data not shown), suggesting polar interactions between the two. The dissociation constant (Kd) and binding capacities (Bmax) of the proteins (Table 1) were calculated from the binding isotherms (Fig. 9B). The wild type protein showed the lowest Kd (0.64 ± 0.10 μm) and highest Bmax (4.68 ± 0.16 μmol of protein/g of chitin), which is well within the range of binding constants reported for other chitin binding proteins (23, 30, 31). The apical domain showed an ∼2-fold increase in Kd (1.14 ± 0.05 μm) and reduction in Bmax (2.95 ± 0.05 μmol of protein/g). The double mutant T347A,S356A, with the lowest oral toxicity to H. armigera larvae, showed an ∼8-fold increase in Kd (4.71 ± 0.76 μm) compared with wild type protein and the lowest Bmax (1. 84 ± 0.02 μmol of protein/g). Mutations T347A and S356A had about an ∼3- and ∼4.6-fold increase in dissociation constants than the wild type protein, respectively (Table 1). The binding capacities of all the mutants were lower than the wild type protein, suggesting their combined role in forming the binding motif. The binding characteristic of the proteins is in good agreement with their lethality to insect larva.
FIGURE 9.
Chitin binding activity of XnGroEL. A, adsorption of XnGroEL and mutant proteins on α-chitin. The reaction mix containing 1 mg/ml α-chitin and 100 μg/ml protein in 50 mm Tris-Cl buffer, pH 7.0, was incubated at room temperature. 500-μl samples were processed at different time points, and free protein (Pfree) in the supernatant was estimated. B, different concentrations of serially diluted XnGroEL and variants were mixed with 0.5 mg/ml of α-chitin and incubated overnight with gentle shaking, and free protein in the supernatant was estimated. The binding isotherms were plotted for wild type (WT), intermediate-apical (Api-int), apical, T347A, S356A, and double mutant (DM). Each point represents the average of values obtained in three independent experiments. All datasets were fitted to the equation for one site binding with nonlinear regression.
TABLE 1.
Binding constants of XnGroEL to α-chitin
| Protein | Kd (±S.D.) | Bmax (±S.D.) |
|---|---|---|
| μm | μmol/g chitin | |
| Wild type | 0.64 ± 0.10 | 4.68 ± 0.16 |
| Apical intermediate | 0.96 ± 0.28 | 3.65 ± 0.38 |
| Apical domain protein | 1.14 ± 0.05 | 2.94 ± 0.05 |
| T347A mutant | 1.87 ± 0.05 | 2.02 ± 0.01 |
| S356A mutant | 2.57 ± 0.20 | 1.78 ± 0.08 |
| Double mutant | 4.71 ± 0.76 | 1.84 ± 0.23 |
DISCUSSION
OMVs released by Gram-negative bacteria are considered as a means of transporting effector proteins outside the cell (16). In an earlier study we reported that OMVs produced by X. nematophila were orally toxic to H. armigera larvae (5). Here we describe a protein present in the OMV preparation of X. nematophila and identified it as XnGroEL. Our study unravels a novel property of the chaperon molecule, which has been investigated largely in the context of protein folding in the past. XnGroEL, although highly homologous to E. coli protein, was secreted in the culture supernatant in association with the OMVs. A number of pathogenic bacteria are known to release GroEL protein in the culture medium (12), but their secretion mechanism is not characterized.
Discovery of oral insecticidal activity of XnGroEL illustrates a unique property of this universally occurring chaperon molecule. The insecticidal activity was not related to its chaperoning activity, as no ATP or GroES was required (data not shown). The insecticidal activity was observed only after oral ingestion of the protein; it had no effect when injected in the hemocoel or when added to cultures of insect hemocytes or Sf21 cells (data not shown), indicating that larval gut is the primary target of the protein. X. nematophila has a large arsenal of potent toxins that work together to kill the larval prey (1–3). Because GroEL happens to be an essential protein for viability of an organism, our efforts to disrupt the gene to determine its role in pathogenicity of X. nematophila were unsuccessful.
Taking a cue from the insect toxic GroEL of E. aerogenes (11), it would be interesting to explore if toxicity of GroEL is a common feature among symbiotic bacterium with conserved substitutions at specific positions. Comparison of GroEL sequences of free-living and symbiotic bacterial species revealed that the sequence is more conserved among the former than the symbionts (data not shown), which have acquired multiple mutations, scattered across the length of the polypeptide. However, the mutations were not conserved across species, suggesting that symbiotic bacteria, which are subjected to diverse kinds of selection pressures in the host, tend to acquire mutations randomly as a means of adaptation that cannot be fitted into a set pattern. Phylogenetic analysis (Fig. 4B) revealed that the X. nematophila protein is closer to a cluster encompassing mostly free-living and a few endosymbionts like Buchnera and Wigglesworthia, whereas most of the endosymbiotic bacteria belonged to evolutionarily distant clades, showing a high degree of variability among themselves. The seven polar substitutions identified in the apical domain of XnGroEL with respect to E. coli were also present in other symbionts, but none of the positions seemed to be conserved (figure not shown) among all the species. Of the two specific residues implicated in toxicity and chitin binding of XnGroEL, Ser-356 seems to be a unique substitution in Xenorhabdus except Sinorhizobium and Wolbachia, whereas a polar substitution at position 347 is seen more commonly.
Toxicity of the recombinant protein in H. armigera larvae was accompanied with specific recognition and binding to the BBMVs through GlcNAc or GalNAc moieties, whereas nontoxic EcGroEL showed no binding with the membranes. Because GlcNAc and GalNAc are known to act as the binding/recognition motifs of glycoprotein receptors in membranes (32), it was inferred that XnGroEL might be interacting with a glycoprotein, glycolipids, or chitinous peritrophic membrane of the gut. Inhibition of protein binding to BBMV with chito-oligosaccharides and its loss after chitinase treatment made a strong case for chitin as the binding moiety.
Specific functions of the three domains of GroEL in protein chaperoning are well characterized (33). To map the minimum bioactive region of XnGroEL, recombinantly produced protein domains were examined. The observation that at equimolar concentration toxicity of the apical domain was one-third and apical-intermediate domain was one-half of the full-length protein (comprising of apical-intermediate-equatorial domains) implied that all the domains were necessary for optimal activity of XnGroEL, in contrast to low activity of full-length EcGroEL. Furthermore, with the addition of successive domains, a progressive increase in binding affinity to α-chitin (decrease in Kd) also emphasized the necessity of an intact protein for optimum functioning. In light of the similarity in binding properties of the apical domain to the full-length protein and the ability to cause larval toxicity, we assume that the former contained the minimum essential region for binding to gut membrane. It is difficult to say whether the equatorial and intermediate domains directly participate in binding or strengthen binding by enabling oligomerization, recruiting the cooperative strength of seven subunits compared with much smaller monomeric apical domain. Thus, larger dimensions of the oligomeric protein can be rationalized as more efficient in physically blocking the membrane surface and interfering with chitin metabolism. Due to noncontiguous domain organization of XnGroEL, we were unable to produce and test independent contribution of intermediate and equatorial domains in binding and toxicity. In addition, the inability to produce monomeric XnGroEL due to structural instability made it difficult to further analyze the importance of oligomerization (34, 35).
Requirement of full protein for optimum insecticidal activity and distribution of amino acid substitutions in the entire length of the protein sequence suggested that alteration in the nature of the residues could be responsible for acquisition of insecticidal activity by XnGroEL vis à vis EcGroEL. It was argued that an increase in surface hydrophilicity of XnGroEL would on the one hand have a positive effect on its stability in aqueous environment and on the other increase propensity of polar interactions with other molecules (36–38). In the insect gut, where the pH ranges from 7 to 9, XnGroEL is likely to remain oligomeric, as it was found to withstand high pH. In such an event the polar residues exposed on the outer surface are more likely to participate in protein-ligand interactions than those facing the cavity due to steric constraints. Among different domains of XnGroEL, the equatorial domain will be less favorably disposed to participate in protein-ligand interactions than apical domain due to intersubunit associations, necessary for the stability of the tetradecameric complex, and as expected, mutations in this domain lead to protein destabilization (this study and Refs. 39–43). The importance of five polar substitutions (Tyr-219, Ser-244, Asn-297, Thr-347, and Ser-356) on the outer surface of the apical domain of XnGroEL was revealed by their mutation to alanine. It needs to be mentioned here that the above residues were different from those involved in chaperoning interactions with denatured proteins (44). Analysis of the results revealed that mutation S356A resulted in reduction of both binding and larval toxicity, whereas the corresponding effect of T347A was relatively low. However, incorporating the mutations together in the double mutant reduced both binding and toxicity of the protein substantially (∼80%, p < 0.001). Based on the above data it was concluded that residue Ser-356 is critical for binding and most likely initiates interaction of the protein with membranes, whereas Thr-347 assists in binding once the primary contact is made. Reciprocal mutation of Ala-347 in the E. coli protein to threonine using two-step energy simulation predicted a very large increase in its energy (45), from -1179 kcal/mol to +12000 kcal/mol, resulted in a highly destabilized variant protein and, hence, was not attempted. Mutation A356S was produced, but no biological activity was observed (data not shown) in the protein. It is interesting to note that the critical Ser-356 is located centrally in a stretch of six polar residues (EESTSD) forming a hairpin loop; considering the fact that binding of XnGroEL with α-chitin was abolished by NaCl, it is conceivable that the protein interacts with the target through this polar loop.
Generally, the insecticidal proteins cause toxicity either by enzymatic action or by cytolysis (46). Because none of the activities was detected in XnGroEL, its mode of action must be different form other toxins. Based on the results of inhibition of protein binding to BBMV by GlcNAc, chito-oligosaccharides, and chitinase treatment, with in vitro chitin binding activity, we propose that XnGroEL binds to chitin in the peritrophic membrane and inhibits chitin metabolism, although interaction with other membrane components also cannot be ruled out at this stage. The primary role of peritrophic membrane is to protect the underlying epithelium against bacterial pathogens and their toxins in the food of the larvae (47). A number of insecticidal proteins with chitinase activity are known; however, not all the chitin-binding proteins have catalytic activity (23, 30, 31); XnGroEL appears to belong to the second category. Another atypical chitin-binding protein has been described earlier in S. marcescens, which interacted with chitin through solvent-exposed polar side chains (23). The common carbohydrate binding architecture of chitin-binding proteins, consisting of a surface groove, cleft, or tunnel lined with aromatic residues, were absent in both S. marcescens and XnGroEL proteins.
Insect growth and development is strictly dependent on the capability to remodel chitinous structures in different developmental stages. The insects constantly synthesize and degrade chitin in a highly controlled manner to allow ecdysis and regeneration of the peritrophic matrices. Because chitin metabolism is crucial for arthropod development, inhibition or interference with chitin metabolism provides a good target for development of insecticides, more so as chitin polymers are absent in vertebrates (47).
In conclusion, findings of this study demonstrate for the first time that XnGroEL is an insecticide with chitin binding property. The correlation between binding to α-chitin/BBMV and toxicity, established by mutational analysis, strongly supports our hypothesis that by virtue of its chitin binding property, XnGroEL interacts with the larval peritrophic lining, leading to cessation of growth and development of the larva. Based on the evidence, we propose that the oligomeric protein is more likely to interact through its heptameric apical face, covering 140 Å (diameter of the heptamer) of membrane surface and 7 binding equivalents, than the equatorial surface, providing only 1–2 subunits. It would be interesting to verify the structural model experimentally.
Acknowledgments
We thank Prof. Jonathan Beckwith for DsbA antibody and Dr. Roberto Spurio for the HNS antibody.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AY184491.
This work was supported in part by the Department of Biotechnology, Government of India. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: OMV, outer membrane vesicle; XnGroEL, X. nematophila GroEL; BBMV, brush border membrane vesicle; LacNAc, N-acetyllactosamine; EcGroEL, E. coli GroEL; kb, kilobase(s); PBS, phosphate-buffered saline.
References
- 1.Herbert, E. E., and Goodrich-Blair, H. (2007) Nat. Rev. Microbiol. 5 634-646 [DOI] [PubMed] [Google Scholar]
- 2.Forst, S., and Nealson, K. (1996) Microbiol. Rev. 60 21-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akhurst, R. J., and Dunphy, G. B. (1993) in Parasites and pathogens of Insects (Beckage, N., Thompson, S., and Federici, B., eds) Vol. 2, pp. 1-23, Academic Press Inc., New York, NY [Google Scholar]
- 4.Sicard, M., Brugirard-Ricaud, K., Pages, S., Lanois, A., Boemare, N. E., Bréhelin, M., and Givaudan, A. (2004) Appl. Environ. Microbiol. 70 6473-6478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khandelwal, P., and Banerjee-Bhatnagar, N. (2003) Appl. Environ. Microbiol. 69 2032-2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khandelwal, P., Bhatnagar, R., Choudhury, D., and Banerjee, N. (2004) Biochem. Biophys. Res. Commun. 314 943-949 [DOI] [PubMed] [Google Scholar]
- 7.Fink, A. L. (1999) Physiol. Rev. 79 425-449 [DOI] [PubMed] [Google Scholar]
- 8.Bukau, B., and Horwich, A. L. (1998) Cell 92 351-366 [DOI] [PubMed] [Google Scholar]
- 9.Braig, K. (1998) Curr. Opin. Struct. Biol. 8 159-165 [DOI] [PubMed] [Google Scholar]
- 10.Sigler, P. B., Xu, Z., Rye, H. S., Burston, S. G., Fenton, W. A., and Horwich, A. L. (1998) Annu. Rev. Biochem. 67 581-608 [DOI] [PubMed] [Google Scholar]
- 11.Yoshida, N., Oeda, K., Watanabe, E., Mikami, T., Fukita, Y., Nishimura, K., Komai, K., and Matsuda, K. (2001) Nature 411 44. [DOI] [PubMed] [Google Scholar]
- 12.Ranford, J. C., and Henderson, B. (2002) Mol. Pathol. 55 209-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fayet, O., Ziegelhoffer, T., and Georgopoulos, C. (1989) J. Bacteriol. 171 1379-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dukan, S., and Nystrom, T. (1999) J. Biol. Chem. 274 26027-26032 [DOI] [PubMed] [Google Scholar]
- 15.Fares, M. A., Barrio, E., Sabater-Munoz, B., and Moya, A. (2002) Mol. Biol. Evol. 19 1162-1170 [DOI] [PubMed] [Google Scholar]
- 16.Wai, S. N., Lindmark, B., Soderblom, T., Takade, A., Westermark, M., Oscarsson, J., Jass, J., Richter, D. A., Mizunoe, Y., and Uhlin, B. E. (2003) Cell 115 25-35 [DOI] [PubMed] [Google Scholar]
- 17.Vashisht, A. A., Pradhan, A., Tuteja, R., and Tuteja, N. (2005) Planta J. 44 76-87 [DOI] [PubMed] [Google Scholar]
- 18.Clarke, A. R., Waldman, A. D., Hart, K. W., and Holbrook, J. J. (1985) Biochem. Biophys. Acta 829 397-407 [DOI] [PubMed] [Google Scholar]
- 19.Khandelwal, P., Choudhury, D., Birah, A., Reddy, M. K., Gupta, G. P., and Banerjee, N. (2004) J. Bacteriol. 186 6465-6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cioffi, M., and Wolfersberger, M. G. (1983) Tissue Cell 15 781-803 [DOI] [PubMed] [Google Scholar]
- 21.Guo, X. F., Kikuchi, K., Matahira, Y., Sakai, K., and Ogawa, K. (2002) J. Carbohydr. Chem. 21 149-161 [Google Scholar]
- 22.Jurat-Fuentes, J. L., and Adang, M. J. (2004) Eur. J. Biochem. 271 3127-3135 [DOI] [PubMed] [Google Scholar]
- 23.Vaaje-Kolstad, G., Huston, D. R., Riemen, A. H. K., Eijsink, V. G. H., and van Alten, D. M. F. (2005) J. Biol. Chem. 280 11313-11319 [DOI] [PubMed] [Google Scholar]
- 24.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Anang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice, P., Longden, I., and Bleaseby, A. (2000) Trends Genet. 16 276-277 [DOI] [PubMed] [Google Scholar]
- 26.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 31 3381-3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comp. Chem. 25 1605-1612 [DOI] [PubMed] [Google Scholar]
- 28.Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, D. R., Kale, L., and Schulten, K. (2005) J. Comp. Chem. 26 1781-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeltins, A., and Schrempf, H. (1997) Eur. J. Biochem. 246 557-564 [DOI] [PubMed] [Google Scholar]
- 31.Goldstein, M. A., Takagi, M., Hashida, S., Shoseyov, O., Roy, H. D., and Segel, I. H. (1993) J. Bacteriol. 175 5762-5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pigott, C. R., and Ellar, D. J. (2007) Microbiol. Mol. Biol. Rev. 71 255-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber, F., Keppel, F., Georgopoulos, C., Hayer-Hartl, M. K., and Hartl, F. U. (1998) Nat. Struct. Biol. 5 977-985 [DOI] [PubMed] [Google Scholar]
- 34.Motojima, F., Chaudhry, C., Fenton, W. A., Farr, G. W., and Horwich, A. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15005-15012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surin, A. K., Kotova, N. V., Marchenkova, S., Marchenkov, V. V., and Semisotnov, G. V. (1999) Bioorg. Khim. 25 358-364 [PubMed] [Google Scholar]
- 36.Ward, E. S., Ellar, D. J., and Chilcott, C. N. (1988) J. Mol. Biol. 202 527-535 [DOI] [PubMed] [Google Scholar]
- 37.Zhang, S., Wei, L., Bastow, K., Zheng, W., Brossi, A., Lee, K. H., and Tropsha, A. (2007) J. Comput. Aided Mol. Des. 21 97-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonet, J., Caltabiano, G., Khan, A. K., Johnston, M. A., Corbí, C., Gómez, A., Rovira, X., Teyra, J., and Villà-Freixa, J. (2006) Proteins Struct. Funct. Genet. 63 65-77 [DOI] [PubMed] [Google Scholar]
- 39.Singh, J., Garber, E., Van Vlijmen, H., Karpusas, M., Hsu, Y. M., Zheng, Z., Naismith, J. H., and Thomas, D. (1998) Protein Sci. 7 1124-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bochkareva, E., Safro, M., and Girshovich, A. (1999) J. Biol. Chem. 274 20756-20758 [DOI] [PubMed] [Google Scholar]
- 41.Boisvert, D. C., Wang, J., Otwinowski, Z., Horwich, A. L., and Sigler, P. B. (1996) Nat. Struct. Biol. 3 170-177 [DOI] [PubMed] [Google Scholar]
- 42.Horovitz, A., Bochkareva, E. S., and Girshovich, A. S. (1993) J. Biol. Chem. 268 9957-9959 [PubMed] [Google Scholar]
- 43.Sigler, P. B., and Horwich, A. L. (1995) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 348 113-119 [DOI] [PubMed] [Google Scholar]
- 44.Fenton, W. A., Kashi, Y., Furtak, K., and Horwich, A. L. (1994) Nature 371 614-619 [DOI] [PubMed] [Google Scholar]
- 45.Wan, S., Coveney, P. V., and Flower, D. R. (2005) J. Immun. 175 1715-1723 [DOI] [PubMed] [Google Scholar]
- 46.Koni, P. A., and Ellar, D. J. (1994) Microbiology 140 1869-1880 [DOI] [PubMed] [Google Scholar]
- 47.Merzendorfer, H., and Zimoh, L. (2003) J. Exp. Biol. 206 4393-4412 [DOI] [PubMed] [Google Scholar]